Abstract
miR-101-3p has been identified as a tumor suppressor in several cancers, but its exact role in gastric adenocarcinoma is still largely unknown. In this study, we found that, compared with the RGM-1 human normal gastric epithelial cells, miR-101-3p was significantly downregulated in all six human gastric adenocarcinoma cell lines, including BGC-823, MNK-45, MGC-803, SGC-7901, AGS, and HGC-27. Overexpression of miR-101-3p suppressed both the proliferation and invasion of AGS gastric adenocarcinoma cells, and knockdown of miR-101-3p displayed the opposite effect. In addition, miR-101-3p could directly target and suppress the expression of the serum response factor (SRF) gene, which is a transcription factor of HOTAIR, a well-characterized tumor promoter lncRNA. miR-101-3p negatively regulated SRF-mediated transcription of HOTAIR. Moreover, silencing of either SRF or HOTAIR could counteract the promotion of gastric adenocarcinoma cell proliferation and invasion by miR-101-3p inhibition. Our findings indicate that miR-101-3p suppresses HOTAIR-induced proliferation and invasion through directly targeting SRF in gastric carcinoma cells.
Key words: miR-101-3p, Gastric carcinoma, Proliferation and invasion, Serum response factor (SRF), HOX transcript antisense RNA (HOTAIR)
INTRODUCTION
Gastric cancer has the third highest mortality of all malignant tumors worldwide, and it is even worse in East Asian countries such as China and Japan1. Although the incidence of gastric cancer has declined slightly in recent years in China, the trend for younger patients with gastric cancer makes it more threatening. Gastric carcinoma, accounting for about 95% of malignant gastric tumors, has posed an enormous clinical challenge because most gastric carcinoma cases are diagnosed in the advanced stages, when prognosis is poor and treatment options are limited2. Moreover, its pathogenesis is still not clear. Therefore, it is urgent to investigate the regulatory networks in the initiation and development of gastric carcinoma and excavate potential therapeutic targets.
Noncoding RNAs (ncRNAs) are a large collection of RNA transcripts that are not able to encode proteins. They are categorized in a few groups, including “age-old” ribosomal RNAs (rRNAs) and transfer RNAs (rRNAs), and recently discovered microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), Piwi-interacting RNAs (piRNAs), and long ncRNAs (lncRNAs). In recent years, the interaction between ncRNAs and coding genes has aroused much interest in researchers studying the pathogenesis of many diseases. Among the ncRNAs, lncRNAs and miRNAs are two of the most representative groups of regulatory ncRNAs. lncRNAs exhibit large differences in length and are famous for their complex regulatory manners3, whereas the molecular size of miRNAs is about 20 nucleotides, and their regulatory manner is relatively simple4. lncRNAs and miRNAs may interact directly or indirectly with each other in the regulatory networks of progression in a number of diseases, including gastric carcinoma5,6.
HOX transcript antisense RNA (HOTAIR) is a relatively well-characterized lncRNA and is the first example of an RNA transcribed from one chromosome but influences transcription on another chromosome7. HOTAIR has been closely linked to cancer progression. It was demonstrated that high levels of HOTAIR were strongly associated with metastasis, poor prognosis, and recurrence in multiple cancers, including breast cancer, esophageal cancer, colorectal cancer, pancreatic cancer, and liver cancer8–12. HOTAIR was also dysregulated, and a higher level of HOTAIR has been linked to more capacity for proliferation, metastasis, and drug resistance in gastric cancer13,14. Overexpression of HOTAIR definitely increases gastric cancer carcinogenesis, invasion, and drug resistance, whereas knockdown of HOTAIR suppresses its proliferation and invasion15–17. HOTAIR has been regarded to be a therapeutic target for gastric cancer.
miR-101-3p has been reported to be a tumor suppressor miRNA in hepatocellular carcinoma, gastric adenocarcinoma, salivary gland adenoid cystic carcinoma, and pancreatic cancer18–21. However, the exact role of miR-101-3p in the progression of gastric adenocarcinoma is almost unknown. In this study, the effect of miR-101-3p on cell proliferation and invasion in gastric adenocarcinoma was explored. Furthermore, we investigated whether this effect is associated with HOTAIR.
