Abstract
Intestinal Paneth cells are professional exocrine cells that play crucial roles in maintenance of homeostatic microbiome, modulation of mucosal immunity, and support for stem cell self-renewal. Dysfunction of these cells may lead to the pathogenesis of human diseases such as inflammatory bowel disease (IBD). Cdk5 activator binding protein Cdk5rap3 (also known as C53 and LZAP) was originally identified as a binding protein of Cdk5 activator p35. Although previous studies have indicated its involvement in a wide range of signaling pathways, the physiological function of Cdk5rap3 remains largely undefined. In this study, we found that Cdk5rap3 deficiency resulted in very early embryonic lethality, indicating its indispensable role in embryogenesis. To further investigate its function in the adult tissues and organs, we generated intestinal epithelial cell (IEC)-specific knockout mouse model to examine its role in intestinal development and tissue homeostasis. IEC-specific deletion of Cdk5rap3 led to nearly complete loss of Paneth cells and increased susceptibility to experimentally induced colitis. Interestingly, Cdk5rap3 deficiency resulted in downregulation of key transcription factors Gfi1 and Sox9, indicating its crucial role in Paneth cell fate specification. Furthermore, Cdk5rap3 is highly expressed in mature Paneth cells. Paneth cell-specific knockout of Cdk5rap3 caused partial loss of Paneth cells, while inducible acute deletion of Cdk5rap3 resulted in disassembly of the rough endoplasmic reticulum (RER) and abnormal zymogen granules in the mature Paneth cells, as well as loss of Paneth cells. Together, our results provide definitive evidence for the essential role of Cdk5rap3 in Paneth cell development and maintenance.
Subject terms: Ubiquitylation, Differentiation
Introduction
The intestinal epithelium consists of a single layer of epithelial cells, and its primary function includes absorbing nutrients and serving as a barrier against luminal pathogens. To maintain the intestinal integrity, intestinal stem cells (ISCs), located in the bases of crypts of Lieberkuhn, rapidly proliferate and differentiate into mature epithelial cells. Among major types of intestinal epithelial cells (IECs), absorptive enterocytes are mainly responsible for nutrient absorption, whereas professional secretory cells such as Paneth and goblet cells play crucial roles in maintaining intestinal homeostasis and mucosal immunity. Interspersed with ISCs at the base of crypts, Paneth cells are distinctive professional exocrine cells containing an extensive rough endoplasmic reticulum (RER) network and many zymogen granules. They synthesize and secrete a large quantity of antimicrobial peptides/proteins and inflammatory cytokines that are essential for shaping healthy gut microbiome and maintaining innate immunity1. Loss or impairment of Paneth cell function is often observed in the patients with inflammatory bowel disease, and contributes to the onset and progression of the disease2–4. Paneth cells also produce niche factors EGF, Wnt3, and Notch ligands to support Lgr5+ intestinal stem cells5. Moreover, Paneth cells can serve as key nutrient sensors to enhance stem cell self-renewal in response to calorie restriction, further highlighting their role as ISC niche to couple organismal nutritional status to stem cell function6. More recently, several studies have demonstrated that Paneth cells possess multipotency to transdifferentiate into other types of IECs and directly contribute to intestinal regeneration under stress conditions7–9. Therefore, elucidation of the molecular and cellular mechanisms that regulate Paneth cell development, function, and plasticity would be crucial for understanding of intestinal homeostasis and disease pathogenesis.
Cdk5 activator binding protein Cdk5rap3 (also known as C53 and LZAP) was originally identified as a binding protein of Cdk5 activator p35, CBP, and ARF protein10–12. It is highly conserved in multi-cellular organisms, and its orthologues are found in vertebrate, invertebrate and plants but not in yeast and bacteria. Previous studies indicate its involvement in a wide variety of signaling pathways, including NF-κB13, ARF/p5311,14, Wnt15–17, STAT318, DNA damage response19,20 and UFMylation21,22. Not surprisingly, it was reported to interact with various proteins, including p3510, CBP/p30012, Rel A13, Chk1/220, PAK423, ARF11, p38MAPK24, UFL1 (also known as RCAD, NLBP and Maxer)25–27, γ-tubulin28, and TIP-129. Clinical studies using cancer patient samples have suggested its tumor suppressing activity in head/neck and gastric cancers13,14,16,17,30, but the conflicting results were reported in liver cancer23,31. Despite its potential involvement in many important signaling pathways, Cdk5rap3’s physiological functions remain poorly defined. In a zebrafish model, Liu et al. have shown that Cdk5rap3 is essential for early zebrafish development32. More recenlty, Yang et al. reported that Cdk5rap3 deficiency in mice led to embyonic lethality possibly due to severe liver hypoplasia22. Hepatocyte-specific Cdk5rap3 knockout mice suffered post-weaning lethality, owing to serious hypoglycemia and impaired lipid metabolism22. These findings have demonstrated an essential role of Cdk5rap3 in both embryogenesis and organ development.
In this study, we found that Cdk5rap3 is highly expressed in intestinal Paneth cells. IEC-specific knockout of Cdk5rap3 led to nearly complete absence of Paneth cells and increased susceptibility to experimentally induced colitis. Cdk5rap3 deficiency impaired the development of intestinal stem cells into Paneth cell lineage. Furthermore, Paneth cell-specific deletion of Cdk5rap3 caused partial loss of Paneth cells, while its acute ablation resulted in disassembly of RER, abnormality of zymogen granules, and loss of mature Paneth cells. Taken together, our results clearly demonstrate that Cdk5rap3 is required in Paneth cell development and maintenance.
Results
Cdk5rap3 knockout leads to early embryonic lethality
To elucidate the physiological function of Cdk5rap3, we generated Cdk5rap3 knockout mice using ES cell clone from the Knockout Mice Project (KOMP). Insertion of the “lacZ-neo” (>20 kb) cassette between exons 5 and 6 disrupted normal RNA splicing, resulting in a knockout allele and a truncated protein product (we designate this allele as “m”) (Supplementary Fig. S1a, b). Crossing of heterozygous mice failed to produce the pups with homozygous knockout alleles, suggesting that Cdk5rap3 deficiency causes embryonic lethality (Supplementary Fig. 1c). Indeed, no Cdk5rap3 deficient embryos were recovered from timed pregnant mice after E8.5, a result that seems to be consistent with the report by Liu et al.32.
IEC-specific deletion of Cdk5rap3 leads to complete depletion of Paneth cells
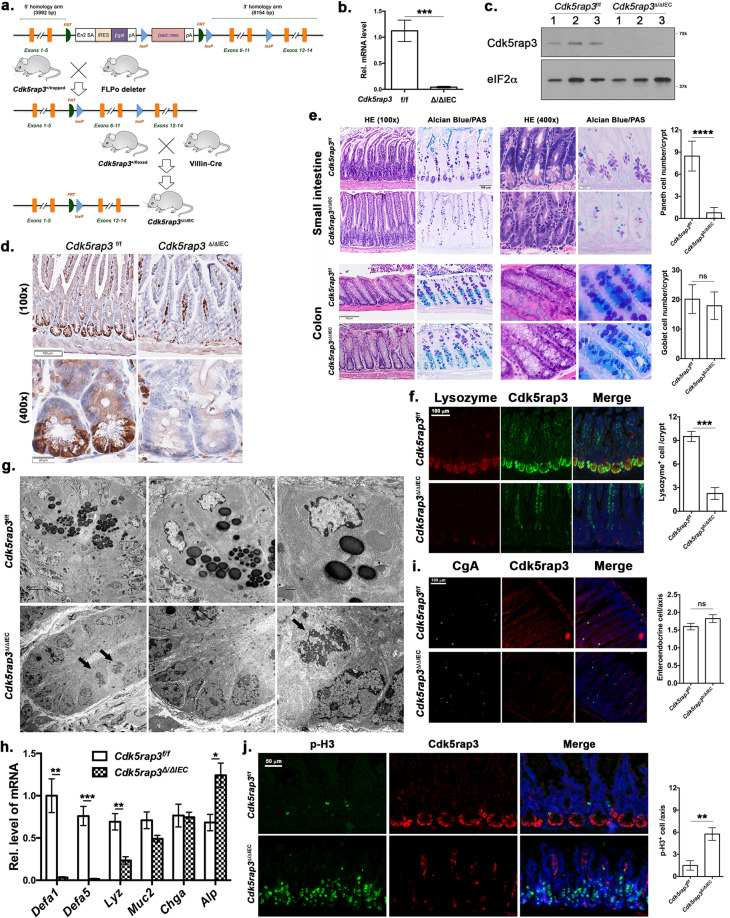
To further investigate Cdk5rap3’s function in the adult animal, we first crossed Cdk5rap3+/m mice with FLPo deletor mice to remove the “lacZ-neo” cassette and create floxed Cdk5rap3 allele. Subsequently, we crossed Cdk5rap3 floxed mice with CAG-CreERT2 transgenic mice to generate the whole-body conditional KO mice. Upon tamoxifen (TAM) administration, Cdk5rap3 deficient mice were viable, but bleeding in the gut was frequently observed. To explore the impact on the intestinal epithelium, we crossed Cdk5rap3 floxed mice with Villin-Cre transgenic mice to generate IEC-specific knockout mice (Fig. 1a). Deficiency of Cdk5rap3 in the intestinal epithelium was confirmed by quantitative RT-PCR (Fig. 1b), immunoblotting (Fig. 1c), and immunohistochemistry (Fig. 1d). Interestingly, we found that Cdk5rap3 was highly expressed in Paneth cells at the bottom of intestinal crypts (Fig. 1d).
