Abstract
Therapeutic payload delivery to the central nervous system (CNS) remains a major challenge in gene therapy. Recent studies using function-driven evolution of adeno-associated virus (AAV) vectors have successfully identified engineered capsids with improved blood-brain barrier (BBB) penetration and CNS tropism in mouse. However, these strategies require transgenic animals and thus are limited to rodents. To address this issue, we developed a directed evolution approach based on recovery of capsid library RNA transcribed from CNS-restricted promoters. This RNA-driven screen platform, termed TRACER (Tropism Redirection of AAV by Cell-type-specific Expression of RNA), was tested in the mouse with AAV9 peptide display libraries and showed rapid emergence of dominant sequences. Ten individual variants were characterized and showed up to 400-fold higher brain transduction over AAV9 following systemic administration. Our results demonstrate that the TRACER platform allows rapid selection of AAV capsids with robust BBB penetration and CNS tropism in non-transgenic animals.
Keywords: Adeno-associated virus, AAV, Brain, CNS, Blood-Brain barrier, Directed evolution, Capsid library
Graphical Abstract
In this article, Nonnenmacher and colleagues establish TRACER, a cell-specific RNA-driven directed evolution platform for functional biopanning of adeno-associated virus (AAV) capsid libraries in animal tissue. Neuron- and glial-specific selection of an AAV9 library in mouse brain identified several AAV variants with unprecedented brain and spinal cord transduction.
Introduction
Clinical applications of gene therapy in the central nervous system (CNS) are currently limited by the poor transduction of brain and spinal cord by adeno-associated virus (AAV) and other viral vectors.1,2 The blood-brain barrier (BBB) represents a formidable obstacle for delivery of AAV into brain tissue following intravenous administration, and even the best-in-class natural BBB-penetrating serotypes, namely AAV9 and other clade F derivatives,3,4 only allow limited brain distribution.1,5,6 This challenge can be partially overcome by using local delivery routes, such as intraparenchymal injection,7,8 intrathecal infusion,9 or cisterna magna administration.10 However, these methods are invasive and only achieve limited distribution and transduction throughout the brain and spinal cord, short of therapeutically desired coverage. These shortcomings could be mitigated by engineered AAV capsids capable of efficiently crossing the BBB via intravascular delivery.
High-throughput mutagenesis and directed evolution of AAV capsids were first described in 200311,12 and are greatly facilitated by the simplicity of the viral genome organization, extensive knowledge of capsid structure,13 and the natural propensity of wild-type AAV to assemble capsids with low mosaicism and high genome-capsid correlation.14, 15, 16 Early designs of AAV-directed evolution were strictly tailored for in vitro selection in cultured cells and used helper adenovirus coinfection to enrich transduction-competent variants.11,12,17,18 Helper-dependent selection is not easily accomplished in vivo, and library selection in the mouse initially relied on indiscriminate PCR amplification of AAV genomes from the tissue of interest.19 Although this approach proved successful to some degree, tremendous effort has been spent on developing new biopanning approaches to efficiently select true positives in vivo.20,21 Over the past decade, functional AAV library screens based on cell-specific sorting,22 in vivo helper virus coinfection,23 or cell-specific Cre-lox selection24, 25, 26 have identified improved capsid variants. In particular, two AAV9 variants, PHP.B and PHP.eB, showed an unprecedented ability to transduce C57BL/6 mouse brain via systemic injection.26,27 Follow-up studies, however, showed that these properties did not translate to other laboratory mouse strains or to non-human primates (NHPs).28, 29, 30
Importantly, Cre-dependent AAV library screening methods24, 25, 26 strictly rely on transgenic animals for specific recovery of transduction-competent variants, which precludes the use of clinically relevant animals such as NHPs. In this study, we describe TRACER (Tropism Redirection of AAV by Cell-type-specific Expression of RNA), an AAV evolution platform based on recovery of bulk capsid library RNA expressed in a cell-type-specific manner from non-transgenic animal tissue. We used TRACER in a directed evolution screen focused on mouse CNS and were able to isolate multiple capsid variants capable of widespread brain transduction via systemic administration. We demonstrate that TRACER is a highly effective AAV evolution platform, with potential applications in a broad range of tissues and non-transgenic animal species.
Results
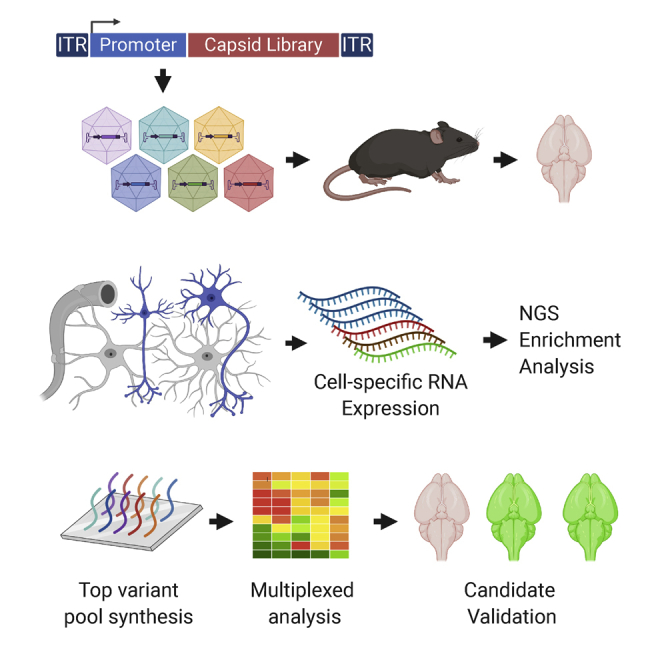
Design and construction of AAV libraries with cell-type-specific expression vectors and biopanning
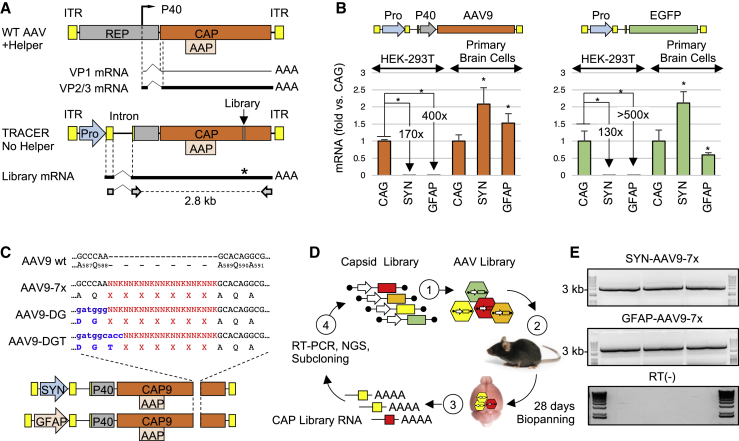
The AAV genome is transcriptionally repressed in the absence of helper virus.31,32 In order to generate transcription-competent libraries for RNA-driven directed evolution, we inserted a non-AAV promoter upstream of the Cap gene, which would confer cell-type expression specificity while retaining the minimal regulatory elements essential for capsid protein expression and stoichiometry (Figure 1A). We determined that a minimal Rep fragment starting at nucleotide 1700 that contains the AAV P40 promoter and splice donor and acceptor sequences was sufficient for efficient virus production, albeit with a lower yield than wild-type AAV (Figure S1).
Figure 1.
Design of RNA recovery strategy for cell-specific biopanning
(A) Map of wild-type AAV (top) and TRACER library vectors (bottom). ITR, inverted terminal repeat. Pro, promoter. Dashed lines indicate AAV intron (top) or synthetic CMV-globin intron (bottom), solid lines represent minor (thin line) and major (thick line) capsid transcripts. Primers used for the recovery of the 2.8-kb capsid library amplicon are indicated at the bottom. (B) Activity of CAG, SYN, and GFAP promoters in TRACER tandem configuration (left panel) or single promoter configuration (right panel). Depicted transgenes were packaged in AAV9 capsid and tested in HEK293T cells or primary mouse brain cells (1e5 VG/cell, n = 3). RNA was quantified by real-time RT-PCR 48 h post-treatment. Values indicate RNA expression normalized to CAG vectors in each cell type (mean ± SD). ∗p < 0.05 (t test). (C) Construction of peptide display libraries. Randomized sequences preceded by AQ, DG, or DGT residues were introduced in AAV9 VP1 at the indicated positions in vectors containing SYN or GFAP promoter. (D) Overview of the in vivo selection process. (1) DNA libraries are used to produce a virus library, (2) virus libraries are injected intravenously (i.v.) into mice (1e12VG per mouse), (3) bulk RNA is recovered from whole brains 28 days post-injection, (4) capsid fragments encoding the peptide library are amplified by RT-PCR, analyzed by next-generation sequencing (NGS), and cloned into TRACER vectors for another round of selection. (E) Example of RT-PCR products obtained from three mice 28 days after injection with SYN-driven and GFAP-driven library (top and middle panel, respectively). The 3-kb band from the molecular weight marker is indicated. Bottom panel: RT-negative controls.
