Abstract
Long noncoding RNA (lncRNA) colon cancer-associated transcript 2 (CCAT2) has been demonstrated to play an important role in diverse tumorigenesis. However, the biological function of lncRNAs in glioma is still unknown. In this study, we found that lncRNA CCAT2 was overexpressed in glioma tissues and cell lines and associated with tumor grade and size. Furthermore, patients with high levels of lncRNA CCAT2 had poorer survival than those with lower levels of lncRNA CCAT2. Knocking down lncRNA CCAT2 expression significantly suppressed the glioma cell growth, migration, and invasion, as well as induced early apoptosis of glioma cells in vitro. Moreover, lncRNA CCAT2 regulated epithelial–mesenchymal transition (EMT)-associated gene expression. In conclusion, lncRNA CCAT2 plays an important role in glioma tumorigenesis and progression and may act as a potential biomarker for therapeutic strategy and prognostic prediction.
Key words: Long noncoding RNAs (lncRNAs), Colon cancer-associated transcript 2 (CCAT2), Glioma, Proliferation, Invasion, Epithelial–mesenchymal transition (EMT)
INTRODUCTION
Glioma is the most prevalent and aggressive primary tumor of the nervous system and accounts for about 80% of primary malignant brain tumors1. Patients suffering from this disease exhibit an extremely poor prognosis with an unsatisfactory 5-year overall survival rate despite the advanced available treatments2. Although intensive research has been performed to detect and validate a number of molecules linked with glioma cell invasion and cell proliferation, only a few molecular mechanisms have been revealed and translated into clinical applications3. Therefore, there is an urgent need to discover novel and effective strategies and establish potential therapeutic targets for the early diagnosis and treatment of human glioma.
Long noncoding RNAs (lncRNAs) are highly conservative across mammalian species4. Anomalous expression of lncRNAs has been reported in a wide variety of human diseases including cancer5,6. A recent study indicated that lncRNA expression in glioma tissue was significantly altered by screening lncRNA expression profile7. lncRNAs, such as HOTAIR, MALAT1, PRNCR1, PCGEM1, CRNDE, and H19, have demonstrated that they play a critical role in the prognosis and progression of glioma8–12. However, the function of lncRNAs in glioma is only partially understood.
Colon cancer-associated transcript 2 (CCAT2), a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer13. lncRNA CCAT2 is overexpressed in tumor tissues and is associated with clinical characteristics and prognosis in various human cancers such as lung, esophageal squamous cell, gastric, cervical, ovarian, and bladder cancers14–19. lncRNA CCAT2 also promotes breast tumor growth by regulating the Wnt signaling pathway, upregulating cell migration, and downregulating chemosensitivity to 5-fluorouracil (5-FU) in a rs6983267-independent manner20,21. In addition, lncRNA CCAT2 functions as an oncogene in hepatocellular carcinoma, regulating cellular proliferation, migration, and apoptosis22. However, the role of lncRNA CCAT2 in the progression of glioma is unclear and needs to be investigated.
Our results showed that lncRNA CCAT2 expression levels were higher in tumor tissues than those in adjacent normal tissues and overexpressed in glioma cell lines. Moreover, the relatively higher expression of lncRNA CCAT2 was significantly associated with malignant status and poor prognosis of glioma patients. Knockdown of lncRNA CCAT2 remarkably inhibited growth, arrested migration and invasion, induced early apoptosis, and reduced the expression of epithelial–mesenchymal transition (EMT)-associated genes in glioma cells.
MATERIALS AND METHODS
Human Tissue Samples
A total of 128 glioma tissues were obtained from the Department of Neurosurgery, Taihe Hospital of the Hubei University of Medicine between January 2010 and May 2011. Nontumor brain tissue obtained during the tumor resection procedures and the adjacent tissue were defined as 2 cm away from the lesions. All specimens were frozen immediately in liquid nitrogen and stored at −80°C until RNA extraction. All glioma samples had confirmed pathological diagnosis and were divided into low grade (grades I–II) and high grade (grades III–IV) according to the WHO classification by neuropathologists. None of the patients from whom the samples were obtained had undergone preoperative chemotherapy or radiotherapy. Informed consents were obtained from all patients, and this study was approved by the Clinical Research Ethics Committee of the Taihe Hospital of Hubei University of Medicine.