Fig. 1. Intestinal epithelial cell (IEC)-specific deletion of Cdk5rap3 leads to ablation of Paneth cells.
a Scheme of mouse breeding to generate IEC-specific KO mice. b Quantitative RT-PCR analysis of Cdk5rap3 mRNA using the primers specific for floxed exons (n = 3 mice per genotype); Total RNA was isolated from intestinal crypts. c Immunoblotting of Cdk5rap3 in the lysate of intestinal epithelial cell lysates. d Immunohistochemistry of Cdk5rap3 in wild-type and Cdk5rap3 KO ileal sections. e Significant loss of Paneth cells in Cdkrap3∆/∆IEC intestine. The number of Paneth and goblet cells were counted in double-blinded fashion from more than 20 crypts, and 4 mice of each phenotypes were scored. ****p < 0.0001 (n = 4 mice per genotype). f Lysozyme staining of ileal sections of wild-type and Cdkrap3∆/∆IEC mice. Lysozyme-positive cells per crypt were scored. ***p < 0.001 (n = 4 mice per genotype). g Electron micrographs of the crypts of wild-type and Cdk5rap3∆/∆IEC mice. Mitotic cells with condensed chromatin were marked by solid arrows. h Quantitative RT-PCR analysis of cell type-specific gene expression. Total RNA was isolated from scraped cells of ileal section. *p < 0.05, **p < 0.01, ***p < 0.001 (n = 4 mice per genotype). i The number of enteroendocrine cells (Chromogranin A-positive) in wild-type and Cdkrap3∆/∆IEC intestine. The number of ChA+ cells per crypt-villus axis was counted and four mice of each phenotypes were scored. j Mitotic cells (phospho-Histone H3 Ser10 staining) in wild-type and Cdk5rap3∆/∆IEC ileal sections. The number of p-H3+ cells per crypt-villus axis was scored. **p < 0.01. (n = 4 mice per genotype).
IEC-specific Cdk5rap3 KO (referred to as Cdk5rap3∆/∆IEC hereafter) mice were born healthy without obvious developmental defects, growth retardation, and comprised gross epithelial organization (Fig. 1e). However, close examination of intestinal sections revealed substantial changes in the crypts. As shown by Alcian blue (AB)/Periodic acid–Schiff (PAS) staining, Cdk5rap3∆/∆IEC mice almost completely lacked Paneth cells at the bottom of the crypts in the small intestine (Fig. 1e). This phenotype was further confirmed by lysozyme staining (Paneth cell marker, Fig. 1f), transmission electron microscopy (TEM) of the crypts (Fig. 1g), and RT-PCR showing significant reduction of Paneth cell-specific gene expression (defensin 1, 5 and lysozyme) (Fig. 1h). In contrast to Paneth cells, the number of goblet cells in both small and large intestine was not significantly altered even though AB/PAS+ granules appeared to be smaller in the small intestine of Cdk5rap3 KO mice (Fig. 1e). In addition, Cdk5rap3 deficiency led to a slight increase in the number of enteroendocrine cells that failed to reach statistical significance (Chromogranin A positive cells) (Fig. 1i). Overall organization of enterocytes was not affected in Cdk5rap3∆/∆IEC mice, yet expression of enterocyte specific intestinal alkaline phosphatase was increased in Cdk5rap3∆/∆IEC intestine (Fig. 1h). Interestingly, the cells with condensed chromatin were frequently observed in Cdk5rap3∆/∆IEC crypts (marked by solid arrows), while such cells were rarely found in wild-type mice (Fig. 1g). These cells appeared to be in mitotic phase because more phospho-Histone H3 positive cells were observed in Cdk5rap3∆/∆IEC crypts than wild-type (Fig. 1j). A similar phenomenon was observed during development of Cdk5rap3 knockdown zebrafish embryos32.
We also took advantage of organoid culture to confirm the findings from the knockout mouse model. Interestingly, unlike Math1 deficient crypts without Paneth cells that could not grow ex vivo organoids in the absence of exogenous Wnt ligand33, Cdk5rap3 deficient crypts were able to proliferate and differentiate into well-formed organoids without exogenous Wnt ligand (Supplementary Fig. S2a). This is reminiscent of recently reported Lsd1 deficient organoid culture34. Cdk5rap3 deficient organoids did not contain Paneth cells (Supplementary Fig. S2b), and the absence of Paneth cells was further confirmed by lysozyme staining (Supplementary Fig. S2c) and RT-PCR analysis (Supplementary Fig. S2d). Therefore, the organoid culture of Cdk5rap3 deficient crypts truthfully recapitulated the phenotype of Cdk5rap3∆/∆IEC intestine. Collectively, our results strongly suggest that Cdk5rap3 is essential for Paneth cell development.
Cdk5rap3 ablation impairs fate specification of Paneth cell lineage
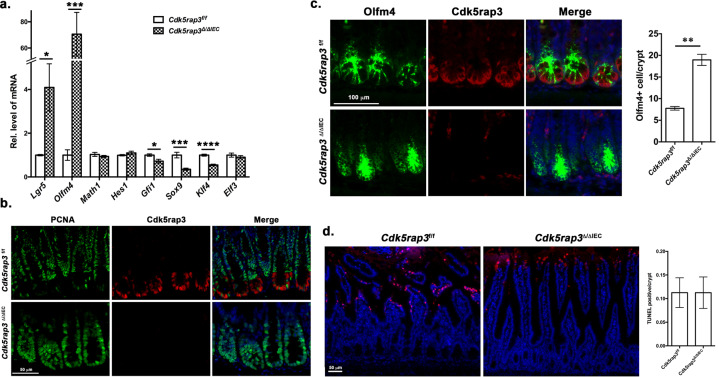
Paneth cells are derived from Math1/Atoh1 positive secretory progenitor cells, and their fate is determined by transcription factors (TFs) such as Gfi1 and Sox935–39. After lineage commitment, progenitor cells further develop into the mature Paneth cells that contain extensive rough ER network and zymogen granules. The Notch-Hes1 signaling represses Math1/Atoh1 and promotes absorptive enterocyte fate40–43. Transcription factor Elf3 is required for differentiation of enterocytes44, while Klf4, along with Elf3, is essential for terminal differentiation of goblet cells45. We examined whether Cdk5rap3 is involved in stem cell differentiation and Paneth cell specification. As showing Fig. 2a, both Gfi1 and Sox9, two critical TFs for Paneth cell lineage, were significantly down-regulated by Cdk5rap3 deficiency. In contrast, expression of Hes1 and Elf3 that are important for enterocyte development were not significantly altered by Cdk5rap3 ablation (Fig. 2a). Expression of Klf4 was also significantly reduced in Cdk5rap3∆/∆IEC mice, and this reduction may contribute to smaller goblet cells observed in Cdk5rap3∆/∆IEC small intestine (Fig. 1e). The result of TF expression profile suggests that Cdk5rap3 is critical for fate determination, and its deficiency impairs Paneth cell lineage specification. Interestingly, expression of Lgr5 and Olfm4, two intestinal stem cell markers, were also dramatically elevated in Cdk5rap3 KO intestine (Fig. 2a). While proliferating (PCNA+) intestinal stem cells (Olfm4+ cells) were located at the bottom of crypts and interspaced between Paneth cells in the wild-type mice, the crypt bases of Cdk5rap3∆/∆IEC intestine were occupied by proliferating (PCNA+) and Olfm4+ cells (Fig. 2b, c). Further study will determine whether Cdk5rap3 deficiency promotes ISC proliferation in a cell autonomous manner. We also examined cell death in Cdk5rap3∆/∆IEC small intestine and found no significant difference in the number of TUNEL positive cells (Fig. 2d).
Fig. 2. Cdk5rap3 deletion impairs fate specification of Paneth cell lineage.
a Quantitative RT-PCR analysis of specific genes involved in intestinal stem cell renewal and differentiation. Total RNA was isolated from intestinal scrapes and used for qRT-PCR analysis. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 (n = 4 mice per genotype). b Proliferating cells at the crypt base of wild-type and Cdk5rap3 deficient small intestine. Ileal sections were so-stained with PCNA and Cdk5rap3 antibodies. c Olfm4 staining of wild-type and Cdk5rap3∆/∆IEC ileal sections. Olfm4+ cells per crypt were scored and compared. **p < 0.01. (n = 4 mice per genotype). d TUNEL staining of wild-type and Cdk5rap3∆/∆IEC ileal sections. TUNEL positive cells per crypt were scored from four mice of each genotype.
