Abstract
miRNAs have been involved in various types of cancer, including T-cell leukemia. In this study, the role of miR-1193 in the proliferation and invasion of T-cell leukemia cells was explored. First, we found that miR-1193 was sharply downregulated in T-cell leukemia cells when compared with normal T cells. miR-1193 markedly decreased the proliferation and invasion in Jurkat human T-cell leukemia cells. Transmembrane 9 superfamily 3 (TM9SF3) was then predicted to be a potential target gene of miR-1193, the levels of which displayed a strongly negative correlation with miR-1193 levels in T-cell leukemia patients. We confirmed that TM9SF3 was a target gene of miR-1193 by luciferase reporter gene assay. Finally, gene overexpression and knockdown experiments in Jurkat cells revealed that TM9SF3 positively regulated cell proliferation and invasion.
Key words: miR-1193, Cell proliferation and invasion, Transmembrane 9 superfamily 3 (TM9SF3), T-cell leukemia
INTRODUCTION
T-cell leukemia represents a minority of lymphocytic leukemia cases, and the clinical manifestations differ greatly among patients. It has a poor prognosis and early relapse, and it is difficult to achieve complete remission after chemotherapy1. Therefore, T-cell leukemia has been regarded to be a research hotspot, and it is urgent that reliable biomarkers are found and effective treatments are developed.
MicroRNAs (miRNAs) are endogenous tiny RNAs with a length of ∼22 nt that play an important regulatory role by targeting specific mRNAs for posttranscriptional degradation or translational repression2. As of this date, more than 6,000 miRNAs have been identified in humans3. Some of them have been well studied, but more need to be investigated. They have been involved in many cellular events, including cell death, proliferation, migration, differentiation, and cell cycle control4. miRNA mutations and dysregulation have been found in various human cancers, and they can function as tumor suppressors and oncogenes5. Increased expression of miRNAs usually represents an additional mechanism of oncogenesis. A few highly abundant miRNAs regulate leukemogenesis through the dominant repression of critical tumor suppressor or promoter genes6. For example, miR-19 is one of the best-characterized miRNAs, and it could promote T-cell leukemia development in Notch1-induced T-cell acute lymphoblastic leukemia in vivo7. Some other miRNAs, such as miR-223, miR-20a, miR-26a, miR-92, miR-193b, and miR-143, have also been found to converge on a set of tumor-related genes and regulate T-cell leukemia cell survival or drug resistance8–11.
Recently, miR-1193 was found to be dysregulated in several pathological processes such as pulmonary fibrosis, acute liver failure, and breast cancer development12–14. However, its expression pattern and exact role in the development of human T-cell leukemia are barely known. In this study, the levels of miR-1193 in normal human T cells and T-cell leukemia cells were first detected. We found that miR-1193 was sharply reduced in T-cell leukemia cells. miR-1193 was also overexpressed and knocked down in Jurkat human T-cell leukemia cells, respectively, and the effect on the proliferation and invasion of T-cell leukemia cells was investigated. Furthermore, we explored the mechanism through which miR-1193 regulated the proliferation and invasion of T-cell leukemia cells.
MATERIALS AND METHODS
Subjects and Ethics Statement
Thirty healthy volunteers (36.5 ± 12.4 years) and 90 peripheral T-cell leukemia patients (35.5 ± 11.3 years, including 30 patients at the chronic phase, 30 patients at the accelerated phase, and 30 patients at blast crisis) were enrolled in this study. Peripheral T cells were isolated by Ficoll-Hypaque density-gradient centrifugation followed by flow cytometry. Informed consent was obtained from all guardians of the patients in accordance with the Declaration of Helsinki and with approval of the medical ethics committee of The Second Affiliated Hospital of Zhengzhou University.
Cell Culture and Transfection
Human normal T-lymphocyte cell line H9 and three human T-cell leukemia cell lines including C8166-CD4, HUT78, and Jurkat were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were cultured with RPMI-1640 medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) plus 100 U/ml penicillin and 0.1 mg/ml streptomycin. The cells were maintained in a 5% CO2 humidified incubator at 37°C.
