Abstract
Breast cancer is the leading cause of cancer deaths in females all over the world, mainly resulting from metastasis. Previous studies have revealed that repressor element-1 (RE-1) silencing transcription (REST) acted as a tumor suppressor in breast cancer. However, the mechanism by which REST is regulated remains unknown, and its role in the metastasis in breast cancer cells remains unclear. In the present study, we showed that the expression of REST was lower in breast cancer samples than that of adjacent samples by immunohistochemical analysis, which may be due to hypermethylation of the REST promoter. Low REST levels are significantly associated with malignant progression in breast cancer patients. Additionally, we elucidated the functions of REST on proliferation and invasion in breast cancer cells. Lentivirus transfection was used to overexpress REST in human breast MDA-MB-231 cells. Then the biologic consequences of overexpressing REST in regard to cell proliferation, apoptosis, and invasion were determined. Furthermore, we also determined matrix metalloproteinase-9 (MMP9) as a target of REST. These results demonstrate that downregulation of REST, a tumor suppressor in breast cancer, is associated with hypermethylation. Induced REST expression is capable of attenuating invasion ability of breast cancer cells, which may be a novel strategy for metastatic breast cancer treatment.
Key words: Breast cancer, Repressor element-1 silencing transcription (REST), Methylation, Metastasis, Matrix metalloproteinase (MMP)
INTRODUCTION
Breast cancer is the leading cause of cancer deaths in females all over the world, with increasing morbidity each year1. With the progress of surgery and neoadjuvant chemotherapy, the overall survival of patients with breast cancer can be effectively increased in the early stages2. However, once metastasis occurs, the mortality of breast cancer is sharply increased3. Thus, it is a necessity to repress cancer cell metastasis if we want to lower the mortality rate of breast cancer.
The repressor element-1 (RE-1) silencing transcription (REST) factor, also known as neuron-restrictive silencing factor (NRSF), is highly expressed in stem cells and nonneural cells, with low expression in neurons and other neural cells4. Previous studies have revealed that REST has a dual role in cancer cell proliferation depending on the type of cancer. It acts as an oncogene in medulloblastomas5, neuroblastomas6, and glioblastomas7, while acting as a tumor suppressor in lung cancer8, colon cancer9, and breast cancer10. In some epithelial cell types, such as human mammary epithelial cells, high REST could prevent cell proliferation. By genetic screening, REST was identified as a tumor suppressor in human mammary carcinoma cells and prevented anchorage-independent proliferation. Deletions and truncations of REST in colon carcinoma lines resulted in rapid proliferation, which was greatly reduced by the transfection of exogenous REST10. Our previous study demonstrated that REST showed low levels in breast carcinoma tissues and cell lines, and downregulation of REST stimulated MCF-7 cell proliferation11. These results suggest that REST as a tumor suppressor plays a role in the development of breast cancer.
However, the mechanism by which REST is regulated remains unknown, and its role in cancer cell metastasis, including breast cancer, is yet unclear. Epigenetic mechanism, including methylation and acetylation, has been implicated in tumor suppressor silence12. Class I histone deacetylases are the key REST effectors13. Valproic acid (VPA), an inhibitor of histone deacetylases, can prevent their invasion into surrounding tissues and may inhibit tumor angiogenesis5. Despite the broad substrate specificity of VPA, these data indicated the close relationship between REST and epigenetic mechanism.
In the present study, we showed that the expression of REST was lower in breast cancer samples than that in adjacent samples by immunohistochemical (IHC) analysis, which may be due to hypermethylation of the REST promoter. Additionally, we elucidated the functions of REST on proliferation and invasion in breast cancer cells. Lentivirus transfection was used to overexpress REST in human breast MDA-MB-231 cells. The biologic consequences of overexpressing REST in regard to cell proliferation, apoptosis, and invasion were then determined. Furthermore, we determined matrix metalloproteinase-9 (MMP9) as a target of REST. These results demonstrate that downregulation of REST is associated with hypermethylation and is a biomarker for evaluating the malignant progression. Induced REST expression is capable of attenuating the invasion ability of breast cancer cells, which may be a novel strategy for metastatic breast cancer treatment.
MATERIALS AND METHODS
Breast Cancer Tissue Samples
A total of 144 tissue samples, including 72 cancer tissues and 72 paired adjacent tumor tissues, were obtained from the Xiangya Hospital according to the Legislation and Ethical Boards of Central South University. Informed consents were signed by all the subjects. Information on the patients providing specimens was obtained from surgical and pathological records. All samples were collected and identified by histopathological evaluation and stored in liquid nitrogen until use.
Cell Culture
The human breast cancer cell lines SK-BR3, T47D, MCF-7, and MDA-MB-231, and the normal human mammary gland cells, HBL100, were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). SK-BR3, MCF-7, and MDA-MB-231 cells were cultured in DMEM (Invitrogen Life Technologies, Carlsbad, CA, USA) supplemented with 10% (v/v) fetal bovine serum (FBS) (Invitrogen Life Technologies). T47D and HBL100 cells were grown in RPMI-1640 (Invitrogen Life Technologies) supplemented with 10% FBS (Invitrogen Life Technologies). All cells were cultured at 37°C in a humidified 5% CO2 incubator.
