Abstract
Tripartite motif 16 (TRIM16), a member of the RING B-box coiled-coil (RBCC)/tripartite motif (TRIM) protein family, has been shown to play a role in tumor development and progression. However, the role of TRIM16 in ovarian cancer has never been revealed. Thus, in this study, we investigated the roles and mechanisms of TRIM16 in ovarian cancer. Our results demonstrated that TRIM16 expression was low in ovarian cancer cell lines. In addition, overexpression of TRIM16 significantly inhibited the migration and invasion in vitro, as well as suppressed the epithelial–mesenchymal transition (EMT) phenotype in ovarian cancer cells. Furthermore, overexpression of TRIM16 greatly inhibited the protein expression levels of Shh, Smo, Ptc, Gli-1, MMP2, and MMP9 in ovarian cancer cells. Taken together, these results strongly suggest that TRIM16 inhibits the migration and invasion via suppressing the Sonic hedgehog signaling pathway in ovarian cancer cells. Thus, TRIM16 may be a novel potential therapeutic target for ovarian cancer.
Key words: Tripartite motif 16 (TRIM16), Ovarian cancer, Cell invasion, Epithelial–mesenchymal transition (EMT)
INTRODUCTION
Ovarian cancer is the most common cause of death in the world among women with gynecological cancer. It is estimated that there are 21,290 new ovarian cancer cases and 14,180 deaths in the US in 20151. Although substantial advances have been made in cancer therapy, the 5-year survival rate of patients with ovarian cancer is less than 30% due to severe invasion and metastasis2–5. Therefore, a thorough investigation into the mechanisms of tumor invasion and metastasis may facilitate potential molecular targets for the treatment of ovarian cancer.
The tripartite motif (TRIM) family of proteins is characterized by three zinc-binding domains, a RING, a B-box type 1, and a B-box type 2, followed by a coiled-coil region6. Many studies have indicated that TRIM proteins play an important role in regulating cell cycle regulation, autophagy, innate immunity, and cell migration7–10. Tripartite motif 16 (TRIM16), a member of the TRIM protein family, has been shown to play a role in tumor development and progression11–13. For example, Li et al. confirmed that the expression of TRIM16 was significantly downregulated in hepatocellular carcinoma (HCC) tissues, and knockdown of TRIM16 promoted epithelial–mesenchymal transition (EMT) in a manner associated with HCC metastasis in vitro and in vivo14. In addition, TRIM16 expression was significantly reduced in squamous cell carcinoma (SCC) and inhibited SCC cell migration in vitro15. However, the role of TRIM16 in ovarian cancer has never been revealed. Thus, in this study, we investigated the roles and mechanisms of TRIM16 in ovarian cancer. Our results show that TRIM16 has a low expression in ovarian cancer cell lines and that TRIM16 suppresses migration and invasion by suppressing the Sonic hedgehog (Shh) signaling pathway in ovarian cancer cells.
MATERIALS AND METHODS
Cell Culture
The human ovarian cancer cell lines (SKOV3, 3AO, and OVCAR3) and human normal ovarian surface epithelial cell line (HOSEpiC) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). All cells were cultured at 37°C in 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, and 1% l-glutamine (Gibco, Rockville, MD, USA).
Construction of the pcDNA3.1-TRIM16 Vector and Cell Transfection
The human TRIM16 expression vector pcDNA3.1-TRIM16 or empty vector was obtained from Shanghai Sangon Co. Ltd. (Shanghai, P.R. China). For in vitro transfection, ovarian cancer cells (5 × 105 cells/well) were transfected with TRIM16 or empty vector using Lipofectamine 2000 (Invitrogen, Victoria, Australia) according to the manufacturer’s protocol. The overexpression efficacy was demonstrated by Western blotting.
RNA Preparation and Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from cells using the PureLink RNA mini extraction kit (Ambion, Victoria, Australia), in accordance with the manufacturer’s instruction. Complementary DNA (cDNA) was synthesized using a first-strand cDNA synthesis kit (TaKaRa, Dalian, P.R. China). RT-PCR was performed with SYBR Premix Ex Taq™ (TaKaRa, Biotech Co., Ltd., Dalian, P.R. China). The following primer pairs were used for PCR amplification: TRIM16, 5′-GTGCATTGGGCTCCTTCCTCCTTA-3′ (forward) and 5′-CTGGCTGACCCAGGCTGGTCTT-3′ (reverse); GADPH, 5′-ATCCCATCACCATCTTCCAG-3′ (forward) and 5′-CCATCACGCCACAGTTTCC-3′ (reverse). Relative expression was standardized using the quantity of β-actin, and data were analyzed accordingto the 2−ΔΔCt formula.