For transfection, Jurkat cells were subgrown in six-well plates at a density of 106/ml until they reach about 70% confluence. To investigate the function of miR-1193, the single-strand oligo miR-1193 mimic, miR-1193 inhibitor, and negative controls (NCs), which were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, P.R. China), were transfected into Jurkat cells with Lipofectamine 3000 reagent (Invitrogen) according to the manufacturer’s instructions. To investigate the function of TM9SF3, pcDNA-TM9SF3 expression vector (Sangon Biotech, Shanghai, P.R. China), TM9SF3 siRNA (Invitrogen), and their corresponding controls were respectively transfected into Jurkat cells with Lipofectamine 3000 reagent.
Real-Time PCR
Total RNA was isolated using TRIzol reagent (Invitrogen), and 1 μg of RNA was applied to synthesizing cDNA using an MMLV Reverse Transcriptase Kit (TaKaRa, Da Lian, P.R. China). Real-time PCR was conducted using SYBR Premix Ex TaqTM Kit (TaKaRa). Specific primers used in this study were designed and synthesized by Sangon Biotech. The reactions were initially denatured at 95°C for 30 s followed by 40 cycles at 95°C for 10 s and 60°C for 60 s in ABI 7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). Abundances of the mRNAs were calculated using the 2−ΔΔCT method. U6 RNA was regarded as the reference gene.
Cell Viability and Apoptosis Assays
Following transfection for 48 h, cell viability was measured using the cell counting kit-8 (CCK-8; Sigma-Aldrich, St. Louis, MO, USA) assay according to the manufacturer’s instructions. Absorbance at the 450-nm wave was measured. Cell apoptosis was detected with Annexin-V/PI Cell Apoptosis Kit (Promega, Madison, WI, USA). The cells were collected and resuspended in 100 μl of flow cytometry binding buffer and then stained with 5 μl of annexin V/FITC and 1 μl of propidium iodide (PI). After cells were incubated in the dark for 15 min and 400 μl of binding buffer was added, the cells were finally measured by FACSCalibur (Becton-Dickinson, San Jose, CA, USA) to calculate the percentage of early apoptotic cells.
Transwell Invasion Assay
Cell invasion was detected with a Transwell invasion assay in the Transwell chamber (Corning, Lowell, MA, USA). The cells that invaded through the membrane were fixed with methanol and stained with crystal violet. Photographs of six random fields of the fixed cells were captured, and the number of invading cells was counted by a countess automatic cell counter (Invitrogen).
Dual-Luciferase Activity Assay
Full-length 3′-untranslated region (3′-UTR) sequence of TM9SF3 was cloned by PCR and inserted into a p-MIR luciferase plasmid. A mutated (MUT) 3′-UTR sequence was produced by MutanBEST Kit (TaKaRa) based on the wild-type (WT) 3′-UTR. It was then inserted into a p-MIR luciferase plasmid. miR-1193 mimic or control mimic was cotransfected with the WT or MUT p-MIR luciferase plasmids (Ambion, Austin, TX, USA) into HEK293 cells. After transfection for 72 h, luciferase activity was measured with a microplate reader (Bio-Rad, Hercules, CA, USA).
Western Blotting
Total protein (30 μg) was separated by SDS-polyacrylamide gel electrophoresis and transferred to the polyvinylidene difluoride (PVDF) membranes (Millipore, Boston, MA, USA). The membranes were then incubated with specific primary antibodies, including anti-caspase 3 (1:300 dilution; Abcam, Cambridge, UK), anti-Bcl-2 (1:500 dilution; Abcam), anti-fibronectin (1:500 dilution; Abcam), anti-MMP-2 (1:200 dilution; Abcam), anti-MMP-9 (1:200 dilution; Abcam), anti-TM9SF3 (1:400 dilution; Abcam), and anti-GAPDH (1:500 dilution; Abcam) overnight. The appropriate horseradish peroxidase-conjugated secondary antibody was added for a 1-h incubation period at room temperature. The immunoreactive proteins were visualized with an Anmobilon Western Chemiluminescent HRP Substrate System (Millipore, Billerica, MA, USA).
