Abstract
The tripartite motif (TRIM) family of proteins is a class of highly conservative proteins that have been implicated in multiple processes. TRIM59, one member of the TRIM family, has now received recognition as a key regulator in the development and progression of human diseases. However, its role in human tumorigenesis has remained largely unknown. In this study, the effects of TRIM59 expression on cell proliferation and migration were investigated in human cervical cancer cells. The expression of TRIM59 in clinical cervical cancer tissues and cervical cancer cells was initially determined by RT-PCR and Western blot. Specific shRNA against TRIM59 was then employed to knock down the expression of TRIM59 in cervical cancer lines HeLa and SiHa. The effects of TRIM59 knockdown on cell proliferation was assessed by MTT assay and colony formation assay. Transwell assay was conducted to reveal cell migration and invasion abilities before and after TRIM59 knockdown. Our results showed that the expression of TRIM59 was significantly elevated in cervical cancers. Knockdown of TRIM59 significantly inhibited cell proliferation and colony formation as well as cell migration and invasion abilities in cervical cancer HeLa and SiHa cells. Cell cycle progression analysis showed that TRIM59-depleted cells preferred to accumulate in the S phase. These data suggest that TRIM59 is a potential target that promotes the progression of cervical cancer.
Key words: TRIM59, Proliferation, Migration, Invasion, Cervical cancer
INTRODUCTION
Cancer of the cervix is the second most common cancer among women worldwide. In Latin America, cervical cancer remains the first cause of mortality among 20- to 40-year-old women, and the third most common cause of cancer-related deaths in females, second only to breast and lung cancers1. In the US, the burden from cervical cancer also remains high, with 12,340 cases and 4,030 deaths in the year 2013, representing a significant economic loss2. Moreover, there are large geographic variations in cervical cancer incidence and mortality rates3,4. Mounting evidence has accumulated to suggest that human papillomavirus (HPV) serves as an important factor in cervix tumorigenesis. However, only a small proportion of patients infected by HPV progress to cervical cancer5–7, indicating that factors other than HPV infection may also contribute to the initiation of cervical cancer. Hence, the tumor biology of this malignancy needs to be elucidated.
The tripartite motif (TRIM) family of proteins are a revolutionarily class of highly conservative proteins that have been implicated in a number of critical processes such as immunity8–10, cell proliferation11,12, tumorigenesis13, cell differentiation14, neuron development15, antiviral activities11, and transcriptional regulation11,16. TRIM proteins are conservative in the N-terminal really interesting new gene (RING) domain, which are E3 ubiquitin ligases and are frequently involved in the ubiquitin–proteasome system and proteolysis17. Members from the TRIM family have recently been implicated in oncogenesis. TRIM13, TRIM19, and TRIM25, for example, were shown to be involved in leukemia, breast, and prostate cancers, respectively18–20, demonstrating the crucial role of TRIM proteins in tumorigenesis.
TRIM59, a novel member of the TRIM family, is a surface molecule with its biological importance significantly implicated in tumors. The oncogenic activity of the TRIM59 gene was initially characterized in mouse cancer models21. Using immunohistochemical detection, TRIM59 was deciphered to be a multiple cancer biomarker22. TRIM59 was remarkably upregulated in gastric tumors and promoted ubiquitination and degradation of p53, by which it promotes tumor growth, cell proliferation, and migration in gastric tumors23. Of note, TRIM59 communicated with the Ras signaling pathway in a transgenic prostate cancer mouse model21, indicating its critical role in tumors from the genital system. However, little is known so far about the role of TRIM59 in other genital tumors, especially in cervical cancer.
This study aimed to investigate the role of TRIM59 in human cervical cancer growth and metastasis. Expression of TRIM59 was initially determined in clinical cervical cancer tissues and a series of cervical cancer cell lines. Furthermore, specific shRNA against TRIM59 (shTRIM59) was employed to knock down the expression of TRIM59. The effects of TRIM59 knockdown on cervical cancer cell proliferation, migration, and invasion will be discussed in this study. To our knowledge, this is the first report that studied the critical roles of TRIM59 in cervical tumorigenesis.