MATERIALS AND METHODS
Cell Culture
RGM-1 human normal gastric epithelial cell line and six human gastric adenocarcinoma cell lines, including BGC-823, MNK-45, MGC-803, SGC-7901, AGS, and HGC-27, were purchased from Shanghai Institutes for Biological Sciences (Shanghai, P.R. China). The cells were cultured in RPMI-1640 medium containing 4.5 g/L glucose and 4 mmol/L l-glutamine (Invitrogen, Carlsbad, CA, USA) and supplemented with 10% fetal bovine serum (FBS; Invitrogen). The cells were incubated in a humidified incubator with an atmosphere of 95% air–5% CO2 at 37°C until adhesion.
Transfection
Oligo miR-101-3p mimics, antagomirs, and relative negative controls (NCs) as well as single-strand siRNAs against serum response factor (SRF) and HOTAIR were synthesized and confirmed to be effective by Ribobio Company (Guangzhou, P.R. China). The pcDNA-SRF expression and pcDNA3.1 empty vectors were constructed by GenScript Company (Nanjing, P.R. China). For transfection, cells were seeded into six-well plates at a density of 105/cm2. On reaching 70% confluence, 60 nM miR-101-3p mimics, 60 nM miR-101-3p antagomirs, 60 nM relative NCs, 40 nM siRNAs, or 2 μg of vectors was transfected into the cells with Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. The medium was changed after 24 h, and after incubation for another 48 h, the cultures were applied in the following assays.
Cell Proliferation and Invasion Analysis
Cell proliferation was evaluated using the cell counting kit-8 (Sigma-Aldrich, St. Louis, MO, USA) assay that was carried out according to the manufacturer’s instructions.
Cell invasion was detected with the Transwell cell invasion assay. Following treatment, cells were incubated in serum-free medium for 24 h, and trypsin was then added to stimulate the retraction of cytoplasm and disconnection of cell-to-cell adhesion. Cells were collected by centrifugation and seeded into the plate at a concentration of 105/well. Transwell chambers (Corning, Corning, NY, USA) precoated with 1:8 Matrigel (Sigma-Aldrich) were filled with serum-free medium (up to 0.2 ml/well). Serum-containing medium was then added into each lower chamber, followed by 12-h incubation at 37°C. Cells on the surface of the upper chamber were removed by scraping with a cotton swab. The cells that invaded through the membrane were fixed in 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet for 30 min. The cells that crossed the membrane were counted under a light microscope (Olympus, Tokyo, Japan).
Real-Time Quantitative PCR
Total RNA was isolated using TRIzol reagent (Invitrogen). Real-time quantitative PCRs (qPCRs) were carried out in a final volume of 25 μl using SYBR Premix Ex Taq (TaKaRa, Dalian, P.R. China), 0.4 mM of each primer, and 200 ng of a cDNA template. miR-101-3p stem-loop primer and quantitative primers, as well as U6 RNA primers, were designed and produced by Ribobio Company. The primers for HOTAIR were designed and produced by GenScript Company. Each individual sample was run in triplicate wells. PCR amplification cycles were performed using iQ™5 Multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The reactions were initially denatured at 95°C for 1 min followed by 35 cycles of 95°C for 15 s, and 60°C for 60 s. The data were calculated using the 2−ΔΔCt method. Enrichment of the RNAs was normalized to the U6 RNA.
3′-UTR Luciferase Reporter Assay
The cDNA fragment corresponding to the 3′-UTR of SRF mRNA that contains the binding site of miR-101-3p was cloned into psiCHECK™-2 Vectors (Promega, Madison, WI, USA) between the XhoI and NotI sites. The constructs were confirmed by sequencing. HEK293T cells (104)/well were seeded into 96-well plates. After 24 h, transfections were performed using Lipofectamine 3000 Transfection Reagent (Invitrogen). Constructs (100 ng) and either 50 nM of miR-101-3p mimics or NC were included in each well. Cells were harvested 48 h after transfection, and luciferase activity was measured using the DualGlo Luciferase Assay System (Promega).
Western Blotting
Protein (25 μg) from each sample was separated by 12% SDS-PAGE and electrotransferred to PVDF membrane (Millipore, Boston, MA, USA) for immunoblotting analysis. The SRF rabbit polyclonal antibody (1:400; Abcam, Cambridge, MA, USA) was used to incubate with the membrane at 4°C overnight. β-Actin was used as the internal reference. After incubation with the appropriate HRP-conjugated secondary antibody, the enrichment of the proteins was detected using a ChemiDoc XRS Imaging System (Bio-Rad).