Western Blot Analysis
Ovarian cancer cell lines were washed twice with 10 ml of ice-cold PBS and whole-cell extracts prepared in 1× lysis buffer (Cell Signaling Technology, Danvers, MA, USA) in sterile water containing a protease inhibitor cocktail and phophatase inhibitor tablets (Roche, Indianapolis, IN, USA). Protein concentration was measured using the BCA-200 Protein Assay kit (Pierce, Rockford, IL, USA). Protein lysates were separated using 12% SDS-PAGE gels and then transferred onto nitrocellulose membranes. After blocking with 5% skim milk solution, the membranes were incubated with various primary antibodies (TRIM16, E-cadherin, N-cadherin, vimentin, Shh, Smo, Ptc, Gli-1, MMP2, MMP9, or GAPDH; Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4°C overnight. Then membranes were washed three times with TBST and incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz Biotechnology) for 1 h at room temperature. Blots were visualized using ECL reagents (Pierce) by a chemiluminescence imaging system (Bio-Rad, Richmond, CA, USA).
Cell Migration and Invasion Assays
Cell migration and invasion were tested using the modified Transwell chamber assay. In brief, for cell migration assay, infected ovarian cancer cells (1 × 105 cell/chamber) in 100 μl of serum-free medium were plated into the upper chamber. The lower compartment of the chamber was supplemented with 10% FBS. After incubation for 24 h at 37°C, the nonmigrating cells were removed from the upper surface of the membrane. The migrated cells were fixed with methanol and stained with 0.1% crystal violet. The number of cells that migrated was quantified by counting the number of stained cells in six individual fields under light microscopy. For invasion assay, the procedures were the same as the migration assay apart from the chamber being replaced with Matrigel-coated filters (BD Bioscience, San Jose, CA, USA).
Statistical Analysis