Cell Culture
Normal human astrocyte (NHA) cells were purchased from Lonza (Basel, Switzerland). U87, U251, A172, and SHG44 cells were purchased from the Chinese Academy of Sciences (Shanghai, P.R. China). They were maintained in DMEM with high glucose and sodium pyruvate, supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin (GIBCO, Grand Island, NY, USA) in humidified air at 37°C with 5% CO2. All cell lines were tested and authenticated by DNA (short tandem repeat genotyping) profiling before use.
Quantitative Real-Time RT-PCR (qRT-PCR) Analysis
Total RNA was isolated from cancerous/noncancerous specimens or cultured cells with TRIzol reagents (Invitrogen, Carlsbad, CA, USA). One microgram of total RNA was reverse transcribed into first-strand cDNA using the PrimeScript RT Reagent Kit (TaKaRa Biotechnology, Dalian, P.R. China). Q-PCR was performed according to the manufacturer’s instructions on the SYBR Premix ExTaq Kit (TaKaRa) on an Mx3005P thermal cycler (Stratagene, San Diego, CA, USA). Results were normalized to GAPDH expression levels. The PCR primers were as follows: CCAT2, 5′-CCCTGGTCAAATTGCTTAACCT-3′ (forward) and 5′-TTATTCGTCCCTCTGTTTTATGGAT-3′ (reverse); E-cadherin, 5′-TGTAGTTACGTATTTATTTTTAGTGGCGTC-3′ (forward) and 5′-CGAATACGATCGAATCGAACCG-3′ (reverse); N-cadherin, 5′-CACTGCTCAGGACCCAGAT-3′ (forward) and 5′-TAAGCCGAGTGATGGTCC-3′ (reverse); β-catenin, 5′-TGCAGTTCGCCTTCACTATG-3′ (forward) and 5′-ACTAGTCGTGGAATGGCACC-3′ (reverse); vimentin, 5′-TGGATTCACTCCCTCTGGTT-3′ (forward) and 5′-GGTCATCGTGATGCTGAGAA-3′ (reverse); Snail, 5′-GAGGCGGTGGCAGACTAG-3′ (forward) and 5′-GACACATCGGTCAGACCAG-3′ (reverse); Twist, 5′-TGAATCTTGCTCAGCTTGTC-3′ (forward) and 5′-CGGGAGTCCGCAGTCTTA-3′ (reverse); GAPDH, 5′-CGCTCTCTGCTCCTCCTGTTC-3′ (forward) and 5′-ATCCGTTGACTCCGACCTTCAC-3′ (reverse). All primers were obtained from Invitrogen (Shanghai, P.R. China). GAPDH was used as internal controls for CCAT2. PCR cycling conditions for mRNA were 2 min at 50°C, 10 min at 95°C, followed by 45 cycles at 95°C for 15 s, and 60°C for 60 s. Conditions for miRNA were 10 min at 95°C followed by 45 cycles at 95°C for 10 s, 60°C for 20 s, and 72°C for 12 s. All qRT-PCRs were performed in triplicate. Relative quantification of gene expression was calculated by the 2−ΔΔCt method.
siRNA Transfection
U87 cells and U251 cells were plated on six-well plates at a density of 2 × 105 cells/well and grown overnight until 30–40% confluency. The cells were transfected with two validated siRNA for CCAT2 (si-CCAT2-1, 5′-GUGCAACUCUGCAAUUUAAUU-3′; si-CCAT2-2, 5′-UUAACCUCUUCCUAUCUCATT-3′) and a scramble negative control si-NC (synthesized by Invitrogen, Shanghai, P.R. China) at a concentration of 100 nM using Lipofectamine 2000 transfection reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. The medium was replaced with standard culture medium 6 h posttransfection. Transfection was repeated 72 h after the first transfection.
Cell Proliferation Assay
Cell proliferation was assayed using a cell proliferation kit, cell counting kit-8 (CCK-8; Dojindo Molecular technologies Inc., Kyushu, Japan) according to the manufacturer’s instructions. Cells were seeded into 96-well tissue culture plates at a density of 2 × 103 cells/well the day before transfection. Approximately 20 μl of the CCK8 reagent was added to each well after transfection and incubated at 37°C for 2 h. Cell growth was analyzed at a wavelength of 450 nm at 24, 48, 72, and 96 h after transfection using EnVision (PerkinElmer Genetics, Bridgeville, PA, USA). Experiments were performed in triplicate.