Cell Treatment
The lentivirus overexpressed REST and MMP9 were designed and purchased from GeneChem (Shanghai, P.R. China). Forty-eight hours after transfection, the expression of REST and MMP9 was detected by Western blotting, and the transfected cells were used for further analysis. 5-Aza (5′-aza-deoxycytidine) was purchased from Sigma-Aldrich (St. Louis, MO, USA). The cells were treated with 4 μmol/L 5-Aza for 48 h and then used for further analysis.
Immunohistochemical (IHC) Staining
The paraffin-embedded tissue samples were serially cut at 4 μm. Slides were deparaffinized in dimethylbenzene and dehydrated in alcohol gradient. Slides were immersed in 3% hydrogen peroxide for inactivating endogenous peroxidase and then retrieved in a citric acid buffer (pH 6.0) by high-pressure repair for 15 min. After cooling, the slices were rinsed with phosphate-buffered saline (PBS) and blocked with normal goat serum and incubated with primary antibody rabbit polyclonal anti-REST antibody (1:100 dilution; Abcam, Cambridge, UK) overnight at 4°C. The sections were then washed with PBS and incubated with anti-rabbit secondary antibody (Zsbio, Beijing, P.R. China) for 2 h at 37°C. The sections were then washed with PBS and stained using a DAB Detection Kit (Zsbio). Finally, the sections were counterstained with hematoxylin.
The staining was scored by two independent pathologists who were blinded to clinicopathologic features. REST staining intensity was scored as 0 (negative, −), 1 (weak, +), 2 (moderate, ++), and 3 (strong, +++). The extent of staining was scored as 0–1.0 (0–100%). The final staining score (0–3) was calculated as the multiplication of the intensity score and extent score. The final score ≥1 was defined as high expression; otherwise it was defined as low expression.
Western Blot
RIPA lysis buffer (Zsbio) was used to extract protein from tissues and indicated cells. BCA Protein Assay Kit (Thermo Scientific, Waltham, MA, USA) was used to measure protein concentration. A total of 50 μg of protein was separated on 10% SDS-PAGE and blotted onto 0.22-μm nitrocellulose membranes. The membranes were blocked for 2 h with 5% nonfat dry milk diluted with tris-buffered saline (TBS) and incubated with primary antibodies [rabbit polyclonal anti-REST (1:100), rabbit polyclonal anti-MMP9 (1:100; Abcam), and mouse monoclonal anti-β-GAPDH (1:2,000; Abcam)] overnight at 4°C. The membranes were washed with TBS containing 0.1% Tween 20 (TBST) and then incubated with appropriate horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit, 1:2,000; goat anti-mouse, 1:2,000; Santa Cruz, Santa Cruz, CA, USA) for 1 h at 37°C. Enhanced chemiluminescence reagent (Merck Millipore, Darmstadt, Germany) was used to detect the signal on the membrane. The data were analyzed via densitometry using the Image-Pro plus software 6.0 (Bio-Rad, Hercules, CA, USA) and normalized to the expression of the internal control (GAPDH).
Quantitative Real-Time Polymerase Chain Reaction (qPCR)
Total RNA was extracted from the tissues and indicated cells using the Ultrapure RNA Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The expression of REST and MMP9 mRNA was detected by real-time PCR using FastLane Cell SYBR® Green Kit (Qiagen). The primers of REST, MMP9, and β-actin are as follows: REST, CCAGCACCCAACTTTACCAC (sense) and TGTTATCCCCAACCGGCATC (antisense); MMP9, CATCCGGCACCTCTAT GGTC (sense) and CATCGTCCACCGGACTCAAA (antisense); β-actin, AGGGGCCGGA CTCGTCATACT (sense) and GGCGGCACCACCATGTACCCT (antisense). The 2−ΔΔCT method was used to analyze the data, and β-actin expression was used as the internal control.
Measurement of REST Promoter CpG Island Methylation Status by Methylation-Specific PCR (MSP)
Genomic DNA was extracted using the Qiagen FFPE DNA Kit (Qiagen). Genomic DNA (1 μg per sample) was modified with bisulfite using the Qiagen DNA Methylation-Gold™ Kit (Qiagen) following the manufacturer’s instructions. MSP was performed on bisulfate-treated DNA. The primers used were unmethylated REST (forward, TATTGTAAATGTTGAATTTTAGGTTTTGA; reverse, AATATATCTTTTTCTATACAATATAAACAA) and methylated REST (forward, ATATTGTAAATGTTGAATTTTAGGTTTC; reverse, AATATATCTTTTTCTATACAATATA AACGA). The annealing temperature was 60°C for methylated PCR, and 55°C for unmethylated PCR, with 27 cycles used for each.
MTT Assay
The indicated cells were seeded into 96-well plates at 1 × 103 cells/well. Cell proliferation was assessed every 24 h by MTT assay. Briefly, cells were exposed to MTT (Sigma-Aldrich) at a final concentration of 5 mg/ml and incubated for an additional 4 h at 37°C. The formazan generated in each well was dissolved in 150 ml of dimethyl sulfoxide (Sigma-Aldrich). Absorbance of each well at 490 nm was read using a microplate reader (Synergy™ Mx; BioTek, Winooski, VT, USA).