All statistical analyses were performed using SPSS 16.0 software (Chicago, IL, USA). The Student’s t-test was used to determine differences between two groups, and one-way ANOVA was used to analyze the differences among multiple groups. A value of p < 0.05 was considered statistically significant.
RESULTS
TRIM16 Has a Low Expression in Ovarian Cancer Cells
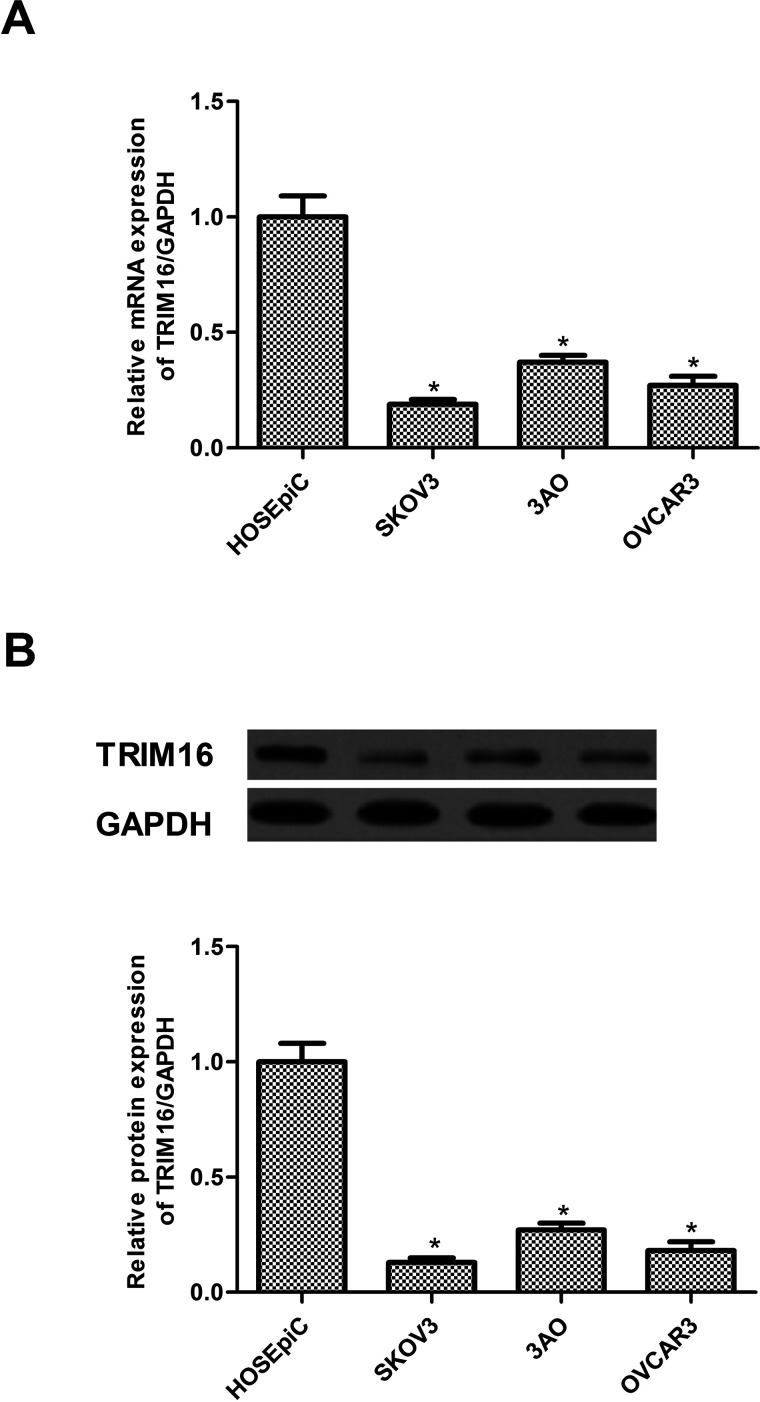
First, we examined the expression of TRIM16 in human ovarian cancer cell lines by RT-qPCR and Western blotting. The mRNA expression of TRIM16 in human ovarian cancer cell lines was lower than that in ovarian epithelial cell lines (Fig. 1A). Consistent with the results of the RT-qPCR analysis, decreased protein expression of TRIM16 was also observed in human ovarian cancer cell lines (Fig. 1B).
Figure 1.
TRIM16 has a low expression in ovarian cancer cells. (A) The mRNA expression levels of TRIM16 were analyzed in four human ovarian cancer cell lines by RT-qPCR. (B) The protein expression levels of TRIM16 were detected in four human ovarian cancer cell lines by Western blot. Results are means ± SD from three independent experiments performed in duplicate. *p < 0.05 compared with HOSEpiC group.
Overexpression of TRIM16 Inhibits the Migration and Invasion in Ovarian Cancer Cells
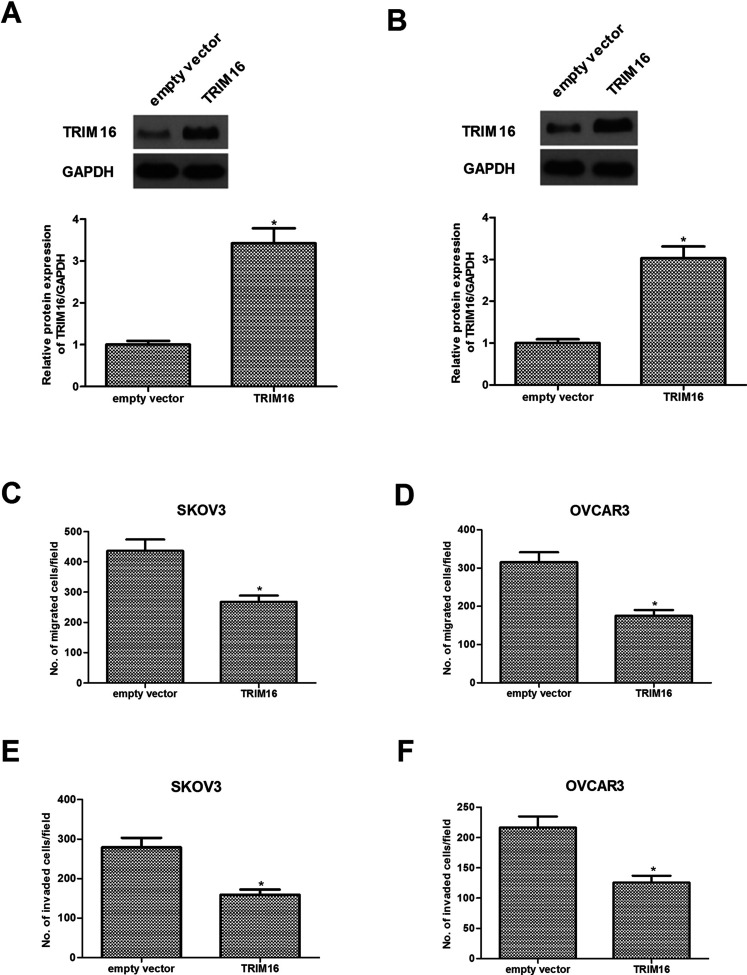
To determine the underlying cellular functions of TRIM16 in ovarian cancer, we overexpressed TRIM16 in two cell lines (SKOV3 and OVCAR3) with lower TRIM16 expression. Western blotting indicated that TRIM16 expression was sharply increased in SKOV3 (Fig. 2A) and OVCAR3 cells (Fig. 2B) after transfection of TRIM16 overexpression. The results of the Transwell migration assay indicated that overexpression of TRIM16 greatly inhibited the cell migration ability of SKOV3 (Fig. 2C) and OVCAR3 cells (Fig. 2D). Furthermore, overexpression of TRIM16 significantly reduced the invasive ability of SKOV3 (Fig. 2E) and OVCAR3 cells (Fig. 2F) compared to cells treated with empty vector.