MATERIALS AND METHODS
Human Samples
This study was approved by the ethics committee of the Affiliate Tumor Hospital of Xinjiang Medical University (Xinjiang, P.R. China). Thirty patients with cervical cancer, who were treated in the Department of Gynecology between 2013 and 2015, were randomly selected. Cervical cancer tissues and their adjacent noncancerous tissues were dissected and frozen in liquid nitrogen prior to total mRNA and total protein extraction. All patients showed their full intention to participate in our study, and written informed consents were obtained.
Cell Culture and Lentivirus Infection
The human cervical cancer cell lines HeLa, C4-1, SiHa, Caski, and C-33A were obtained from the American Type Culture Collection (ATCC; Rockville, MD, USA). Human umbilical vein endothelial cell ECV304 was included as an internal control. All these cells were incubated in the recommended culture medium supplemented with 10% fetal bovine serum (FBS; Gibco, NY, USA) at 37°C in a humidified 5% CO2 incubator. Specific shRNA against TRIM59 was designed and chemically synthesized by GenePharma Co. (Shanghai, P.R. China). Lentivirus was packaged and testified by GenePharma Co.
mRNA Extraction and Real-Time PCR
Total RNAs from both clinical tissues and cultured cells were extracted by a QuickPrep mRNA Purification Kit (GE Healthcare, Piscataway, NJ, USA) according to the manufacturer’s protocols. The first-strand cDNA was produced by a synthesis kit (TaKaRa, Dalian, P.R. China) for reverse transcriptions. Real-time PCR was performed with the SYBR Premix Ex Taq Kit (TaKaRa) in an ABI PRISM 7900 Real-Time System. The protocol is briefly listed here: denaturation (95°C for 5 min), annealing (35 repetition of 95°C for 30 s, 60°C for 45 s, and 72°C for 60 s), and extension (72°C for 10 min). The primers were synthesized by JieLi Co. (Shanghai, P.R. China). TRIM59: 5′-TACGAGAGCAGCAGCTTGAA-3′ (forward) and 5′-ACGGGTTGAACCTCAGGAAG-3′ (reverse). GAPDH: 5′-GTGGACATCCGCAAAGAC-3′ (forward) and 5′-AAAGGGTGTAACGCAACTA-3′ (reverse). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. All experiments were performed at least three times in triplicate.
Western Blot Analysis
Western blotting analysis was performed according to the manufacturer’s protocols. Total proteins from both human tissues and cultured cells were extracted with lysis buffer, and the protein quality and quantity were detected by the Bradford dye-binding protein assay (Bio-Rad Laboratories, Hercules, CA, USA). A total of 50 μg of protein was loaded onto a 10% SDS polyacrylamide gel, after which PVDF membrane was used to transfer the proteins. The primary antibody against TRIM59 was purchased from Abcam (Cambridge, MA, USA), and primary antibody against GAPDH and secondary antibodies were commercially purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Cell Proliferation Assay
Cervical cancer cell proliferation was assessed using cell counts and bromodeoxyuridine (BrdU) incorporation with a commercial kit (EMD Millipore, Billerica, MA, USA) according to the manufacturer’s protocols. Briefly, cells were cultured with serum-free medium for 24 h and trypsinized. An equal number (5,000) of HeLa and SiHa cells from each group were seeded into 96-well plates and allowed to grow in culture medium (10% FBS) supplied with 10 μM BrdU at the indicated times. BrdU incorporation was examined by additional peroxidase substrates. Spectrophotometric detection was explored at a wavelength of 450 nm using a microplate reader (TECAN, Morrisville, NC, USA). Each assay was repeated at least three times in triplicate.
Colony Formation Assay
For the colony formation assay, both HeLa and SiHa cells (500 per/well) were seeded into six-well culture plates and treated with specific shRNA against TRIM59. After incubation for 2 to 4 weeks, the colonies were fixed in a 4% paraformaldehyde solution, stained with crystal violet, imaged, and calculated in five randomly selected fields.