Flow Cytometric Analysis of the Cell Apoptosis
Following the indicated treatment, cells were trypsinized and washed with ice-cold PBS. The cell suspensions were incubated for 15 min in the dark using annexin V/propidium iodide (PI) detection kit (Transgen Inc., Beijing, P.R. China) according to the manufacturer’s instructions. Then cell apoptosis was analyzed by flow cytometry (Becton, Dickinson and Company, San Jose, CA, USA). The experiments were performed in triplicate.
Transwell Assay
The indicated cells were starved for 24 h, resuspended in serum-free medium, and added to the upper chamber. The lower chamber was filled with medium containing 10% FBS. Following 48 h in the culture, cells attached to the bottom were fixed and stained with crystal violet for 45 min and air dried. The optical density (OD) at 570 nm of crystal violet dissolved by 10% acetic acid was detected by an enzyme immunoassay analyzer (Synergy™ Mx; BioTek).
Luciferase Reporter Gene Assays
The MMP9 promoter region was ligated into the pGL3-enhancer vector (Promega, Madison, WI, USA) generating pGL3-enhancer MMP9 (MMP9 promoter) performed by GeneCopoeia (Guangzhou, P.R. China). All vector sequences were confirmed by DNA sequencing. Cells were transfected with the MMP9 promoter or pGL3-enhancer reporter plasmid together with expression vectors for REST. At 48 h after transfection, cells were collected and lysed. The supernatants (10 ml) were used to detect luciferase activities using the luciferase reporter gene assay kit (BioVision, Milpitas, CA, USA) on a microplate luminometer.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5 software (GraphPad Software Inc., La Jolla, CA, USA), and the data are presented as the mean ± standard deviation. An unpaired two-tailed Student’s t-test or one-way analysis of variance (ANOVA) with Bonferroni t posttest was used to analyze the data depending on conditions. Chi-square test was used to analyze the association between REST expression and clinicopathologic characteristics in breast cancer. Multivariate prognostic factors were examined using Cox’s proportional hazards model. A value of p < 0.05 was considered to indicate a statistically significant difference.
RESULTS
REST Is Downregulated in Breast Cancer, Which May Be Induced by Hypermethylation of Promoter
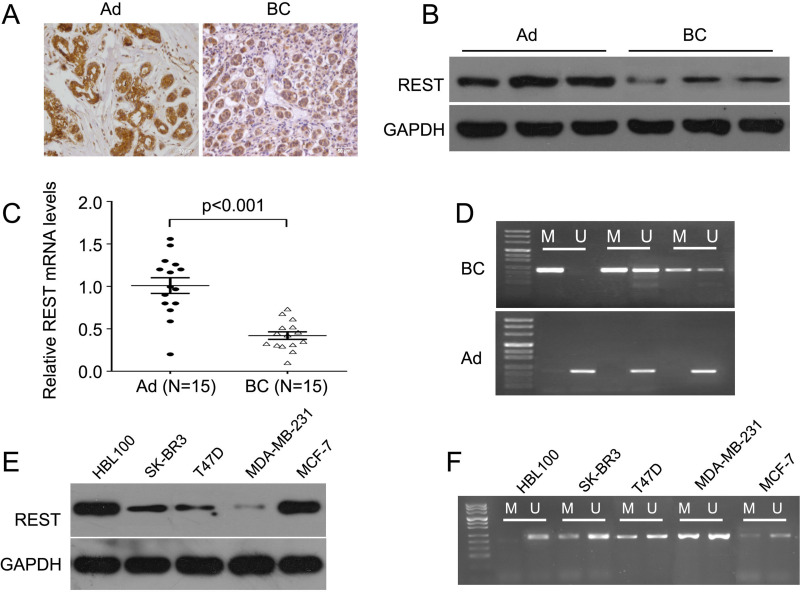
To investigate the role of REST in breast cancer, we detected the expression level of REST in 72 breast cancer tissues and the paired adjacent tissues by IHC staining. We found that REST expression was significantly lower in breast cancer samples compared with adjacent controls, as indicated by the immunostaining (Fig. 1A). In addition, the Western blot results also showed that REST was significantly downregulated in breast cancer tissues compared to adjacent control (Fig. 1B). In addition, we randomly selected 15 pairs of breast cancer tissues and the adjacent tissues to analyze the mRNA levels of REST by qPCR. The results showed that the mRNA levels of REST were significantly reduced in breast cancer tissues compared with the adjacent controls (Fig. 1C). In light of this, we further investigated the role of epigenetic mechanisms that may be involved in the silencing of REST. MSP was used to detect methylation of REST in three pairs of breast cancer tissues and adjacent tissues. A significantly higher level of methylation was observed in breast cancer tissues, but not in adjacent tissues. All the normal tissues presented as unmethylated (Fig. 1D). Additionally, we detected the expression of REST in four breast cancer cell lines. The results showed that REST expression was lower in breast cancer cell lines, especially in MDA-MB-231 cells, than in normal human mammary gland cells, HBL100 (Fig. 1E). Importantly, in line with the results in breast cancer tissues, we also observed a higher level of methylation of REST in the breast cancer cell lines, especially in the MDA-MB-231 cells (Fig. 1F).
Figure 1.