We performed a series of in vitro experiments to test possible interference between the P40 promoter and the non-AAV promoters placed in tandem configuration. We tested three promoters in the context of AAV9 TRACER vectors: the ubiquitous cytomegalovirus (CMV) enhancer-chicken β-actin (CAG) promoter, the neuron-specific human synapsin 1 (SYN) promoter,33 and the astrocyte-specific gfaABC1D (GFAP) promoter.34 All constructs were used to produce AAV9 capsids, and the resulting virions were added to HEK293T cells or cultured primary mouse brain cells. As expected, RNA expression from SYN and GFAP promoters was strongly repressed in non-CNS HEK293T cells (170-fold and 400-fold lower than CAG, respectively), but not in primary mouse brain cells, where both SYN and GFAP showed a strength similar to CAG promoter (Figure 1B, left panel). A similar trend was observed with GFP vectors without P40 sequence (Figure 1B, right panel), indicating that the presence of the P40 element has little or no impact on the regulation of upstream tandem promoters used for biopanning.
Since TRACER constructs lack a full-length functional REP reading frame, REP proteins were provided in trans by a separate plasmid during virus production. We first tested a Rep2 plasmid missing most of the Cap gene, but higher titers were obtained with a Rep2-Cap9Δ plasmid containing a CAP C terminus deletion and in-frame stop codons to eliminate VP1-3 translation (Figure S1).
Capsid libraries were generated by inserting 7-mer randomized peptides between residues 588 and 589 in the hypervariable surface loop VIII35 of AAV9. Random peptides were N-terminally flanked by the original AAV9 residues AQ(587, 588) or by the PHP.eB-derived residues DG(587, 588) or DGT(587, 588, 589) (Figure 1C).27 To avoid the loss of variants resulting from bacterial transformation, library DNA assembled with SYN or GFAP vectors was amplified in vitro by rolling circle amplification (RCA) and protelomerase end cleavage joining (Figure S1).36 This technique generated large amounts of transfection-ready DNA with a diversity beyond the capacity of our next-generation sequencing (NGS) analysis (>108 unique variants) and without obvious sequence bias. Viral libraries were produced in HEK293T cells using low-DNA-input conditions to minimize capsid mosaicism and cross-packaging14 and were administered intravenously to adult C57BL/6 mice (n = 6). Whole-brain RNA was isolated after 28 days and capsid library sequences were recovered by RT-PCR. Amplified pools were re-cloned into SYN or GFAP TRACER vectors for a second round of selection (Figure 1D). Abundant capsid amplicons were recovered from all brain samples regardless of the promoter driving the expression (Figure 1E), indicating that some variants achieve high transduction in the CNS, a tissue with one of the lowest AAV biodistributions.37
Selection of AAV9 capsid variants with enhanced BBB penetration and CNS transduction
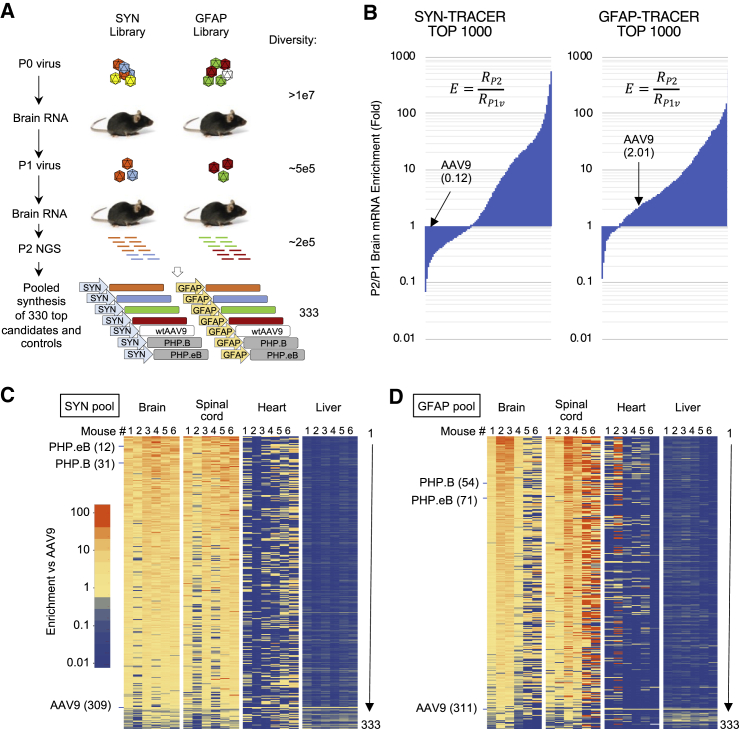
NGS analysis was performed after each step of in vivo selection to estimate variant diversity and enrichment (Figure 2A). The first round of selection eliminated approximately 95% of variants (from ≥10 million to 500,000 unique sequences), and the second round removed 60% of the remaining variants (from 500,000 to 200,000 unique sequences). Enrichment analysis performed after the second round of biopanning showed that hundreds of capsid variants displayed high enrichment scores compared to AAV9 (Figure 2B).
Figure 2.
NGS-driven evolution of TRACER libraries in C57BL/6 mice
(A) TRACER workflow and library diversity through successive rounds of evolution and pooled synthesis. Values indicate the number of unique variants detected by NGS. (B) Enrichment analysis of P2 brain RNA. Enrichment score E indicates the relative RNA abundance of each variant (RP2) normalized to P1 virus stock (RP1v). Top 1,000 variants of SYN and GFAP libraries are depicted. (C and D) Fitness analysis of SYN-driven (C) and GFAP-driven (D) pool of 330 capsid candidates plus AAV9, PHP.B, and PHP.eB controls. Heatmaps represent relative RNA enrichment score in brain and spinal cord and DNA enrichment score in heart and liver. Values are normalized to AAV9 control. Numbered columns represent individual animals (n = 6). Values represent the average of two codon variants for each mutant and are ranked according the average of 6 brains. Ranking of control capsids is indicated.
Bioinformatics analysis based on absolute read numbers, enrichment scores, cross-animal consistency, and collapsing of pseudo-variants from sequencing errors led us to select 330 capsid candidates with a favorable CNS enrichment profile (see Materials and methods). Phylogenetic analysis of this brain-enriched variant pool identified several conserved families of variants harboring 9-mer peptides with striking sequence similarities (Figure S2). The largest families clustered in three dominant groups: the most prominent (107 variants) shared the consensus motif DGTxxxxGW, a second group (68 variants) displayed the motif DGTxxxP(F/P)(K/R) reminiscent of the PHP.eB capsid and herein referred to as “PHP-like,” and a third group (43 variants) displayed the motif DGTxxxLSS (Figure S2). Smaller groups displayed the motif (DG/AQ)xxxxYD(A/S) or AQxxxxxRW (8 and 9 variants, respectively). Of note, we also observed a cluster of 9 variants sharing the motif AQWxxxxGY, similar to the recently identified PHP.C2,38 and also one single variant (AQFVVGQQY) closely related to the CNS-trophic AAV-F capsid (AQFVVGQSY).25
A synthetic library containing these 330 candidates plus AAV9, PHP.B, and PHP.eB, each encoded as two distinct codon versions (666 nucleotide variants total), was produced de novo using pooled primer synthesis as recently described38 and cloned in SYN or GFAP vectors (Figure 2A). Each AAV pool was injected to C57BL/6 mice (n = 6), and NGS enrichment analysis was performed 28 days post-injection using RNA from the brain and spinal cord, as well as DNA from heart and liver tissues. Overall, 300 variants showed a brain transduction superior to AAV9, and 92 variants outperformed AAV9 by more than 10-fold (Figures 2C and 2D). By contrast, whereas AAV9 was among the lowest performers in the brain and spinal cord, it showed the highest score in the heart and liver, consistent with CNS cell-type-specific selection conferred by the TRACER platform used in this study (Figures 2C and 2D). The alternative codon versions of each variant showed highly correlated behavior in DNA, virus, and tissue samples, supporting the robustness of the assay (Figure S3). SYN- and GFAP-driven versions of each variant behaved very consistently at the DNA level but diverged at the RNA level (Figure S3), which was not unexpected, since the DNA enrichment is not subject to variations in promoter activity among cell types. Consistent enrichment scores were observed among animals injected with the SYN-driven pool, whereas the GFAP-driven library showed a higher inter-animal variability (Figures 2C and 2D). As expected, both PHP.eB- and PHP.B-positive controls showed a high CNS enrichment score in both screens (Figures 2C and 2D; Data S1). Taken together, the high inter-animal reproducibility, the strong codon variant correlation, and the precise calibration from internal controls allowed us to identify multiple capsids with improved CNS fitness with a very high degree of confidence. Strikingly, despite the absence of effort to select variants with low liver and heart tropism in our screen, the majority of CNS-trophic capsids showed substantial de-targeting from both tissues.