Cell Cycle Assay
Each group of HeLa and SiHa cells was seeded at a concentration of 3 × 105 cells/well in six-well plates and treated with shRNA for 72 h when 85% confluence was reached. After being washed with PBS three times, cells were collected by low-speed centrifugation (1,000 rpm, 4°C, 5 min). Cell pellets were resuspended with 1 ml of PBS solution, fixed with 75% ice-cold ethanol, and stored in −20°C for 2 days. Prior to FCM analysis, cells were centrifuged, washed twice with PBS, and resuspended in propidium iodide (PI) staining solution (50 μl/ml PI and 250 μl/ml RNase A). The cell mixture was incubated at 4°C for 30 min in darkness and analyzed with FACS (Beckman, Germany).
Transwell Assay
Cell migration and invasion were explored with Transwell chambers (pore size: 8 μm; Corning, NY, USA). For cell migration assay, a total of 5 × 104 HeLa or SiHa cells were seeded into the upper chamber in serum-free DMEM, and 600 μl of DMEM supplied with 10% FBS was added into the lower chamber. After incubating in a 37°C incubator for 24 h, cells were washed with PBS, fixed with ice-cold methanol, and stained with crystal violet for 5 min. Cells on the upper surface of the membrane were removed by cotton swabs, and those on the bottom surface were photographed and counted under a light microscope (Nikon, Minato, Tokyo, Japan) in five random fields. For the invasion assay, the membranes on the upper chamber were precoated with Matrigel (BD Biosciences, Woburn, MA, USA) for 6 h at 37°C.
Wound-Healing Assay
For wound-healing scratch assay, HeLa and SiHa cells were preinfected with shTRIM59 for 48 h and seeded in six-well plates. After 24 h of incubation, a wound was scratched with 10-μl pipette tips across the bottom of the plates on the monolayer of the confluent cells. Afterward, cells were washed gently with PBS and then incubated in DMEM supplemented with 10% FBS. The same area of the gap in each well was imaged at 20× magnification by a light microscope equipped with a digital camera (Nikon) at 0 and 24 h after scratching and quantified by calculating the difference between the initial and final areas.
Statistical Analysis
All data were presented as the means ± SD unless otherwise indicated. Student’s t-test was used to assess the statistical significance between variables. All statistical analyses were carried out with SPSS PASW Statistics 18.0 software (Chicago, IL, USA), and a value of p < 0.05 was considered as significant.
RESULTS
TRIM59 Is Upregulated in Cervical Cancer
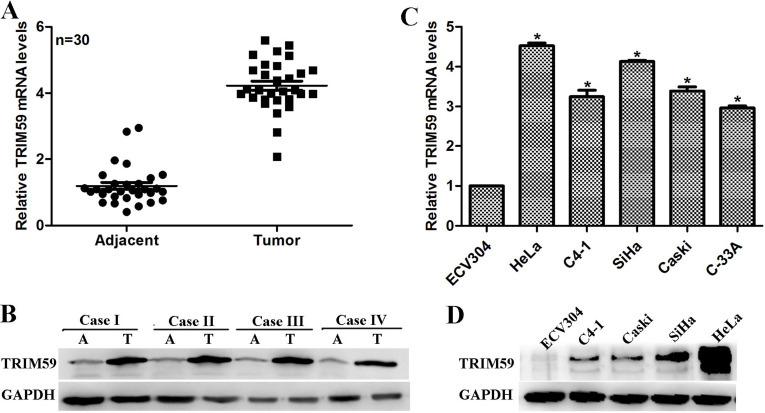
The expression of TRIM59 was initially determined in 30 clinical cervical cancer tissues. qRT-PCR analysis showed that, compared with adjacent noncancerous tissues, the relative mRNA of TRIM59 had an approximately fourfold increase in the cancerous tissues (Fig. 1A). Likewise, the protein level of TRIM59 was remarkably high in the cancerous tissues compared with their counterparts (Fig. 1B). These data suggest increased expression of TRIM59 in clinical cervical cancer. Furthermore, compared with the normal epithelial cell ECV304, the mRNA level of TRIM59 was revealed to be upregulated in cervical cancer cell lines, with its increase extent varied among the selected cancer cells including HeLa, C4-1, SiHa, Caski, and C-33A. HeLa and SiHa cells were the two most increased cell lines, which exhibited a 4.5- and 4-fold increase, respectively (Fig. 1C). Western blot analysis further verified that expression of TRIM59 was mostly remarkable in HeLa cells and SiHa cells (Fig. 1D). These in vitro assessments suggested a differentially expressed profile of TRIM59 and also verified the clinical observation that TRIM59 was upregulated in cervical cancer.