Statistical Analysis
All data were presented as mean ± SD. Statistics were calculated with SPSS statistics v19.0 software. Multiple comparisons were assessed by one-way ANOVA followed by Dunnett’s tests. The correlation between miR-1193 and TM9SF3 levels was evaluated by Pearson nonparametric correlation coefficient assay. A value of p < 0.05 was considered statistically significant.
RESULTS
miR-1193 Was Downregulated in T-Cell Leukemia Cells
The expression of miR-1193 in peripheral blood T cells from 30 healthy volunteers and 90 patients with peripheral T-cell leukemia at different stages was measured. Our results showed that miR-1193 was decreased in patients with chronic-phase T-cell leukemia, and further decreased in patients with accelerated-phase and blast crisis T-cell leukemia (Fig. 1A). The expression of miR-1193 in human normal T-lymphocyte cell line H9 and three human T-cell leukemia cell lines including C8166-CD4, HUT78, and Jurkat was then measured. The results showed that the miR-1193 level was decreased by more than 80% in all three T-cell leukemia cell lines (Fig. 1B). It is worth mentioning that miR-1193 expression was reduced by more than 90% in Jurkat T-cell leukemia cells (Fig. 1B). These data suggest that miR-1193 may play a role in the progression of T-cell leukemia.
Figure 1.
miR-1193 was downregulated in T-cell leukemia cells. (A) miR-1193 was decreased in the peripheral blood T cells in patients with T-cell leukemia. Peripheral blood T cells were isolated from 30 healthy volunteers and 90 patients with peripheral T-cell leukemia at different stages. Total RNA was isolated from the T cells, and miR-1193 expression was detected with qPCR. (B) miR-1193 was downregulated in human T-cell leukemia cell lines. The expression of miR-1193 in human normal T-lymphocyte cell line H9 and three human T-cell leukemia cell lines including C8166-CD4, HUT78, and Jurkat. *p < 0.05 versus Control, #p < 0.05 versus Chronic phase. **p < 0.01 versus H9.
miR-1193 Suppressed the Proliferation and Invasion of Jurkat T-Cell Leukemia Cells
To explore the role of miR-1193 in the progression of T-cell leukemia, the miR-1193 mimic at concentrations of 5, 10, 20, 40, and 80 nM and the miR-1193 antagomir at concentrations of 20, 40, and 80 nM were transfected into Jurkat T-cell leukemia cells. The miR-1193 level was increased by the miR-1193 mimic in a dose-dependent manner, and the miR-1193 mimic at concentrations of 20, 40, and 80 nM increased the miR-1193 level by about sixfold (Fig. 2A). The cell viability assay showed that the miR-1193 mimic decreased cell viability in a dose-dependent manner. The miR-1193 mimic at 10 nM reduced the viability of Jurkat cells by about 30%, and the miR-1193 mimic at concentrations of 20, 40, and 80 nM reduced cell viability by more than 50% (Fig. 2B). The cell apoptosis assay showed that 10 nM of the miR-1193 mimic and the miR-1193 mimic at concentrations of 20 nM modestly increased apoptosis, while 40 and 80 nM dramatically increased the apoptosis of Jurkat cells (Fig. 2C). The Transwell invasion assay showed that 10 nM of the miR-1193 mimic significantly suppressed the invasive ability of Jurkat cells, and the miR-1193 mimic at concentrations of 20, 40, and 80 nM further reduced this ability (Fig. 2D). Simultaneously, Western blotting analysis showed that 10 nM of the miR-1193 mimic increased the expression of apoptotic protein caspase 3 and 20 nM of the miR-1193 mimic further increased this expression (Fig. 2E). The miR-1193 mimic at 10 nM reduced the expression of antiapoptotic protein Bcl-2 and several invasion marker proteins, including fibronectin, MMP-2, and MMP-9, and 20 nM of the miR-1193 mimic further reduced this expression (Fig. 2E). In contrast to the results from the miR-1193 mimic transfection, the miR-1193 antagomir decreased the miR-1193 level in a dose-dependent manner in Jurkat cells. The miR-1193 antagomir at 20 nM decreased the miR-1193 level by about 50%, and the miR-1193 antagomir at concentrations of 40 and 80 nM decreased the miR-1193 level by about 80% (Fig. 3A). The miR-1193 antagomir at 20 nM did not affect the viability, apoptosis, or invasion of Jurkat cells, whereas the miR-1193 antagomir at concentrations of 40 and 80 nM slightly increased cell viability and invasion, as well as suppressed cell apoptosis (Fig. 3B–D). These data indicated that miR-1193 suppressed the proliferation and invasion of Jurkat T-cell leukemia cells.