Statistical Analysis
Data were obtained from four independent experiments in every assay. Values were expressed as means ± SEM. Multiple comparisons were assessed by one-way ANOVA followed by Dunnett’s tests with SPSS 22.0 software. Differences between groups were considered to be statistically significant with a value of p < 0.05.
RESULTS
miR-101-3p Was Downregulated in Human Gastric Adenocarcinoma Cell Lines
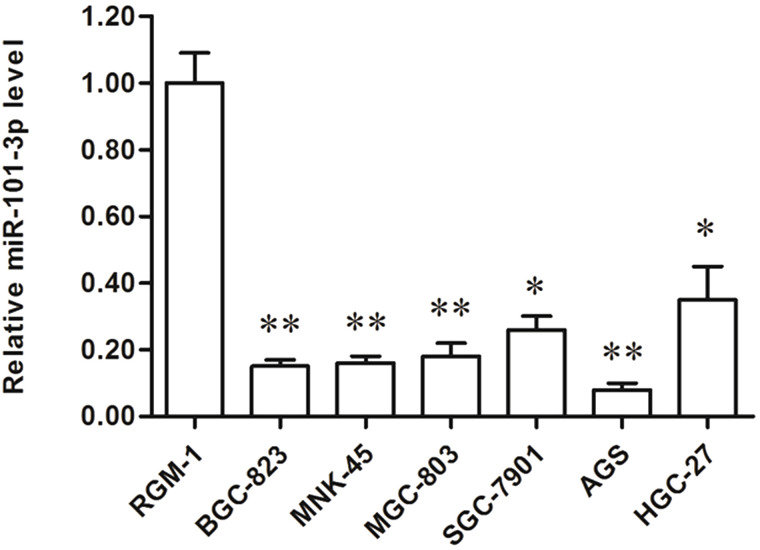
To investigate the role of miR-101-3p in the progression of human gastric adenocarcinoma, we first detected the levels of miR-101-3p in the RGM-1 human normal gastric epithelial cell line and in six human gastric adenocarcinoma cell lines, including BGC-823, MNK-45, MGC-803, SGC-7901, AGS, and HGC-27. Real-time qPCR analysis showed that the levels of miR-101-3p in the six gastric adenocarcinoma cell lines were all decreased by more than 60% compared with that in the RGM-1 cells (Fig. 1). It is worth mentioning that the decrease in miR-101-3p level was nearly 90% in AGS cells (Fig. 1). These results suggest that miR-101-3p might be negatively associated with the progression of gastric adenocarcinoma.
Figure 1.
miR-101-3p was significantly downregulated in human gastric adenocarcinoma cell lines. The expression of miR-101-3p was detected in RGM-1 human normal gastric epithelial cell line and in six human gastric adenocarcinoma cell lines, including BGC-823, MNK-45, MGC-803, SGC-7901, AGS, and HGC-27. *p < 0.05, **p < 0.01 versus RGM-1.
miR-101-3p Directly Targeted SRF and Negatively Regulated the Proliferation and Invasion in Human Gastric Adenocarcinoma Cells
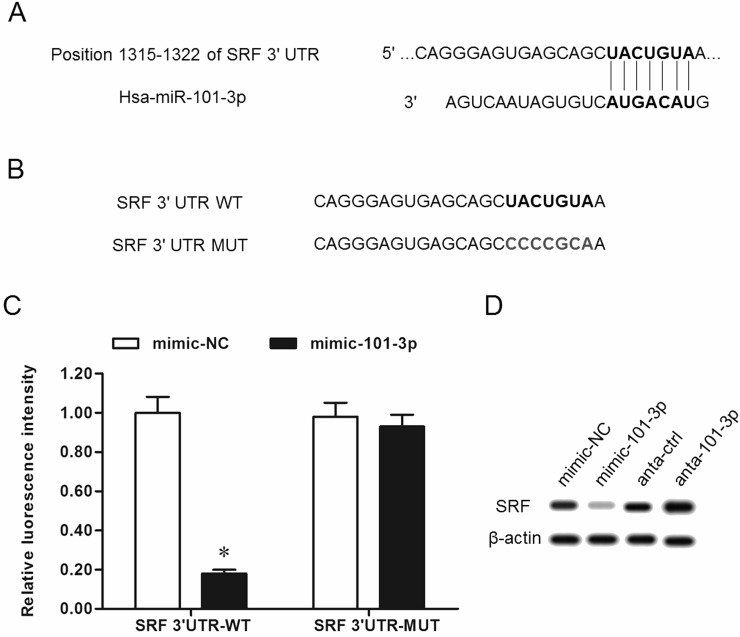