Figure 2.
Overexpression of TRIM16 inhibits the migration and invasion in ovarian cancer cells. SKOV3 and OVCAR3 cells were transfected with TRIM16 or empty vector for 24 h. The corresponding transfection efficiency was detected by Western blot in SKOV3 (A) and OVCAR3 cells (B). (C, D) Cell migration was analyzed by the Transwell assay. (E, F) Cell invasion was determined by the Matrigel invasion assay. Results are means ± SD from three independent experiments performed in duplicate. *p < 0.05 compared with empty vector group.
Overexpression of TRIM16 Inhibits the EMT Phenotype in Ovarian Cancer Cells
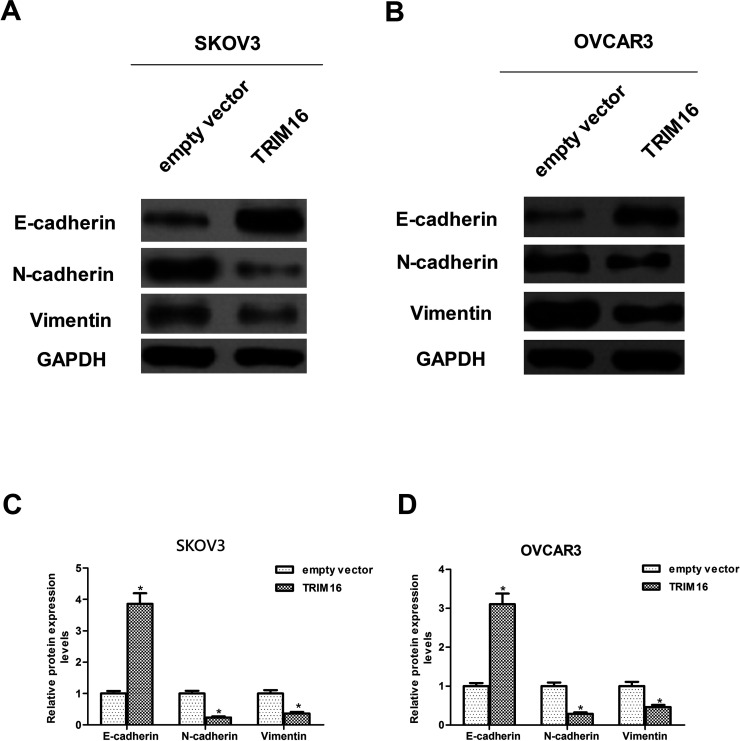
Next, we examined the effects of TRIM16 on the expression profile of EMT markers, including E-cadherin, vimentin, and N-cadherin in SKOV3 and OVCAR3 cells. The results demonstrated that E-cadherin, the epithelial marker, was significantly upregulated by overexpression of TRIM16. Meanwhile, the mesenchymal markers, including vimentin and N-cadherin, were downregulated by overexpression of TRIM16 in SKOV3 cells (Fig. 3A and C). Similarly, a significant increase in E-cadherin expression and a significant decrease in vimentin and N-cadherin expression were observed after overexpression of TRIM16 in OVCAR3 cells when compared to the empty vector group (Fig. 3B and D).
Figure 3.