Figure 2.
miR-1193 overexpression suppressed the proliferation and invasion of Jurkat T-cell leukemia cells. (A) The overexpression efficiency of the miR-1193 mimic at different concentrations. (B) miR-1193 overexpression suppressed the proliferation of Jurkat cells. (C) miR-1193 overexpression promoted the apoptosis of Jurkat cells. (D) miR-1193 overexpression suppressed the invasion of Jurkat cells. (E) miR-1193 overexpression regulated the expression of apoptosis and invasion-related proteins. The miR-1193 mimic at concentrations of 5, 10, 20, 40, and 80 nM was respectively transfected into Jurkat cells. After incubation for 48 h, the miR-1193 level was detected with qPCR, cell proliferation was detected with the cell counting kit-8 (CCK-8) assay, cell apoptosis was detected with flow cytometry, cell invasion was detected with Transwell invasion assay, and the levels of caspase 3, Bcl-2, fibronectin, MMP-2, and MMP-9 were detected with Western blotting. *p < 0.05 versus Ctrl mimic, #p < 0.05 versus 5 nM, &p < 0.05 versus 10 nM.
Figure 3.
miR-1193 knockdown promoted the proliferation and invasion of Jurkat T-cell leukemia cells. (A) The knockdown efficiency of the miR-1193 antagomir at different concentrations. (B) miR-1193 knockdown promoted the proliferation of Jurkat cells. (C) miR-1193 knockdown suppressed the apoptosis of Jurkat cells. (D) miR-1193 knockdown promoted the invasion of Jurkat cells. The miR-1193 antagomir at concentrations of 20, 40, and 80 nM was respectively transfected into Jurkat T-cell leukemia cells. After incubation for 48 h, the miR-1193 level, cell proliferation, cell apoptosis, and cell invasion were detected. *p < 0.05 versus Ctrl-anta, #p < 0.05 versus 20 nM.
TM9SF3 Is a Target Gene of miR-1193
To investigate the underlying mechanism from which miR-1193 suppressed proliferation and invasion of T-cell leukemia cells, TargetScan online bioinformatic software was used to search the target gene of miR-1193. The results showed that TM9SF3 was a potential target gene of miR-1193 (Fig. 4A). miR-1193 significantly decreased the luciferase activity of the WT TM9SF3-3′UTR luciferase reporter gene plasmid but had no effect on the MUT luciferase reporter gene plasmid (Fig. 4B and C). Furthermore, we measured the effect of miR-1193 on the expression of TM9SF3 protein. Western blotting analysis showed that the miR-1193 mimic sharply decreased and the miR-1193 antagomir significantly increased TM9SF3 protein expression (Fig. 4D). These results demonstrated that TM9SF3 is a target gene of miR-1193.
Figure 4.