Correlation of variant origin and genotype with in vivo and in vitro properties
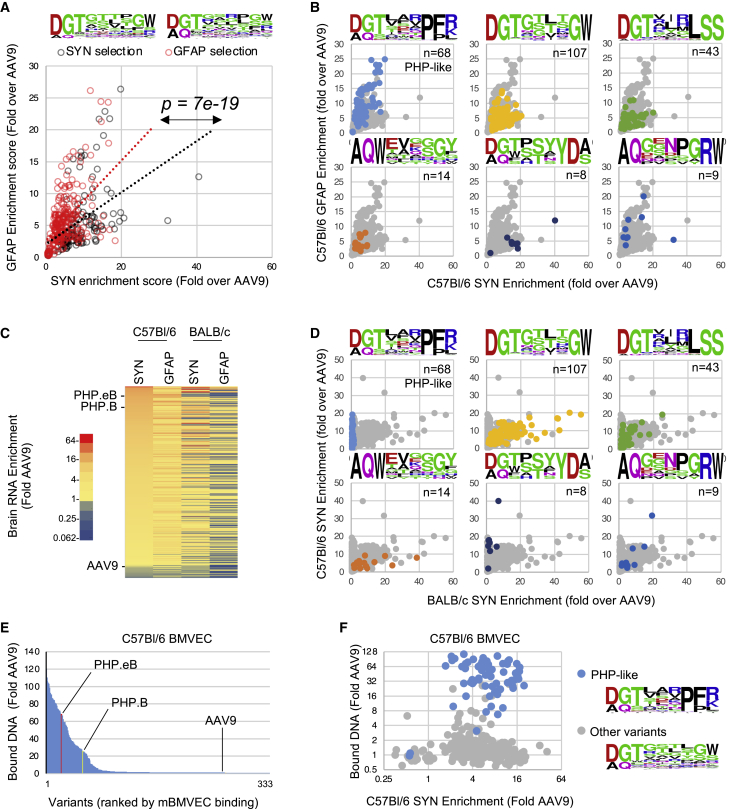
Our synthetic library of 330 brain-enriched variants contained 120 candidates originating from SYN-driven library evolution and 210 candidates from GFAP-driven evolution (Data S1). We asked if the capsid evolution path would favor variants with a bias toward neurons (SYN) or astrocytes (GFAP). When brain RNA enrichment data were stratified according to the library selection method, we observed a significant, albeit modest, transduction bias in favor of the cell type where the library screen was performed (Figure 3A). On average, capsids selected with the GFAP-TRACER library showed a 2-fold increased fitness for astrocyte versus neuron than capsids evolved with the SYN-TRACER library. This relatively modest cell-type specificity likely reflects the absence of negative selection in our screen, combined with the ability of most variants to transduce both neurons and astrocytes, at least to some extent.
Figure 3.
Genotype-to-phenotype analysis of synthetic capsid pool from C57BL/6 CNS biopanning
(A) Comparative neuron and astrocyte fitness of the capsid variants originating from SYN- or GFAP-driven library biopanning (black and red circles, respectively). Each data point represents the average neuron (SYN-driven) and astrocyte (GFAP-driven) RNA enrichment score in i.v.-dosed C57BL/6 mice (n = 6), normalized to AAV9. Linear regression trendline of each population is indicated. p value indicates the statistical difference between the average GFAP-to-SYN score ratio of each subpopulation (unpaired t test). Frequency plots of peptides from SYN- and GFAP-evolved subpools are indicated on top. (B) Enrichment scores of each capsid sequence family in GFAP- (y axis) and SYN-driven RNA assays (x axis). The frequency plots and number of variants in each group are indicated. (C) Comparative brain RNA enrichment of 330 variants in C57BL/6 mice (n = 6) and BALB/c mice (n = 6) following i.v. injection. Color scale indicates the average RNA enrichment score normalized to AAV9. Variants are ranked by SYN-driven RNA enrichment score in C57BL/6 mice. (D) Comparative SYN-driven RNA enrichment score of distinct capsid families in C57BL/6 and BALB/c mice. The frequency plots and number of variants of each group are indicated. (E) Multiplexed binding assay of synthetic capsid pool to C57BL/6 mouse primary brain microvascular endothelial cells (BMVECs). Values indicate bound viral DNA enrichment score relative to AAV9. Ranking of reference PHP.eB, PHP.B, and AAV9 capsids is indicated. (F) Scatterplot presenting the correlation between virus binding to mouse BMVECs and C57BL/6 brain RNA enrichment scores. The PHP-like capsid variants are indicated by blue dots, all other variants by gray dots.
We next investigated the impact of capsid sequence on cellular tropism by clustering the neuron/astrocyte fitness of variants with high sequence homology. Although most variants showed improved transduction in both cell types, the PHP-like DGTxxxPF(K/R) group and the AQxxxxxRW group showed the highest transduction in astrocytes (Figure 3B). By contrast, variants belonging to the DGTxxxxGW sequence group showed no obvious preference for either cell type, and variants belonging to the (DG/AQ)xxxxYD(A/S) group displayed a marked neuronal bias. This observation could suggest that while multiple variants show an increased ability to cross the BBB, the displayed peptide sequence could also modulate the transduction efficiency of different cell types in the brain parenchyma.
We next examined brain transduction in BALB/c mice, since some engineered CNS-trophic capsids have shown substantial variation between C57BL/6 and BALB/c strains due to the usage of a polymorphic receptor on the BBB lumen.39, 40, 41 Brain RNA enrichment analysis showed that a fraction of variants did not transduce BALB/c brain efficiently, while others showed strong enrichment in both strains (Figure 3C). Strikingly, all PHP-like variants were strictly restricted to C57BL/6 mice and did not outperform AAV9 in BALB/c mice, consistent with previous data obtained with PHP.B and PHP.eB capsids (Figure 3D).29,41,42 Similarly, low CNS transduction in BALB/c mice was observed with the capsids from the (DG/AQ)xxxxYD(A/S) group. By contrast, other capsid families showed similar performance across strains (Figure 3D), which strongly suggest the use of different mechanisms to cross the BBB endothelium.
We evaluated the capacity of our 330 variants to bind brain microvascular endothelial cells from C57BL/6 mice (mBMVECs) in culture, a property demonstrated by PHP.B and PHP.eB capsids.41 Out of the 333 capsids present in the library, 69 showed 10-fold or more binding relative to AAV9 (Figure 3E; Data S1). PHP.B and PHP.eB capsids showed 25- and 68-fold improvement over AAV9, respectively, in agreement with published data.41 Strikingly, the capsids with high affinity for mBMVECs almost exclusively belonged to the PHP-like cluster (Figure 3F), suggesting that the recapitulation of capsid-receptor interaction in BMVECs is unique to the LY6a-binding variants.41,42 Interestingly, capsids from the (DG/AQ)xxxxYD(A/S) group, which share the C57BL/6-specific transduction phenotype with PHP-like capsids, did not bind mBMVECs, suggesting a different receptor usage. Overall, correlation between the in vivo performance of capsids and the mBMVEC binding was limited to a unique sequence group (Figure 3F), which clearly indicates that monolayer cultures of BMVECs are a poor predictor of in vivo transport across the BBB. Taken together, our data strongly suggest that the TRACER platform identified a broad population of diverse capsid families with distinct BBB transport and brain cell transduction mechanisms, as is evident from their distinct tropism across cell types and mouse strains.