Overexpression of TRIM16 inhibits the EMT phenotype in ovarian cancer cells. SKOV3 and OVCAR3 cells were transfected with TRIM16 or empty vector for 24 h. Western blot assays were performed to detect the expression of N-cadherin, E-cadherin, and vimentin in SKOV3 cells (A, C). Western blot assays were performed to detect the expression of N-cadherin, E-cadherin, and vimentin in OVCAR3 cells (B, D). Results are means ± SD from three independent experiments performed in duplicate. *p < 0.05 compared with empty vector group.
Overexpression of TRIM16 Inhibits the Activation of Sonic Hedgehog Signaling Cascade in Ovarian Cancer Cells
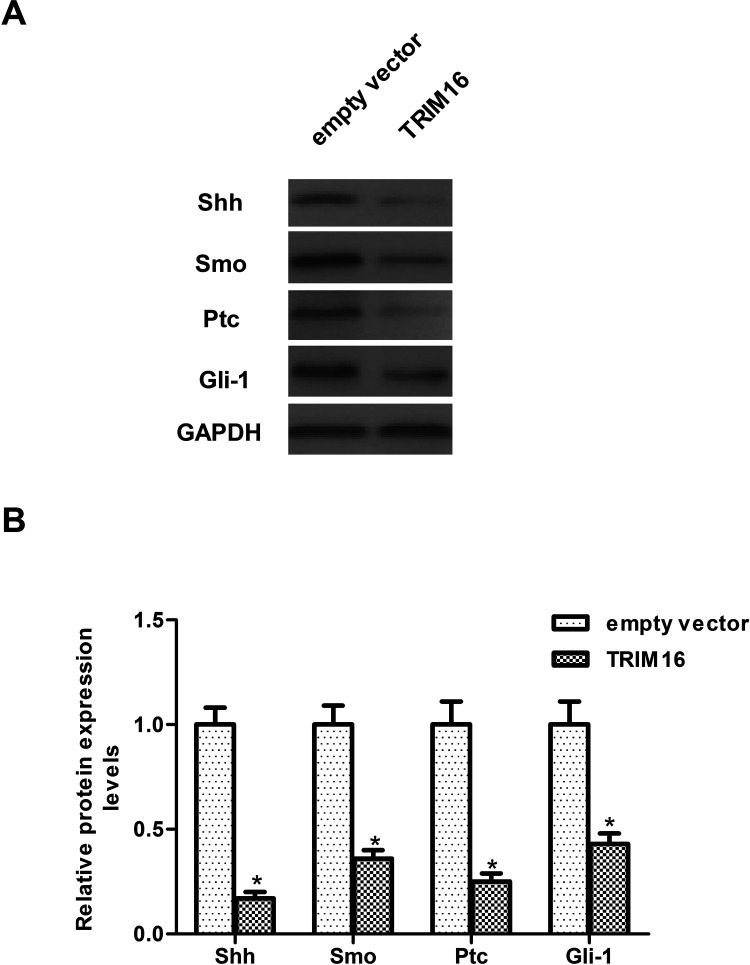
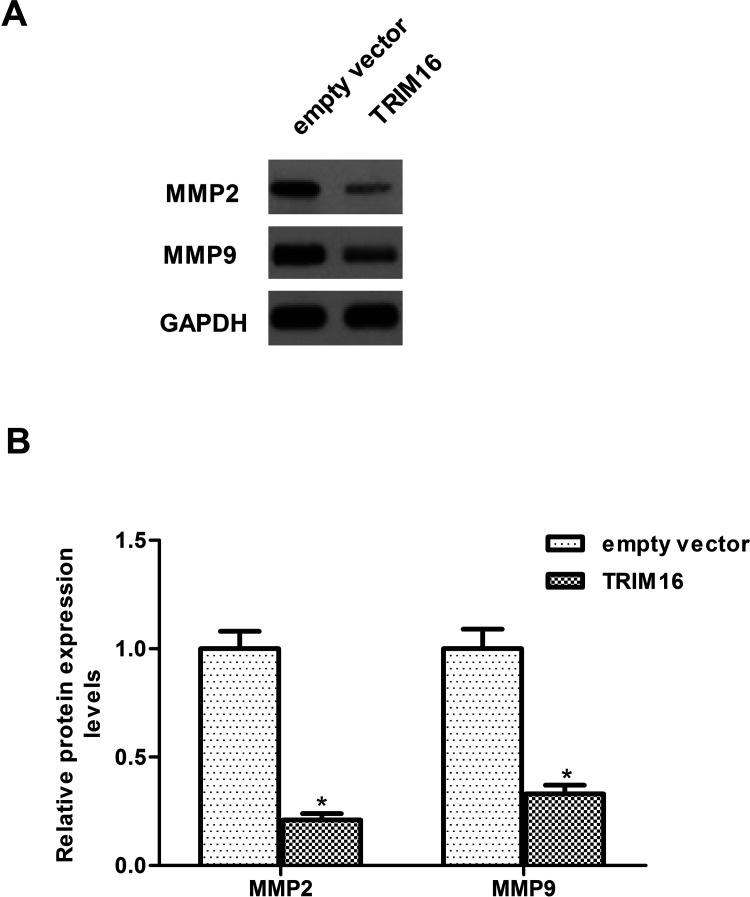
To further explore the molecular mechanism responsible for the function of TRIM16 in ovarian cancer, we examined the effect of TRIM16 on the activation of Shh signaling cascade in SKOV3 cells. Overexpression of TRIM16 dramatically downregulated the protein expression levels of Shh as well as Smo, Ptc, and Gli-1 in SKOV3 cells, compared with the empty vector group (Fig. 4). Furthermore, we examined whether TRIM16 affects the expression of MMP2 and MMP9 in SKOV3 cells. The results of the Western blot analysis showed that overexpression of TRIM16 obviously inhibited the expression of MMP2 and MMP9 in SKOV3 cells (Fig. 5).
Figure 4.
Overexpression of TRIM16 inhibits the activation of Sonic hedgehog signaling cascade in ovarian cancer cells. SKOV3 cells were transfected with TRIM16 or empty vector for 24 h. (A) Western blot assays were performed to detect the expression of Shh, Smo, Ptc, and Gli-1 in SKOV3 cells. (B) The relative protein expression levels of Shh, Smo, Ptc, and Gli-1 were quantified using Image-Pro Plus 6.0 software and normalized to GAPDH. Results are means ± SD from three independent experiments performed in duplicate. *p < 0.05 compared with empty vector group.
Figure 5.