Clonogenic Assay
Tumor cells were plated in six-well plates at a density of 800–1,200 cells per well. When the colonies were visible by the naked eye after 10–14 days, the cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Colonies were counted by ImageJ software (NIH, Bethesda, MD, USA).
Annexin V/Propidium Iodide (PI) Staining for Detecting Apoptotic Cells
The cell apoptosis was determined by an annexin V/PI assay according to the protocol of the Annexin-V–FITC apoptosis kit (BD Biosciences, Carlsbad, CA, USA). The Annexin-V–FITC and PI fluorescence levels were measured by flow cytometry (BD Biosciences, FACSCalibur). The annexin V+ cells (both PI− and PI+) were defined as apoptotic cells.
Migration and Invasion Assay
BD 24-well Transwell chambers (Costar, Corning, NY, USA) with or without Matrigel coating were used to measure cell migration or invasion according to the manufacturer’s instructions. Cells (1 × 105 cells per well) suspended in 0.5 ml of serum-free medium were added to the upper compartment of the inserts in the 24-well plate, and medium supplemented with 10% FBS was applied to the lower compartment. After incubating for 24 h at 37°C in an atmosphere of 5% CO2, the cells attached to the lower membrane surface were fixed with 4% formaldehyde and stained with 1% crystal violet in PBS. The number of cells that invaded through the Matrigel-coated membrane was determined in five randomly selected high-power fields across the center and the periphery of the membrane. Experiments were performed in triplicate.
Western Blot Analysis
Cells were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris base, 5 mM EDTA, 1% NP-40, 0.25% deoxycholate, pH 7.4) with protease and phosphatase inhibitors (Roche, Complete Mini). Protein concentrations were measured by the BCA protein assay (23227; Thermo Fisher Scientific, Waltham, MA, USA). Equal amounts (30–50 μg) of the protein were electrophoresed by SDS-PAGE, transferred to NC membranes, and incubated with the following primary antibodies: anti-β-actin antibody (Cell Signaling Technology, Danvers, MA, USA), rabbit polyclonal anti-E-cadherin (A01589; GenScript, Edison, NJ, USA), mouse monoclonal anti-N-cadherin (BD Transduction, San Jose, CA, USA), rabbit polyclonal anti-vimentin (A01189; GenScript), rabbit monoclonal anti-β-catenin (8480; Cell Signaling Technology), rabbit monoclonal anti-Snail (3879; Cell Signaling Technology), and rabbit monoclonal anti-Twist (ab50581; Abcam, Cambridge, MA, USA). The primary antibody incubation was followed by incubation with an HRP-conjugated secondary antibody. The bound antibodies were detected using enhanced chemiluminescence reagent (32109; Thermo Fisher Scientific).
Statistical Analysis
The quantitative data are presented as the means ± standard deviation (SD). The chi-square test or Student’s t-test was used to assess the association between CCAT2 expression and clinicopathological characteristics. The Kaplan–Meier method with log-rank test was performed to estimate overall survival (OS). Two-tailed Student’s t-test was used for comparisons of two independent groups. One-way analysis of variance (ANOVA) was used to identify significant differences in multiple comparisons in vitro analyses. Statistical analysis was performed using SPSS 18.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA, USA). Statistically significant differences were defined as p < 0.05.
RESULTS
Expression of lncRNA CCAT2 Is Overexpressed in Glioma Tissues and Cell Lines
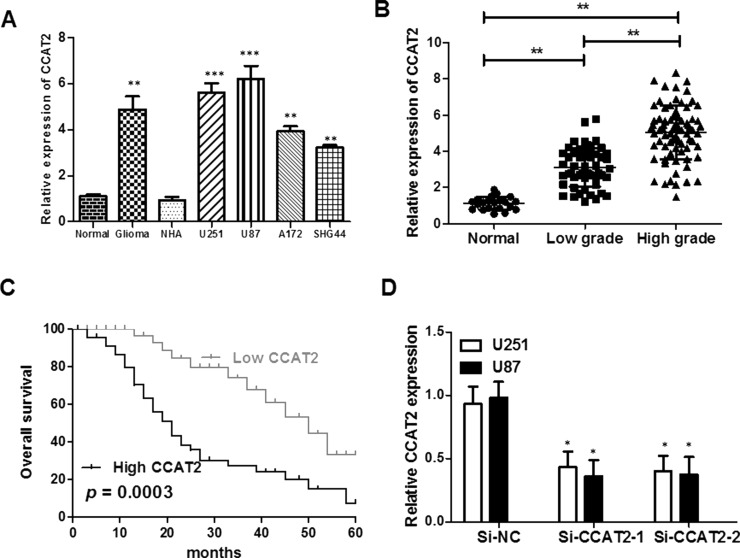
We detected the expression levels of lncRNA CCAT2 in glioma tissues and cell lines by real-time PCR. The lncRNA CCAT2 expression level was markedly increased in glioma samples compared with paired adjacent normal tissue samples (p < 0.01) (Fig. 1A and B). Additionally, the expression of lncRNA CCAT2 was increased in glioma cell lines (U87, U251, A172, and SHG44) compared with NHAs (p < 0.001). Furthermore, the expression levels of lncRNA CCAT2 were upregulated with the rising pathological grades of gliomas. The results suggested that lncRNA CCAT2 overexpression may be significant in glioma carcinogenesis.