TM9SF3 is a direct target gene of miR-1193. (A) TargetScan online bioinformatic software reveals that there is a binding site of miR-1193 in TM9SF3 3′-untranslated region (3′-UTR). (B) Details of wild-type (WT) and mutant (MUT) TM9SF3 3′-UTR sequences inserted in luciferase reporter gene plasmids. (C) Luciferase activity assay for WT and MUT luciferase reporter gene plasmids in HEK293 cells. (D) The effect of miR-1193 overexpression and knockdown on the expression levels of TM9SF3 protein. **p < 0.01.
TM9SF3 Was Upregulated in T-Cell Leukemia Cells, and it Positively Regulated the Proliferation and Invasion of Jurkat T-Cell Leukemia Cells
The exact role of TM9SF3 in the regulation of carcinogenesis has not been revealed. The expression of TM9SF3 in peripheral blood T cells from 30 healthy volunteers and 90 patients with peripheral T-cell leukemia at different stages was measured. The results showed that TM9SF3 mRNA and protein were upregulated in patients with chronic-phase T-cell leukemia, and further increased in patients with accelerated-phase and blast crisis T-cell leukemia (Fig. 5A and C). In cultured cell lines, levels of TM9SF3 mRNA and protein were increased by more than fourfold in all three T-cell leukemia cell lines, and they were increased by more than sixfold in Jurkat cells (Fig. 5B and D). Pearson nonparametric correlation coefficient assay revealed that the TM9SF3 protein levels had a strongly negative correlation with miR-1193 levels in 22 patients randomly selected from the 90 patients (Fig. 5E).
Figure 5.
TM9SF3 was upregulated in T-cell leukemia cells, and its levels were negatively related with miR-1193 levels. (A) TM9SF3 mRNA was upregulated in the peripheral blood T cells in patients with T-cell leukemia. (B) TM9SF3 mRNA was upregulated in human T-cell leukemia cell lines. (C) TM9SF3 protein was upregulated in the peripheral blood T cells in patients with T-cell leukemia. (D) TM9SF3 protein was upregulated in human T-cell leukemia cell lines. Peripheral blood T cells were isolated from 30 healthy volunteers and 90 patients with peripheral T-cell leukemia at different stages. Total RNA and total protein were isolated from the T cells. TM9SF3 mRNA and protein levels in peripheral blood T cells, human normal T-lymphocyte cell line H9, and three human T-cell leukemia cell lines including C8166-CD4, HUT78, and Jurkat were detected with qPCR and Western blotting. (E) TM9SF3 levels were negatively related with miR-1193 levels in 22 patients randomly selected. *p < 0.05 versus control, #p < 0.05 versus Chronic phase, **p < 0.01 versus H9.
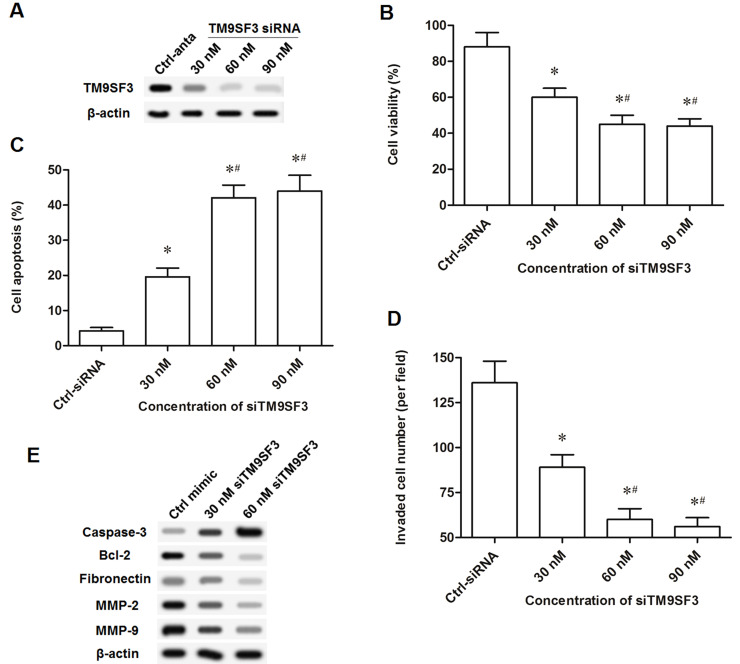