Overexpression of TRIM16 inhibits the expression of MMPs in ovarian cancer cells. SKOV3 cells were transfected with TRIM16 or empty vector for 24 h. (A) Western blot assays were performed to detect the expression of MMP2 and MMP9 in SKOV3 cells. (B) The relative protein expression levels of MMP2 and MMP9 were quantified using Image-Pro Plus 6.0 software and normalized to GAPDH. Results are means ± SD from three independent experiments performed in duplicate. *p < 0.05 compared with empty vector group.
DISCUSSION
The main findings of the present study can be summarized as follows: (1) TRIM16 has a low expression in ovarian cancer cell lines; (2) overexpression of TRIM16 inhibits the migration and invasion of ovarian cancer cells; (3) overexpression of TRIM16 inhibits the EMT phenotype in ovarian cancer cells; (4) overexpression of TRIM16 dramatically downregulated the protein expression levels of Shh, Smo, Ptc, Gli-1, MMP2, and MMP9 in SKOV3 cells.
Previous studies have shown that reduced TRIM16 expression in a range of human primary malignant tissues correlates with increased malignant potential13. Sutton et al. confirmed that the protein levels of TRIM16 were markedly reduced in human melanoma cell lines, compared with normal human epidermal melanocytes16. Yao et al. reported that TRIM16 had a low expression in breast cancers17. Consistent with these results, we observed that TRIM16 has a low expression in ovarian cancer cell lines, suggesting that TRIM16 may act as a tumor suppressor in the development and progression of ovarian cancer.
EMT is a dynamic process mediating cancer metastasis. It is characterized by loss of epithelial markers, acquisition of mesenchymal molecules, and enhancement of cell mobility18. In cancer, EMT allows neoplastic cells to become motile and invasive, thereby contributing to the metastatic potential of tumors19. It has been reported that upregulation of TRIM16 obviously inhibited EMT and metastasis of non-small cell lung cancer (NSCLC) cells. In contrast, silencing TRIM16 greatly promoted the EMT and metastasis of NSCLC cells20. Similarly, we observed that overexpression of TRIM16 inhibited the migration and invasion of ovarian cancer cells, as well as suppressed the EMT phenotype in ovarian cancer cells. These data suggest that TRIM16-mediated inhibition of migration and invasion likely operates through its suppression effect on EMT.
