Abstract
Telithromycin has been reported to possess robust in vitro antibacterial activity against many species of gram-positive bacteria, and telithromycin is also effective against Staphylococcus aureus biofilms. However, the in vitro antimicrobial susceptibility of telithromycin against clinical enterococci isolates in China is rarely reported and the impacts of telithromycin on the biofilm formation and eradication of enterococci remain elusive. Therefore, this study aimed to explore the inhibitory effects of telithromycin on planktonic cells and biofilms of Enterococcus strains. A total of 280 Enterococcus faecalis and 122 Enterococcus faecium isolates were collected from individual inpatients in China. The 50% minimum inhibitory concentration (MIC50) values of telithromycin against the E. faecalis and E. faecium strains carrying erythromycin-resistant methylase (erm) genes such as the ermA, ermB, or ermC, were 2 and 4 μg/mL, respectively. In addition, these isolates were typed using multilocus sequence typing (MLST) based on housekeeping genes. The predominant sequence types (STs) of E. faecalis were ST16, ST30, and ST179, and the main STs of E. faecium isolates were ST18, ST78, and ST80. Among these major STs, 87.1% (135/158) of E. faecalis and 80.4% (41/51) of E. faecium carried erm genes. Furthermore, at the subinhibitory concentrations (1/4 and 1/8 × MIC) of telithromycin, the biofilm formation of 16 E. faecalis isolates were inhibited by approximately 35%. Moreover, treatment with 8 × MIC of telithromycin or ampicillin led to an almost 40% reduction in the established biofilms of E. faecalis isolates, whereas vancomycin or linezolid with 8 × MIC had minimal effects. The combination of telithromycin and ampicillin resulted in an almost 70% reduction in the established biofilms of E. faecalis. In conclusion, these results revealed that telithromycin significantly decreased the planktonic cells of both E. faecalis and E. faecium. In addition, the data further demonstrated that telithromycin has the robust ability to inhibit E. faecalis biofilms and the combination of telithromycin and ampicillin improved antibiofilm activity. These in vitro antibacterial and antibiofilm activities suggest that telithromycin could be a potential candidate for the treatment of enterococcal infections.
Keywords: telithromycin, Enterococcus faecalis, Enterococcus faecium, erythromycin-resistance, MIC, MLST, biofilm
Introduction
Enterococci are gram-positive cocci which are commonly found in the gastrointestinal tracts of nearly all land animals, including humans (Fiore et al., 2019). Although a core member of the microbiome, enterococci are capable of resulting in various infectious diseases, such as urinary tract infections, wound infections, intra-abdominal and pelvic regions infections, and bloodstream infections (Murray and Weinstock, 1999; Richards et al., 2000; Mohamed and Huang, 2007). Enterococci are now the third most common nosocomial pathogen. Statistics showed that enterococci caused almost 15% of hospital-acquired infections in the United States between 2011 and 2014 (Weiner et al., 2016), an increase of 12% from 2006 to 2007 (Hidron et al., 2008). In addition, enterococci are also responsible for 5–20% of cases of infective endocarditis (Megran, 1992). Enterococcus faecalis, the most common species of Enterococcus in the clinical setting, causes 85 to 90% of human enterococcal infections, while Enterococcus faecium is responsible for 5 to 10% of the remainder (Jett et al., 1994; Jones et al., 2004). Because E. faecalis and E. faecium usually carry a range of intrinsic and acquired resistance genes, these Enterococcus strains are frequently resistant to many commonly used antibiotics, such as glycopeptides (vancomycin and teicoplanin), beta-lactams (ampicillin, penicillin), aminoglycoside (gentamicin or streptomycin), and macrolides (van Harten et al., 2017; Ch’ng et al., 2019). Of note, the widespread emergence of vancomycin-resistant enterococci (VRE) has caused further concern due to the high mortality rate (Eliopoulos, 1997). In 2013, almost 70% of clinical E. faecium isolates in the United States displayed vancomycin resistance, while this was up to 20% in Europe (Mendes et al., 2016). In contrast, E. faecalis isolates are less frequently resistant to vancomycin (<10%) (Sievert et al., 2013).