Figure 1.
TRIM59 is upregulated in cervical cancer. (A, B) The expression of TRIM59 was determined by qRT-PCR analysis and Western blot, respectively. A total of 30 clinical cervical cancer cases were collected and subjected to qRT-PCR analysis of the mRNA level of TRIM59. Results representing the high protein level of TRIM59 in tumor tissues (T) instead of adjacent noncancerous tissues (A) were also shown. (C, D) A series of cell lines, including normal epithelial ECV304 cells and cervical cancer cells HeLa, C4-1, SiHa, Caski, and C-33A, were cultured. The total RNAs and proteins were collected from the cultured cells and subjected to detection of TRIM59 mRNA and protein levels. Compared with the normal epithelial ECV304 cells, TRIM59 was upregulated in cervical cancer cell lines at both mRNA and protein levels.
Knockdown of TRIM59 Inhibits Cell Viability and Colony Formation in Cervical Cancer Cells
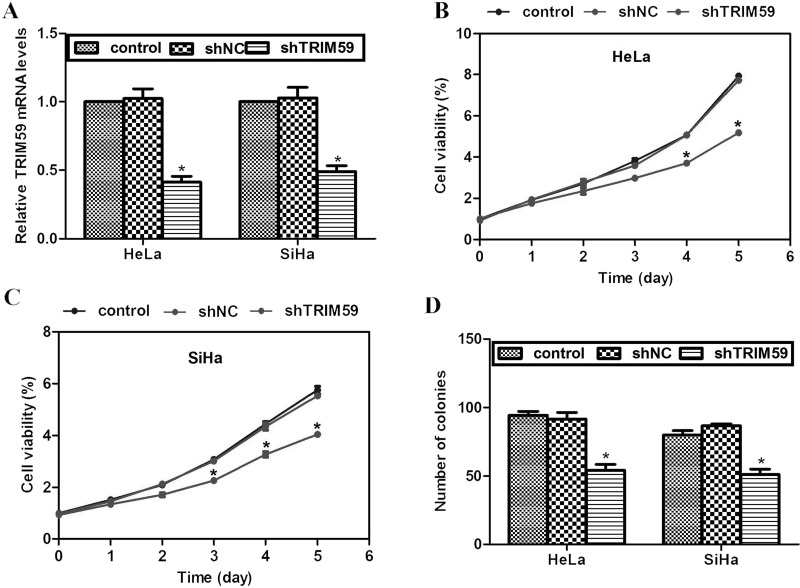
Next, a specific shRNA against TRIM59 (shTRIM59) was employed to knock down the expression of TRIM59. HeLa cells and SiHa cells were selected because these two cell lines had the highest expression of TRIM59. As shown in Figure 2A, transfection of shTRIM59 into either HeLa cells or SiHa cells caused significant decreases of TRIM59 mRNA levels, while a scramble negative control shRNA (shNC) barely had any effect on TRIM59 expression (Fig. 2A), suggesting the high effectiveness of our synthesized shRNA. With the specific shTRIM59, HeLa cells with or without transfection of shTRIM59 were assayed for cell viability detection. Cell viabilities among the groups were not significantly different for the former 3 monitored days. However, by day 4, cells from the shTRIM59-transfected group exhibited significantly less viability. This was even true by day 5 when shTRIM59-transfected HeLa cells were only 56.2% active compared with that from control groups (Fig. 2B). Comparable results were also observed in SiHa cells, where knockdown of TRIM59 caused cell viability decrease by up to 30% by day 5 (Fig. 2C). In the colony formation assay, it was observed that control cells formed an average of 100 colonies; however, shTRIM59-transfected cells only formed approximately 50 colonies, representing a 50% decrease in colony formation ability (Fig. 2D). These data suggest that knockdown of TRIM59 inhibits cervical cancer cell proliferation in vitro.
Figure 2.