The hedgehog (Hh) pathway plays an important role in the progression of various human carcinomas, including ovarian cancer21–23. Shh and Indian Hh (Ihh) are secreted proteins in the Hh pathway. Shh signaling activates Gli transcription factors, which possess oncogenic characteristics and are deregulated in malignant cancers resulting in invasion and metastasis, chemoresistance, and EMT24. It was reported that overexpression of Patched and Gli-1 protein in ovarian cancers correlated with poor survival of patients, and Gli-1 upregulation promoted the EMT, proliferation, invasion, and migration abilities of ovarian cancer cells25,26. Furthermore, inhibition of the Hh signaling pathway can promote extensive cell death and reduce tumor growth in vivo in mouse xenograft models of human ovarian cancer27. Interestingly, Huo et al. reported that downregulation of TRIM16 activated the Shh pathway in NSCLC cells20. Thus, the Hh signaling pathway is a possible molecular target for new treatment strategies for ovarian carcinoma. In this study, we found that overexpression of TRIM16 dramatically downregulated the protein expression levels of Shh as well as Smo, Ptc, and Gli-1 in SKOV3 cells. Previous studies reported that suppression of Shh decreased the invasive phenotype of cancer cells through inhibition of MMP2 and MMP9 expression28,29. Herein we observed that overexpression of TRIM16 also inhibited the expression of MMP2 and MMP9 in SKOV3 cells. These data suggest that TRIM16 inhibits the migration and invasion in ovarian cancer cells by downregulating MMP2 and MMP9 expression mediated by the Shh signaling pathway.
In conclusion, our study is the first to show that TRIM16 inhibits migration and invasion via suppressing the Shh signaling pathway in ovarian cancer cells. These findings reveal that TRIM16 plays an important role in the regulation of ovarian cancer progression; thus, TRIM16 may be a potential therapeutic target for the treatment of ovarian cancer.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. [DOI] [PubMed] [Google Scholar]
- 2. Marcus CS, Maxwell GL, Darcy KM, Hamilton CA, McGuire WP. Current approaches and challenges in managing and monitoring treatment response in ovarian cancer. J Cancer 2014;5:25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lin JJ, Egorova N, Franco R, Prasad-Hayes M, Bickell NA. Ovarian cancer treatment and survival trends among women older than 65 years of age in the United States, 1995–2008. Obstet Gynecol. 2016;127:81–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Banerjee S, Kaye SB. New strategies in the treatment of ovarian cancer: Current clinical perspectives and future potential. Clin Cancer Res. 2013;19:961–8. [DOI] [PubMed] [Google Scholar]
- 5. Schmid BC, Oehler MK. New perspectives in ovarian cancer treatment. Maturitas 2014;77:128–36. [DOI] [PubMed] [Google Scholar]
- 6. Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer 2011;11:792–804. [DOI] [PubMed] [Google Scholar]
- 8. Ozato K, Shin DM, Chang TH, Morse HC. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kosaka Y, Inoue H, Ohmachi T, Yokoe T, Matsumoto T, Mimori K, Tanaka F, Watanabe M, Mori M. Tripartite motif-containing 29 (TRIM29) is a novel marker for lymph node metastasis in gastric cancer. Ann Surg Oncol. 2007;14:2543–29. [DOI] [PubMed] [Google Scholar]
- 10. Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C, Silvestri G, Münch J, Kirchhoff F, Simonsen A. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev Cell 2014;30:394–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marshall G, Bell J, Koach J, Tan O, Kim P, Malyukova A, Thomas W, Sekyere E, Liu T, Cunningham A. TRIM16 acts as a tumour suppressor by inhibitory effects on cytoplasmic vimentin and nuclear E2F1 in neuroblastoma cells. Oncogene 2010;29:6172–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bell JL, Malyukova A, Kavallaris M, Marshall GM, Cheung BB. TRIM16 inhibits neuroblastoma cell proliferation through cell cycle regulation and dynamic nuclear localization. Cell Cycle 2013;12:889–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim PY, Rahmanto AS, Tan O, Norris MD, Haber M, Marshall GM, Cheung BB. TRIM16 overexpression induces apoptosis through activation of caspase-2 in cancer cells. Apoptosis 2013;18:639–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li L, Dong L, Qu X, Jin S, Lv X, Tan G. Tripartite motif 16 inhibits hepatocellular carcinoma cell migration and invasion. Int J Oncol. 2016;48:1639–49. [DOI] [PubMed] [Google Scholar]