REST expression is downregulated and hypermethylated in breast cancer tissues and cell lines. (A) Representative images of immunostaining with REST in adjacent (left) and breast cancer tissue (right). The intensity of adjacent tissues was higher than in the breast cancer tissue. Scale bars: 50 μm. (B) Western blot detected REST protein expression levels. REST expression was significantly lower in breast cancer tissues than in adjacent tissues. (C) qPCR detected REST mRNA expression levels in breast cancer and adjacent tissues (15 cases for each). (D) MSP test results for three breast cancer tissues and the paired adjacent controls. (E) Western blot detected REST protein expression levels. REST expression was significantly lower in breast cancer cell lines than in normal cell line. (F) MSP test results for breast cancer cell lines. AD, adjacent; BC, breast cancer; M, methylation; U, unmethylated.
Decreased REST Levels Are Associated With T Stage, Clinical Stage, and Lymph Node Metastasis in Breast Cancer
We further studied the association between the REST levels and the clinical characteristics of breast cancer patients. All breast cancer patients were divided into two groups: high levels of REST and low levels of REST, according to the staining scores of REST. As demonstrated in Table 1, the REST levels were not correlated with age (p = 0.24), pathological grade (p = 0.80), distant metastasis (p = 0.3), Her2 expression (p = 0.24), or p53 status (p = 0.79). However, it was significantly associated with the T grade (p = 0.016), clinical stage (p = 0.002), lymph node metastasis (p = 0.008), ER expression (p = 0.018), PR expression (p = 0.008), and Ki-67 expression (p = 0.017). Thus, our data suggest that the REST levels may be used as a biomarker for evaluating the malignant progression of breast cancer.
Table 1.
Association Between REST Expression and Clinicopathologic Characteristics in Breast Cancer
| Variables | REST Expression | χ2 Test p | |
|---|---|---|---|
| Low (Score <1) (n = 42) | High (Score ≥1) (n = 30) | ||
| Age | 0.24 | ||
| <50 | 23 | 12 | |
| ≥50 | 19 | 18 | |
| Pathological grade | 0.80 | ||
| G1–2 | 28 | 21 | |
| G3 | 14 | 8 | |
| T stage | 0.016 | ||
| T1–2 | 22 | 23 | |
| T3–4 | 20 | 7 | |
| Lymph nodal metastasis | 0.008 | ||
| Yes | 28 | 10 | |
| No | 14 | 20 | |
| Distant metastasis | 0.30 | ||
| Yes | 10 | 11 | |
| No | 32 | 19 | |
| Clinical stage | 0.002 | ||
| I–II | 15 | 22 | |
| III–IV | 27 | 8 | |
| ER expression | 0.018 | ||
| Positive | 18 | 21 | |
| Negative | 26 | 9 | |
| PR expression | 0.008 | ||
| Positive | 28 | 10 | |
| Negative | 14 | 20 | |
| HER2 expression | 0.24 | ||
| Positive | 23 | 12 | |
| Negative | 19 | 18 | |
| Ki-67 expression | 0.017 | ||
| <14% | 25 | 26 | |
| ≥14% | 17 | 4 | |
| p53 status | 0.79 | ||
| Wild type | 29 | 22 | |
| Mutant | 13 | 8 | |
We further investigated the factors that could predict the prognosis of breast patients using the univariate and multivariate analyses. Univariate analysis data indicated that the level of REST (p = 0.004), as well as the T stage (p = 0.018), lymph node metastasis (p = 0.007), and clinical stage (p = 0.0008) were significantly associated with survival (Table 2). Moreover, as demonstrated in Table 3, the levels of REST (p = 0.015), lymph node metastasis (p = 0.031), and clinical stage (p = 0.008) were found to be independent factors for predicting the prognosis of breast cancer patients.
Table 2.
Univariate Analysis of Prognostic Factors of Breast Cancer
| Factors | Hazard Ratio | p Value |
|---|---|---|
| Age (≥50/<50) | 1.15 | 0.845 |
| Pathological grade (G3/G1–2) | 1.73 | 0.135 |
| T stage (T3–4/T1–2) | 2.83 | 0.018 |
| Lymph node metastasis (yes/no) | 3.75 | 0.007 |
| Distant metastasis (yes/no) | 1.31 | 0.224 |
| Clinical stage (III–IV/I–II) | 5.82 | 0.0008 |
| REST protein levels (low/high) | 4.88 | 0.004 |
Table 3.