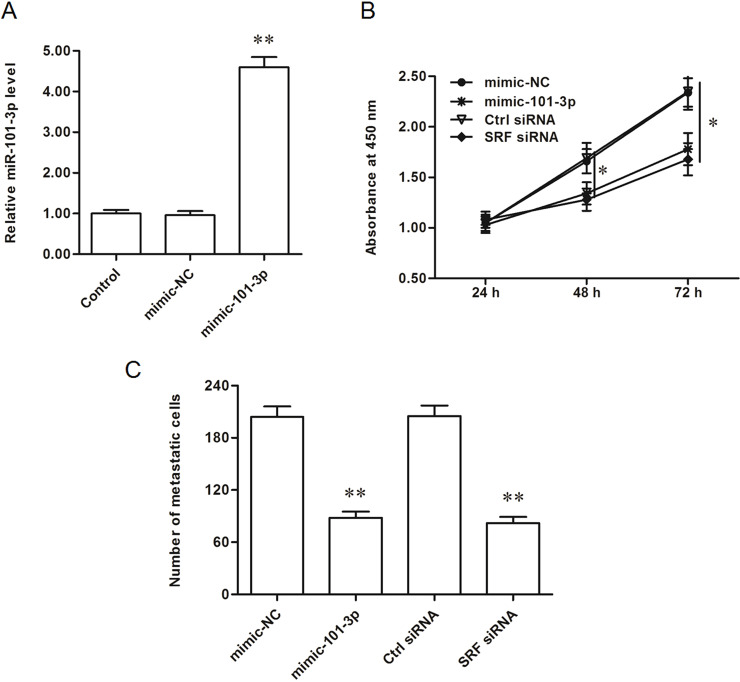
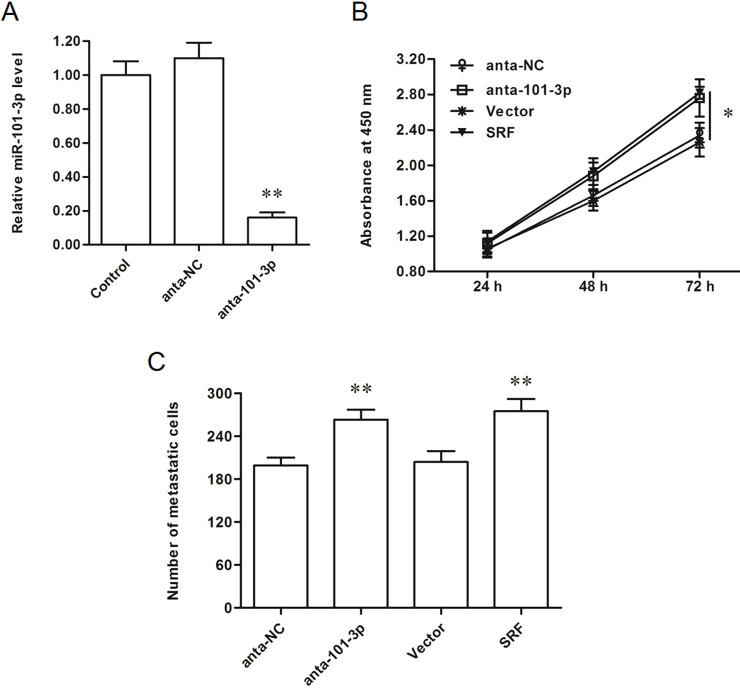
We searched the target genes of miR-101-3p, and the output from the online bioinformatic tool TargetScan showed that the seed sequence of miR-101-3p completely matched with seven continuous bases at the 3′-UTR of SRF mRNA (Fig. 2A). Wild-type (WT) SRF-3′-UTR and mutated (MUT) luciferase reporter vectors were, respectively, constructed (the mutated site is shown in Fig. 2B) and applied in the following luciferase reporter gene assay. The results showed that miR-101-3p significantly suppressed the luciferase activity of the WT SRF-3′-UTR luciferase reporter gene but could not change that of the MUT SRF-3′-UTR (Fig. 2C). Western blotting analysis showed that overexpression of miR-101-3p dramatically decreased and knockdown of miR-101-3p increased the expression of SRF protein (Fig. 2D). SRF was suggested to promote the progression of gastric cancer via multiple pathways, so miR-101-3p is likely to play a negative role in gastric adenocarcinoma progression. Gain- and loss-of-function experiments were performed in AGS gastric adenocarcinoma cells. The results showed that miR-101-3p mimics mediated overexpression of miR-101-3p and suppressed the proliferation and invasion of AGS cells (Fig. 3A–C). Similar to the effect miR-101-3p mimics, siRNA-mediated SRF silencing suppressed the proliferation and invasion of AGS cells (Fig. 3B and C). In contrast, miR-101-3p antagomirs mediated the inhibition of miR-101-3p and promoted the proliferation and invasion of AGS cells (Fig. 4A–C), which was consistent with the results from SRF overexpression (Fig. 4B and C).