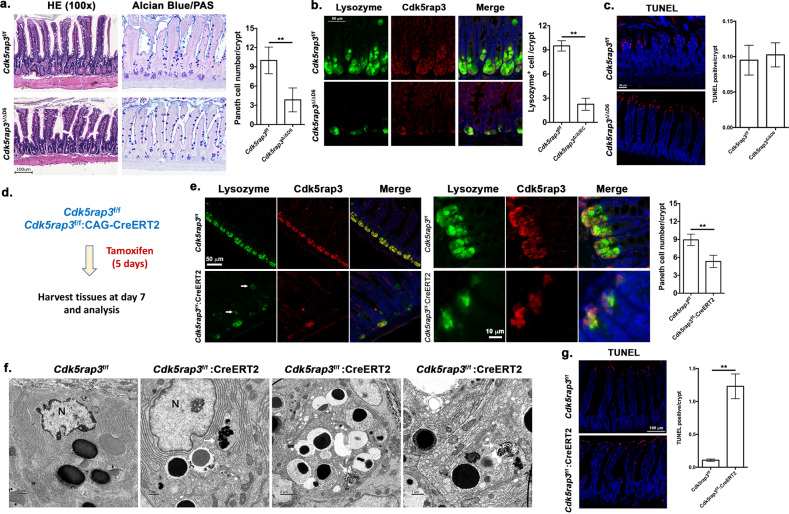
Deletion of Cdk5rap3 in mature Paneth cells causes their loss and abnormality
As shown in Fig. 1d, Cdk5rap3 is highly expressed in Paneth cells, indicating its potential role in maintenance of mature cells. To address this possibility, we generated Paneth cell-specific KO model of Cdk5rap3 using Defensin 6-Cre (D6-Cre) transgenic line2. Cdk5rap3 deletion (Cdk5rap3∆/∆D6) led to significant loss of Paneth cells (Fig. 3a, b) even though the reduction was not as dramatic as in Cdk5rap3∆/∆IEC mice (Fig. 1e). This may be caused by variable penetrance of Cre transgene expression and efficiency of Cre-mediated deletion of Cdk5rap3 allele because most Lysozyme+ cells in Cdk5rap3∆/∆D6 intestine were also Cdk5rap3+ (Fig. 3b). We also examined cell death, and no significant increase of TUNEL-positive cells was detected in the Cdk5rap3∆/∆D6 crypts (Fig. 3c). The result of this KO model suggests that Cdk5rap3 is also important for mature Paneth cells. Further investigation using the mice with tracing reporter will determine whether Cdk5rap3 deficient Paneth cells undergo cell death or trans-differentiate to other types of cells.
Fig. 3. Paneth cell-specific deletion of Cdk5rap3 causes loss of Paneth cell.
a H&E and Alcian blue/PAS staining of ileal sections of wild-type and Cdk5rap3 Paneth cell-specific Cdk5rap3 KO mice (Cdk5rap3∆/∆D6). The number of Paneth cells per crypt was scored. **p < 0.01. (n = 4 mice per genotype). b Lysozyme staining of ileal sections of wild-type and Cdkrap3∆/∆D6 mice. Lysozyme-positive cells per crypt were scored. **p < 0.01. (n = 4 mice per genotype). c TUNEL staining of ileal sections of wild-type and Cdkrap3∆/∆D6 mice. TUNEL positive cells per crypt were scored (n = 4 mice per genotype). d Experimental procedure to analyze the effect of acute deletion of Cdk5rap3. Mice were treated with daily i.p. injection of tamoxifen (75 mg/kg) for 5 consecutive days, and the tissues were harvested at day 7 for fixation and analysis. e Lysozyme staining of ileal sections of wild-type and Cdkrap3 deficient mice. Lysozyme-positive cells per crypt were scored. **p < 0.01. (n = 4 mice per genotype). f TEM analysis of the crypts of wild-type and Cdkrap3 deficient small intestine. The nucleus was labeled as “N”, and autolysosomes were marked by white arrows. g TUNEL staining of ileal sections of wild-type and Cdkrap3 deficient mice. TUNEL positive cells per crypt were scored. **p < 0.01. (n = 4 mice per genotype).
We also examined the effect of acute deletion of Cdk5rap3 by treating Cdk5rap3f/f:CAG-CreERT2 mice with TAM. The mice were treated with TAM for 5 days and analyzed at day 7 after the first injection (Fig. 3d). TAM-induced acute deletion of Cdk5rap3 resulted in fewer lysozyme+ Paneth cells, and more metaplastic Paneth cells outside of the crypts (Fig. 3e, marked by white arrows), while zymogen granules appeared to be disorganized (Fig. 3e). TEM analysis showed that Cdk5rap3 abolition led to local disassembly of the rough ER network, increase of autophagosomes and deformed zymogen granules, a phenotype that is similar to the one of Mist1 deficient Paneth cells (Fig. 3f)46. Compared to Cdk5rap3f/f mice, the number of TUNEL-positive cells was significantly elevated in TAM-treated Cdk5rap3f/f:CAG-CreERT2 intestine (Fig. 3g). Our results suggest that Cdk5rap3 is critical for either Paneth cell maturation or maintenance of mature Paneth cells.
Cdk5rap3∆/∆IEC mice are more susceptible to dextran sulfate sodium (DSS)-induced colitis
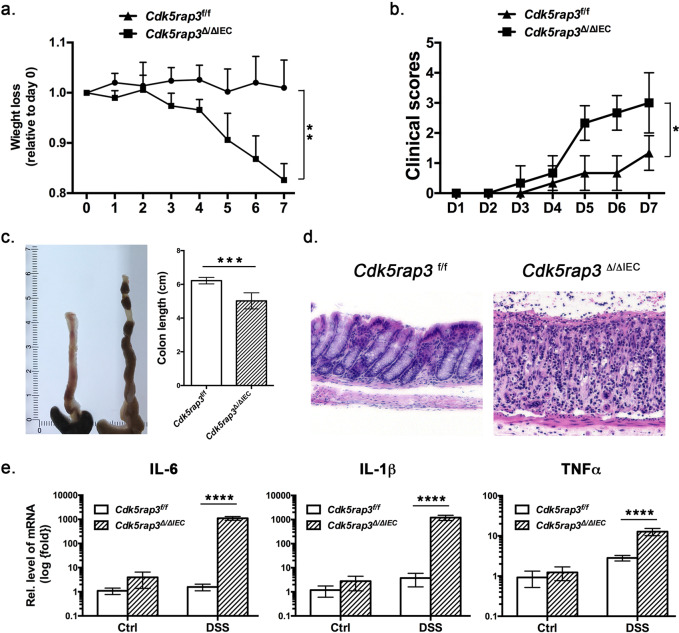
Paneth cells play a pivotal role in maintaining homeostasis of intestinal microbiota and innate immunity, and impairment or loss of their function may lead to dysbiosis and inflammatory response. It has been shown that dysfunction of Paneth cells in mice leads to dysbiotic microbiota and loss of intestinal homeostasis, thereby conferring susceptibility to experimentally induced colitis47. To address whether Cdk5rap3 deficiency predisposes susceptibility to inflammatory colitis, we tested IEC-specific Cdk5rap3 deficient mice in the DSS (dextran sulfate sodium)-induced colitis model. In comparison to wild-type mice, Cdk5rap3∆/∆IEC mice exhibited accelerated weight loss (Fig. 4a), deteriorated clinical scores (Fig. 4b), and colon shrinkage (Fig. 4c). In addition, Cdk5rap3∆/∆IEC mice possessed severe epithelial damage including significant loss of basal crypts, massive infiltration of immune cells (Fig. 4d), and elevated expression of inflammatory cytokines IL-6, IL-1β and TNFα (Fig. 4e). Altogether, these results demonstrate that Cdk5rap3∆/∆IEC mice are more susceptible to DSS-induced colitis.
Fig. 4. Cdk5rap3∆/∆IEC mice are susceptible to experimentally induced colitis.
Mice of each genotype were treated with 2.5% DSS in drinking water for 5 consecutive days and monitored for weight loss and clinical symptoms such as diarrhea and blood in stools. a Weight loss of DSS-treated mice **p < 0.01 (n = 5 mice per genotype). b Clinical scores of diarrhea of DSS-treated mice. Diarrhea (0 = no diarrhea, 1 = soft stool, 2 = very soft stool, 3 = liquid stool, 4 = severe diarrhea). *p < 0.05 (n = 5 mice per genotype). c Colon length. ***p < 0.001 (n = 5 mice per genotype). d H&E staining of colonic sections of wild-type and Cdk5rap3 deficient mice after DSS treatment. e Elevated expression of inflammatory cytokines in Ufbp1 deficient mice after DSS treatment. Total colonic RNA was subjected to quantitative RT-PCR analysis. ****p < 0.0001 (n = 5 mice per genotype).