Besides antibiotic resistance, enterococci are also known for their ability to form biofilms which is a population of cells growing on a surface and surrounded by a matrix of macromolecules like polysaccharides, proteins, lipids, and extracellular DNA (Jakubovics and Burgess, 2015). The term biofilm was introduced into medicine in 1982 by Costerton as Staphylococcus aureus biofilms were observed on a cardiac pacemaker lead (Marrie et al., 1982). Bacteria that are not innately resistant to antibiotics can also become resistant by forming persistent biofilms that lead to chronic infections (Lebeaux et al., 2014). In today’s healthcare environment, the diversity of biofilm-associated infections has risen with time, it is suggested that biofilms are present in more than 65% of all bacterial infections (Costerton et al., 1999; Lewis, 2001). Especially, due to a sharp increase in the number of patients receiving implanted medical devices in recent years, the rates of infection are 40% for ventricular assist devices, 10% for ventricular shunts, and 4% for mechanical heart valves, pacemakers, and defibrillators (Darouiche, 2004). Biofilms serve as a new nidus for bacterial dissemination and as a reservoir for antimicrobials resistant genes. Additionally, biofilms protect bacteria from detergent solutions, antimicrobial agents, environmental stress, and effectively make bacteria 10 to 1000-fold more resistant to antibiotic treatment, making their eradication extremely difficult (Lewis, 2001). Therefore, biofilm infections are becoming increasingly difficult to effectively treat due to the decreasing efficacy of antibiotics (Paganelli et al., 2012; van Harten et al., 2017). Enterococci frequently cause biofilm-associated infections such as catheter-related bloodstream infections, urinary tract infections, and infective endocarditis (Arias and Murray, 2012). Among Enterococcus species, E. faecalis isolates usually have a higher capacity of producing biofilms than E. faecium isolates and the prevalence of E. faecalis biofilms varies in different regions (Mohamed and Huang, 2007). For example, in Sardinia, biofilm production was identified among 87% of E. faecalis clinical isolates and 16% of E. faecium clinical isolates (Duprè et al., 2003). In Rome, 80% of E. faecalis and 48% of E. faecium isolates from infected patients were able to form biofilms (Baldassarri et al., 2001). Other study showed similar results and indicated that E. faecalis (95%) isolates produced biofilms more often than E. faecium (29%) (Di Rosa et al., 2006). In contrast with planktonic cells, biofilm-embedded enterococci are involved in multidrug resistance and may even be untreatable with conventional antibiotics (Lewis, 2001). Therefore, there is an urgent need to identify novel treatments for enterococcal infections.
Telithromycin (HMR 3647), a semi-synthetic derivative of erythromycin, belongs to a new chemical class of antibiotics called ketolides that have been added to the macrolide-lincosamide-streptogramin class of antibiotics (Douthwaite and Champney, 2001). Telithromycin has been approved by the U.S. Food and Drug Administration (FDA) to treat several infectious diseases, such as community-acquired pneumonia, acute exacerbations of chronic bronchitis, and acute maxillary sinusitis (Nguyen and Chung, 2005). The antimicrobial action of telithromycin, and probably that of the other 14-member-ring macrolides, is compounded by binding to 23S rRNA and blocking protein synthesis at the early stages (Menninger, 1995). Macrolides resistance is dominantly explained by the prevalence of erm genes in a wide spectrum of gram-positive bacteria (Westh et al., 1995). erm gene classes can encode a series of methyltransferase that specifically methylate the N-6 position of adenosine 2058 (A2058) or neighboring nucleotides in domain V of 23S rRNA within the large ribosomal subunit (Weisblum, 1995; Vester and Douthwaite, 2001), this prevents interaction with macrolides. In addition, erythromycin-resistant enterococci often exhibit macrolide-lincosamide-streptogramin B (MLSB) antibiotic resistance phenotypes due to the presence of erm genes. However, compared with MLSB antibiotics like erythromycin, telithromycin has an additional target site at position A752 in domain II of 23S rRNA. U747 methylation promotes G748 methylation, resulting in the increased binding of telithromycin to ribosomes and enhanced telithromycin susceptibility (Shoji et al., 2015). Telithromycin has been found to conquer erm-mediated resistance in S. pneumonia and S. aureus (Van Laethem and Sternon, 2003; Sun et al., 2018), and it also showed less efficacy against erythromycin-resistant Enterococcus isolates from different regions (TEL, MIC ≥ 4 μg/mL) (Singh et al., 2000; Min et al., 2011). However, the antimicrobial impacts of telithromycin on Enterococcus carrying erm genes remain less clear in China. Moreover, a previous study reported that telithromycin exhibited effective antibiofilm activity against S. aureus in vitro (Zheng et al., 2020). Therefore, the effect of telithromycin on Enterococcus biofilms should be further investigated.