Multivariate Analysis of Independent Prognostic Factors of Breast Cancer
| Factors | Hazard Ratio | p Value |
|---|---|---|
| Pathological grade | 1.53 | 0.079 |
| Lymph node metastasis | 2.73 | 0.031 |
| Clinical stage | 4.22 | 0.008 |
| REST protein levels | 3.68 | 0.015 |
Upregulation of REST Represses Cell Proliferation, Induces Apoptosis, and Inhibits Cell Invasion in MDA-MB-231 Cells
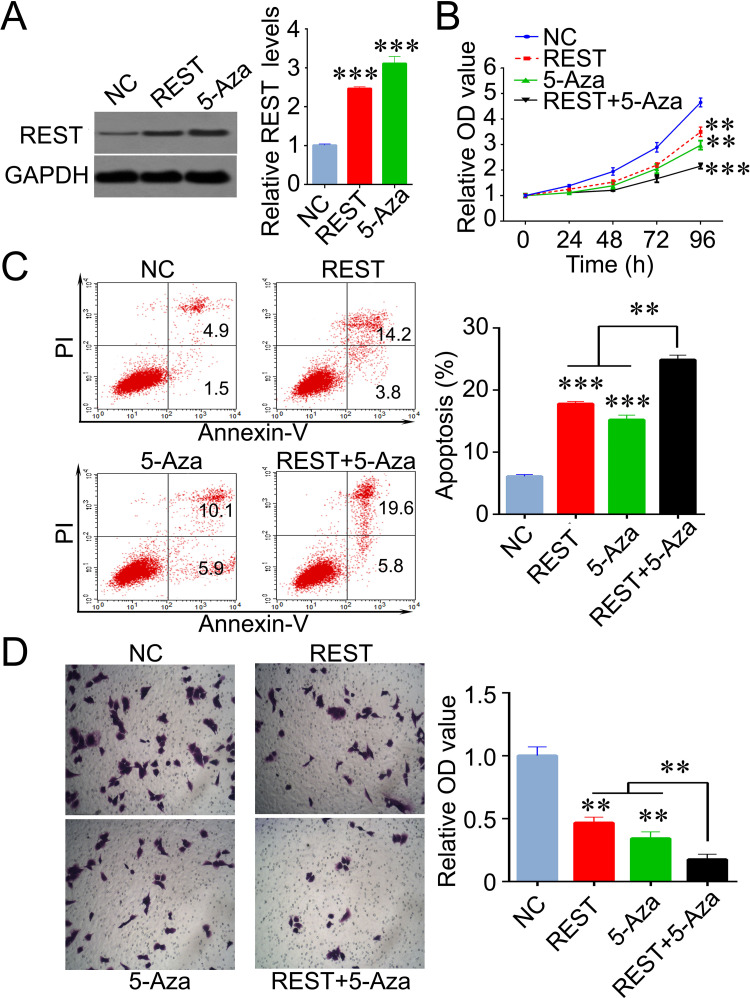
To investigate the role of REST on breast cancer cells, we induced the expression of REST by transfecting lentivirus and treating with 5-Aza in MDA-MB-231 cells. We found that the expression of REST was significantly increased after these treatments, indicating that hypermethylation might be, at least partially, the reason for REST downregulation in breast cancer (Fig. 2A). Next, with a battery of cell biology experiments, we found that induced REST expression resulted in inhibition of proliferation, increased apoptosis, and decreased invasive ability in MDA-MB-231 cells. Additionally, REST showed synthetic effects with 5-Aza in the inhibition of cell proliferation and induction of cell apoptosis in breast cancer cells (Fig. 2B–D).
Figure 2.
Induced REST exhibits an inhibitory role in breast cancer cells. (A) REST expression levels were significantly increased by REST lentivirus transfection and epigenetic drug (5-Aza) treatment in MDA-MB-231 cells. (B) MDA-MB-231 cell proliferation was inhibited by 5-Aza and REST. (C) 5-Aza and REST lentivirus transfection induced an increased cell apoptosis. (D) Invasive ability of cells was reduced by 5-Aza and REST lentivirus transfection. Data are expressed as mean ± standard deviation from three independent experiments. **p < 0.01, ***p < 0.001 versus negative control. NC, negative control; 5-Aza, 5′-aza-deoxycytidine.
REST Downregulates the Expression of MMP9 mRNA and Protein by Directly Binding to the MMP9 Promoter
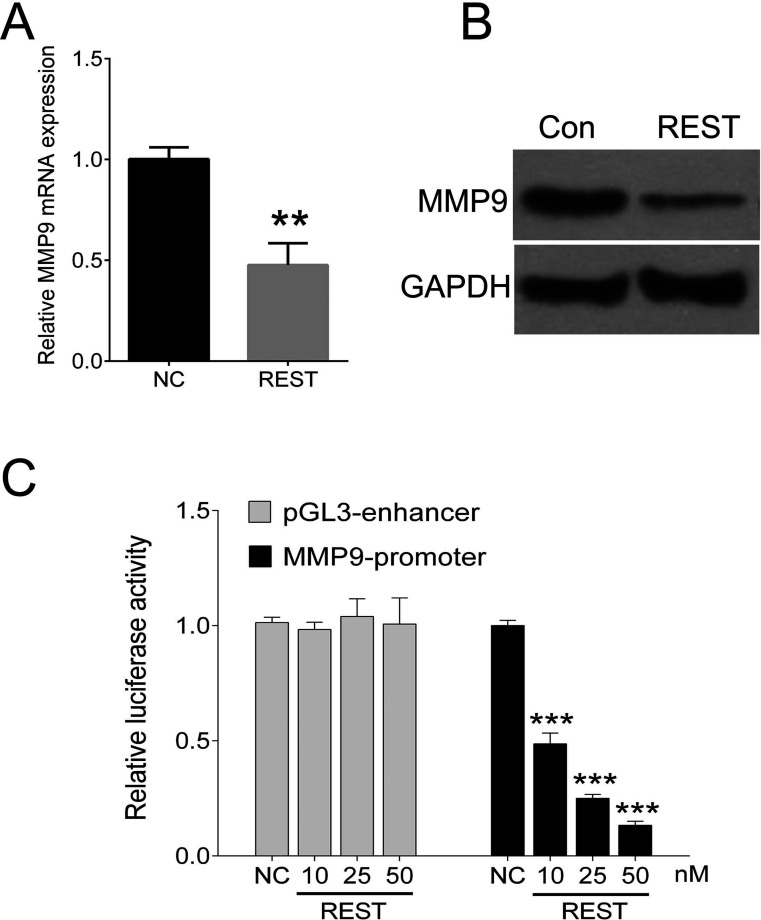
MMP9 is a putative target gene for REST that is involved in invasion and metastasis of breast cancer. We investigated endogenous MMP9 gene expression and transduced MDA-MB-231 cells with a lentivirus encoding for REST. The mRNA and protein levels of MMP9 were sigificantly reduced in cells expressing REST (Fig. 3A and B). To investigate whether REST directly binds to the MMP9 promoter, we cotransfected MDA-MB-231 cells with different amounts of a REST expression vector, together with the reporter plasmid MMP promoter or empty vector (pGL3 enhancer). Clearly, cotransfection of REST concentration dependently reduced luciferase expression from the MMP9 promoter (Fig. 3C).