Figure 1.
Expression of lncRNA CCAT2 is increased in glioma tissues and cell lines and associates with overall survival in glioma patients. (A) Expression of CCAT2 in human glioma cancer tissues and cell lines; expression of CCAT2 in human glioma tissues was higher than in noncancerous tissues, and lncRNA CCAT2 expression is increased in glioma cancer cell lines compared with normal glioma cell line. (B) Expression of CCAT2 in high-grade gliomas was higher than in low-grade gliomas. (C) High expression of lncRNA CCAT2 predicts a poor prognosis in glioma cancer patients. The relationship between lncRNA CCAT2 and glioma cancer patient survival was estimated using the Kaplan–Meier method and the log-rank test. (D) lncRNA CCAT2 expression is decreased by small interfering RNA (si-CCAT2) in U87 and U251 cells. *p < 0.05 compared with normal control; **p < 0.01 compared with normal control; ***p < 0.001 compared with normal control.
lncRNA CCAT2 Overexpression Is Associated With Malignant Status and Poor Prognostic Factor in Glioma Patients
We further analyzed the association between the expression of lncRNA CCAT2 and clinicopathological characteristics of glioma cancer patients. The 128 glioma patients were classified into the low-CCAT2 expression group (n = 65, CCAT2 expression ratio ≤ median ratio) and the high-CCAT2 expression group (n = 63, CCAT2 expression ratio ≥ median ratio) according to the median ratio of relative CCAT2 expression in all glioma samples. This classification was based on published studies by Ma et al.12 and Xie et al.23. The high-CCAT2 expression group was obviously correlated with tumor grade (low grades I–II vs. high grades III–IV, p = 0.006) and tumor size (<3 vs. ≥3 cm, p = 0.001). However, there were no significant associations between lncRNA CCAT2 expression and gender (p = 0.291), age (p = 0.608), family history of cancer (p = 0.461), or tumor location (p = 0.410). We used Kaplan–Meier analysis and log-rank test to explore the effects of lncRNA CCAT2 expression and the clinicopathological factors on OS. The level of lncRNA CCAT2 expression was remarkably correlated with the OS of glioma patients, as patients with higher levels of lncRNA CCAT2 expression had a shorter survival than those with lower levels of lncRNA CCAT2 expression (p = 0.0003) (Fig. 1C). Taken together, these data indicate that the lncRNA CCAT2 expression level is an independent risk factor for glioma patients.