In vivo characterization of selected AAV capsid candidates
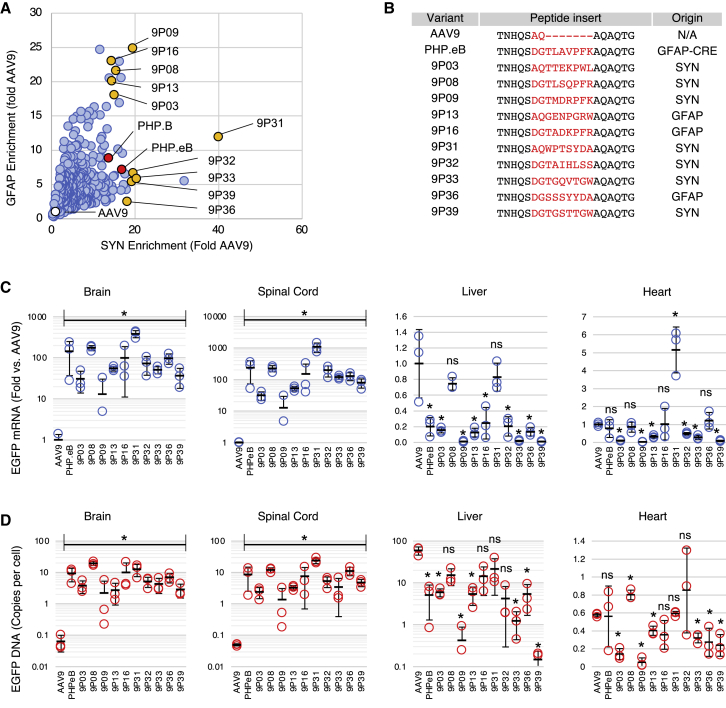
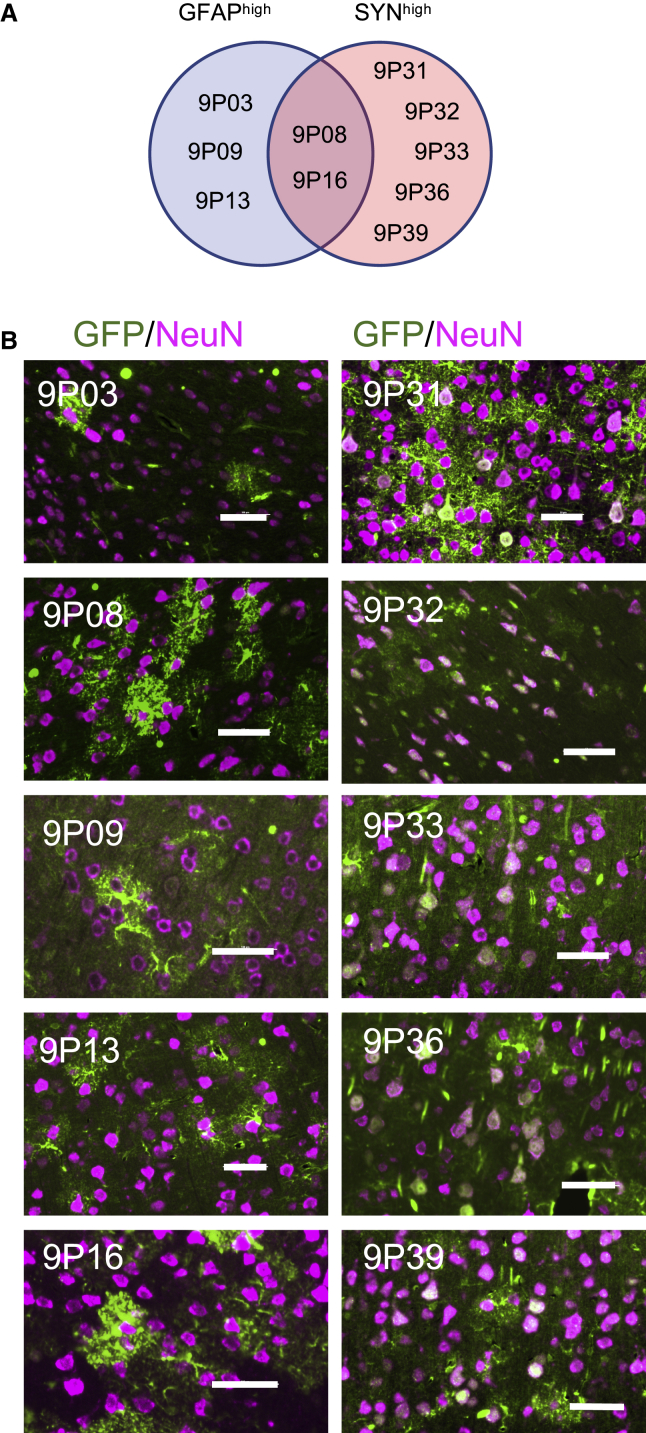
Ten capsid candidates were selected for individual evaluation, based on high CNS enrichment scores in either the SYN or GFAP screen (Figure 4A) and sequence divergence (Figure 4B). Each capsid was used to produce recombinant AAV containing a self-complementary43 EGFP reporter driven by the ubiquitous CAG promoter and administered intravenously to C57BL/6 mice at a dose of 4e11 viral genomes (VG) per animal. AAV9 and PHP.eB were used as references. Relative EGFP mRNA expression was measured 4 weeks post-injection. All 10 candidates largely outperformed AAV9 in the brain and spinal cord (Figure 4C). When compared to AAV9, EGFP expression in the CNS was increased from 13-fold (9P09) to 385-fold (9P31). In the spinal cord, 9P31 transduction was more than 1,000-fold higher than AAV9. By comparison, PHP.eB transduction was 144-fold higher than AAV9 in the brain and 236-fold higher in the spinal cord, in agreement with the published data.27 Of note, the qPCR scores from individual capsids showed a high correlation with the SYN screen, but not with the GFAP screen, indicating that SYN-driven expression was more predictive of global brain transduction (Figure S4). As suggested by our NGS analysis, no capsid variant transduced the liver better than AAV9; instead, some variants (9P09, 9P33, and 9P39) were de-targeted by approximately 100-fold. In the heart, no capsid variants outperformed AAV9 with the exception of 9P31, which showed a 5-fold increase in RNA expression (Figure 4C, right panel).
Figure 4.
Individual characterization of TRACER capsid candidates
(A) Brain RNA enrichment score of pooled synthetic variants in the SYN- and GFAP-driven assay. Values are normalized to AAV9. Yellow dots indicate candidates chosen for individual testing; red and white dots indicate the PHP and AAV9 control capsids, respectively. (B) Sequence of capsids selected for individual characterization. The right column indicates the biopanning method used to evolve each variant. (C) Real-time RT-PCR analysis of EGFP transgene RNA expression in the brain, spinal cord, liver, and heart 28 days after i.v. injection of each capsid in C57BL/6 mice (4e11 VG per mouse, n = 3). Values indicate mean ± SD (n = 3), normalized to AAV9. ∗p < 0.05 relative to AAV9 (unpaired t test); ns, not significant. (D) AAV genome biodistribution measured by qPCR. Values indicate mean ± SD (n = 3) EGFP copies per diploid cell. ∗p < 0.05 relative to AAV9 (unpaired t test); ns, not significant. All brain and spinal cord samples were statistically different from AAV9.
Biodistribution of AAV genomes showed a pattern similar to RNA expression (Figure 4D). The CNS biodistribution of novel variants ranged from 2 VG per cell (9P03 and 9P09) to 20–25 VG per cell (9P08 and 9P31). Biodistribution of PHP.eB (8.6 and 9.3 VG per cell in the brain and spinal cord, respectively) was in the expected range.26,27 By comparison, mice injected with AAV9 showed less than 0.1 VG per cell in the brain and spinal cord (Figure 4D). Consistent with the RNA data, 9P09 and 9P39 showed a very low DNA biodistribution in the liver (<1 VG per cell) relative to AAV9 (∼60 VG per cell).
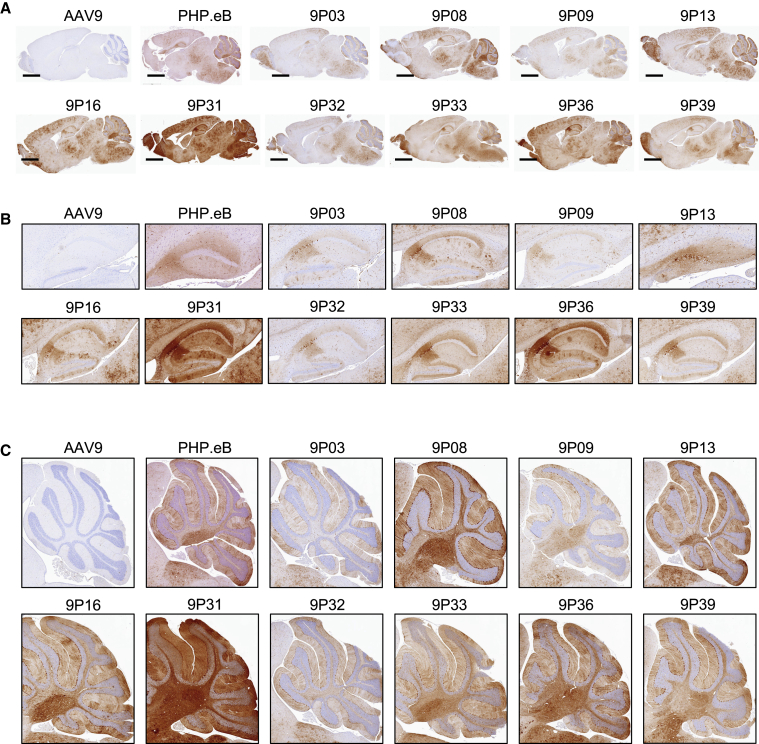
We then analyzed EGFP protein expression in brain sagittal sections by immunohistochemistry. All the TRACER-evolved capsids showed strong and widespread EGFP expression throughout the entire brain (Figure 5A). As expected from the RNA quantitation data, capsids 9P03 and 9P09 showed the lowest transduction; capsids 9P08, 9P16, 9P33, and 9P36 were similar to PHP.eB; and capsid 9P31 showed a strikingly high EGFP expression in all brain regions (Figure 5A). Similar results were obtained by observing native EGFP fluorescence in frozen sections from the same animals (Figure S5). Despite different transgene expression levels, all capsid variants showed a similar spatial distribution characterized by preferential transduction of the cerebellum, brain stem, medulla, pons, hippocampus, thalamus, cortex, anterior olfactory nucleus, and olfactory bulb (Figure 5A; Figure S6). Notably, all capsid variants showed a strong transduction of the CA2 region of the hippocampus (Figure 5B) and of the Purkinjie and molecular layers of the cerebellum (Figure 5C).