Finally, to investigate the exact role of TM9SF3 in the proliferation and invasion of T-cell leukemia cells, TM9SF3 was overexpressed via the pcDNA-TM9SF3 expression vector or knocked down via the TM9SF3 siRNA. The pcDNA-TM9SF3 expression vector at concentrations of 2 and 4 μg/ml markedly increased the level of TM9SF3 protein (Fig. 6A). Similar to the effect of the miR-1193 antagomir on Jurkat cells, the pcDNA-TM9SF3 vector at concentrations of 2 and 4 μg/ml slightly increased cell viability and invasion and suppressed cell apoptosis (Fig. 6B–D). Additionally, 30 nM TM9SF3 siRNA significantly decreased the TM9SF3 protein level, and the TM9SF3 siRNA at concentrations of 60 and 90 nM dramatically decreased TM9SF3 protein expression (Fig. 7A). Similar to the effect of the miR-1193 mimic on Jurkat cells, TM9SF3 siRNA at concentrations of 60 and 90 nM dramatically decreased cell viability and invasion and promoted cell apoptosis (Fig. 7B–D). TM9SF3 siRNA at 30 nM increased caspase 3 expression and reduced the levels of Bcl-2, fibronectin, MMP-2, and MMP-9; 60 nM of TM9SF3 siRNA further increased caspase 3 expression and further reduced the levels of Bcl-2, fibronectin, MMP-2, and MMP-9 (Fig. 7E). These data indicated that TM9SF3 positively regulated the proliferation and invasion of T-cell leukemia cells.
Figure 6.
TM9SF3 overexpression promoted the proliferation and invasion of Jurkat cells. (A) The overexpression efficiency of the pcDNA-TM9SF3 vector at different concentrations. (B) TM9SF3 overexpression promoted the proliferation of Jurkat cells. (C) TM9SF3 overexpression suppressed the apoptosis of Jurkat cells. (D) TM9SF3 overexpression promoted the invasion of Jurkat cells. The pcDNA-TM9SF3 vector at concentrations of 1, 2, and 4 μg/ml was respectively transfected into Jurkat T-cell leukemia cells. After incubation for 48 h, the TM9SF3 protein level, cell proliferation, cell apoptosis, and cell invasion were detected. *p < 0.05 versus Vector.
Figure 7.
TM9SF3 knockdown suppressed the proliferation and invasion of Jurkat cells. (A) The knockdown efficiency of the TM9SF3 siRNA at different concentrations. (B) TM9SF3 knockdown suppressed the proliferation of Jurkat cells. (C) TM9SF3 knockdown promoted the apoptosis of Jurkat cells. (D) TM9SF3 knockdown suppressed the invasion of Jurkat cells. (E) TM9SF3 knockdown regulated the expression of apoptosis and invasion-related proteins. The TM9SF3 siRNA at concentrations of 30, 60, and 90 nM was respectively transfected into Jurkat cells. After incubation for 48 h, the protein TM9SF3 level, cell proliferation, cell apoptosis, cell invasion, and the protein levels of caspase 3, Bcl-2, fibronectin, MMP-2, and MMP-9 were detected. *p < 0.05 versus Ctrl-anta, #p < 0.05 versus 30 nM.
DISCUSSION
miR-1193 was discovered in human melanoma tissues in 200015. There were rare reports on the function of miR-1193. Quite recently, Li et al. reported that miR-1193 was sharply reduced in breast cancer tissues and cell lines, and it suppressed the proliferation and invasion of breast cancer cells16. In this study, we also found that, compared with healthy volunteers and a normal T-lymphocyte cell line, miR-1193 was sharply reduced in peripheral blood T cells of T-cell leukemia patients and cell lines. Moreover, gain- and loss-of-function experiments revealed that miR-1193 was able to inhibit proliferation and invasion.