Cdk5rap3 knockout results in defective ufmylation pathway and activation of Unfolded Protein Response (UPR)
In attempt to gain insight into the molecular mechanism of Cdk5rap3’s function, we examined gene expression profiles in wild-type and Cdk5rap3∆/∆IEC intestine. Among 2674 DEGs (differentially expressed genes) (cutoff 1.3-fold, Padj < 0.05), 1400 genes were upregulated and 1274 genes were downregulated in Cdk5rap3∆/∆IEC small intestine (Supplementary Tables S1 and S2, A1-A3 for Cdk5rap3f/f, and A4-A8 for Cdk5rap3∆/∆IEC). Consistent with our results described above (Fig. 1h), RNA-seq analysis confirmed down-regulation of Paneth cell-specific defensin genes (Supplementary Table S2 and Supplementary Fig. S3a). Furthermore, RNA-Seq result validated the under-expression of Gfi1 and Sox9 and up-regulation of Olfm4 induced by Cdk5rap3 deletion (Fig. 2d, Supplementary Tables S1 and S2 and Supplementary Fig. S3b).
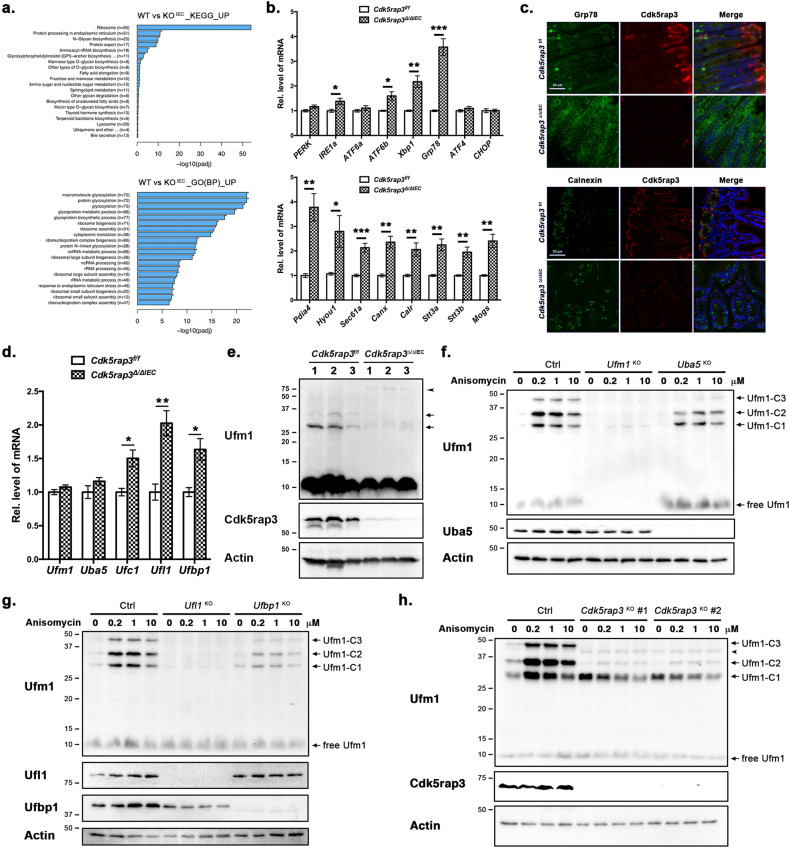
We then performed Gene Ontology (GO) of Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of upregulated DEGs. As shown in Fig. 5a, the top hits included ribosome biogenesis, protein translation, glycosylation and processing in the ER, cellular response to ER stress, and other metabolic pathways such as lipid metabolism (Fig. 5a). Cdk5rap3 was identified as an interacting protein of Ufl1, a Ufm1-specific E3 ligase25,27. The Ufm1 conjugation system is a novel ubiquitin-like modification system that consists of Uba5 (Ufm1-specific E1 enzyme), Ufc1 (Ufm1-specific E2 enzyme), and Ufl1/Ufbp1 complex (Ufm1-specific E3 ligase)48,49. Previous studies have demonstrated its indispensable role in mouse embryogenesis and erythropoiesis50–52. We have recently reported that Ufl1 and Ufbp1 is essential for maintenance of intestinal homeostasis, and knockout of Ufl1 and its co-factor Ufbp1 led to complete ablation of Paneth cells and partial loss of goblet cells47. Elevated ER stress and UPR activation has been observed in both Ufbp1 KO intestine and B cells as well as several UFMylation-deficient cell lines21,47,53. Therefore, we examined UPR activation first. As shown in Fig. 5b, Cdk5rap3 deletion caused significant up-regulation of ER stress sensors IRE1α and ATF6β, but not PERK and ATF6α (Fig. 5b). Moreover, expression of IRE1α downstream effector Xbp-1 and its target Grp78/Bip were also significantly increased (Fig. 5b), indicating that Cdk5rap3 ablation activates the IRE1α-Xbp-1 branch of the UPR. In contrast, expression of ATF4 and CHOP, two downstream targets of the PERK branch, was not altered by Cdk5rap3 KO, suggesting that the PERK branch may not be activated in Cdk5rap3 KO intestine. Furthermore, representative genes in glycosylation (Stt3a, Stt3b, and Mogs), protein folding and chaperones (Pdia4, Hyou1, Canx, and Calr), and co-translational translocation (Sec61a) were also upregulated in Cdk5rap3 KO intestine (Fig. 5b). Activation of the UPR in Cdk5rap3 deficient cells was further confirmed by enhanced immunostaining of Grp78/Bip and Calnexin (Fig. 5c). Taken together, our results suggest that like Ufl1 and Ufbp1, Cdk5rap3 deficiency may lead to activation of the UPR, especially the IRE1α-Xbp-1 branch.
Fig. 5. Cdk5rap3 knockout results in defective ufmylation pathway and activation of Unfolded Protein Response (UPR).
a GO and KEGG enrichment analysis of up-regulated DEGs in Cdkrap3∆/∆IEC intestine. b Quantitative RT-PCR confirmation of up-regulated genes that are involved in UPR and ER-related protein glycosylation and folding. *p < 0.05, **p < 0.01, ***p < 0.001 (n = 4 mice per genotype). c Immunofluorescence staining of Grp78/Bip and Calnexin in ileal section of wild-type and Cdkrap3∆/∆IEC mice. d Up-regulation of the components of the Ufm1 conjugation system induced by Cdk5rap3 knockout. Total RNA was isolated from small intestinal scrapes. *p < 0.05, **p < 0.01 (n = 4 mice per genotype). e The Ufm1 conjugates in wild-type and Cdk5rap3 KO IECs. Cell lysates of isolated crypts were subjected to immunoblotting of Ufm1. Endogenous of Ufm1 conjugates in wild-type cells were marked by arrows, while a new Ufm1 conjugate in Cdk5rap3 KO cells was indicated by an arrowhead. f UFMylation in wild-type, Ufm1 KO and Uba5 KO 293 T cells in the absence and presence of anisomycin. Cells were treated with anisomycin for 1 h. Ufm1-C1, 2, 3 are endogenous Ufm1 conjugates. g UFMylation in wild-type, Ufl1 KO and Ufbp1 KO 293T cells with and without anisomycin. h UFMylation in wild-type and Cdk5rap3 KO 293T cells with and without anisomycin. A new Ufm1 conjugate in Cdk5rap3 KO cells was indicated by an arrowhead (around 45 kD).
As an interacting protein of Ufm1-specific E3 Ligase Ufl1, Cdk5rap3 has been shown to modulate the UFMylation pathway21,22,54. Interestingly, loss of Cdk5rap3 in IECs led to elevated expression of other components of the Ufm1 system, including Ufc1 (Ufm1 E2 enzyme), Ufl1 and Ufbp1 (Ufm1 E3 ligase) (Fig. 5d), indicating a possible compensatory response to Cdk5rap3 deficiency. Knockout of Cdk5rap3 also altered UFMylation of endogenous targets, as evidenced by a reduction of several endogenous Ufm1 conjugates and appearance of new Ufm1 conjugate (around 80 kD, Fig. 5e). To further explore the impact of Cdk5rap3 on the UFMylation pathway, we examined Ufm1 conjugation in HEK293T cells under various conditions. It has been shown that low concentration of translation elongation inhibitor anisomycin causes ribosome stalling and collision, resulting in increased Ufm1 conjugation of ribosomal protein RPL2655. As shown in Fig. 5f, Ufm1 KO completely abolished basal and anisomycin-induced UFMylation, while Uba5 KO (Ufm1 E1 ligase) partially blocked UFMylation. Similarly, UFMylation was almost completely eliminated by Ufl1 KO and attenuated by Ufbp1 deletion (Fig. 5g). In comparison to parental cells, Cdk5rap3 KO attenuated basal and anisomycin-induced multi-UFMylation (Ufm1 conjugates C2 and C3) but enhanced basal mono-UFMylation (Ufm1-C1) (Fig. 5h). Mono-UFMylated form in Cdk5rap3 KO cells was slightly reduced by anisomycin treatment, possibly due to anisomycin-induced multi-UFMylation of this mono-form (Fig. 5h). Collectively, our data strongly suggest that Cdk5rap3 plays a pivotal role in the UFMylation pathway, and alteration of UFMylation caused by Cdk5rap3 deficiency may lead to activation of UPR and loss of Paneth cells.