Figure 5.
Brain transduction profile of TRACER capsid candidates in adult mice
EGFP was detected by immunohistochemistry (IHC) from formalin-fixed paraffin-embedded (FFPE) sections in the brain of adult C57BL/6 mice 28 days after i.v. infusion with 4e11 VG per mouse. (A–C) Representative images of whole-brain sagittal sections (A), hippocampus (B), and cerebellum (C) are shown. Scale bar in (A), 2 mm.
All engineered capsids demonstrated a predominant tropism for neuronal cells in the hippocampus, as demonstrated by NeuN immunostaining (Figure S7). By contrast, differences in cellular tropism were observed in the cortex and the thalamus regions, where some variants showed an obvious neuronal preference (9P31, 9P32, 9P33, 9P36, and 9P39), whereas others (9P03, 9P08, 9P09, 9P13, and 9P16) transduced mostly NeuN-negative cells with distinctive highly branched morphological features (Figure 6; Figure S7). Capsids with marked neuronal preference were also the strongest performers in the SYN-driven NGS RNA enrichment assay (Figure 6A). Intriguingly, capsids with high performance in the GFAP-driven RNA assay did not appear to efficiently transduce GFAP-positive astrocytes in the cortex, thalamus, or hippocampus, possibly suggesting a preferred tropism for immature or protoplasmic astrocytes rather than mature GFAP+ astrocytes. Similarly, despite a strong transduction of the entire cerebellum, all variants appeared to be restricted to Purkinje cells and did not transduce the GFAP+ Bergmann glia (Figure S8).
Figure 6.
Cortical neuron transduction by TRACER capsid candidates in adult mice
(A) Relative fitness of TRACER capsids in the GFAP-driven and SYN-driven library NGS assay. (B) EGFP (green) and NeuN (magenta) were detected by IHC from FFPE sections in the brain of adult C57BL/6 mice 28 days after intravascular infusion with 4e11 VG per mouse. Representative images of cortex are shown. Bar, 50 μm.
Discussion
Here, we developed a transcription-dependent platform that allowed rapid selection of AAV capsid variants with high CNS tropism. Our platform does not require the use of transgenic animals and is, therefore, compatible with virtually all in vivo models, including higher mammals.
Directed evolution strategies relying on total viral DNA recovery can suffer from the confounding effects of virus productivity bias or the carryover of inert particles accumulated in the tissue of interest.20,21 In our case, using RNA expression as an obligatory step allowed a reliable selection of true positives, attested by the improvement of all our individually tested candidates by at least one order of magnitude over AAV9. In addition, the use of cell-type-specific promoters presumably reduced the recovery of capsids from undesired cell types and contributed to the rapid emergence of variants with high neuronal or glial tropism after only two rounds of screening. It is noteworthy that our screen did not uncover capsids with a strict tropism for brain endothelial cells, which constituted the dominant species in a previous DNA-based brain library screen.44 The TRACER system can be adapted to any promoter sequence of less than 2 kb, to accommodate the size limit allowed by AAV packaging, and may be of great value in numerous applications where selective recovery from rare cell types is required.
Another key aspect of our study was the identification of classes of conserved variants with similar properties, in agreement with a recent report.38 This is important when considering the undersampling of AAV libraries. Consistent with previous observations,14 our AAV libraries contained about 2e7 unique variants regardless of the initial DNA library diversity. For a 7-mer random library (1.28e9 theoretical variants), the probability for any given variant to be present in the original pool is therefore less than 2%. However, if one or more amino acid substitutions is tolerated, the likelihood for recovering a biochemically similar capsid increases dramatically. We identified, among many others, more than 60 variants harboring a peptide motif similar to PHP.B and PHP.eB capsids, as well as close homologs to the recently identified AAV-F and PHP.C2 capsids.25,38 The fact that our screen “re-discovered” close homologs of multiple capsids previously identified using several independent Cre-Lox selections, in addition to novel variants with unprecedented CNS transduction, suggests that the sequence space coverage and the robustness of the TRACER platform are equivalent or possibly superior to the existing transgenic evolution platforms.
The SYN-driven NGS analysis data were highly predictive of individual capsid properties and identified multiple variants with a high tropism for neurons. By comparison, capsids with high enrichment score in the GFAP-driven assay showed lower inter-animal and inter-assay consistency and did not transduce GFAP-positive astrocytes efficiently. These observations could suggest a disconnect between the cellular tropism of certain capsid variants and the specificity of the synthetic gfaABC1D promoter used to drive library mRNA expression. If capsids predominantly transduce non-astrocytic cells, nonspecific transcription from the gfaABC1D promoter45 could result in positive but inconsistent RNA recovery. In addition, the recently described neuronal preference of the CBA promoter46 (similar to the CAG used in our individual capsid study) could lead to an underestimation of capsid tropism for non-neuronal cells. Regardless, the high success ratio of individual capsid candidates (10/10 candidates with at least 10-fold improvement over AAV9) argues for the robustness of our cell-type-specific NGS platform.
Variants from different sequence clusters showed qualitative and quantitative variation in their cellular tropism and mouse strain restriction, which strongly suggest the use of multiple mechanisms for transport across the BBB. Brain tissue examination, however, showed that all candidates had a strikingly similar tropism for specific brain regions such as the hippocampus CA2 region, the cerebellum, or the thalamus. This repeated pattern, also shared by PHP.B and AAV-F capsids,25,26 suggest that while the 7-mer peptide insertion is critical for transport across the BBB, other AAV9 capsid domains also contribute to the regional distribution of the virus regardless of the BBB translocation mechanism. This finding underlies the multifactorial nature of in vivo transduction and could have important implications for future library designs.
Materials and methods
TRACER vectors construction
Library shuttle vectors harboring an AAV capsid expression cassette under the control of the human Synapsin 1 promoter33 or the gfaABC1D promoter34 were assembled using standard molecular biology techniques, and full sequences are available in the Supplemental information. These plasmids contain full-length inverted terminal repeats (ITRs) as well as minimal AAV cis sequences required for capsid mRNA expression and splicing during virus production. The capsid fragment extending from the hypervariable loop VIII to the stop codon was removed and replaced by a unique BsrGI restriction site used for library insertion. TelN protelomerase palindromic recognition sites (TATCAGCACACAATTGCCCATTATACGCGCGTATAATGGACTATTGTGTGCTGATA) were inserted outside of the ITRs to allow concatemer resolution and end joining after cloning-free library amplification (see below). The helper vector pREP-3stop encoding the REP protein in trans was generated by MscI digestion and self-ligation of a pREP2-CAP9 plasmid, in order to delete nucleotides 1514–2030 of AAV9 VP1. Artificial stop codons were added downstream of the start codons of VP1, VP2, and VP3. The amino acid sequence of the assembly-activating protein (AAP) was kept unchanged. The full sequence of this construct is available in the Supplemental information.
AAV library construction and virus production
Primers 9L8-F24, 9DGL8-F24, and 9GDTL8-F24 containing hand-mixed randomized NNK codons flanked by conserved sequences (Table S1) were generated by Integrated DNA Technologies (IDT, Coralville, IA, USA) and used together with the CAP9-StopR23 primer to generate library amplicons containing a randomized loop embedded in a fragment covering nucleotides 1735–2211 of AAV9 VP1. A gBlock fragment (IDT) was used as a template to prevent carryover from plasmid material in subsequent reactions. PCR was performed for 15 cycles using Q5 polymerase (New England Biolabs [NEB], Ipswich, MA, USA). The resulting amplicons were gel purified, and 500 ng of each amplicon was assembled with 2 μg of BsrGI-digested TRACER vectors using 100 μL 2× NEBuilder mix (NEB). Assembled products were treated with T5 exonuclease to eliminate unassembled DNA and purified on DNA Clean and Concentrator-5 columns (Zymo Research, Irvine, CA, USA). The resulting products were quantified by nanodrop, and the entire reaction (routinely 500–600 ng) was used in a 900 μL rolling circle amplification reaction performed at 30°C overnight with TruePrime RCA kit (4Basebio, Madrid, Spain). The reaction was heat-inactivated 10 min at 65°C, diluted 1:5 in 1× ThermoPol buffer (NEB), and treated at 30°C for 1 h with 50 μL of protelomerase (NEB) to obtain linear closed-end DNA monomers. After heat inactivation of protelomerase at 70°C for 10 min, a 5 μL aliquot (∼1:1,000 of the total reaction) was run on an agarose gel to confirm amplification and complete concatemer resolution. Qiaprep 2.0 columns (QIAGEN) were used for final DNA purification. The procedure routinely yielded ∼200 μg of transfection-ready dogbone DNA.