Figure 3.
REST regulates MMP9 expression by repressing its promoter activity. (A) Induced REST in MDA-MB-231 cells reduced MMP9 mRNA levels. (B) Induced REST in MDA-MB-231 cells reduced MMP9 protein levels. (C) Luciferase reporter gene assay using the reporter vector containing the promoter of MMP9 gene. MDA-MB-231 cells were cotransfected with MMP9 promoter together with different amounts of a REST expression lentivirus. REST reduces luciferase expression in a concentration-dependent manner in MDA-MB-231 cells. Data are expressed as mean ± standard deviation from three independent experiments. **p < 0.01, ***p < 0.001 versus negative control. Con, control; NC, negative control.
Restoration of MMP9 Blocked the Inhibitory Effects of REST on Breast Cancer Cells
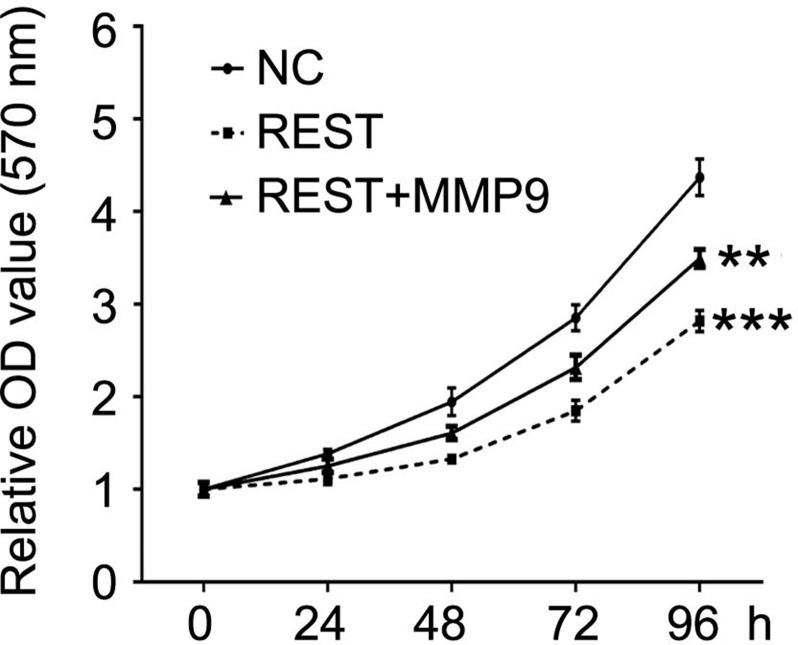
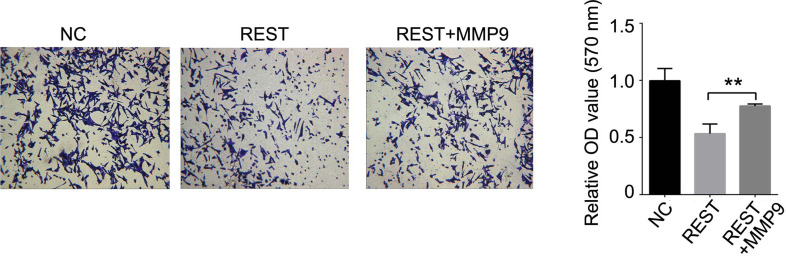
The previous results demonstrate that REST reduces target gene expression by inhibiting the activity of the MMP9 promoter. Consequently, we investigated whether induced MMP9 expression affected the role of REST in MDA-MB-231 cells. MDA-MB-231 cells were transfected with REST expression lentivirus alone or cotransfected REST lentivirus together with MMP9 expression lentivirus or empty vector. MTT results showed that the inhibitory effects of REST on cell proliferation were mostly blocked by induced expression of MMP9 (Fig. 4). In addition, Transwell assays revealed that restoration of MMP9 increased the invasion ability that was repressed by the upregulation of REST (Fig. 5). These results demonstrated that restoration of MMP9 blocked the inhibitory effects of REST on breast cancer cells.
Figure 4.