Knockdown of TRIM59 inhibits cell proliferation and colony formation in cervical cancer cells. (A) Specific shRNA against TRIM59 (shTRIM59) was employed to knock down TRIM59 in cervical cancer HeLa cells and SiHa cells. (B, C) Effects of TRIM59 knockdown on cell proliferation were assessed in HeLa cells and SiHa cells. This assay was monitored for 5 consecutive days. (D) Colony formation assay was conducted to assess effects of TRIM59 knockdown on cell growth.
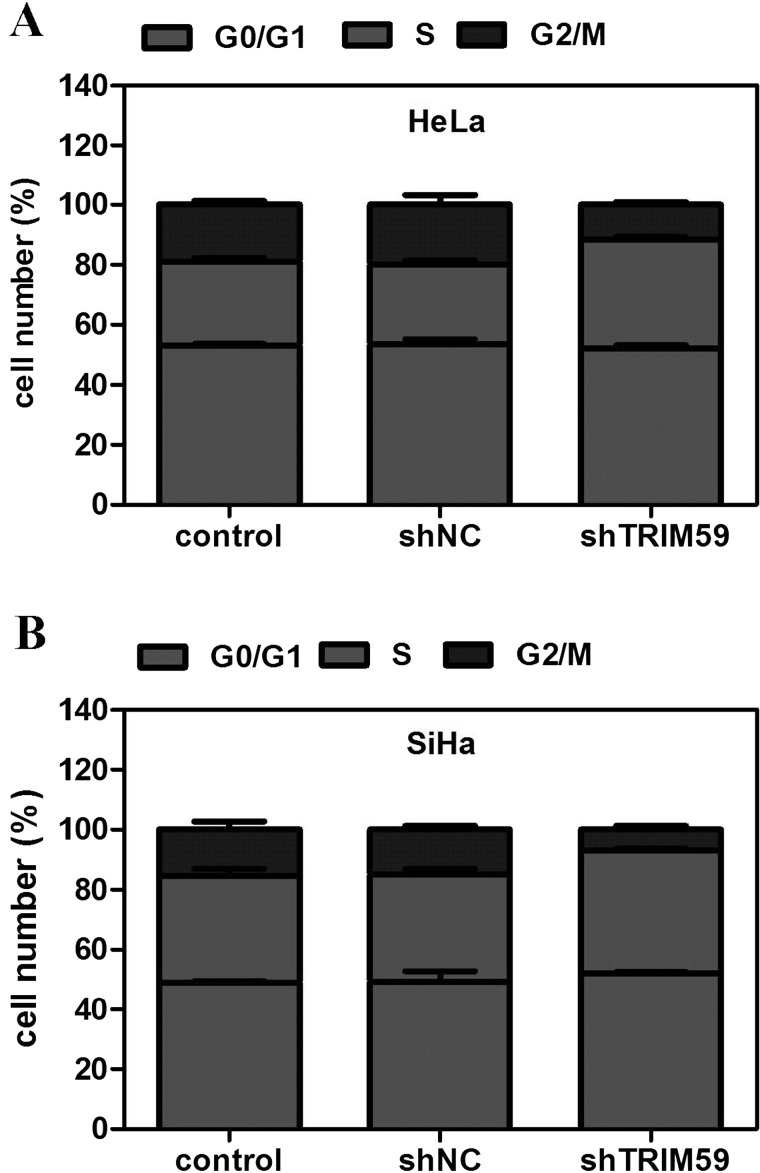
Knockdown of TRIM59 Arrests Cell Cycle in the S Phase
Cell proliferation depends largely on cell cycle progression. Hence, the effect of TRIM59 knockdown on cell cycle progression was also assessed using flow cytometry. In HeLa cells, it was observed that after knockdown of TRIM59, cells were proportionally accumulated in the S phase with cell proportion in the G2/M phase largely decreased (Fig. 3A). Similarly, cells in the G2/M phase were significantly decreased, while cells in the S phase were increased in TRIM59-depleted SiHa cells (Fig. 3B). These data suggest that knockdown of TRIM59 interrupts cell cycle progression.
Figure 3.