In this study, the in vitro antibacterial activity of telithromycin was tested against enterococcal clinical isolates from inpatients in Shenzhen Nanshan People’s Hospital, and then the antibacterial activity of telithromycin was further compared with that of other antimicrobials. In addition, the multilocus sequence types (MLSTs) and erm genes expression in enterococci were detected by PCR assay. Moreover, the effect of telithromycin on the biofilms of E. faecalis isolates was further explored.
Materials and Methods
Bacterial Isolates and Growth Conditions
A total of 280 E. faecalis and 122 E. faecium strains were isolated from individual patients at Shenzhen Nanshan People’s Hospital from 2011 to 2015. Bacterial isolates were determined by standard methods using a VITEK 2 system (Biomérieux, Marcy l’Etoile, France). E. faecalis ATCC 29212 and OG1RF (ATCC 47077) were tested as quality control strains. The isolates were cultured overnight in tryptic soy broth (TSB) (Oxoid, Basingstoke, United Kingdom) at 37°C with a shaker at 220 rpm. Telithromycin (TEL), erythromycin (ERY), ampicillin (AMP), vancomycin (VAN), tetracycline (TET), doxycycline (DOX), minocycline (MIN), ciprofloxacin (CIP), nitrofurantoin (NIT), linezolid (LZD), tedizolid (TED), and tetracycline (TEC) were purchased from MCE (Princeton, NJ, United States).
Antimicrobials Susceptibility Testing
Antimicrobial susceptibilities of Enterococcus to several clinical antibiotics, including TEL, ERY, AMP, VAN, TET, DOX, MIN, CIP, NIT, LZD, TED, and TEC were tested with the VITEK 2 system. The MICs of AMP, VAN, TEL, and ERY were determined by the broth macrodilution method in cation-adjusted Mueller-Hinton broth (CAMHB) according to the 2019 Clinical and Laboratory Standards Institute (CLSI) guidelines1. The isolates were cultured overnight in TSB at 37°C with a shaker at 220 rpm, then the strains were diluted at 1:200 [2.0–3.0 × 107 colony-forming units (CFU) ml–1], and inoculated into 96 polystyrene microtiter plates with 200 μL of CAMHB containing the indicated concentrations of antibiotics. As the MIC breakpoint of telithromycin against enterococci has not been established, the MIC value of telithromycin against S. aureus (≤1, 2, ≥4 μg/mL) was based on the 2019 CLSI guidelines. Four MIC levels were thus employed for telithromycin in the antimicrobial susceptibility analysis (≤1, 2, 4, ≥8 μg/mL). The used concentrations of indicated antibiotics are given in the figure legends and Supplementary Table 4. All experiments were performed at least three times.
Detection of ERY Resistance Genes
DNA was extracted from all clinical enterococcal isolates with lysis buffer as templates for PCR according to the manufacturer’s instructions (Takara Bio Inc., Japan). As described previously (Bai et al., 2018), PCR analysis was performed to detect ermA, ermB, and ermC genes, and the primers used for PCR amplification were as follows:
ermA: sense: 5′- TCTAAAAAGCATGTAAAAGAAA-3′ and antisense: 5′- CGATACTTTTTGTAGTCCTTC-3′; ermB: sense: 5′-CCGTTTACGAAATTGGAACAGGTAAAGGGC-3′ and antisense: 5′-GAATCGAGACTTGAGTGTGC-3′; ermC: sense: 5′-GCTAATATTGTTTAAATCGTCAATTCC-3′ and antisense: 5′- GGATCAGGAAAAGGACATTTTAC -3′.