Discussion
In this report, we present the genetic evidence to demonstrate the pivotal role of Cdk5rap3 protein in Paneth cell development and intestinal homeostasis. IEC-specific deletion of Cdk5rap3 caused nearly complete loss of Paneth cells (Fig. 1). Gene expression analysis revealed that Cdk5rap3 deficiency resulted in under-expression of key TFs that are important for Paneth cell lineage specification (Fig. 2 and Supplementary Table S2), suggesting a crucial role of Cdk5rap3 in cell fate decision. Moreover, Paneth cell-specific Cdk5rap3 knockout caused partial loss of Paneth cells, and TAM-induced acute deletion of Cdk5rap3 led to abnormal Paneth cells and subcellular abnormalities (Fig. 3), indicating its important role in mature Paneth cells. Consequently, Cdk5rap3 deficient mice manifested increased susceptibility to DSS-induced colitis (Fig. 4). Taken together, our results establish Cdk5rap3 as an important regulator of Paneth cell development.
The intestinal epithelium is one of the fastest renewing tissues, and the renewal process is driven by intestinal stem cell under stringent control of multiple signaling pathways such as Wnt, Notch and BMP pathways56. Under physiological condition, the Wnt pathway is the dominant force to drive proliferation of Lgr5+ intestinal stem cells. Aberrant activation of this pathway results in intestinal hyperplasia and colorectal cancers, whereas its inhibition leads to loss of crypts and altered lineage development57,58. In comparison, the Notch pathway is not only essential for maintenance of stem cell compartment, but also controls absorptive versus secretory cell fate specification. Inhibition of Notch signaling drives stem cells towards secretory lineages59. Paneth cell development is tightly controlled by both Wnt and Notch pathways and their downstream transcription factors60. The combination of Wnt activation and Notch inhibition promotes Lgr5+ ISCs to differentiate to Paneth cells in organoid culture61. Deletion of Lgr4, a positive regulator of Wnt signaling, significantly impaired Paneth cell formation62. Math1/Atoh1 is a master regulator of secretory cells and essential for development of all secretory lineages, including both exocrine (Paneth and goblet) and enteroendocrine lineages35,37. Sox-9, a Wnt/β-catenin target, is essential for Paneth cell fate decision and differentiation38,39. Gfi1 is important for fate decision of exocrine cell lineage, while Sox9 is specifically required for differentiation of Paneth cells36,38,39. Interestingly, β-catenin activity is required for Paneth cell maturation63. Yet, how these signaling pathways and transcription factors coordinate to control differentiation, maturation and plasticity of Paneth cells remains poorly understood.
Interestingly, our result shows that Cdk5rap3 is critical for both fate decision and development of Paneth cells, a feature that distinguishes Cdk5rap3 from other important regulators of Paneth cell development. On the one hand, Cdk5rap3 is critical for Paneth cell specification. Cdk5rap3 knockout led to down-regulation of Gfi1 and Sox9 (Fig. 2a), and Cdk5rap3 deficient intestine was similar to the ones of Gfi1 and Sox9 KO mice. Yet there are several phenotypic differences among these KO mouse models. Cdk5rap3 KO mice exhibited neither significant increase of enteroendocrine cells as manifested in Gfi1 KO mice nor apparent crypt enlargement as observed in Sox9 KO mice36,38,39. On the other hand, Cdk5rap3 is also important for mature Paneth cells, and Paneth cell-specific deletion led to Paneth cell loss (Fig. 3a–c). Taken together, our data strongly suggest that Cdk5rap3 represents a novel and unique regulator of Paneth cell biology.
What is the molecular mechanism underlying Cdk5rap3’s function? One of possible mechanisms is its role in the UFMylation pathway. The Ufm1 conjugation system is involved in multiple signaling pathways and cellular processes, including transcriptional regulation64, p53 stability65, DNA damage response66,67, autophagy68,69 and UPR70,71. Notably, the Ufm1-specific E3 ligase consisting of Ufl1/Ufbp1 is highly expressed in intestinal exocrine cells and vital for their survival and function47. IEC-specific knockout of either Ufl1 and Ufbp1 leads to almost complete loss of Paneth cells and partial loss of goblet cells47. Cdk5rap3 is an interacting partner of Ufl1 and Ufbp125–27. Therefore, it is plausible that Cdk5rap3 may affect Paneth cell development and function through modulating the UFMylation pathway. Indeed, its deficiency alters basal and anisomycin-induced UFMylation of endogenous targets (Fig. 5), a result that is consistent with previous observations21,22. Moreover, similar to Ufl1 and Ufbp1, Cdk5rap3 deletion causes preferentially activation of the IRE1α branch of UPR (Fig. 5). Although how UFMylation deficiency increases ER stress remains elusive, elevated ER stress and UPR activation may affect many aspects of intestinal homeostasis and physiology, including reduced stemness of ISC and impaired development and survival of exocrine cells72. Further studies will be conducted to explore the underlying mechanism of cross-talk between the ER and the UFMylation pathway and its impact on Paneth cell development.
Despite phenotypic similarities shared by Cdk5rap3 and Ufl1/Ufbp1 KO mice, there are also several notable differences. Cdkr5rap3 KO embryos die much earlier than Ufl1 and Ufbp1 null embryos52. Additionally, no severe anemia was observed in adult Cdk5rap3 conditional KO mice (our unpublished observation), suggesting that unlike other components of the Ufm1 system, Cdk5rap3 is not essential for red blood cell development. Interestingly, Cdk5rap3 appears to have a distinct effect on UFMylation. One of the principal Ufm1 targets is RPL26, a ribosomal protein whose UFMylation is enhanced by ribosome stalling21,55. Ufm1-C1 and C2 conjugates represent mono- and di-UFMylation of RPL26 (Fig. 5)21,55. As shown in Fig. 5f and g, Ufl1 KO nearly abolishes RPL26 UFMylation, while Uba5 or Ufbp1 KO significantly attenuates its UFMylation. Interestingly, Cdk5rap3 KO cells exhibited a distinct UFMylation pattern. Specifically, multi-UFMylation was dramatically diminished by Cdk5rap3 KO while basal RPL26 mono-UFMylation is substantially increased (Fig. 5h). Additional Ufm1 conjugate was also observed in Cdk5rap3 KO cells (Fig. 5h). It remains to be determined whether the differential influence on UFMylation contributes to the phenotypic difference of Cdk5rap3 and Ufl1/Ufbp1 KO mice.
In addition to the UFMylation pathway, Cdk5rap3 has been implicated in other signaling pathways. Several studies suggest that Cdk5rap3 may directly modulate transcription by interacting with TFs such as RelA and CBP/p30011,12. Therefore, it is plausible that Cdk5rap3 may directly regulate expression of key TFs such as Gfi1 and Sox9 to control lineage allocation. Alternatively, cytosolic Cdk5rap3 may modulate WNT signaling to affect intestinal development. Previous studies have indicated that Cdk5rap3 is a negative regulator of WNT signaling by stabilizing GSK-3β15,16. More studies will be conducted to determine whether Cdk5rap3 deficiency results in altered output of WNT signaling and whether Cdk5rap3 is directly involved in regulation of WNT signaling in the intestine. Furthermore, Cdk5rap3 deficiency also affected cell cycle progression of proliferating IECs (Fig. 1j) and ribosome biogenesis (Fig. 5a), indicating its involvement in multiple cellular processes. Given a long list of its potential interacting proteins, further mechanistic investigations will establish whether Cdk5rap3 acts in a unified molecular mechanism or it is a truly multi-faceted protein with pleotropic functions in different type of cells and tissues.
Materials and methods
Generation of Cdk5rap3 knockout mice and genotyping protocol
ES cell clone HEPD0516_2_A06 (JM8.N4 with C57BL/6N background) containing trapped Cdk5rap3 allele was purchased from the EUCOMM (European Conditional Mouse Mutagenesis) team (supplemental Fig. 1). The ES cells were injected into the blastocysts of C57BL/6 mice (Northwestern University Transgenic and Targeted Mutagenesis Laboratory). Chimeric mice were crossed with B6(Cg)-TyrC-2J/J albino mice, and heterozygous offspring with germ-line transmission were confirmed by genotyping.