Figure 2.
miR-101-3p directly targeted serum response factor (SRF) mRNA at the 3′-UTR region and suppressed SRF expression. (A) The output of the TargetScan online server about the binding of miR-101-3p with the 3′-UTR region of SRF mRNA. The bold letters represent the miR-101-3p seed sequence and its binding site. (B) Wild-type (WT) and mutated (MUT) SRF mRNA 3′-UTR sequences. The bold letters represent the WT and MUT sites. (C) miR-101-3p mimic suppressed the luciferase activity of the WT vector but not the MUT vector. The mimic-NC and mimic-101-3p were, respectively, cotransfected with a WT or mutant 3′-UTR luciferase vector into HEK293 cells. After incubation for 48 h, the luciferase activity in each group was measured. (D) miR-101-3p negatively regulated the expression of SRF. The 60 nM miR-101-3p mimics, miR-101-3p antagomirs, or relative negative controls were transfected into AGS cells. After incubation for 48 h, the level of SRF protein was detected with Western blotting. *p < 0.05.
Figure 3.
miR-101-3p overexpression decreased the proliferation and invasion of AGS gastric adenocarcinoma cell lines. (A) miR-101-3p expression was distinctly increased by the miR-101-3p mimic transfection. (B, C) The miR-101-3p mimics and SRF siRNA both reduced the proliferation and invasion of AGS cells. The 60 nM mimic-NC or miR-101-3p mimics were transfected into AGS human gastric adenocarcinoma cell lines. As a comparison, a ctrl-siRNA and a specific siRNA against SRF were also transfected into AGS cells. After incubation for 48 h, miR-101-3p expression was detected with real-time quantitative PCR (qPCR). Cell proliferation was detected with the cell counting kit-8 (CCK-8) assay, and cell invasion was detected with the Transwell invasion assay. *p < 0.05, **p < 0.01 versus mimic-NC or ctrl-siRNA.
Figure 4.
Inhibition of miR-101-3p increased the proliferation and invasion of AGS cells. (A) miR-101-3p level was sharply decreased by the anta-miR-101-3p transfection. (B, C) The anta-101-3p and SRF overexpression both significantly promoted the proliferation and invasion of AGS cells. The 60 nM control antagomir or miR-101-3p antagomirs were transfected into AGS human gastric adenocarcinoma cell lines. As a comparison, an empty pcDNA3.1 vector and a pcDNA-SRF expression vector were also transfected into AGS cells. After incubation for 48 h, miR-101-3p expression was detected with real-time qPCR. Cell proliferation was detected with the CCK-8 assay, and cell invasion was detected with the Transwell invasion assay. *p < 0.05, **p < 0.01 versus anta-NC or Vector.