Knockdown of lncRNA CCAT2 Inhibits Glioma Cell Proliferation and Promotes Apoptosis In Vitro
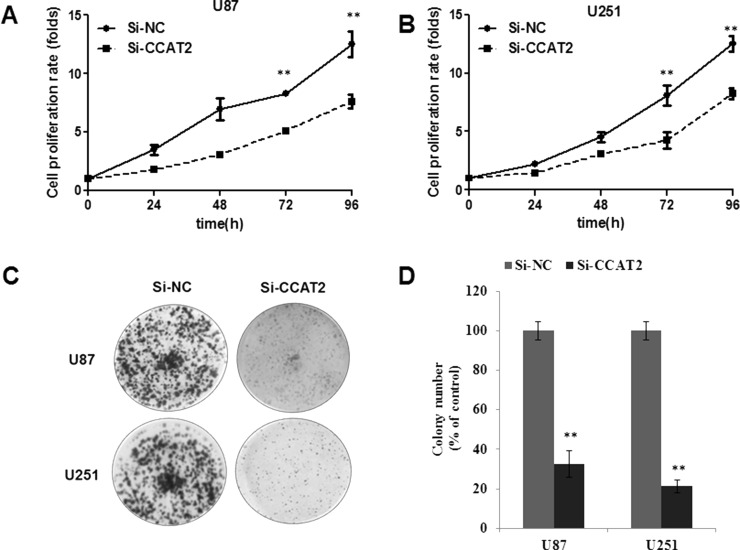
To assess the biological role of lncRNA CCAT2 in glioma, we investigated the effect of targeted knockdown of lncRNA CCAT2 on cell proliferation and apoptosis. lncRNA CCAT2 expression was relatively higher in U87 and U251 glioma cell lines than in A172 and SHG44 glioma cell lines (Fig. 1A). Therefore, we chose the U87 and U251 cell lines for the following loss-of-function studies. We induced downregulation of lncRNA CCAT2 expression induced by siRNA in U87 and U251, and these efficiencies were confirmed by qRT-PCR (Fig. 1D). Subsequently, we investigated the effect of knockdown lncRNA CCAT2 expression on glioma cell growth in vitro. The growth curves determined by CCK-8 assay showed that suppressing lncRNA CCAT2 significantly inhibited glioma cell viability (Fig. 2A and B). Meanwhile, the results of the CCK-8 assay were also consistent with clonogenicity tests as knockdown lncRNA CCAT2 has a remarkably reduced effect on the number of colonies compared to the si-NC group over a 10-day period (Fig. 2C and D). In addition, we performed flow cytometry to examine whether glioma cancer cell proliferation was induced by cell apoptosis. The results revealed that knockdown of lncRNA CCAT2 could obviously induce cell apoptosis. The proportion of apoptotic cells following lncRNA CCAT2 siRNA treatment was significantly increased in U87 and U251 cells (Fig. 3A and B), suggesting that CCAT2-mediated promotion of glioma cell proliferation seems to be mediated by regulation of apoptosis.
Figure 2.
Knockdown of lncRNA CCAT2 inhibits the growth of glioma cancer cells. (A, B) si-CCAT2 decreases viability of U87 and U251 cells through CCK-8 assay in vitro. (C, D) Knockdown lncRNA CCAT2 expression inhibits the proliferative ability of U87 and U251 cells through colony formation assay. Data represent the mean ± SD from three independent experiments. **p < 0.01 compared with the si-NC.
Figure 3.
Knockdown of lncRNA CCAT2 increases apoptosis of U87 and U251 cells. (A) Knockdown of CCAT2 induced apoptosis in U87 and U251 cells. (B) Quantification of the rate of apoptosis assay is presented. Representative images and data based on three independent experiments. Data are mean ± SD. *p < 0.05 compared with si-NC; **p < 0.01 compared with si-NC.
Knockdown of lncRNA CCAT2 Inhibits Glioma Cell Migration and Invasion
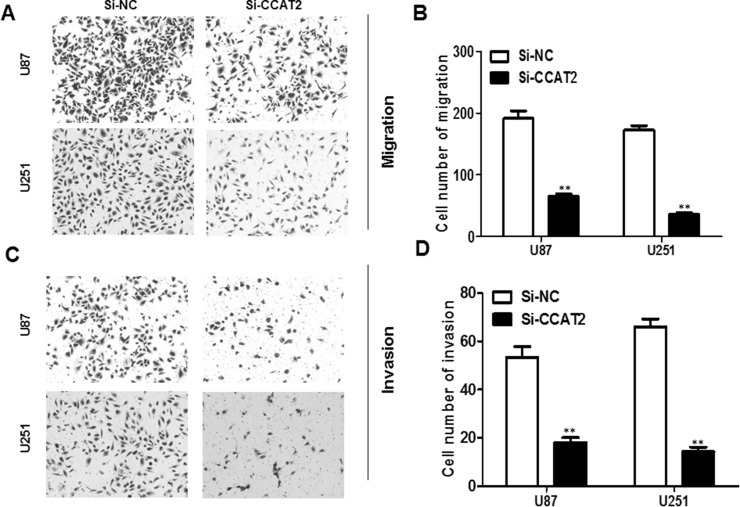
To examine the effect of lncRNA CCAT2 on cell migration, after 24 h of transfection the number of migrated cells in both si-CCAT2 glioma cell groups was significantly less than that in the si-NC glioma cells (for both p < 0.01) (Fig. 4A and B). Using a Boyden chamber coated with Matrigel, we determined changes in cell invasiveness after 24 h of transfection. Compared with the si-NC glioma cells, the si-CCAT2 glioma cells showed significantly reduced invasiveness (for both p < 0.01) (Fig. 4C and D).