Multilocus Sequence Type
The genotypes of the enterococcal isolates were analyzed by MLST. Seven pairs of housekeeping genes: gdh, gyd, pstS, gki, aroE, xpt, and yqiL for E. faecalis, and atpA, ddl, gdh, purK, gyd, pstS, and adk for E. faecium were amplified by PCR. As previously reported (Zheng et al., 2017), the purified PCR products were sequenced, and the results were submitted to the MLST database2 for comparison, and the sequence types (STs) of enterococci were determined. The primers used for PCR amplification are listed in Supplementary Tables 5, 6.
Inhibition and Eradication of E. faecalis Biofilms
A detailed protocol has been previously reported (Zheng et al., 2020). As for the inhibition experiments, E. faecalis isolates were cultured overnight in TSB at 37°C with a shaker at 220 rpm, then the strains were diluted at 1:200 [2.0–3.0 × 107 colony-forming units (CFU) ml–1], and inoculated into 96 polystyrene microtiter plates with 200 μL of TSBG (TSB with 0.5% glucose) containing the indicated concentration of antibiotics. TSBG without antimicrobials was used as an untreated control. After incubation for 24 h, the biofilm biomasses were washed three times with ddH2O before and after crystal violet staining, the stained biofilms were detected by optical density (OD570). Solithromycin was used as a positive control (Wang et al., 2020). As for the eradication assays of the established biofilms, E. faecalis isolates were cultivated in tryptic TSB medium at 37°C for 24 h to form matured biofilms, then they were treated with antimicrobials (8 × MIC) for 48 h with the medium replaced daily. TSBG without antimicrobials was used as an untreated control. Biofilm biomasses were stained and then detected. The used concentrations of indicated antibiotics are given in the figure legends and Supplementary Table 4. All data are representative of three independent experiments.
Detecting the Adherent Cells in the Established Biofilms
The adherent cells in the established biofilms of E. faecalis were identified by the CFU numbers as described previously (Zheng et al., 2019). Briefly, the E. faecalis isolates were inoculated into 24 polystyrene microtiter plates with TSBG and formed mature biofilms after 24 h of static incubation. The supernatant was discarded and plates were washed, fresh TSBG containing antimicrobials was added, and TSBG without antimicrobials was used as an untreated control. After 48 h of static incubation with the medium being replaced daily, the supernatant was discarded and the remaining adherent cells in the established biofilms were collected by scratching the wall of the wells with a flat end toothpick. Finally, the bacteria were centrifuged and the numbers of CFU were determined. The used concentrations of indicated antibiotics were given in the figure legends and Supplementary Table 4. All data are representative of three independent experiments.
Time-Kill Curve Assay
Two E. faecalis 16C3 and 16C6 isolates were cultured in TSB at 37°C for 16 h, then diluted 200 times with TSB and antibiotics were added to make the final concentrations at 4 × MIC. A colony count was performed after 0, 1, 3, and 24 h. Data are representative of three independent experiments.