Knockdown of TRIM59 arrests cell cycle in the S phase. (A, B) Effects of TRIM59 knockdown on cell cycle progression were assessed in HeLa and SiHa cells by flow cytometry. The cell proportions in each phase are shown in bars.
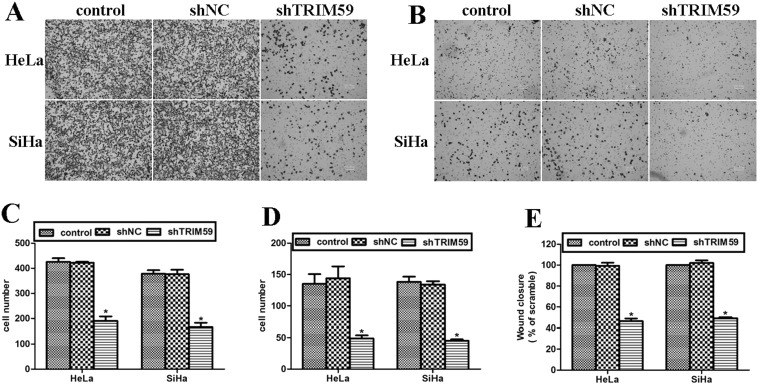
Knockdown of TRIM59 Inhibits Cell Migration and Invasion Abilities in Cervical Cancer
Cell migration and invasion abilities were assessed using Transwell assay. It was shown that knockdown of TRIM59 caused a significantly decreased number of cells that migrated to the lower surface of the chamber (Fig. 4A). Cells that migrated through the Matrigel were also sparse under the microscopic views (Fig. 4B). By counting the number of migrated cells, it was shown that knockdown of TRIM59 decreased the migrated cells by up to 57.1% in HeLa cells and 52.6% in SiHa cells (Fig. 4C). HeLa cells that invaded through the Matrigel were also decreased by up to 75.8%, while the decrease was up to 74.7% in SiHa cells (Fig. 4D). Wound-healing assay, which reflects cell migration ability, was also conducted. The percentage of wound closure was calculated 24 h after the wound scratch. It was observed that TRIM59-depleted cells were significantly inhibited from wound recovery based on the half wound closure rate compared with control groups (Fig. 4E). All these observations suggest that knockdown of TRIM59 significantly inhibits cell migration and invasion abilities in HeLa and SiHa cells.
Figure 4.
Knockdown of TRIM59 inhibits cell migration and invasion abilities in cervical cancer. (A, B) Cell migration (A) and invasion abilities (B) were assessed using Transwell assay. After each treatment, cells were allowed to migrate for 8 h before proceeding to crystal violet staining. (C, D) After staining, three randomly selected fields were photographed under microscopy. The cell numbers in the three images were then averaged and shown in bars. Cell migration (C) and invasion (D) were shown separately in the histograms. (E) Wound-healing assay, which reflects cell migration ability, was conducted. The percentage of wound closure in HeLa and SiHa cells was calculated 24 h after the wound scratch.
DISCUSSION
Cervical cancer remains one of the most common cancers affecting women worldwide. Two complementary approaches have been launched for the prevention of cervical cancer: primary prevention through vaccination to prevent HPV infection and secondary prevention through screening to detect and treat cervical precancerous lesions before they become invasive24. However, substantial numbers of patients are newly diagnosed and suffering from this malignancy. It is estimated that 500,000 cases are newly diagnosed, and nearly 200,000 deaths are attributable to the disease around the world annually25. Novel biomarkers are hence desperately needed for early diagnosis and treatment of this malignancy.