Figure 4.
Knockdown of lncRNA CCAT2 inhibits glioma cell migration and invasion. (A, B) Downregulated lncRNA CCAT2 expression dramatically decreased the ability of U87 and U251 cell migration in vitro. (C, D) Suppressed lncRNA CCAT2 expression inhibited invasion of U87 and U251 cells. Data represent the mean ± SD from three independent experiments. **p < 0.01 compared with the si-NC.
lncRNA CCAT2 Regulates the Expression of EMT-Associated Genes in Glioma Cells
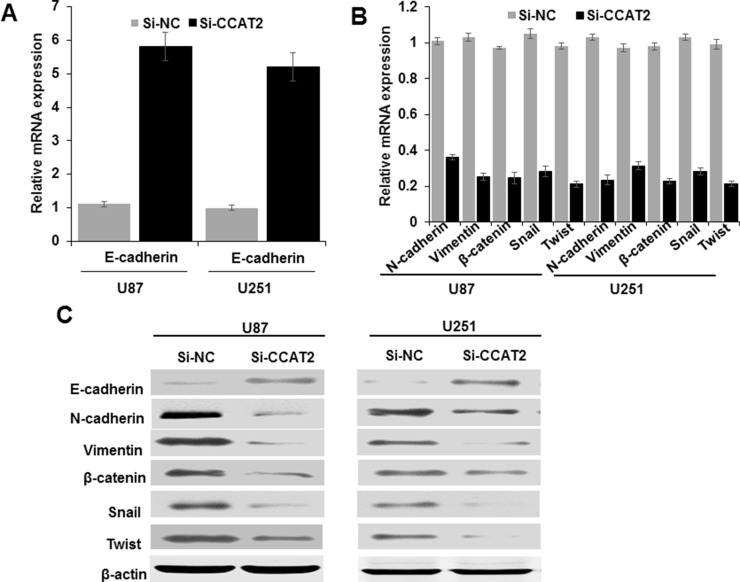
To further explore the mechanism by which lncRNA CCAT2 regulates cell migration and invasion, we examined protein levels of metastasis- and EMT-associated genes in glioma cells with knockdown of lncRNA CCAT2 expression by qRT-PCR and Western blotting. We found that suppressing lncRNA CCAT2 expression increased the expression of epithelial marker genes, including E-cadherin, and decreased the expression of mesenchymal marker genes including vimentin, N-cadherin, Twist, β-catenin, and SNAIL, suggesting that knockdown of lncRNA CCAT2 could inhibit the EMT process (Fig. 5A–C). Collectively, the dysregulation of the expression of EMT-related genes partially explains the involvement of lncRNA CCAT2 in glioma cell migration and invasion.
Figure 5.
Effect of lncRNA CCAT2 on EMT-related gene expression in glioma cells. U87 and U251 cells were transfected with CCAT2-specific si-RNA and si-NC for 48 h. The expression of EMT-associated genes including E-cadherin, N-cadherin, vimentin, β-catenin, SNAIL, and TWIST was analyzed by (A, B) qRT-PCR and (C) Western blotting. Each assay was performed in triplicate. Data are mean ± SD.
DISCUSSION
lncRNAs have a crucial effect on the development of the malignant phenotype of glioma cells, including cell survival, proliferation, differentiation, tumor angiogenesis, and stem cell generation24. Detection of the clinical and biological significance of lncRNA dysregulation in glioma cells not only contributes to identifying mechanistic components underlying the pathogenesis of the disease but also provides a novel insight for the role of lncRNA in tumorigenesis and cancer progression7,25.
In our study, we detected the lncRNA CCAT2 expression in glioma and further explored the functional role and possible mechanism of lncRNA CCAT2. Despite great progress that has been made in early diagnosis, surgical techniques, and chemotherapy, the prognosis of patients with glioma cancer is still poor. Aberrant expression of lncRNAs has been found to be associated with glioma progression. So there is an urgent need for the identification of novel biomarkers, especially glioma-associated lncRNAs, in order to realize early diagnosis of glioma and to work out effective methods with precise targets, improving patients’ prognosis.