Statistical Analysis
The SPSS software (version 19.0) and GraphPad Prism software (version 5.0) were used for statistical analysis. For multiple comparisons, one-way analysis of variance (ANOVA) and a post hoc-Dunnett test were applied to analyze the data. All experiments were repeated at least three times. Sample size, n, for each experiment is given in the figure legends. Results are shown as mean ± SEM. Value differences were considered significant when ∗p < 0.05 (not significant p > 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Results
In vitro Antibacterial Activity of Telithromycin Against Enterococci Isolates
Enterococcus faecalis isolates (n = 280) and E. faecium isolates (n = 122) were retrospectively collected from different clinical specimens in China, including urine, wound secretions, blood, bile, phlegm, and other sources (Supplementary Figures 1A,B). The in vitro antibacterial activity of telithromycin against clinical isolates of E. faecalis and E. faecium are summarized in Tables 1, 2. As expected, these clinical isolates of enterococci showed a high frequency of resistance to erythromycin (ERY, MIC ≥ 8 μg/mL) and some commonly used tetracycline antibiotics, including tetracycline (TET, MIC ≥ 16 μg/mL), doxycycline (DOX, MIC ≥ 16 μg/mL), and minocycline (MIN, MIC ≥ 16 μg/mL), whereas they remained highly susceptible to vancomycin (VAN, MIC ≤ 4 μg/mL), nitrofurantoin (NIT, MIC ≤ 32 μg/mL), and linezolid (LZD, MIC ≤ 2 μg/mL).
TABLE 1.
In vitro antibacterial activity of TEL compared with that of various antibiotics against E. faecalis isolates.
| Antibiotic | No. isolates | Resistance rate (%) | MIC (μg/mL) breakpoints | NO. | TEL MIC (μg/mL) |
ERY MIC (μg/mL) |
||||||||
| ≤0.5 | 1 | 2 | 4 | ≥8 | MIC50/MIC90 | ≤0.5 | 1–4 | ≥8 | MIC50/MIC90 | |||||
| Total | 280 | – | – | – | 116 | 7 | 27 | 97 | 33 | 2/8 | 10 | 58 | 212 | >256/>256 |
| Ampicillin | 275 | 0.4 | ≤8 | 274 | 130 | 6 | 26 | 83 | 29 | 2/8 | 8 | 55 | 211 | >256/>256 |
| ≥16 | 1 | 0 | 0 | 0 | 1 | 0 | 4/4 | 0 | 0 | 1 | – | |||
| Vancomycin | 280 | 0 | ≤4 | 278 | 115 | 7 | 27 | 96 | 33 | 2/8 | 10 | 58 | 210 | >256/>256 |
| 8–16 | 2 | 1 | 0 | 0 | 1 | 0 | 0.125/4 | 0 | 0 | 2 | >256/>256 | |||
| ≥32 | 0 | 0 | 0 | 0 | 0 | 0 | – | 0 | 0 | 0 | – | |||
| Tetracycline | 276 | 83.7 | ≤4 | 39 | 33 | 3 | 1 | 2 | 0 | 0.06/1 | 7 | 17 | 15 | 4/>256 |
| 8 | 6 | 3 | 0 | 0 | 2 | 1 | 0.06/4 | 1 | 1 | 4 | 128/>256 | |||
| ≥16 | 231 | 79 | 4 | 25 | 91 | 32 | 4/8 | 2 | 39 | 190 | >256/>256 | |||