miR-101-3p Decreased Expression of SRF-Mediated HOTAIR Expression
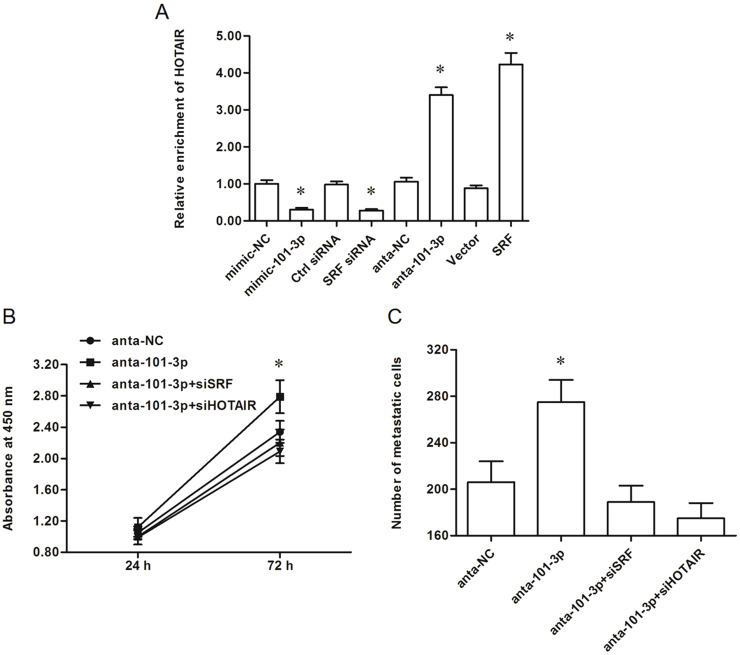
It was recently revealed that SRF is a transcription factor of lncRNA-HOTAIR. We subsequently investigated the effect of miR-101-3p on HOTAIR expression. The results showed that miR-101-3p overexpression and SRF knockdown suppressed HOTAIR expression in AGS cells (Fig. 5A). In contrast, miR-101-3p inhibition and SRF overexpression could both increase HOTAIR expression (Fig. 5A). These data indicated that miR-101-3p functions as a negative regulator in HOTAIR expression via suppression of SRF in AGS cells.
Figure 5.
The suppression of miR-101-3p on gastric adenocarcinoma cell proliferation and invasion was associated with SRF-mediated HOTAIR expression. (A) miR-101-3p negatively regulated SRF-mediated HOTAIR expression. Upon reaching 70% confluence, AGS cells were transfected with 60 nM miR-101-3p mimics, 60 nM miR-101-3p antagomirs, 60 nM relative NCs, 40 nM siRNAs, or 2 μg of vectors. After incubation for 48 h, HOTAIR expression was detected with real-time qPCR. (B, C) Silencing of either SRF or HOTAIR counteracted the promotion of gastric adenocarcinoma cell proliferation and invasion by the miR-101-3p antagomirs. The miR-101-3p antagomirs were transfected alone or cotransfected with SRF siRNA or HOTAIR siRNA into AGS cells. After incubation for 48 h, cell proliferation and invasion were detected. *p < 0.05 versus mimic-NC, ctrl-siRNA, anta-NC, or Vector.
Silencing SRF or HOTAIR Can Counteract the Promotion of Gastric Adenocarcinoma Cell Proliferation and Invasion by miR-101-3p Antagomirs
Finally, we researched whether the suppressive effect of miR-101-3p on gastric adenocarcinoma cell proliferation and invasion was associated with HOTAIR. The miR-101-3p antagomirs were cotransfected with SRF-siRNA or HOTAIR-siRNA. Cell proliferation and invasion assays showed that silencing of either SRF or HOTAIR could counteract the promotion of gastric adenocarcinoma cell proliferation and invasion by the miR-101-3p antagomirs (Fig. 5B and C). These data suggest that the suppressive effect of miR-101-3p on gastric adenocarcinoma progression is associated with SRF-mediated HOTAIR expression.
DISCUSSION
miR-101-3p has been identified as a tumor suppressor in several cancers, but its exact role in gastric adenocarcinoma is still largely unknown. A recent study revealed that miR-101 expression in gastric cancer tissues was significantly downregulated compared to matched normal tissues, and miR-101 exhibited a proapoptotic function in gastric cancer cells22. In our study, we found that miR-101-3p was significantly downregulated in all six human gastric adenocarcinoma cell lines compared with the normal gastric epithelial cells. Overexpression of miR-101-3p suppressed both the proliferation and invasion of AGS gastric adenocarcinoma cells, and knockdown of miR-101-3p displayed the opposite effect. Our findings indicate that miR-101-3p is also a tumor suppressor in gastric adenocarcinoma.
The function of miRNAs is basically dependent on the roles of their target genes. Existing reports indicate that miR-101-3p is able to directly target several tumor-promoter genes. For example, in triple negative breast cancer, mir-101-3p directly targets adenosine 5′-monophosphate (AMP)-activated protein kinase (AMPK) and regulates tumor metabolism19. mir-101-3p could also directly target and suppress the expression of the proto-oncogene enhancer of zeste homolog 2 (EZH2)20 and premature initiation of mitosis (Pim-1)21. In this study, SRF was identified as a novel target gene of miR-101-3p. SRF is a widely expressed transcription factor that has been involved in the progression of multiple cancers. In gastric cancer, high levels of SRF were associated with invasion and metastasis23. In addition, SRF could promote gastric cancer metastasis via a couple of pathways24,25. In this study, we also showed that SRF overexpression and miR-101-3p antagomir-mediated upregulation of SRF both promoted the proliferation and invasion of gastric adenocarcinoma cells, while SRF knockdown and miR-101-3p-mediated downregulation of SRF displayed the opposite effect.