Virus production was performed in HEK293T maintained in Dulbecco’s modified Eagle’s medium with GlutaMAX, penicillin/streptomycin (all from Gibco) and 5% fetal bovine serum (Corning). Calcium phosphate transfection was performed using 15 μg pAdDeltaF6 adenovirus helper plasmid, 10 μg pREP-3stop, and 1 μg library DNA per dish. These conditions have been previously shown to allow optimal virus yields, library diversity, and DNA-capsid correlation.14 Cells and culture medium were harvested 72 h after transfection by scraping. Cells were pelleted by low-speed centrifugation and lysed by addition of 0.1% Triton X-100 (Thermo Scientific), while supernatant was precipitated on ice with 1:10th volume of a 40% PEG-8000, 2.5 M NaCl solution followed by centrifugation. Lysate and supernatant fractions were pooled and fractionated on two successive rounds of iodixanol gradients as previously described.47 Buffer exchange was performed on Amicon-100 columns (Millipore) with phosphate-buffered saline containing 200 mM total NaCl and 0.001% Pluronic F-68 (Gibco), and the final virus samples were analyzed by real-time PCR using a REP-specific primer/probe set (Table S1). Final virus preparations were tested by silver stain of PAGE gels and Endosafe endotoxin assay (Charles River Laboratories, Wilmington, MA, USA).
Animals
Adult male C56Bl/6J mice (22–30 g; 7–8 weeks; stock #027) were purchased from the Charles River Laboratory. Adult male BALB/C mice (22–30 g; 7–8 weeks; #000651) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Animals were housed in a 12-h light:12-h dark environment and provided food and water ad libitum. All animal protocols were approved by the Voyager Therapeutics (Cambridge, MA, USA) Institutional Animal Care and Use Committee (IACUC). For all experiments, animals were maintained in the animal colony for 1 week before dosing, and all animals were euthanized using a ketamine/xylazine cocktail.
Library screening by in vivo selection
AAV libraries were injected to C57BL/6 mice (n = 3) at a dose of 1e12 VG per animal into the lateral tail vein. For multiplexed analysis of synthetic libraries, 5e11 VG were injected to each C57BL/6 (n = 6) and BALB/c mouse (n = 6). Animals were euthanized 28 days post-injection and perfused with cold PBS. Brain, spinal cord, heart, and liver were promptly collected, snap-frozen in liquid nitrogen, and stored at −80°C.
Whole-brain RNA was extracted using the RNeasy plus universal kit (QIAGEN) following manufacturer’s instructions, and mRNA was purified using Oligotex beads (QIAGEN). Reverse transcription was performed with the gene-specific CAP-RT primer (Table S1) using Superscript IV first-strand synthesis kit (Life Technologies). Full-length spliced capsid cDNA was then amplified for 25 cycles using SpliceF6 and CAP-RT primers in multiple 50 μL PCR reactions containing 4 μL of cDNA and 25 μL Q5 HotStart high-fidelity 2× master mix (NEB). Gel-purified amplicons were used in nested PCR reactions for cloning into TRACER SYN or GFAP vectors (using CAP9-L8F and CAP9-StopR23 primers) or for generation of NGS templates (using 9∗NGS-F and 9∗NGS-R primers).
In vivo characterization of TRACER AAV variants
REP-CAP plasmids containing various capsid candidates were generated from an ITR-less REP2-CAP9-BsrGI plasmid, and virus production was performed by co-transfection of HEK293T with 15 μg pAdDeltaF6 adenovirus helper plasmid, 10 μg REP-CAP plasmid, and 5 μg self-complementary AAV CAG-EGFP plasmid (Supplemental information). Cell harvesting and lysis, iodixanol gradient purification, and buffer exchange were performed as described for libraries. Final virus titers were determined by TaqMan real-time PCR using a EGFP primer/probe set (Life Technologies).
Twelve capsid variants including AAV9 and PHP.eB were injected, respectively, into the lateral tail vein (4e11 vg per animal). Three mice were used for mRNA extraction and three mice for immunohistochemistry staining. Animals were euthanized 28 days post-injection. Animals assigned for mRNA extraction were perfused with cold PBS, and tissues were snap-frozen in liquid nitrogen. Animals assigned for immunohistochemistry were perfused with cold PBS and 4% paraformaldehyde (PFA), and tissues were immersion-fixed in 10% neutral formalin and cryo-preserved in sucrose. Brains were sectioned in half following the sagittal midline. One hemisphere was frozen in OCT and sectioned with a cryostat for EGFP direct observation and whole-slide scanning, and the other hemisphere was processed for paraffin embedding.
Immunohistochemistry and imaging
Frozen right brain hemispheres were sectioned at 10 μm thickness on a cryostat (Leica Biosystems) and mounted on glass slides with Prolong Gold anti-fade mountant (Thermo Fisher Scientific, cat. P36934). Left hemispheres were fixed in 4% paraformaldehyde at room temperature for 24 h and processed for paraffin embedding and 5 μm thickness sectioning. Antigen retrieval was performed at 95°C for 40 min with CC1 (Ventana). Rabbit anti-GFP (A-11122, Invitrogen) was incubated for 60 min at 1:250 dilution in antibody diluent (PBS with 10% normal goat serum and 0.01% Triton X-100), followed by 16 min incubation with OmniMap anti-Rabbit HRP (Ventana #760-4311). Signal was detected with 3,3′-diaminobenzidine ChromoMap DAB kit (Ventana), and counterstaining was performed with hematoxylin II and bluing reagent (Ventana) before slides were dehydrated and mounted in Cytoseal 60 (Thermo Scientific). Immunofluorescence for NeuN-EGFP or EGFP-GFAP was performed on a Ventana Discovery Ultra autostainer. All the incubations were at 37°C unless otherwise stated. Antigen retrieval was performed at 95°C for 40 min with Ventana CC1. Rabbit anti-NeuN (ABN78, Millipore), rabbit anti-GFAP (Z0334, Dako) and rabbit anti-GFP (see above) were used at 1:3,000, 1:400, and 1:250 dilution, respectively. This was followed by 16 min incubation with OmniMap anti-rabbit horseradish peroxidase (HRP) and signal detection with Cy5, rhodamine, or fluorescein isothiocyanate (FITC) discovery reagents (Ventana). Slides mounted in Cytoseal 60 were imaged on a Nikon Eclipse Ti-2 epifluorescence microscope.
NGS and bioinformatics analysis
NGS amplicon libraries were generated by 15 cycles of nested PCR with Q5 High-Fidelity DNA Polymerase (NEB) using 9∗NGS-F and 9∗NGS-R primers. PCR products were separated on 3.0% agarose gel and purified with Zymo Gel DNA Recovery Kit (Zymo Research). Final NGS libraries were quantified with Qubit dsDNA HS kit (Life Technology). Samples were spiked with 20% PhiX libraries (Illumina), denatured with 0.2 N NaOH at room temperature for 5 min, further diluted with HT1 buffer, and subjected to Illumina Nextseq500 analysis with 75-cycle or 150-cycle high-output kit.
A custom AAV amplicon-sequencing pipeline was developed to process the NGS raw data from round 1 and round 2 screens. Briefly, raw reads were first quality filtered, and a sequential trimming (https://sourceforge.net/projects/bbmap/) was applied to map invariable flanking sequences allowing a 10% error rate. Variable library inserts were length filtered (27 nt) for peptide translation. Inserts with nucleotide sequences within 1 Levenshtein distance to the relatively abundant ones were collapsed to reduce sequencing and PCR errors.48 T/G filtering of every third position of NNK codons was also applied to filter out sequencing errors.
For round 2 enrichment analysis, the reads per million (rpm) of each capsid variant in brain RNA recovery was divided by the rpm of this variant in P1 virus pool. The list of 330 candidates were selected based on: (1) raw count of the variant in virus input (>10), and (2) fold change between P2 brain RNA recovery and P1 virus stock (>20). In total, 330 unique capsid variants were kept for primer pool synthesis and round 3 characterization studies. A mammalian NNK and NNM codon table was applied to generate non-rare codon variants of each peptide. To process the data from round 3 synthetic library analysis, NNK and NNM codon sequences of 330 capsid variants plus PHP.eB and PHP.B were built by bowtie index as reference genome.49 The minimal hamming distance50 of the 664 codon sequences was 2. Sequence reads of inserts were aligned to bowtie references with 1 mismatch allowed.