To generate conditional KO mice, we crossed Cdk5rap3Trap-F/+ mice with FLPo deleter mice (stock # 011065, The Jackson Laboratory, Bar harbor, ME) to remove the gene trap cassette. The Cdk5rap3 floxed mice were crossed with various Cre strain to generate tissue and cell-type specific KO mice. For tamoxifen-induced whole body KO, CAG-CreERT2 mice (stock# 004453, The Jackson Laboratory) was crossed with Cdk5rap3 floxed mice, and Cdk5rap3 deletion was induced by tamoxifen injection. Tamoxifen (20 mg/ml in corn oil, Sigma, St. Louis, MO) was administrated by 5-day IP injection with an approximate dose of 75 mg tamoxifen/kg body weight. For IEC-specific KO, Cdk5rap3 floxed mice were crossed with Villin-Cre transgenic mouse that was originally from Dr. Sylvie Robine’s laboratory73. Mice were housed in the animal facility of Augusta University, and all animal procedures were approved by AU IACUC. All mice used in this study were at the age of 8–12 weeks, and randomly chosen for the experiments.
The following primers were used for PCR genotyping of Cdk5rap3 trapped allele: F1 (CACAACGGGTTCTTCTGTTAGTCC), R1 (GATTGGCAGGAGATCGTAAGCCTG) and R2: (GCAGTACGCACTACCTCCCCAAGG). A 35-cycle (92 °C, 45 s, 62 °C, 45 s, 72 °C 45 s), 260 bp PCR product is for wild-type allele, while 425 bp is for trapped allele. For Cdk5rap3 floxed allele, two primers are used: P1 (TAG CTC GGG GCT CAG ACG CTC TGA) and P2 (TTA TCT GCT CTT CCC GCT AGA ATA). The floxed allele generates a 356 bp PCR product while wild-type allele gives 330 bp product. Genotyping of Cre-ERT2 mice was performed according to the standard protocol of the Jackson Laboratory, while Villin-Cre and D6-Cre mice were genotyped as previously described41.
Isolation and in vitro organoid culture of intestinal crypts
Crypts were isolated from the ileum of small intestine according to Sato et al. with minor modifications74. Briefly, the ileum of small intestine was harvested and opened lengthwise, and then washed multiple times with cold PBS. After removal of the villi with a cover glass, the intestine was cut into 2–3 large pieces, and then washed with cold PBS (10–20 ml) for more than ten times. Subsequently the fragments were incubated in 25 ml of 2 mM EDTA on ice for 15 min with a gentle shaking. After removal of the supernatant, the tissue was washed with cold PBS and then pipetted up and down 3–5 times. Released crypts were passed through a 70 μM cell strainer and collected by 3-min centrifugation at 100 × g. Isolated crypts were counted and resuspended in Growth Factor Reduced Matrigel (Corning Life sciences, Corning, NY) and cultured in the complete medium: Advanced DMEM/F12 supplemented with 1× B27, 1× N2 (Thermo Fisher Scientific, Waltham, MA), 1 mM N-acetyle-L-cysteine, HEPES (10 mM, pH 7.4), murine EGF (50 ng/ml, PeproTech, Rocky Hill, NJ), murine Noggin (100 ng/ml, PeproTech), 1/10th volume of R-Spondin-1 conditional medium (Trevigen, Gaitherburg, MD), 1 × GlutaMax, 1 × Penicillin/Streptomycin (ThermoFisher Scientific) and 100 μg/ml Normocin (InvivoGen, San Diego, CA),
RNA-seq, differential, and enrichment analysis
The cells were scraped off from ileal section of small intestine with slide glass. Total RNAs from three wild-type and four Cdk5rap3 KO mice were isolated from the scrapes with TRIzol (ThermoFisher Scientific) and Zymo Direct-zol RNA Miniprep Plus kit (R2072, Zymo Research, Irvine, CA). RNA-seq and analyses were conducted by Novogene (Beijing, China). Raw data were obtained by Illumina platform. After data filtering, the genes were mapped to the genome with STAR software, and quantification was determined by HTSeq software. DESeq2 was used for differential analysis, and ClusterProfiler was used for enrichment analysis (GO, KEGG and Reactome). Gene Set Enrichment Analysis was performed online at https://www.gsea-msigdb.org/gsea/index.jsp.
Histology, immunohistochemistry, immunofluorescent staining, and immunoblotting
HE and PAS/Alcian blue staining was performed by the Histology core of Augusta University, while TEM was conducted by the EM core of Augusta University according to the standard procedures. Immunohistochemistry, immunofluorescent staining, and immunoblotting were performed as described previously45. Bright field and Epifluorescence images were obtained using Zeiss Observer D1 with AxioVision 4.8 software (Carl Zeiss Microscopy GmbH, Jena, Germany) and Keyence BZ-X700 fluorescent microscope with its corresponding software (Keyence America, Itasca, IL, USA). All histopathological analysis and quantification were performed blindly by lab personnel who had no prior information on the genotypes of animals and tissues.
The antibodies used in this study included: Cdk5rap3 rat polyclonal antibody (Li lab), Uba5 (Li Lab), Ufl1 (Li lab), Ufbp1 (21445-1-AP, Proteintech, Rosemont, IL), phospho-Histone H3 (ser10) (#9706, Cell Signaling), PCNA (#2586, Cell Signaling), Olfm4 (#39141, Cell Signaling), Calnexin (#2679, Cell Signaling), Ufm1 (Ab109305, Abcam), Lysozyme (A0099, Agilent Dako, Santa Clara, CA), Grp78/Bip (#3177, Cell Signaling), Chromogranin A (Ab15160, Abcam, Cambridge, MA), and β-Actin (#3700, Cell Signaling). All affinity-purified and species-specific HRP- and fluorophore-conjugated secondary antibodies were obtained from Jackson ImmunoResearch (West Grove, PA).
Generation of CRISPR/cas-9-mediated knockout cell lines
The plasmids containing sgRNAs were constructed by cloning sgRNA-containing oligos into pLentiCRISPR V2 vector (Addgene # 52961). Lentiviral particles expressing sgRNA and cas-9 were prepared by standard 293T cell transfection, and then used for infection of parental 293T cells. Knockout cell clones were isolated by puromycin selection, limiting dilution, and immunoblotting. Sequences of sgRNAs were listed below:
| Gene (human) | sgRNA |
|---|---|
| Cdk5rap3 sgRNA-1 | GTTGACATTCCGAACCAGG |
| Cdk5rap3 sgRNA-2 | GATTATAGCTCTGTATGAGA |
| Ufm1 sgRNA-1 | TCACGCTGACGTCGGACCCA |
| Ufm1 sgRNA-2 | CTTTAAGATCACGCTGACGT |
| Uba5 sgRNA-1 | TCCCGAGGAGCGGCGACGGA |
| Uba5 sgRNA-2 | GCTGGAGCGGGAACTTGCCC |
| Ufl1 sgRNA-1 | CCAGCGGGCGCAGTTCGCCG |
| Ufl1 sgRNA-2 | GGAAGAGATTAGGCGGTTGG |
| Ufbp1 sgRNA-1 | GTAGCGGCGGCTCTGCTAGT |
Quantitative real-time PCR
Total RNA was isolated with TRIzol (ThermoFisher Scientific) and Zymo Direct-zol RNA Miniprep Plus kit (R2072, Zymo Research, Irvine, CA), and then reversely transcribed with High-Capacity cDNA Reverse Transcription kit according to the manufacturer’s instruction (ThermoFisher Scientific). Quantitative RT-PCR was performed using the iTaq Universal SYBR Green Supermix kit (BIO-RAD, Hercules, CA) with 40 cycles of 95 °C for 15 s and 60 °C for 1 min on StepOnePlus Real-Time PCR System (ThermoFisher Scientific). The results were analyzed by StepOne Software (Version 2.1, Life Technologies). Relative expression of each transcript was normalized to murine beta-actin by using the 2^(-delta delta Ct) method. The following is the list of primers used in this study:
| Gene (mouse) | Forward primer | Reverse primer |
|---|---|---|
| Actin | GACCTCTATGCCAACACAGT | AGTACTTGCGCTCAGGAGGA |
| Cdk5rap3 (exon6-12) | TGGAAGCTGTTTCCGACTCT | AGCCTCAGTTCCTGTCTCCA |
| Lysozyme 2 | ATGGAATGGCTGGCTACTATGG | ACCAGTATCGGCTATTGATCTGA |
| Defcr1 | AAGAGACTAAAACTGAGGAGCAGC | CGACAGCAGAGCGTGTA |
| Defcr5 | AGGCTGATCCTATCCACAAAACAG | TGAAGAGCAGACCCTTCTTGGC |
| Intestinal ALP | CACAGCTTACCTGGCACTGA | GGTCTCTGACGACAGGGGTA |
| Chromogranin A | CCAATACCCAATCACCAACC | TTGTAGCCTGCATGGAAGTG |
| Mucin 2 | GCCTGTTTGATAGCTGCTATGTGCC | GTTCCGCCAGTCAATGCAGACAC |
| Lgr5 | ACCCGCCAGTCTCCTACATC | GCATCTAGGCGCAGGGATTG |
| Olfm4 | TGGCCCTTGGAAGCTGTAGT | ACCTCCTTGGCCATAGCGAA |
| Math1 | GAGTGGGCTGAGGTAAAAGAGT | GGTCGGTGCTATCCAGGAG |
| Hes1 | CCAGCCAGTGTCAACACGA | AATGCCGGGAGCTATCTTTCT |
| Gfi1 | AGGAACGCAGCTTTGACTGT | CCTGTGTGGATGAAGGTGTG |
| Sox9 | GAGGAAGTCGGTGAAGAACG | CCCTCTCGCTTCAGATCAAC |
| Klf4 | CTGAACAGCAGGGACTGTCA | GTGTGGGTGGCTGTTCTTTT |
| Elf3 | GCATGTCCTTCCAAGAGAGC | ACATCACTTCCACCGGAGTC |
| IL-6 | TAGTCCTTCCTACCCCAATTTCC | TTGGTCCTTAGCCACTCCTTC |
| IL-1β | GCCCATCCTCTGTGACTCAT | AGGCCACAGGTATTTTGTCG |
| TNFα | CCGATGGGTTGTACCTTGTC | TGGAAGACTCCTCCCAGGTA |