| Doxycycline | 280 | 78.9 | ≤4 | 40 | 36 | 1 | 2 | 1 | 0 | 0.06/0.125 | 7 | 19 | 14 | 2/>256 |
| 8 | 19 | 7 | 2 | 3 | 6 | 1 | 2/4 | 1 | 2 | 16 | 128/>256 | |||
| ≥16 | 221 | 73 | 4 | 22 | 90 | 32 | 4/8 | 2 | 37 | 182 | >256/>256 | |||
| Minocycline | 280 | 73.2 | ≤4 | 43 | 37 | 2 | 2 | 2 | 0 | 0.06/2 | 7 | 21 | 15 | 2/>256 |
| 8 | 32 | 11 | 0 | 9 | 9 | 3 | 2/4 | 1 | 5 | 26 | >256/>256 | |||
| ≥16 | 205 | 68 | 5 | 16 | 86 | 30 | 4/8 | 2 | 32 | 171 | >256/>256 | |||
| Ciprofloxacin | 252 | 26.6 | ≤1 | 151 | 68 | 3 | 11 | 51 | 18 | 2/8 | 5 | 25 | 121 | >256/>256 |
| 2 | 34 | 22 | 1 | 2 | 8 | 1 | 0.125/4 | 1 | 12 | 21 | 128/>256 | |||
| ≥4 | 67 | 24 | 3 | 14 | 20 | 6 | 2/8 | 2 | 12 | 53 | >256/>256 | |||
| Nitrofurantoin | 254 | 1.2 | ≤32 | 247 | 102 | 6 | 26 | 84 | 29 | 2/8 | 9 | 52 | 186 | >256/>256 |
| 64 | 4 | 2 | 0 | 0 | 2 | 0 | 0.5/4 | 0 | 0 | 4 | >256/>256 | |||
| ≥128 | 3 | 3 | 0 | 0 | 0 | 0 | 0.03/0.25 | 0 | 2 | 1 | 4/>128 | |||
| Linezolid | 280 | 5.4 | ≤2 | 214 | 87 | 6 | 21 | 75 | 25 | 2/8 | 9 | 44 | 161 | >256/>256 |
| 4 | 51 | 23 | 1 | 6 | 17 | 4 | 2/4 | 1 | 11 | 39 | >256/>256 | |||
| ≥8 | 15 | 6 | 0 | 0 | 5 | 4 | 4/8 | 0 | 3 | 12 | >256/>256 | |||
MIC, minimum inhibitory concentration; TEL, telithromycin; ERY, erythromycin; MIC50/MIC90, the MIC values for 50% or 90% of bacterial growth inhibition.
TABLE 2.
In vitro antibacterial activity of TEL compared with that of various antibiotics against E. faecium isolates.
| Antibiotic | No. isolates | Resistance rate (%) | MIC (μg/mL) breakpoints | No. | TEL MIC (μg/mL) | ERY MIC (μg/mL) | ||||||||
| ≤0.5 | 1 | 2 | 4 | ≥8 | MIC50/MIC90 | ≤0.5 | 1–4 | ≥8 | MIC50/MIC90 | |||||
| Total | 122 | – | – | – | 25 | 2 | 12 | 34 | 49 | 4/8 | 6 | 12 | 104 | >256/>256 |
| Ampicillin | 112 | 87.5 | ≤8 | 14 | 11 | 0 | 0 | 3 | 0 | 0.06/4 | 1 | 6 | 7 | 8/>128 |
| ≥16 | 98 | 13 | 2 | 12 | 29 | 42 | 4/8 | 5 | 6 | 87 | >128/>256 | |||
| Vancomycin | 120 | 0.0 | ≤4 | 116 | 23 | 2 | 12 | 31 | 48 | 4/8 | 6 | 11 | 99 | 128/>256 |
| 8–16 | 4 | 2 | 0 | 0 | 2 | 0 | 0.06/4 | 0 | 1 | 3 | 8/>256 | |||
| ≥32 | 0 | 0 | 0 | 0 | 0 | 0 | – | 0 | 0 | 0 | – | |||
| Teicoplanin | 115 | 0.9 | ≤8 | 114 | 24 | 2 | 12 | 33 | 43 | 4/8 | 6 | 12 | 96 | >128/>256 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | – | 0 | 0 | 0 | – | |||
| ≥32 | 1 | 0 | 0 | 0 | 0 | 1 | 8 | 0 | 0 | 1 | >256/>256 | |||
| Tetracycline | 119 | 46.2 | ≤4 | 45 | 11 | 1 | 1 | 16 | 16 | 4/8 | 1 | 6 | 38 | >128/>256 |
| 8 | 19 | 3 | 0 | 1 | 3 | 12 | 8/8 | 2 | 0 | 17 | >256/>256 | |||
| ≥16 | 55 | 10 | 1 | 10 | 14 | 20 | 4/8 | 3 | 5 | 47 | >256/>256 | |||