Vector mRNA and DNA quantification in mouse tissue
For transgene mRNA quantification, total RNA was extracted from 100–200 mg of tissue using RNeasy plus universal kit (QIAGEN), and reverse transcription was performed with 1 μg RNA using the Quantitect kit (QIAGEN). Spliced EGFP transcripts were quantified by TaqMan PCR using a primer-probe set specific for CMV-globin exon-exon junction (Table S1). Murine TATA box-binding protein (TBP) RNA was used as a housekeeping control in all experiments. Relative expression levels were calculated from the ΔCt values and normalized to AAV9 samples.
For vector DNA quantification, 20 mg tissue was processed using the Blood and Tissue DNeasy kit (QIAGEN). Concentration was adjusted to 40 ng/μL in all samples, and 100 ng was used for TaqMan PCR quantification with a probe/primer set specific for EGFP (Life Technologies). Normalization was performed with a TaqMan set specific for the murine TERT gene (Life Technologies; Table S1).
In vitro binding and transduction assays
Recombinant AAV9 containing CAG-, SYN-, or GFAP-driven EGFP, as well as AAV9 TRACER vectors containing CAG-, SYN-, or GFAP-driven CAP, were generated as described above. Primary mixed neuronal culture was prepared with brains from embryonic day 17 CD1 mice (Charles River Laboratories). Dissociated hippocampal neurons were plated onto poly-D-lysine coated plates at 100,000 cells/well and cultured in neurobasal media supplemented with B27, GlutaMAX, and penicillin/streptomycin (Gibco). Cells were transduced at day in vitro 4. HEK293T cells and primary mixed neuronal cells were transduced 48 h with 1e5 VG per cell. Total RNA was extracted using RNeasy mini columns (QIAGEN), and reverse transcription was performed with a Quantitect kit (QIAGEN). Expression levels of EGFP transcripts or CAP transcripts were measured with the same TaqMan primer/probe set specific for the CMV-globin exon-exon junction (Table S1). Murine TBP and human Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used as housekeeping controls in mouse primary brain cells and HEK293T cells, respectively. Primary murine C57BL/6-derived BMVECs were obtained from Cell Biologics (Chicago, IL, USA) and cultured with endothelial media from the same source. Synthetic AAV library was added in culture medium for 2 h at 37°C before extensive PBS washes. Cells were then lysed directly in the flask, and low-molecular-weight DNA was extracted using Zyppy miniprep columns (Zymo Research). Viral DNA was amplified for 20 cycles with primers containing Illumina adapters and gel purified for NGS sequencing.
Statistical analysis
Unpaired two-tailed t tests were performed in Excel and are reported in figure legends. A p value < 0.05 was considered significant. Results are reported as mean ± SD. Correlation and R2 values were calculated in Excel.
Acknowledgments
This study was funded by Voyager Therapeutics.
Author contributions
M.N. designed and performed experiments, analyzed data, prepared figures, and wrote the manuscript. W.W. developed pipelines and performed bioinformatics analysis of NGS data, helped with data analysis, and assisted with manuscript preparation. M.A.C. and A.Z.R. performed experiments, virus production, and characterization. X.-Q.R. performed NGS runs. C.H., Q.C., and J.T. performed in vivo dosing and tissue processing. N.P., K.B., and K.T. performed tissue processing and IHC imaging. C.H.-Y.C. performed experiments with mouse primary cells. S.M.P. provided support and oversight. J.H. helped with study design, data analysis, and manuscript preparation and supervised the project. All authors approved the manuscript.
Declaration of interests
M.N., W.W., M.A.C., X.-Q.R., C.H., A.Z.R., J.T., K.B., K.T., N.P., C.H.-Y.C., and J.H. are paid employees of Voyager Therapeutics Inc. Voyager has filed a patent application related to the subject matter of this paper: WO2020072683. S.M.P. currently serves on the board of Voyager Therapeutics, Sage Therapeutics, Karuna Therapeutics, and Alnylam Pharmaceuticals.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.12.006.
Contributor Information
Mathieu Nonnenmacher, Email: mnonnenmacher@vygr.com.
Jay Hou, Email: jhou@vygr.com.
Supplemental information
References
- 1.Deverman B.E., Ravina B.M., Bankiewicz K.S., Paul S.M., Sah D.W.Y. Gene therapy for neurological disorders: progress and prospects. Nat. Rev. Drug Discov. 2018;17:641–659. doi: 10.1038/nrd.2018.110. [DOI] [PubMed] [Google Scholar]
- 2.Gray S.J., Woodard K.T., Samulski R.J. Viral vectors and delivery strategies for CNS gene therapy. Ther. Deliv. 2010;1:517–534. doi: 10.4155/tde.10.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 4.Ellsworth J.L., Gingras J., Smith L.J., Rubin H., Seabrook T.A., Patel K., Zapata N., Olivieri K., O’Callaghan M., Chlipala E. Clade F AAVHSCs cross the blood brain barrier and transduce the central nervous system in addition to peripheral tissues following intravenous administration in nonhuman primates. PLoS One. 2019;14:e0225582. doi: 10.1371/journal.pone.0225582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hocquemiller M., Giersch L., Audrain M., Parker S., Cartier N. Adeno-Associated Virus-Based Gene Therapy for CNS Diseases. Hum. Gene Ther. 2016;27:478–496. doi: 10.1089/hum.2016.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taymans J.-M., Vandenberghe L.H., Van den Haute C., Thiry I., Deroose C., Mortelmans L. Comparative analysis of adeno-associated viral vector serotypes 1, 2, 5, 7, and 8 in mouse brain. Hum. Gene Ther. 2007;18:195–206. doi: 10.1089/hum.2006.178. [DOI] [PubMed] [Google Scholar]
- 8.Cearley C.N., Wolfe J.H. Transduction characteristics of adeno-associated virus vectors expressing cap serotypes 7, 8, 9, and Rh10 in the mouse brain. Mol. Ther. 2006;13:528–537. doi: 10.1016/j.ymthe.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 9.Hinderer C., Bell P., Katz N., Vite C.H., Louboutin J.P., Bote E., Yu H., Zhu Y., Casal M.L., Bagel J. Evaluation of Intrathecal Routes of Administration for Adeno-Associated Viral Vectors in Large Animals. Hum. Gene Ther. 2018;29:15–24. doi: 10.1089/hum.2017.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinderer C., Bell P., Vite C.H., Louboutin J.P., Grant R., Bote E., Yu H., Pukenas B., Hurst R., Wilson J.M. Widespread gene transfer in the central nervous system of cynomolgus macaques following delivery of AAV9 into the cisterna magna. Mol. Ther. Methods Clin. Dev. 2014;1:14051. doi: 10.1038/mtm.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perabo L., Büning H., Kofler D.M., Ried M.U., Girod A., Wendtner C.M., Enssle J., Hallek M. In vitro selection of viral vectors with modified tropism: the adeno-associated virus display. Mol. Ther. 2003;8:151–157. doi: 10.1016/s1525-0016(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 12.Müller O.J., Kaul F., Weitzman M.D., Pasqualini R., Arap W., Kleinschmidt J.A., Trepel M. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat. Biotechnol. 2003;21:1040–1046. doi: 10.1038/nbt856. [DOI] [PubMed] [Google Scholar]
- 13.Xie Q., Bu W., Bhatia S., Hare J., Somasundaram T., Azzi A., Chapman M.S. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nonnenmacher M., van Bakel H., Hajjar R.J., Weber T. High capsid-genome correlation facilitates creation of AAV libraries for directed evolution. Mol. Ther. 2015;23:675–682. doi: 10.1038/mt.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmit P.F., Pacouret S., Zinn E., Telford E., Nicolaou F., Broucque F., Andres-Mateos E., Xiao R., Penaud-Budloo M., Bouzelha M. Cross-Packaging and Capsid Mosaic Formation in Multiplexed AAV Libraries. Mol. Ther. Methods Clin. Dev. 2019;17:107–121. doi: 10.1016/j.omtm.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Körbelin J., Hunger A., Alawi M., Sieber T., Binder M., Trepel M. Optimization of design and production strategies for novel adeno-associated viral display peptide libraries. Gene Ther. 2017;24:470–481. doi: 10.1038/gt.2017.51. [DOI] [PubMed] [Google Scholar]