- 15. Cheung BB, Koach J, Tan O, Kim P, Bell JL, D’andreti C, Sutton S, Malyukova A, Sekyere E, Norris M. The retinoid signalling molecule, TRIM16, is repressed during squamous cell carcinoma skin carcinogenesis in vivo and reduces skin cancer cell migration in vitro. J Pathol. 2012;226:451–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sutton SK, Koach J, Tan O, Liu B, Carter DR, Wilmott JS, Yosufi B, Haydu LE, Mann GJ, Thompson JF. TRIM16 inhibits proliferation and migration through regulation of interferon beta 1 in melanoma cells. Oncotarget 2014;5:10127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yao J, Xu T, Tian T, Fu X, Wang W, Li S, Shi T, Suo A, Ruan Z, Guo H. Tripartite motif 16 suppresses breast cancer stem cell properties through regulation of Gli-1 degradation via the ubiquitin-proteasome pathway. Oncol Rep. 2016;35:1204–12. [DOI] [PubMed] [Google Scholar]
- 18. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yao D, Dai C, Peng S. Mechanism of the mesenchymal-epithelial transition and its relationship with metastatic tumor formation. Mol Cancer Res. 2011;9:1608–20. [DOI] [PubMed] [Google Scholar]
- 20. Huo X, Li S, Shi T, Suo A, Ruan Z, Yao Y. Tripartite motif 16 inhibits epithelial-mesenchymal transition and metastasis by down-regulating sonic hedgehog pathway in non-small cell lung cancer cells. Biochem Biophys Res Commun. 2015;460:1021–8. [DOI] [PubMed] [Google Scholar]
- 21. Sabol M, Car D, Musani V, Ozretic P, Oreskovic S, Weber I, Levanat S. The hedgehog signaling pathway in ovarian teratoma is stimulated by sonic hedgehog which induces internalization of patched. Int J Oncol. 2012;41:1411–18. [DOI] [PubMed] [Google Scholar]
- 22. Huang RL, Gu F, Kirma NB, Ruan J, Chen CL, Wang HC, Liao YP, Chang CC, Yu MH, Pilrose JM. Comprehensive methylome analysis of ovarian tumors reveals hedgehog signaling pathway regulators as prognostic DNA methylation biomarkers. Epigenetics 2013;8:624–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schmid S, Bieber M, Zhang F, Zhang M, He B, Jablons D, Teng NN. Wnt and hedgehog gene pathway expression in serous ovarian cancer. Int J Gynecol Cancer 2011;21:975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Inaguma S, Riku M, Hashimoto M, Murakami H, Saga S, Ikeda H, Kasai K. GLI1 interferes with the DNA mismatch repair system in pancreatic cancer through BHLHE41-mediated suppression of MLH1. Cancer Res. 2013;73:7313–23. [DOI] [PubMed] [Google Scholar]
- 25. Liao X, Siu MK, Au CW, Wong ES, Chan HY, Ip PP, Ngan HY, Cheung AN. Aberrant activation of hedgehog signaling pathway in ovarian cancers: Effect on prognosis, cell invasion and differentiation. Carcinogenesis 2009;30:131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ke Z, Caiping S, Qing Z, Xiaojing W. Sonic hedgehog-Gli1 signals promote epithelial–mesenchymal transition in ovarian cancer by mediating PI3K/AKT pathway. Med Oncol. 2015;32:1–9. [DOI] [PubMed] [Google Scholar]
- 27. Chen Q, Xu R, Zeng C, Lu Q, Huang D, Shi C, Zhang W, Deng L, Yan R, Rao H. Down-regulation of Gli transcription factor leads to the inhibition of migration and invasion of ovarian cancer cells via integrin β4-mediated FAK signaling. PLoS One 2014;9:e88386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suyama K, Onishi H, Imaizumi A, Shinkai K, Umebayashi M, Kubo M, Mizuuchi Y, Oda Y, Tanaka M, Nakamura M, Katano M. CD24 suppresses malignant phenotype by downregulation of SHH transcription through STAT1 inhibition in breast cancer cells. Cancer Lett. 2016;374:44–53. [DOI] [PubMed] [Google Scholar]
- 29. Wang YH, Sui XM, Sui YN, Zhu QW, Yan K, Wang LS, Wang F, Zhou JH. BRD4 induces cell migration and invasion in HCC cells through MMP-2 and MMP-9 activation mediated by the Sonic hedgehog signaling pathway. Oncol Lett. 2015;10:2227–32. [DOI] [PMC free article] [PubMed] [Google Scholar]