Pervasive studies showed that lncRNA CCAT2 is aberrantly expressed and associated with tumor development and prognosis in various cancers, indicating that CCAT2 is tissue specific and may function as an oncogene in cancer. qRT-PCR first confirmed that CCAT2 expression was strikingly increased in tumor tissues from 128 patients with glioma and a panel of glioma cell lines compared with matched adjacent normal tissues and an NHA cell line. Meanwhile, Kaplan–Meier analysis and log-rank test showed that the prognosis of glioma patients with a high expression level of CCAT2 was poorer than those with a low expression level of CCAT2, and univariate and multivariate Cox regression analysis demonstrated that high expression of CCAT2 was a significant prognostic factor for poor OS of glioma patients. This was similar to a report from Zhang et al.26 that showed a high level of lncRNA CCAT2 to be significantly associated with TNM stages, lymph node metastasis (LNM), and poorer OS in esophageal squamous cell carcinoma. In addition, Huang et al.18 reported that higher CCAT2 expression levels were associated with a shorter OS and disease-free survival in ovarian cancer patients. Thus, our results suggest that CCAT2 may serve as a marker for poor prognosis in glioma.
In order to explore the biological function of lncRNA CCAT2 in glioma, we performed a series of experiments to investigate the role of CCAT2 in glioma cell lines. U87 and U251 cells were chosen to be performed in the following experiment due to the top two expression levels of CCAT2. A series of functional analysis was done after silencing CCAT2 expression in U87 and U251 cells. It has been observed that knockdown of lncRNA CCAT2 inhibited cell proliferation of U87 and U251 cells, promoted cell apoptosis, and impaired metastatic ability. EMT is a main mechanism involved in cell migration and invasion27. We then investigated the expression of EMT-associated genes. Interestingly, CCAT2 knockdown upregulated E-cadherin expression while it downregulated N-cadherin, vimentin, β-catenin, SNAIL, and TWIST expression. These illuminated that CCAT2 affects glioma cell proliferation and metastasis partly through the EMT. In non-small cell lung cancer, Qiu et al.14 demonstrated that knockdown of lncRNA CCAT2 inhibited the proliferation of tumor cells and reduced their ability to migrate and invade. Similarly, knockdown of lncRNA CCAT2 reduced bladder cancer cell proliferation, invasion, and migration19. Thus, our study suggested that lncRNA CCAT2 is an important regulator in controlling glioma cell metastasis. This study advances our understanding of the role of CCAT2 as a regulator of glioma pathogenesis.
In summary, we demonstrated that lncRNA CCAT2 was obviously elevated in glioma tissues and cells lines and correlated with the malignant status and prognosis in glioma patients. Furthermore, inhibition of lncRNA CCAT2 could remarkably decrease glioma cancer cell proliferation and migration and regulate EMT-associated gene expression. Because lncRNA CCAT2 plays this pivotal role in the progression of glioma, it holds great promise as a potential therapeutic target for glioma.
ACKNOWLEDGMENTS
The present study was supported by the National Natural Science Foundation of China (Grant No. 81401447) and the Key Discipline Project of Hubei University of Medicine.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Lefranc F, Rynkowski M, DeWitte O, Kiss R. Present and potential future adjuvant issues in high-grade astrocytic glioma treatment. Adv Tech Stand Neurosurg. 2009;34:3–35. [DOI] [PubMed] [Google Scholar]
- 3. Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. [DOI] [PubMed] [Google Scholar]
- 4. Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–61. [DOI] [PubMed] [Google Scholar]
- 7. Zhang XQ, Leung GK. Long non-coding RNAs in glioma: Functional roles and clinical perspectives. Neurochem Int. 2014;77:78–85. [DOI] [PubMed] [Google Scholar]
- 8. Park JY, Lee JE, Park JB, Yoo H, Lee SH, Kim JH. Roles of long non-coding RNAs on tumorigenesis and glioma development. Brain Tumor Res Treat. 2014;2:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiang X, Yan Y, Hu M, Chen X, Wang Y, Dai Y, Wu D, Wang Y, Zhuang Z, Xia H. Increased level of H19 long noncoding RNA promotes invasion, angiogenesis, and stemness of glioblastoma cells. J Neurosurg. 2016;124:129–36. [DOI] [PubMed] [Google Scholar]