| Doxycycline | 120 | 38.3 | ≤4 | 62 | 15 | 1 | 4 | 22 | 20 | 4/8 | 4 | 8 | 50 | >128/>256 |
| 8 | 12 | 1 | 0 | 4 | 1 | 6 | 4/8 | 0 | 0 | 12 | >256/>256 | |||
| ≥16 | 46 | 9 | 1 | 4 | 8 | 24 | 8/8 | 2 | 4 | 40 | >128/>256 | |||
| Minocycline | 120 | 27.5 | ≤4 | 66 | 16 | 1 | 4 | 22 | 23 | 4/8 | 4 | 8 | 54 | >128/>256 |
| 8 | 21 | 3 | 1 | 6 | 4 | 7 | 2/8 | 1 | 1 | 19 | >256/>256 | |||
| ≥16 | 33 | 6 | 0 | 2 | 7 | 18 | 8/8 | 1 | 3 | 29 | >128/>256 | |||
| Ciprofloxacin | 113 | 9.7 | ≤1 | 29 | 10 | 1 | 7 | 8 | 3 | 2/4 | 1 | 5 | 23 | >256/>256 |
| 2 | 4 | 1 | 0 | 0 | 1 | 2 | 4/8 | 1 | 0 | 3 | >256/>256 | |||
| ≥4 | 80 | 13 | 1 | 5 | 24 | 37 | 4/8 | 3 | 7 | 70 | >128/>256 | |||
| Nitrofurantoin | 114 | 57.0 | ≤32 | 11 | 2 | 0 | 3 | 2 | 4 | 4/8 | 1 | 1 | 9 | >256/>256 |
| 64 | 38 | 6 | 1 | 5 | 12 | 14 | 4/8 | 1 | 4 | 33 | >256/>256 | |||
| ≥128 | 65 | 16 | 1 | 4 | 18 | 26 | 4/8 | 4 | 7 | 54 | >128/>256 | |||
| Linezolid | 122 | 2.4 | ≤2 | 116 | 22 | 2 | 12 | 32 | 48 | 4/8 | 6 | 10 | 100 | >128/>256 |
| 4 | 3 | 2 | 0 | 0 | 1 | 0 | 0.06/4 | 0 | 1 | 2 | 8/>256 | |||
| ≥8 | 3 | 1 | 0 | 0 | 1 | 1 | 4/8 | 0 | 1 | 2 | >256/>256 | |||
MIC, minimum inhibitory concentration; TEL, telithromycin; ERY, erythromycin; MIC50/MIC90, the MIC values for 50% or 90% of bacterial growth inhibition.
In addition, as shown in Tables 1, 2, both E. faecalis and E. faecium isolates had a high MIC50/MIC90 (the MIC values for 50% or 90% of bacterial growth inhibition) of ERY (>256/>256 μg/mL). However, the MIC50/MIC90 values of telithromycin against the E. faecalis and E. faecium strains were 2/8 μg/mL and 4/8 μg/mL, respectively. Moreover, 75.7% (212/280) of E. faecalis isolates were shown with ERY MIC ≥ 8 μg/mL, whereas only 11.8% (33/280) of E. faecalis isolates were shown with telithromycin MIC ≥ 8 μg/mL (Table 1). Similarly, among 122 E. faecium isolates, 85.2% (104/122) of strains had a high resistance to ERY (MIC ≥ 8 μg/mL), whereas 40% (49/122) of strains were shown with telithromycin MIC ≥ 8 μg/mL (Table 2), suggesting that Enterococcus isolates were more susceptible to telithromycin than ERY. Interestingly, it was found that there was a high susceptibility rate of 99.6% (274/275) for E. faecalis isolates toward ampicillin (MIC ≤ 8 μg/mL), whereas there was a high resistant rate of 87.5% (98/112) for E. faecium isolates to ampicillin (MIC ≥ 16 μg/mL). These results indicate that the antibacterial activity of telithromycin against Enterococcus was better than that of ERY.