The regulation of gene expression by miRNAs is complex. Many mRNAs contain binding sites for multiple miRNAs and, likewise, most miRNAs potentially target a number of genes17. In breast cancer cells, miR-1193 binds the insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) mRNA at the 3′-UTR region and suppresses the activation of the ERK and PI3K/Akt proto-oncogene pathways at the downstream IGF2BP2 gene16. Our results showed that miR-1193 targeted the TM9SF3 mRNA at the 3′-UTR region and suppressed expression of TM9SF3 and TM9SF3-mediated proliferation and invasion in T-cell leukemia cells.
TM9SFs are a family of nonaspanin proteins, characterized by a large noncytoplasmic domain and nine putative transmembrane domains18. The family is highly conserved through evolution and comprises four members in mammals (TM9SF1–TM9SF4). Genetic studies revealed that TM9SF members played an irreplaceable role in adhesion and phagocytosis of immune cells19. It was shown that TM9SFs were transcriptionally regulated by TNF receptor-associated factor 2 (TRAF2), which is closely linked to immune development and tumor progression20. Recently, TM9SF3 was found to affect dysregulation in several types of cancer, suggesting that it might play an important role in carcinogenesis21. A serial analysis of gene expression showed that TM9SF3 was upregulated in gastric cancer tissues22. TM9SF3 expression was significantly correlated with depth of invasion and tumor stage in scirrhous-type gastric cancer, indicating that TM9SF3 may serve as a prognostic factor. In addition, oligonucleotide microarray analysis identified TM9SF3 and 27 other genes that are correlated with the resistance of paclitaxel and SAHA in breast cancer23. However, the exact role of TM9SF3 in the regulation of carcinogenesis has not been revealed. In this study, we found that TM9SF3 was significantly upregulated in peripheral blood T cells of T-cell leukemia patients and cell lines. TM9SF3 positively regulated the proliferation and invasion of T-cell leukemia cells.
In conclusion, the present study proved that miR-1193 suppresses proliferation and invasion in human T-cell leukemia cells through directly targeting the newly identified proto-oncogene TM9SF3.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Iwanaga M, Watanabe T, Yamaguchi K. Adult T-cell leukemia: A review of epidemiological evidence. Front Microbiol. 2012;3:322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116(2):281–97. [DOI] [PubMed] [Google Scholar]
- 3. Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2011;223(2):102–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell 2009;136(2):215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Esquelakerscher A, Slack FJ. Oncomirs—MicroRNAs with a role in cancer. Nat Rev Cancer 2006;6(4):259–69. [DOI] [PubMed] [Google Scholar]
- 6. Sanghvi VR, Mavrakis KJ, Van dMJ, Boice M, Wolfe AL, Carty M, Mohan P, Rondou P, Socci ND, Benoit Y, Taghon T, Van Vlierberghe P, Leslie CS, Speleman F, Wendel HG. Characterization of a set of tumor suppressor microRNAs in T cell acute lymphoblastic leukemia. Sci Signal. 2014;7(352):ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS, Parker JS, Paddison PJ, Tam W, Ferrando A, Wendel HG. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol. 2010;12(4):372–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumar V, Palermo R, Talora C, Campese AF, Checquolo S, Bellavia D, Tottone L, Testa G, Miele E, Indraccolo S, Amadori A, Ferretti E, Gulino A, Vacca A, Screpanti I. Notch and NF-kB signaling pathways regulate miR-223/FBXW7 axis in T-cell acute lymphoblastic leukemia. Leukemia 2014;28(12):2324–35. [DOI] [PubMed] [Google Scholar]
- 9. Akao Y, Nakagawa YA, Naoe T. Role of microRNA-143 in Fas-mediated apoptosis in human T-cell leukemia Jurkat cells. Leuk Res. 2009;33(11):1530–8. [DOI] [PubMed] [Google Scholar]