The TRIM family of proteins has been implicated in multiple critical processes including tumorigenesis20. Of the members of the TRIM family, TRIM59 is a novel molecule that has been observed to be highly elevated in gastric cancer and promotes gastric cancer progression by degrading p53 in a ubiquitination-dependent manner23. In fact, TRIM59 has been reported to be critically involved in prostate cancer. TRIM59 communicated with the Ras signaling pathway in a transgenic prostate cancer mouse model21. Upregulation and hyperphosphorylation of TRIM59 protein were initiated in the prostate cytoplasm in early tumorigenesis from prostate intraepithelial neoplasia (PIN)21. Phosphorylation of Ser/Thr sites (p-Ser/Thr) of TRIM59 correlated with tumorigenesis, while p-Tyr-TRIM59 proteins correlated with advanced prostate cancer26. All of this research suggests that TRIM59 might be involved in prostate cancer. However, as a cancer also involving the genital system, the role of TRIM59 in cervical cancer has never been revealed. There is so far only one pioneering study reporting that TRIM59 was upregulated in cervix adenocarcinoma. Its staining intensity in IHC analysis was moderate–strong22. Our present study has provided evidence that TRIM59 is significantly increased in clinical cervical cancer tissues, which is consistent with the previous report22. With the shRNA depleting TRIM59 expression in HeLa and SiHa cells, cell proliferation and colony formation were consistently inhibited. Cell mobility was also suppressed by knockdown of TRIM59. Our data strongly suggest that TRIM59 plays a critical role in the progression of cervical cancer. Hence, this is the first report to our knowledge that identifies TRIM59 as a key mediator of cell proliferation and migration in cervical cancer cells.
One interesting question would be the mechanism(s) of how TRIM59 functions to mediate the progression of cervical cancer. Structurally, TRIM59 contains the RING domain, which acts as E3 ubiquitin ligases and is involved in the ubiquitin–proteasome system in the regulation of numerous cellular processes including cell cycle progression, gene transcription, and signal transduction11. p53, the widely known tumor suppressor, is negatively regulated by TRIM59 in a ubiquitination manner, by which TRIM59 promotes tumor growth, cell proliferation, and migration in gastric cancer23. Hence, the SV40 Tag/p53/pRB routes have been considered as one mechanism for TRIM59 functioning in the process of tumorigenesis. In addition, previous studies have suggested that the Ras pathway is also involved in TRIM59 function. The initial functional targets of TRIM59 were on the Ras signal pathway as an early and rapid signal transmitter27. Upregulation of TRIM59 was correlated with BRAF, an early signal effector of the Ras signal pathway27. Hence, the signal pathways for TRIM59 function may be linked with (1) the Ras/Raf pathway and (2) the p53/pRB routes21. In our study, cell cycle progression was arrested in the S phase after knockdown of TRIM59. In fact, either pathway (Ras or p53) is associated with cell cycle arrest. Hence, the suggested two pathways may be involved in TRIM59-mediated cell cycle arrest in cervical cancer HeLa cells and SiHa cells. Further work concerning exactly which pathway contributes to TRIM59-mediated biological activities in cervical cancer is still needed.
In all, the present study identified a novel protein, TRIM59, as a key mediator of cell proliferation and migration in cervical cancer cells. Knockdown of TRIM59 caused a significant decrease in proliferating cells, formed colonies, and migrated cells. Our data may provide novel evidence that target against TRIM59 may be a potential therapeutic strategy for the detection and treatment of cervical cancer in clinics.
REFERENCES
- 1. Capote Negrin LG. Epidemiology of cervical cancer in Latin America. Ecancermedicalscience 2015;9:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Siegel R, Naishadham D, Jemal A. Cancer Statistics, 2013. CA Cancer J Clin. 2013;63:11–30. [DOI] [PubMed] [Google Scholar]
- 3. Drain PK, Holmes KK, Hughes JP, Koutsky LA. Determinants of cervical cancer rates in developing countries. Int J Cancer 2002;100:199–205. [DOI] [PubMed] [Google Scholar]