| Perk | GAAACGGCTTTCAGTTGAGC | CTGGCCATATCCACCAGAGT |
| Ire1α | CAGATCTGCGCAAATTCAGA | CTCCATGGCTTGGTAGGTGT |
| Atf6α | AGCCGACTGTGGTTCAACTT | CCCATACTTCTGGTGGCACT |
| Atf6β | TTTGACAGCAGCTCTCTGGA | CATCTTCACATGCAGCACCT |
| Xbp1 | ACACGCTTGGGAATGGACAC | CCATGGGAAGATGTTCTGGG |
| Grp78/Bip | ACTTGGGGACCACCTATTCCT | ATCGCCAATCAGACGCTCC |
| Atf4 | GAAACCTCATGGGTTCTCCA | AGCTCATCTGGCATGGTTTC |
| Chop | GCATGAAGGAGAAGGAGCAG | ATGGTGCTGGGTACACTTCC |
| Pdia4 | TGCTGACACACCCTGAGAAG | TGCTGTACCGCTTAGCATCA |
| Hyou1 | GCCCACTTTAACCTGGATGA | GGCTCTCCTCTTCCTCCTGT |
| Sec61α | CTATTTCCAGGGCTTCCGAGT | AGGTGTTGTACTGGCCTCGGT |
| Canx | ACAAGAGCGATTGGATGGAA | TGCTTGTGAATGGAGCAGTC |
| Calr | AGGCTCCTTGGAGGATGATT | TCCCACTCTCCATCCATCTC |
| Stt3a | TATCTCCCTGGTTGGCTTTG | TGGTGCTCAGAAACAGATGC |
| Stt3b | TGGAGGACAGCAGTGATGAG | AAGGACCACACTTGGACTGG |
| Mogs | GGACCTAGCTTTGCCTACCC | TTCAGTCTCCCCAAGCTGTT |
| Ufm1 | CCGTTCACAGCAGTGCTAAA | CAGCTTCCAACTCGGTCTCT |
| Uba5 | CAAGCTATGTTCACGGCAGA | AGTTGTTTTGCCCACCACTC |
| Ufc1 | AACTGCACTTCCGCAGTTTT | CTCCAGTCGGAACCAATCAT |
| Ufl1 | AGCAAACAGGCCTCAACTGT | TTTCTGGTGCATCAGCTCAC |
| Ufbp1 | GAAGCCAGCAGAAGTTCACC | GAAGCCGTTCCTCTTCCTTC |
Statistical analysis
All statistical analyses were performed using Graph Prism 7 software. P values were determined by unpaired t-tests between two set of data. A p value < 0.05 was considered to be significant.
TUNEL staining
TUNEL staining was performed using in situ cell death detection kit (TMR Red, Roche, Basel, Switzerland) according to the manufacturer’s instruction.
Supplementary information
Funding statement
This work was supported by NIH/NIDDK (1R01DK113409) and AU IGP award to H.L., NIH/NIDDK (R01DK088199) to R.B., NIH/NIAID (1R01AI55774) to N.S., NSFC (31970413) to Y.C., NSFC (81760595 and 81560529) to H.X., and CSC (201908360205) to Y.X.
Author contributions
M.Q., S.L., Y.X., Y.H., Y.Z., G.L., Y.C., and H.X. contribute to acquiring and analyzing the data; M.Q., L.H., N.S., R.B., Y.C., H.X., and H.L. contribute to designing the work and analyzing the data; M.Q., S.L., and H.L. contribute to drafting the manuscript; and all authors contribute to commenting and revising the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
The authors state that all animal research complied with USDA Animal Welfare Act and Regulations, and all experimental procedures were approved by AU IACUC.
Footnotes
Edited by A. Stephanou
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Michaela Quintero, Siyang Liu
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-021-03401-8.
References
- 1.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 2.Adolph TE, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu TC, Stappenbeck TS. Genetics and pathogenesis of inflammatory bowel disease. Annu Rev. Pathol. 2016;11:127–148. doi: 10.1146/annurev-pathol-012615-044152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salzman NH, Bevins CL. Dysbiosis-a consequence of Paneth cell dysfunction. Semin. Immunol. 2013;25:334–341. doi: 10.1016/j.smim.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Sato T, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yilmaz OH, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitt M, et al. Paneth cells respond to inflammation and contribute to tissue regeneration by acquiring stem-like features through SCF/c-Kit signaling. Cell Rep. 2018;24:2312–2328 e2317. doi: 10.1016/j.celrep.2018.07.085. [DOI] [PubMed] [Google Scholar]
- 8.Yu S, et al. Paneth Cell Multipotency Induced by Notch Activation following Injury. Cell Stem Cell. 2018;23:46–59 e45. doi: 10.1016/j.stem.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones JC, et al. Cellular plasticity of Defa4(Cre)-expressing paneth cells in response to notch activation and intestinal injury. Cell Mol. Gastroenterol. Hepatol. 2019;7:533–554. doi: 10.1016/j.jcmgh.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ching YP, Qi Z, Wang JH. Cloning of three novel neuronal Cdk5 activator binding proteins. Gene. 2000;242:285–294. doi: 10.1016/S0378-1119(99)00499-0. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, He X, Luo Y, Yarbrough WG. A novel ARF-binding protein (LZAP) alters ARF regulation of HDM2. Biochem. J. 2006;393:489–501. doi: 10.1042/BJ20050960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin X, Warner DR, Roberts EA, Pisano MM, Greene RM. Novel interaction between nuclear co-activator CBP and the CDK5 activator binding protein - C53. Int. J. Mol. Med. 2005;16:251–C256. [PubMed] [Google Scholar]
- 13.Wang J, An H, Mayo MW, Baldwin AS, Yarbrough WG. LZAP, a putative tumor suppressor, selectively inhibits NF-kappaB. Cancer Cell. 2007;12:239–251. doi: 10.1016/j.ccr.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Mak GW, et al. CDK5RAP3 is a novel repressor of p14ARF in hepatocellular carcinoma cells. PLoS One. 2012;7:e42210. doi: 10.1371/journal.pone.0042210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin KY, et al. Tumor suppressor Lzap suppresses Wnt/beta-catenin signaling to promote zebrafish embryonic ventral cell fates via the suppression of inhibitory phosphorylation of glycogen synthase kinase 3. J. Biol. Chem. 2015;290:29808–29819. doi: 10.1074/jbc.M115.669309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang JB, et al. CDK5RAP3 acts as a tumor suppressor in gastric cancer through inhibition of beta-catenin signaling. Cancer Lett. 2017;385:188–197. doi: 10.1016/j.canlet.2016.10.024. [DOI] [PubMed] [Google Scholar]
- 17.Zheng CH, et al. CDK5RAP3 suppresses Wnt/beta-catenin signaling by inhibiting AKT phosphorylation in gastric cancer. J. Exp. Clin. Cancer Res. 2018;37:59. doi: 10.1186/s13046-018-0716-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egusquiaguirre SP, et al. CDK5RAP3 is a co-factor for the oncogenic transcription factor STAT3. Neoplasia. 2020;22:47–59. doi: 10.1016/j.neo.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang H, Luo S, Li H. Cdk5 activator-binding protein C53 regulates apoptosis induced by genotoxic stress via modulating the G2/M DNA damage checkpoint. J. Biol. Chem. 2005;280:20651–20659. doi: 10.1074/jbc.M413431200. [DOI] [PubMed] [Google Scholar]
- 20.Jiang H, Wu J, He C, Yang W, Li H. Tumor suppressor protein C53 antagonizes checkpoint kinases to promote cyclin-dependent kinase 1 activation. Cell Res. 2009;19:458–468. doi: 10.1038/cr.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walczak CP, et al. Ribosomal protein RPL26 is the principal target of UFMylation. Proc. Natl Acad. Sci. USA. 2019;116:1299–1308. doi: 10.1073/pnas.1816202116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang, R. et al. CDK5RAP3, a UFL1 substrate adaptor, is crucial for liver development. Development146 (2019). [DOI] [PubMed]
- 23.Mak GW, et al. Overexpression of a novel activator of PAK4, the CDK5 kinase-associated protein CDK5RAP3, promotes hepatocellular carcinoma metastasis. Cancer Res. 2011;71:2949–2958. doi: 10.1158/0008-5472.CAN-10-4046. [DOI] [PubMed] [Google Scholar]
- 24.Wamsley JJ, et al. LZAP is a novel Wip1 binding partner and positive regulator of its phosphatase activity in vitro. Cell Cycle. 2017;16:213–223. doi: 10.1080/15384101.2016.1261767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Lei G, Mei M, Tang Y, Li H. A novel C53/LZAP-interacting protein regulates stability of C53/LZAP and DDRGK domain-containing Protein 1 (DDRGK1) and modulates NF-kappaB signaling. J. Biol. Chem. 2010;285:15126–15136. doi: 10.1074/jbc.M110.110619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon J, et al. A novel LZAP-binding protein, NLBP, inhibits cell invasion. J. Biol. Chem. 2010;285:12232–12240. doi: 10.1074/jbc.M109.065920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shiwaku H, et al. Suppression of the novel ER protein Maxer by mutant ataxin-1 in Bergman glia contributes to non-cell-autonomous toxicity. EMBO J. 2010;29:2446–2460. doi: 10.1038/emboj.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horejsi B, et al. Nuclear gamma-tubulin associates with nucleoli and interacts with tumor suppressor protein C53. J. Cell Physiol. 2012;227:367–382. doi: 10.1002/jcp.22772. [DOI] [PubMed] [Google Scholar]