- 10. Mets E, Van der Meulen J, Van Peer G, Boice M, Mestdagh P, Van de Walle I, Lammens T, Goossens S, De Moerloose B, Benoit Y, Van Roy N, Clappier E, Poppe B, Vandesompele J, Wendel HG, Taghon T, Rondou P, Soulier J, Van Vlierberghe P, Speleman F. MicroRNA-193b-3p acts as a tumor suppressor by targeting the MYB oncogene in T-cell acute lymphoblastic leukemia. Leukemia 2014;29(4):798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamada N, Noguchi S, Kumazaki M, Shinohara H, Miki K, Naoe T, Akao Y. Epigenetic regulation of microRNA-128a expression contributes to the apoptosis-resistance of human T-cell leukaemia jurkat cells by modulating expression of fas-associated protein with death domain (FADD). Biochim Biophys Acta 2014;1843(3):590–602. [DOI] [PubMed] [Google Scholar]
- 12. Vemuganti R, Silva VR, Mehta SL, Hazell AS. Erratum to: Acute liver failure-induced hepatic encephalopathy is associated with changes in microRNA expression profiles in cerebral cortex of the mouse. Metab Brain Dis. 2015;30(1):247. [DOI] [PubMed] [Google Scholar]
- 13. Zhang L, Li X, Dong W, Sun C, Guo D, Zhang L. Mmu-miR-1894-3p inhibits cell proliferation and migration of breast cancer cells by targeting Trim46. Int J Mol Sci. 2016;17(4):609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lei GS, Kline HL, Lee CH, Wilkes DS, Zhang C. Regulation of collagen V expression and epithelial-mesenchymal transition by miR-185 and miR-186 during idiopathic pulmonary fibrosis. Am J Pathol. 2016;186(9):2310–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stark MS, Tyagi S, Nancarrow DJ, Boyle GM, Cook AL, Whiteman DC, Parsons PG, Schmidt C, Sturm RA, Hayward NK. Characterization of the melanoma miRNAome by deep sequencing. PLoS One 2012;5(3):e9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li X, Li Y, Hong L. MiR-1193 suppresses proliferation and invasion of human breast cancer cells through directly targeting IGF2BP2. Oncol Res. 2017;25(4):579–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pillai RS. MicroRNA function: Multiple mechanisms for a tiny RNA? 2006;11(12):1753–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Benghezal M, Cornillon S, Gebbie L, Alibaud L, Brückert F, Letourneur F, Cosson P. Synergistic control of cellular adhesion by transmembrane 9 proteins. Mol Biol Cell 2003;14(7):2890–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tang X, Parisi D, Spicer B, Morré DM, Morré DJ. Molecular cloning and characterization of human age-related NADH oxidase (arNOX) proteins as members of the TM9 superfamily of transmembrane proteins. Adv Biol Chem. 2013;03(2):187–97. [Google Scholar]
- 20. Pruvot B, Laurens V, Salvadori F, Solary E, Pichon L, Chluba J. Comparative analysis of nonaspanin protein sequences and expression studies in zebrafish. Immunogenetics 2010;62(10):681–99. [DOI] [PubMed] [Google Scholar]
- 21. Oue N, Sentani K, Sakamoto N, Yasui W. Clinicopathologic and molecular characteristics of gastric cancer showing gastric and intestinal mucin phenotype. Cancer Sci. 2015;106(8):951–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oo HZ, Sentani K, Sakamoto N, Anami K, Naito Y, Oshima T, Yanagihara K, Oue N, Yasui W. Identification of novel transmembrane proteins in scirrhous-type gastric cancer by the Escherichia coli ampicillin secretion trap (CAST) method: TM9SF3 participates in tumor invasion and serves as a prognostic factor. Pathobiology 2014;81(81):138–48. [DOI] [PubMed] [Google Scholar]
- 23. Chang H, Jeung HC, Jung JJ, Kim TS, Rha SY, Chung HC. Identification of genes associated with chemosensitivity to SAHA/taxane combination treatment in taxane-resistant breast cancer cells. Breast Cancer Res Treat. 2011;125(1):55–63. [DOI] [PubMed] [Google Scholar]