Restoration of MMP9 blocked growth inhibition induced by REST in MDA-MB-231 cells. MTT assay was used to measure the cell proliferation after indicated treatment. Data are expressed as mean ± standard deviation from three independent experiments. **p < 0.01, ***p < 0.001 versus negative control. NC, negative control.
Figure 5.
Restoration of MMP9 reversed REST-mediated suppression of invasive ability in MDA-MB-231 cells. Transwell was used to measure the ability of invasion after indicated treatment. Data are expressed as mean ± standard deviation from three independent experiments. **p < 0.01 versus negative control. NC, negative control.
DISCUSSION
Previous studies revealed that REST, combined with its cofactors, could maintain transcriptional silencing of a range of neuronal genes in differentiated nonneuronal cells by altering chromatin structure and regulating transcription through histone deacetylation, chromatin remodeling, and methylation14. Recently, REST has been implicated in several types of cancer, such as glioblastomas7 and breast cancer10, as either an oncogene or a tumor suppressor. Our previous study showed that REST expression in breast cancer was lower compared with normal breast tissue, and knocking down REST by shRNA in MCF-7 could stimulate cell proliferation and reduce the sensitivity of MCF-7 cells to 5-FU [5-fluoro-2,4(1h, 3h) pyrimidinedione]11. In line with our previous results, here we confirmed that REST expression was lower compared with normal breast tissue at mRNA and protein levels. The level of REST was differentially expressed in breast cancer cell lines, especially in MDA-MB-231 cells, which is more malignant than other cell lines. Importantly, in the present study, we found that the promoter of REST was hypermethylated in breast cancer and in breast cancer cell lines, especially in MDA-MB-231 cells. Schoofs et al. showed that REST-binding sites identified in embryonic stem cells were preferentially DNA hypermethylated in primary acute promyelocytic leukemia cells, which was associated with the leukemia phenotype15. In small-cell lung cancer (SCLC) cell lines, loss of REST expression regulated by hypermethylation was linked to malignant progression. Reduced REST expression significantly enhanced cell proliferation and, rescued, decreased the potential of cells to grow anchorage independently, indicating that REST acted as an important modulator of malignant progression in SCLC16. We found that REST expression was increased by 5-Aza, a demethylation agent. Combined with hypermethylation of REST promoter, mentioned above, this result suggests that loss of REST expression in breast cancer was due, at least partially, to epigenetic mechanism. In addition, increased REST by lentivirus transfection and 5-Aza significantly inhibited MDA-MB-231 cell proliferation, induced apoptosis, and reduced invasion ability. Taken together, loss of REST by hypermethylation contributes to malignant progression in breast cancer.
Furthermore, we investigated the downstream target gene regulated by REST. Our data demonstrated that upregulation of REST induced a significant reduction of MMP9 at the mRNA and protein levels. Matrix metalloproteinases (MMPs) have been implicated in diverse roles in breast cancer development and progression. MMP9 is most highly expressed in breast cancer of high histological grade and in triple-negative tumors, with significant association with a higher incidence of metastasis and relapse17. Mehner et al. showed that human breast cancer cell-produced MMP9 was required for invasion in a cell culture and for pulmonary metastasis in a mouse orthotopic model of basal-like breast cancer, and silencing of MMP9 expression led to a less malignant phenotype, suggesting that it contributes to metastatic progression18. Park et al. found that the absence of MMP9 delayed tumor onset in mice19. Thus, inactivation of MMP9 in breast cancer would result in a decrease in proliferation and invasion abilities. By luciferase assay, we determined that REST inactivated the MMP9 promoter in a concentration-dependent manner, consequently resulting in the loss of MMP9 expression. By cotransfecting REST and MMP9 into MDA-MB-231 cells, we found that upregulation of MMP9 blocked the inhibitory effects of REST on breast cancer cells. REST can recruit corepressors to modify histones methyltransferases and repress transcription. Zheng et al. revealed that REST mediated the alternations of histone acetylations and methylations, such as H3K27, H3K9, and H3K4, which may be REST dependent20. Hwang et al. showed that REST could bind to the miR-132 promoter and silence miR-132 expression in selectively vulnerable hippocampal CA1 neurons by epigenetic remodeling21. Reddy et al. also demonstrated that REST bound to the 5′-untranslated region of the oncogenic gene promoter, preprotachykinin-A, and suppresses its expression in breast cancer cells22. It is reasonable to conclude that REST is likely to recruit corepressors, such as histone deacetylases and methyltransferases, to repress MMP9 transcription.