As a representative lncRNA, HOTAIR emerges with a wealth of regulatory manners. It can bind with poly r(C)-binding protein (PCBP) 1 to form an RNA–protein complex26, or function as a competing endogenous RNA (ceRNA) to interact with several tumor suppressor miRNAs27–30. However, the regulation of HOTAIR expression in cancer progression has been poorly studied. Quite recently, it was reported that SRF regulated the transcription of lncRNA-HOTAIR through binding the CArG-box at the promoter region of HOTAIR gene in breast cancer cells31. Our data show that SRF overexpression and miR-101-3p antagomir-mediated upregulation of SRF both promoted the expression of HOTAIR in gastric adenocarcinoma cells, while SRF knockdown and miR-101-3p-mediated downregulation of SRF suppressed HOTAIR expression. Moreover, miR-101-3p antagomir-mediated promotion of gastric adenocarcinoma cell proliferation and invasion could both be counteracted by SRF and HOTAIR knockdown.
In conclusion, miR-101-3p suppresses the expression of HOTAIR and promotes the proliferation and invasion in gastric carcinoma cells through directly targeting SRF.
ACKNOWLEDGMENTS
This research was funded by the National Science Foundation of China (Nos. 81402523 and 81672990 to Xiaoyu Wu, and No. 81201797 to Gang Li), Jiangsu Province ‘‘Six Talents’’ High Peak Plan (No. 2015-WSN-052 to Xiaoyu Wu), the Project of Jiangsu Provincial Bureau of Traditional Chinese Medicine (No. JD201510 to Xiaoyu Wu), the Six “1” Project of Jiangsu Province (No. LGY2016012 to Xiaoyu Wu), Youth Projects of Jiangsu Provincial Health Department (No. H201065 to Gang Li), and the Natural Science Foundation of Jiangsu Province (No. BK2009445 to Gang Li).
REFERENCES
- 1. Fock KM. Review article: The epidemiology and prevention of gastric cancer. Aliment Pharmacol Ther. 2014;40:250–60. [DOI] [PubMed] [Google Scholar]
- 2. Carcas LP. Gastric cancer review. J Carcinog. 2014;13:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shahandeh A. Molecular mechanisms of oncogenic long non-coding RNAs. Biosci Res. 2013;10:38–54. [Google Scholar]
- 4. Giovannetti E, Erozenci A, Smit J, Danesi R, Peters GJ. Molecular mechanisms underlying the role of microRNAs (miRNAs) in anticancer drug resistance and implications for clinical practice. Criti Rev Oncol Hematol. 2012;81:103–22. [DOI] [PubMed] [Google Scholar]
- 5. Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochimic Biophys Acta 2016;1859:169–76. [DOI] [PubMed] [Google Scholar]
- 6. Wan X, Ding X, Chen S, Song H, Jiang H, Fang Y, Li P, Guo J. The functional sites of miRNAs and lncRNAs in gastric carcinogenesis. Tumor Biol. 2015;36:521–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu ZL, Zhou GZ, Cao G, Jin L, Xie HW, Wang CM, Lv J, De W, Wu M, Cao XF. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52:908–15. [DOI] [PubMed] [Google Scholar]
- 9. Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6. [DOI] [PubMed] [Google Scholar]
- 10. Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–50. [DOI] [PubMed] [Google Scholar]
- 11. Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013;32:1616–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, Li S, Zhao JC, Yu J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 2015;35:2746–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gan L, Xu M, Zhang Y, Zhang X, Guo W. Focusing on long noncoding RNA dysregulation in gastric cancer. Tumor Biol. 2015;36:129–41. [DOI] [PubMed] [Google Scholar]