- 29.Han M, Wang H, Zhang HT, Han Z. Expression of TIP-1 confers radioresistance of malignant glioma cells. PLoS One. 2012;7:e45402. doi: 10.1371/journal.pone.0045402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin JX, et al. Low expression of CDK5RAP3 and DDRGK1 indicates a poor prognosis in patients with gastric cancer. World J. Gastroenterol. 2018;24:3898–3907. doi: 10.3748/wjg.v24.i34.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao JJ, et al. Identification of LZAP as a new candidate tumor suppressor in hepatocellular carcinoma. PLoS One. 2011;6:e26608. doi: 10.1371/journal.pone.0026608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu D, et al. Tumor suppressor Lzap regulates cell cycle progression, doming, and zebrafish epiboly. Dev. Dyn. 2011;240:1613–1625. doi: 10.1002/dvdy.22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durand A, et al. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1) Proc. Natl Acad. Sci. USA. 2012;109:8965–8970. doi: 10.1073/pnas.1201652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwiggelaar RT, et al. LSD1 represses a neonatal/reparative gene program in adult intestinal epithelium. Sci. Adv. 2020;6:eabc0367. doi: 10.1126/sciadv.abc0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 36.Shroyer NF, Wallis D, Venken KJ, Bellen HJ, Zoghbi HY. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005;19:2412–2417. doi: 10.1101/gad.1353905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shroyer NF, et al. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 38.Mori-Akiyama Y, et al. SOX9 is required for the differentiation of paneth cells in the intestinal epithelium. Gastroenterology. 2007;133:539–546. doi: 10.1053/j.gastro.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 39.Bastide P, et al. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J. Cell Biol. 2007;178:635–648. doi: 10.1083/jcb.200704152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki K, et al. Hes1-deficient mice show precocious differentiation of Paneth cells in the small intestine. Biochem. Biophys. Res. Commun. 2005;328:348–352. doi: 10.1016/j.bbrc.2004.12.174. [DOI] [PubMed] [Google Scholar]
- 41.van Es JH, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 42.Fre S, et al. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 43.Stanger BZ, Datar R, Murtaugh LC, Melton DA. Direct regulation of intestinal fate by Notch. Proc. Natl Acad. Sci. USA. 2005;102:12443–12448. doi: 10.1073/pnas.0505690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ng AY, et al. Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology. 2002;122:1455–1466. doi: 10.1053/gast.2002.32990. [DOI] [PubMed] [Google Scholar]
- 45.Katz JP, et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dekaney CM, King S, Sheahan B, Cortes JE. Mist1 expression is required for Paneth cell maturation. Cell Mol. Gastroenterol. Hepatol. 2019;8:549–560. doi: 10.1016/j.jcmgh.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai Y, et al. Indispensable role of the Ubiquitin-fold modifier 1-specific E3 ligase in maintaining intestinal homeostasis and controlling gut inflammation. Cell Discov. 2019;5:7. doi: 10.1038/s41421-018-0070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komatsu M, et al. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 2004;23:1977–1986. doi: 10.1038/sj.emboj.7600205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tatsumi K, et al. A novel type of E3 ligase for the Ufm1 conjugation system. J. Biol. Chem. 2010;285:5417–5427. doi: 10.1074/jbc.M109.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang M, et al. RCAD/Ufl1, a Ufm1 E3 ligase, is essential for hematopoietic stem cell function and murine hematopoiesis. Cell Death Differ. 2015;22:1922–1934. doi: 10.1038/cdd.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tatsumi K, et al. The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat. Commun. 2011;2:181. doi: 10.1038/ncomms1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai Y, et al. UFBP1, a key component of the ufm1 conjugation system, is essential for ufmylation-mediated regulation of erythroid development. PLoS Genet. 2015;11:e1005643. doi: 10.1371/journal.pgen.1005643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu H, et al. Ufbp1 promotes plasma cell development and ER expansion by modulating distinct branches of UPR. Nat. Commun. 2019;10:1084. doi: 10.1038/s41467-019-08908-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Zhang M, Wu J, Lei G, Li H. Transcriptional regulation of the Ufm1 conjugation system in response to disturbance of the endoplasmic reticulum homeostasis and inhibition of vesicle trafficking. PLoS One. 2012;7:e48587. doi: 10.1371/journal.pone.0048587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, et al. UFMylation of RPL26 links translocation-associated quality control to endoplasmic reticulum protein homeostasis. Cell Res. 2020;30:5–20. doi: 10.1038/s41422-019-0236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 57.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 58.Mah AT, Yan KS, Kuo CJ. Wnt pathway regulation of intestinal stem cells. J. Physiol. 2016;594:4837–4847. doi: 10.1113/JP271754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Demitrack ES, Samuelson LC. Notch regulation of gastrointestinal stem cells. J. Physiol. 2016;594:4791–4803. doi: 10.1113/JP271667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noah TK, Donahue B, Shroyer NF. Intestinal development and differentiation. Exp. Cell Res. 2011;317:2702–2710. doi: 10.1016/j.yexcr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yin X, et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat. Methods. 2014;11:106–112. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mustata RC, et al. Lgr4 is required for Paneth cell differentiation and maintenance of intestinal stem cells ex vivo. EMBO Rep. 2011;12:558–564. doi: 10.1038/embor.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Es JH, et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat. Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 64.Yoo HM, et al. Modification of ASC1 by UFM1 is crucial for ERalpha transactivation and breast cancer development. Mol. Cell. 2014;56:261–274. doi: 10.1016/j.molcel.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 65.Liu J, et al. UFMylation maintains tumour suppressor p53 stability by antagonizing its ubiquitination. Nat. Cell Biol. 2020;22:1056–1063. doi: 10.1038/s41556-020-0559-z. [DOI] [PubMed] [Google Scholar]
- 66.Wang Z, et al. MRE11 UFMylation promotes ATM activation. Nucleic Acids Res. 2019;47:4124–4135. doi: 10.1093/nar/gkz110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qin B, et al. UFL1 promotes histone H4 ufmylation and ATM activation. Nat. Commun. 2019;10:1242. doi: 10.1038/s41467-019-09175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeJesus R, et al. Functional CRISPR screening identifies the ufmylation pathway as a regulator of SQSTM1/p62. Elife. 2016;5:e17290. doi: 10.7554/eLife.17290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liang JR, et al. A genome-wide ER-phagy screen highlights key roles of mitochondrial metabolism and ER-resident UFMylation. Cell. 2020;180:1160–1177 e1120. doi: 10.1016/j.cell.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li J, et al. Ufm1-specific ligase Ufl1 regulates endoplasmic reticulum homeostasis and protects against heart failure. Circ. Heart Fail. 2018;11:e004917. doi: 10.1161/CIRCHEARTFAILURE.118.004917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu J, et al. A critical role of DDRGK1 in endoplasmic reticulum homoeostasis via regulation of IRE1alpha stability. Nat. Commun. 2017;8:14186. doi: 10.1038/ncomms14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coleman OI, Haller D. ER stress and the UPR in shaping intestinal tissue homeostasis and immunity. Front. Immunol. 2019;10:2825. doi: 10.3389/fimmu.2019.02825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.el Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 74.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.