In conclusion, we provided evidence to reveal that the loss of REST expression in breast cancer is linked to hypermethylation, and low REST levels are significantly associated with malignant progression in breast cancer patients. The inhibitory effects of REST in breast cancer cells are achieved, at least partially, by repressing MMP9 transcription via inactivating its promoter activity. Our work represents a novel therapeutic target for breast cancer treatment.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Hart CD, Migliaccio I, Malorni L, Guarducci C, Biganzoli L, Leo AD. Challenges in the management of advanced, ER-positive, HER2-negative breast cancer. Nat Rev Clin Oncol. 2015;12:541–52. [DOI] [PubMed] [Google Scholar]
- 2. De Iuliis F, Taglieri L, Salerno G, Lanza R, Scarpa S. Taxane induced neuropathy in patients affected by breast cancer: Literature review. Crit Rev Oncol Hematol. 2015;96:34–45. [DOI] [PubMed] [Google Scholar]
- 3. Colwell AS. Current strategies with 1-stage prosthetic breast reconstruction. Gland Surg. 2015;4:111–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Negrini S, Prada I, D’Alessandro R, Meldolesi J. REST: An oncogene or a tumor suppressor? Trends Cell Biol. 2013;23:289–95. [DOI] [PubMed] [Google Scholar]
- 5. Taylor P, Fangusaro J, Rajaram V, Goldman S, Helenowski IB, MacDonald T, Hasselblatt M, Riedemann L, Laureano A, Cooper L, Gopalakrishnan V. REST is a novel prognostic factor and therapeutic target for medulloblastoma. Mol Cancer Ther. 2012;11:1713–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang Z, Bao S. Ubiquitination and deubiquitination of REST and its roles in cancers. FEBS Lett. 2012;586:1602–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Conti L, Crisafulli L, Caldera V, Tortoreto M, Brilli E, Conforti P, Zunino F, Magrassi L, Schiffer D, Cattaneo E. REST controls self-renewal and tumorigenic competence of human glioblastoma cells. PLoS One 2012;7:e38486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kreisler A, Strissel PL, Strick R, Neumann SB, Schumacher U, Becker CM. Regulation of the NRSF/REST gene by methylation and CREB affects the cellular phenotype of small-cell lung cancer. Oncogene 2010;29:5828–38. [DOI] [PubMed] [Google Scholar]
- 9. Coulson JM. Transcriptional regulation: Cancer, neurons and the REST. CurrBiol. 2005;15:R665–68. [DOI] [PubMed] [Google Scholar]
- 10. Wagoner MP, Gunsalus KT, Schoenike B, Richardson AL, Friedl A, Roopra A. The transcription factor REST is lost in aggressive breast cancer. PLoS Genet. 2010;6:e1000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lv H, Pan G, Zheng G, Wu X, Ren H, Liu Y, Wen J. Expression and functions of the repressor element 1 (RE-1)-silencing transcription factor (REST) in breast cancer. J Cell Biochem. 2010;110:968–74. [DOI] [PubMed] [Google Scholar]
- 12. Noh KM, Hwang JY, Follenzi A, Athanasiadou R, Miyawaki T, Greally JM, Bennett MV, Zukin RS. Repressor element-1 silencing transcription factor (REST)-dependent epigenetic remodeling is critical to ischemia-induced neuronal death. Proc Natl Acad Sci USA 2012;109:E962–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baiula M, Carbonari G, Dattoli SD, Calienni M, Bedini A, Spampinato S. REST is up-regulated by epidermal growth factor in HeLa cells and inhibits apoptosis by influencing histone H3 acetylation. Biochim Biophys Acta 2012;1823:1252–63. [DOI] [PubMed] [Google Scholar]
- 14. Zheng D, Zhao K, Mehler MF. Profiling RE1/REST-mediated histone modifications in the human genome. Genome Biol. 2009;10:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schoofs T, Rohde C, Hebestreit K, Klein HU, Gollner S, Schulze I, Lerdrup M, Dietrich N, Agrawal-Singh S, Witten A, Stoll M, Lengfelder E, Hofmann WK, Schlenke P, Buchner T, Hansen K, Berdel WE, Rosenbauer F, Dugas M, Muller-Tidow C. DNA methylation changes are a late event in acute promyelocytic leukemia and coincide with loss of transcription factor binding. Blood 2013;121:178–87. [DOI] [PubMed] [Google Scholar]
- 16. Kreisler A, Strissel PL, Strick R, Neumann SB, Schumacher U, Becker CM. Regulation of the NRSF/REST gene by methylation and CREB affects the cellular phenotype of small-cell lung cancer. Oncogene 2010;29:5828–38. [DOI] [PubMed] [Google Scholar]
- 17. Yousef EM, Tahir MR, St-Pierre Y, Gaboury LA. MMP-9 expression varies according to molecular subtypes of breast cancer. BMC Cancer 2014;14:609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mehner C, Hockla A, Miller E, Ran S, Radisky DC, Radisky ES. Tumor cell-produced matrix metalloproteinase 9 (MMP-9) drives malignant progression and metastasis of basal-like triple negative breast cancer. Oncotarget 2014;5:2736–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park JH, Rasch MG, Qiu J, Lund IK, Egeblad M. Presence of insulin-like growth factor binding proteins correlates with tumor-promoting effects of matrix metalloproteinase 9 in breast cancer. Neoplasia 2015;17:421–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng D, Zhao K, Mehler MF. Profiling RE1/REST-mediated histone modifications in the human genome. Genome Biol. 2009;10:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hwang JY, Kaneko N, Noh KM, Pontarelli F, Zukin RS. The gene silencing transcription factor REST represses miR-132 expression in hippocampal neurons destined to die. J Mol Biol. 2014;426:3454–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reddy BY, Greco SJ, Patel PS, Trzaska KA, Rameshwar P. RE-1-silencing transcription factor shows tumor-suppressor functions and negatively regulates the oncogenic TAC1 in breast cancer cells. Proc Natl Acad Sci USA 2009;106:4408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]