- 10. Zhou X, Ren Y, Zhang J, Zhang C, Zhang K, Han L, Kong L, Wei J, Chen L, Yang J, Wang Q, Zhang J, Yang Y, Jiang T, Li M, Kang C. HOTAIR is a therapeutic target in glioblastoma. Oncotarget 2015;6:8353–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367:122–8. [DOI] [PubMed] [Google Scholar]
- 12. Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P, Wu JW. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. 2015;36:3355–9. [DOI] [PubMed] [Google Scholar]
- 13. Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, Ivan C, Barbarotto E, De Vries I, Zhang X, Ferracin M, Churchman M, van Galen JF, Beverloo BH, Shariati M, Haderk F, Estecio MR, Garcia-Manero G, Patijn GA, Gotley DC, Bhardwaj V, Shureiqi I, Sen S, Multani AS, Welsh J, Yamamoto K, Taniguchi I, Song MA, Gallinger S, Casey G, Thibodeau SN, Le Marchand L, Tiirikainen M, Mani SA, Zhang W, Davuluri RV, Mimori K, Mori M, Sieuwerts AM, Martens JW, Tomlinson I, Negrini M, Berindan-Neagoe I, Foekens JA, Hamilton SR, Lanza G, Kopetz S, Fodde R, Calin GA. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L, Yin R. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol. 2014;35:5375–80. [DOI] [PubMed] [Google Scholar]
- 15. Wang J, Qiu M, Xu Y, Li M, Dong G, Mao Q, Yin R, Xu L. Long noncoding RNA CCAT2 correlates with smoking in esophageal squamous cell carcinoma. Tumour Biol. 2015;36:5523–8. [DOI] [PubMed] [Google Scholar]
- 16. Wang CY, Hua L, Yao KH, Chen JT, Zhang JJ, Hu JH. Long non-coding RNA CCAT2 is up-regulated in gastric cancer and associated with poor prognosis. Int J Clin Exp Pathol. 2015;8:779–85. [PMC free article] [PubMed] [Google Scholar]
- 17. Wu L, Jin L, Zhang W, Zhang L. Roles of long non-coding RNA CCAT2 in cervical cancer cell growth and apoptosis. Med Sci Monit. 2016;22:875–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang S, Qing C, Huang Z, Zhu Y. The long non-coding RNA CCAT2 is up-regulated in ovarian cancer and associated with poor prognosis. Diagn Pathol. 2016;11:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li J, Zhuang C, Liu Y, Chen M, Zhou Q, Chen Z, He A, Zhao G, Guo Y, Wu H, Cai Z, Huang W. shRNA targeting long non-coding RNA CCAT2 controlled by tetracycline-inducible system inhibits progression of bladder cancer cells. Oncotarget 2016;7:28989–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cai Y, He J, Zhang D. Long noncoding RNA CCAT2 promotes breast tumor growth by regulating the Wnt signaling pathway. Onco Targets Ther. 2015;8:2657–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, Buiga R, Pop V, Irimie A, Fodde R, Bedrosian I, Martens JW, Foekens JA, Berindan-Neagoe I, Calin GA. CCAT2, a novel long non-coding RNA in breast cancer: Expression study and clinical correlations. Oncotarget 2013;4:1748–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou N, Si Z, Li T, Chen G, Zhang Z, Qi H. Long non-coding RNA CCAT2 functions as an oncogene in hepatocellular carcinoma, regulating cellular proliferation, migration and apoptosis. Oncol Lett. 2016;12:132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xie T, Liu P, Chen L, Chen Z, Luo Y, Chen X, Feng Y, Luo X. MicroRNA-15a down-regulation is associated with adverse prognosis in human glioma. Clin Transl Oncol. 2015;17:504–10. [DOI] [PubMed] [Google Scholar]
- 24. Kiang KM, Zhang XQ, Leung GK. Long non-coding RNAs: The key players in glioma pathogenesis. Cancers 2015;7:1406–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramos AD, Attenello FJ, Lim DA. Uncovering the roles of long noncoding RNAs in neural development and glioma progression. Neurosci Lett. 2016;625:70–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang X, Xu Y, He C, Guo X, Zhang J, He C, Zhang L, Kong M, Chen B, Zhu C. Elevated expression of CCAT2 is associated with poor prognosis in esophageal squamous cell carcinoma. J Surg Oncol. 2015;111:834–9. [DOI] [PubMed] [Google Scholar]
- 27. Ombrato L, Malanchi I. The EMT universe: Space between cancer cell dissemination and metastasis initiation. Crit Rev Oncog. 2014;19:349–61. [DOI] [PubMed] [Google Scholar]