- 14. Emadiandani E, Nikpour P, Emadibaygi M, Bidmeshkipour A. Association of HOTAIR expression in gastric carcinoma with invasion and distant metastasis. Adv Biomed Res. 2014;3:135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Endo H, Shiroki T, Nakagawa T, Yokoyama M, Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, Iijima K, Shimosegawa T, Sugamura K, Satoh K. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS One 2013;8:e77070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L, Yu PF, Cheng XD. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci. 2013;9:587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Na KL, Lee JH, Chan HP, Yu D, Yong CL, Cheong JH, Noh SH, Lee SK. Long non-coding RNA HOTAIR promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochem Biophys Res Commun. 2014;451:171–8. [DOI] [PubMed] [Google Scholar]
- 18. Sheng Y, Li J, Zou C, Wang S, Cao Y, Zhang J, Huang A, Tang H. Downregulation of miR-101-3p by hepatitis B virus promotes proliferation and migration of hepatocellular carcinoma cells by targeting Rab5a. Arch Virol. 2014;159:2397–410. [DOI] [PubMed] [Google Scholar]
- 19. Liu P, Ye F, Xie X, Li X, Tang H, Li S, Huang X, Song C, Wei W, Xie X. mir-101-3p is a key regulator of tumor metabolism in triple negative breast cancer targeting AMPK. Oncotarget 2016;7:35188–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu D, Li Y, Luo G, Xiao X, Tao D, Wu X, Wang M, Huang C, Wang L, Zeng F, Jiang G. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 2016;18:281–91. [DOI] [PubMed] [Google Scholar]
- 21. Liu X, Liu Z, He H, Zhang C, Wang Y. MicroRNA-101-3p suppresses cell proliferation, invasion and enhances chemotherapeutic sensitivity in salivary gland adenoid cystic carcinoma by targeting Pim-1. Am J Cancer Res. 2015;5:3015–29. [PMC free article] [PubMed] [Google Scholar]
- 22. He XP, Shao Y, Li XL, Xu W, Chen GS, Sun HH, Xu HC, Xu X, Tang D, Zheng XF, Xue YP, Huang GC, Sun WH. Downregulation of miR-101 in gastric cancer correlates with cyclooxygenase-2 overexpression and tumor growth. FEBS J. 2012;279:4201–12. [DOI] [PubMed] [Google Scholar]
- 23. Zhao M, Dingjie XU, Hong XU. The clinical significance of SRF, E-cadherin, and β-catenin expression in gastric carcinoma. Chin J Clin Oncol. 2011. DOI: 10.3969/j.issn.1000-8179.2011.22.008 [Google Scholar]
- 24. Qiao J, Liu Z, Yang C, Gu L, Deng D. SRF promotes gastric cancer metastasis through stromal fibroblasts in an SDF1-CXCR4-dependent manner. Oncotarget 2016;7:46088–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao X, He L, Li T, Lu Y, Miao Y, Liang S, Guo H, Bai M, Xie H, Luo G, Zhou L, Shen G, Guo C, Bai F, Sun S, Wu K, Nie Y, Fan D. SRF expedites metastasis and modulates the epithelial to mesenchymal transition by regulating miR-199a-5p expression in human gastric cancer. Cell Death Differ. 2014;21:1900–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang ZZ, Shen ZY, Shen YY, Zhao EH, Wang M, Wang CJ, Cao H, Xu J. HOTAIR long noncoding RNA promotes gastric cancer metastasis through suppression of poly r(C)-binding protein (PCBP) 1. Mol Cancer Ther. 2015;14:1162–70. [DOI] [PubMed] [Google Scholar]
- 27. Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, De W, Wang KM, Wang ZX. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer 2014;13:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song B, Guan Z, Liu F, Sun D, Wang K, Qu H. Long non-coding RNA HOTAIR promotes HLA-G expression via inhibiting miR-152 in gastric cancer cells. Biochem Biophys Res Commun. 2015;464:807–13. [DOI] [PubMed] [Google Scholar]
- 29. Yan J, Dang Y, Liu S, Zhang Y, Zhang G. LncRNA HOTAIR promotes cisplatin resistance in gastric cancer by targeting miR-126 to activate the PI3K/AKT/MRP1 genes. Tumor Biol. 2016;37:16345–55. [DOI] [PubMed] [Google Scholar]
- 30. Liu Y, Sun M, Xia R, Zhang E, Liu X, Zhang Z, Xu T, De W, Liu B, Wang Z. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He H, Wei Z, Du F, Meng C, Zheng D, Lai Y, Yao H, Zhou H, Wang N, Luo X, Ma W, Zhang T. Transcription of HOTAIR is regulated by RhoC-MRTF-A-SRF signaling pathway in human breast cancer cells. Cell Signal. 2017;31:87–95. [DOI] [PubMed] [Google Scholar]