- 4. Gatta G, Ciccolallo L, Kunkler I, Capocaccia R, Berrino F, Coleman MP, De Angelis R, Faivre J, Lutz JM, Martinez C, and others. Survival from rare cancer in adults: A population-based study. Lancet Oncol. 2006;7:132–40. [DOI] [PubMed] [Google Scholar]
- 5. Galloway TJ, Ridge JA. Management of squamous cancer metastatic to cervical nodes with an unknown primary site. J Clin Oncol. 2015;33:3328–37. [DOI] [PubMed] [Google Scholar]
- 6. Biglia N, Bounous VE, Sgro LG, D’Alonzo M, Gallo M. Treatment of climacteric symptoms in survivors of gynaecological cancer. Maturitas 2015;82:296–8. [DOI] [PubMed] [Google Scholar]
- 7. Yan J, Zhang Y, Ren C, Shi W, Chen L. Involvement of nuclear protein C23 in activation of EGFR signaling in cervical cancer. Tumour Biol. 2016;37:905–10. [DOI] [PubMed] [Google Scholar]
- 8. Ozato K, Shin DM, Chang TH, Morse HC 3rd. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci USA 2007;104:6200–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Keeble AH, Khan Z, Forster A, James LC. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc Natl Acad Sci USA 2008;105:6045–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007;446:916–20. [DOI] [PubMed] [Google Scholar]
- 12. Short KM, Cox TC. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J Biol Chem. 2006;281:8970–80. [DOI] [PubMed] [Google Scholar]
- 13. Wang L, Heidt DG, Lee CJ, Yang H, Logsdon CD, Zhang L, Fearon ER, Ljungman M, Simeone DM. Oncogenic function of ATDC in pancreatic cancer through Wnt pathway activation and beta-catenin stabilization. Cancer Cell 2009;15:207–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 2009;136:913–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balastik M, Ferraguti F, Pires-da Silva A, Lee TH, Alvarez-Bolado G, Lu KP, Gruss P. Deficiency in ubiquitin ligase TRIM2 causes accumulation of neurofilament light chain and neurodegeneration. Proc Natl Acad Sci USA 2008;105:12016–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lerner M, Corcoran M, Cepeda D, Nielsen ML, Zubarev R, Ponten F, Uhlen M, Hober S, Grander D, Sangfelt O. The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol Biol Cell 2007;18:1670–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. [DOI] [PubMed] [Google Scholar]
- 18. de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 1991;66:675–84. [DOI] [PubMed] [Google Scholar]
- 19. Cambiaghi V, Giuliani V, Lombardi S, Marinelli C, Toffalorio F, Pelicci PG. TRIM proteins in cancer. Adv Exp Med Biol. 2012;770:77–91. [DOI] [PubMed] [Google Scholar]
- 20. Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer 2011;11:792–804. [DOI] [PubMed] [Google Scholar]
- 21. Valiyeva F, Jiang F, Elmaadawi A, Moussa M, Yee SP, Raptis L, Izawa JI, Yang BB, Greenberg NM, Wang F, Xuan JW. Characterization of the oncogenic activity of the novel TRIM59 gene in mouse cancer models. Mol Cancer Ther. 2011;10:1229–40. [DOI] [PubMed] [Google Scholar]
- 22. Khatamianfar V, Valiyeva F, Rennie PS, Lu WY, Yang BB, Bauman GS, Moussa M, Xuan JW. TRIM59, a novel multiple cancer biomarker for immunohistochemical detection of tumorigenesis. BMJ Open 2012;2(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou ZC, Ji ZZ, Wang Y, Li J, Cao H, Zhu HLH, Gao WQ. TRIM59 is up-regulated in gastric tumors, promoting ubiquitination and degradation of p53. Gastroenterology 2014;147:1043–54. [DOI] [PubMed] [Google Scholar]
- 24. El-Zein M, Richardson L, Franco EL. Cervical cancer screening of HPV vaccinated populations: Cytology, molecular testing, both or none. J Clin Virol. 2016;76(Suppl 1):S62–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Z, Wang H, Wang Z, Cai H. MiR-195 inhibits the proliferation of human cervical cancer cells by directly targeting cyclin D1. Tumour Biol. 2016;37:6457–63. [DOI] [PubMed] [Google Scholar]
- 26. Xuan JW, Bygrave M, Valiyeva F, Moussa M, Izawa JI, Bauman GS, Klibanov A, Wang F, Greenberg NM, Fenster A. Molecular targeted enhanced ultrasound imaging of flk1 reveals diagnosis and prognosis potential in a genetically engineered mouse prostate cancer model. Mol Imaging 2009;8:209–20. [PubMed] [Google Scholar]
- 27. Coleman ML, Marshall CJ, Olson MF. RAS and RHO GTPases in G1-phase cell-cycle regulation. Nat Rev Mol Cell Biol. 2004;5:355–66. [DOI] [PubMed] [Google Scholar]