- 17.Grimm D., Lee J.S., Wang L., Desai T., Akache B., Storm T.A., Kay M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maheshri N., Koerber J.T., Kaspar B.K., Schaffer D.V. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat. Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- 19.Michelfelder S., Kohlschütter J., Skorupa A., Pfennings S., Müller O., Kleinschmidt J.A., Trepel M. Successful expansion but not complete restriction of tropism of adeno-associated virus by in vivo biopanning of random virus display peptide libraries. PLoS ONE. 2009;4:e5122. doi: 10.1371/journal.pone.0005122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Körbelin J., Sieber T., Michelfelder S., Lunding L., Spies E., Hunger A., Alawi M., Rapti K., Indenbirken D., Müller O.J. Pulmonary Targeting of Adeno-associated Viral Vectors by Next-generation Sequencing-guided Screening of Random Capsid Displayed Peptide Libraries. Mol. Ther. 2016;24:1050–1061. doi: 10.1038/mt.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Körbelin J., Trepel M. How to Successfully Screen Random Adeno-Associated Virus Display Peptide Libraries In Vivo. Hum. Gene Ther. Methods. 2017;28:109–123. doi: 10.1089/hgtb.2016.177. [DOI] [PubMed] [Google Scholar]
- 22.Dalkara D., Byrne L.C., Klimczak R.R., Visel M., Yin L., Merigan W.H., Flannery J.G., Schaffer D.V. In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci. Transl. Med. 2013;5:189ra76. doi: 10.1126/scitranslmed.3005708. [DOI] [PubMed] [Google Scholar]
- 23.Lisowski L., Dane A.P., Chu K., Zhang Y., Cunningham S.C., Wilson E.M., Nygaard S., Grompe M., Alexander I.E., Kay M.A. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ojala D.S., Sun S., Santiago-Ortiz J.L., Shapiro M.G., Romero P.A., Schaffer D.V. In Vivo Selection of a Computationally Designed SCHEMA AAV Library Yields a Novel Variant for Infection of Adult Neural Stem Cells in the SVZ. Mol. Ther. 2018;26:304–319. doi: 10.1016/j.ymthe.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanlon K.S., Meltzer J.C., Buzhdygan T., Cheng M.J., Sena-Esteves M., Bennett R.E., Sullivan T.P., Razmpour R., Gong Y., Ng C. Selection of an Efficient AAV Vector for Robust CNS Transgene Expression. Mol. Ther. Methods Clin. Dev. 2019;15:320–332. doi: 10.1016/j.omtm.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.L., Yang B., Huber N., Pasca S.P., Gradinaru V. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan K.Y., Jang M.J., Yoo B.B., Greenbaum A., Ravi N., Wu W.L., Sánchez-Guardado L., Lois C., Mazmanian S.K., Deverman B.E., Gradinaru V. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017;20:1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuzaki Y., Konno A., Mochizuki R., Shinohara Y., Nitta K., Okada Y., Hirai H. Intravenous administration of the adeno-associated virus-PHP.B capsid fails to upregulate transduction efficiency in the marmoset brain. Neurosci. Lett. 2018;665:182–188. doi: 10.1016/j.neulet.2017.11.049. [DOI] [PubMed] [Google Scholar]
- 29.Hordeaux J., Wang Q., Katz N., Buza E.L., Bell P., Wilson J.M. The Neurotropic Properties of AAV-PHP.B Are Limited to C57BL/6J Mice. Mol. Ther. 2018;26:664–668. doi: 10.1016/j.ymthe.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liguore W.A., Domire J.S., Button D., Wang Y., Dufour B.D., Srinivasan S., McBride J.L. AAV-PHP.B Administration Results in a Differential Pattern of CNS Biodistribution in Non-human Primates Compared with Mice. Mol. Ther. 2019;27:2018–2037. doi: 10.1016/j.ymthe.2019.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stutika C., Gogol-Döring A., Botschen L., Mietzsch M., Weger S., Feldkamp M., Chen W., Heilbronn R. A Comprehensive RNA Sequencing Analysis of the Adeno-Associated Virus (AAV) Type 2 Transcriptome Reveals Novel AAV Transcripts, Splice Variants, and Derived Proteins. J. Virol. 2015;90:1278–1289. doi: 10.1128/JVI.02750-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mouw M.B., Pintel D.J. Adeno-associated virus RNAs appear in a temporal order and their splicing is stimulated during coinfection with adenovirus. J. Virol. 2000;74:9878–9888. doi: 10.1128/jvi.74.21.9878-9888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoch S., Cibelli G., Thiel G. Neuron-specific Gene Expression of Synapsin I. J. Biol. Chem. 1996;271:3317–3323. doi: 10.1074/jbc.271.6.3317. [DOI] [PubMed] [Google Scholar]
- 34.Lee Y., Messing A., Su M., Brenner M. GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia. 2008;56:481–493. doi: 10.1002/glia.20622. [DOI] [PubMed] [Google Scholar]
- 35.DiMattia M.A., Nam H.-J., Van Vliet K., Mitchell M., Bennett A., Gurda B.L., McKenna R., Olson N.H., Sinkovits R.S., Potter M. Structural insight into the unique properties of adeno-associated virus serotype 9. J. Virol. 2012;86:6947–6958. doi: 10.1128/JVI.07232-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deneke J., Ziegelin, G., Lurz R., Lanka E. The protelomerase of temperate Escherichia coli phage N15 has cleaving-joining activity. Proc. Natl. Adac. Sci. U S A. 2000;97:7721–7726. doi: 10.1073/pnas.97.14.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zincarelli C., Soltys S., Rengo G., Rabinowitz J.E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 38.Ravindra Kumar S., Miles T.F., Chen X., Brown D., Dobreva T., Huang Q., Ding X., Luo Y., Einarsson P.H., Greenbaum A. Multiplexed Cre-dependent selection yields systemic AAVs for targeting distinct brain cell types. Nat. Methods. 2020;17:541–550. doi: 10.1038/s41592-020-0799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hordeaux J., Wang Q., Katz N., Buza E.L., Bell P., Wilson J.M. The Neurotropic Properties of AAV-PHP.B Are Limited to C57BL/6J Mice. Mol. Ther. 2018;26:664–668. doi: 10.1016/j.ymthe.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hordeaux J., Yuan Y., Clark P.M., Wang Q., Martino R.A., Sims J.J., Bell P., Raymond A., Stanford W.L., Wilson J.M. The GPI-Linked Protein LY6A Drives AAV-PHP.B Transport across the Blood-Brain Barrier. Mol. Ther. 2019;27:912–921. doi: 10.1016/j.ymthe.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Q., Chan K.Y., Tobey I.G., Chan Y.A., Poterba T., Boutros C.L., Balazs A.B., Daneman R., Bloom J.M., Seed C., Deverman B.E. Delivering genes across the blood-brain barrier: LY6A, a novel cellular receptor for AAV-PHP.B capsids. PLoS ONE. 2019;14:e0225206. doi: 10.1371/journal.pone.0225206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hordeaux J., Yuan Y., Clark P.M., Wang Q., Martino R.A., Sims J.J., Bell P., Raymond A., Stanford W.L., Wilson J.M. The GPI-Linked Protein LY6A Drives AAV-PHP.B Transport across the Blood-Brain Barrier. Mol. Ther. 2019;27:912–921. doi: 10.1016/j.ymthe.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z., Ma H.I., Li J., Sun L., Zhang J., Xiao X. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003;10:2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- 44.Körbelin J., Dogbevia G., Michelfelder S., Ridder D.A., Hunger A., Wenzel J., Seismann H., Lampe M., Bannach J., Pasparakis M. A brain microvasculature endothelial cell-specific viral vector with the potential to treat neurovascular and neurological diseases. EMBO Mol. Med. 2016;8:609–625. doi: 10.15252/emmm.201506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taschenberger G., Tereshchenko J., Kügler S. A MicroRNA124 Target Sequence Restores Astrocyte Specificity of gfaABC1D-Driven Transgene Expression in AAV-Mediated Gene Transfer. Mol. Ther. Nucleic Acids. 2017;8:13–25. doi: 10.1016/j.omtn.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Powell S.K., Samulski R.J., McCown T.J. AAV Capsid-Promoter Interactions Determine CNS Cell-Selective Gene Expression In Vivo. Mol. Ther. 2020;28:1373–1380. doi: 10.1016/j.ymthe.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crosson S.M., Dib P., Smith J.K., Zolotukhin S. Helper-free Production of Laboratory Grade AAV and Purification by Iodixanol Density Gradient Centrifugation. Mol. Ther. Methods Clin. Dev. 2018;10:1–7. doi: 10.1016/j.omtm.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zorita E., Cuscó P., Filion G.J. Starcode: sequence clustering based on all-pairs search. Bioinformatics. 2015;31:1913–1919. doi: 10.1093/bioinformatics/btv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Loo M.P.J. The stringdist package for approximate string matching. R J. 2014;6:111–122. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.