Telithromycin Against the Enterococcus Clinical Isolates Harboring erm Genes
Next, this study further examined the effects of E. faecalis and E. faecium isolates carrying erm genes on the sensitivity of telithromycin. At first, the presence of ermA, ermB, or ermC in Enterococcus isolates was detected by PCR assays. As shown in Table 3, the rates of E. faecalis strains harboring ermA and ermB genes were 3.9 and 67.1%, respectively. However, there were no ermC-positive E. faecalis isolates. Additionally, among 122 E. faecium isolates, the rates of E. faecium isolates carrying ermA, ermB, and ermC gene were 9.8, 32.8, and 45.9%, respectively (Table 4). Furthermore, telithromycin MIC50/MIC90 values of ermA and ermB-positive E. faecalis strains or E. faecium strains both were 4/8 μg/mL, and ermC-positive E. faecium strains had a telithromycin MIC50/MIC90 of 8/8 μg/mL (Tables 3, 4). As mentioned, the MIC50/MIC90 values of telithromycin against E. faecalis and E. faecium strains were 2/8 and 4/8 μg/mL, respectively (Tables 1, 2), suggesting that the presence of erm genes slight impacted telithromycin susceptibility in the Enterococcus isolates.
TABLE 3.
In vitro activity of TEL against E. faecalis isolates with ERY-specific resistant genes.
| Erythromycin resistance genes | No. (%) | TEL MIC (μg/mL) |
ERY MIC (μg/mL) |
|||||||||
| ≤0.5 | 1 | 2 | 4 | ≥8 | MIC50/MIC90 | ≤0.5 | 1–4 | ≥8 | MIC50/MIC90 | |||
| Total | 280 | 116 | 7 | 27 | 97 | 33 | 2/8 | 10 | 58 | 212 | >256/>256 | |
| ermA | + | 11 (3.9) | 1 | 0 | 0 | 6 | 4 | 4/8 | 0 | 0 | 11 | >256/>256 |
| − | 269 (96.1) | 115 | 7 | 27 | 91 | 29 | 2/8 | 10 | 58 | 201 | >256/>256 | |
| ermB | + | 188 (67.1) | 39 | 4 | 23 | 92 | 30 | 4/8 | 1 | 8 | 179 | >256/>256 |
| − | 92 (32.9) | 77 | 3 | 4 | 5 | 3 | 0.06/2 | 9 | 50 | 33 | 2/>256 | |
| ermC | + | 0 (0) | 0 | 0 | 0 | 0 | 0 | – | 0 | 0 | 0 | – |
| − | 280 (100) | 116 | 7 | 27 | 97 | 33 | 2/8 | 10 | 58 | 212 | >256/>256 | |
MIC, minimum inhibitory concentration; TEL, telithromycin; ERY, erythromycin; +, positive; −, negative; MIC50/MIC90, the MIC values for 50% or 90% of bacterial growth inhibition.
TABLE 4.
In vitro activity of TEL against E. faecium isolates with ERY-specific resistant genes.
| No. (%) | TEL MIC (μg/mL) |
ERY MIC (μg/mL) |
|||||||||||
| Erythromycin resistance genes | ≤0.5 | 1 | 2 | 4 | ≥8 | MIC50/MIC90 | ≤0.5 | 1–4 | ≥8 | MIC50/MIC90 | |||
| Total | 122 | 25 | 2 | 12 | 34 | 49 | 4/8 | 6 | 12 | 104 | ≥0.25–256 | ||
| ermA | + | 12 (9.8) | 2 | 0 | 2 | 3 | 5 | 4/8 | 0 | 2 | 10 | 128/>256 | |
| − | 110 (90.2) | 23 | 2 | 10 | 31 | 44 | 4/8 | 6 | 10 | 94 | >128/>256 | ||
| ermB | + | 40 (32.8) | 2 | 0 | 3 | 21 | 14 | 4/8 | 0 | 2 | 38 | 128/>256 | |
| − | 82 (67.2) | 23 | 2 | 9 | 13 | 35 | 4/8 | 6 | 10 | 66 | >128/>256 | ||
| ermC | + | 56 (45.9) | 5 | 2 | 7 | 10 | 32 | 8/8 | 0 | 1 | 55 | >256/>256 | |
| − | 66 (54.1) | 20 | 0 | 5 | 24 | 17 | 4/8 | 6 | 11 | 49 | >128/>256 | ||
MIC, minimum inhibitory concentration; TEL, telithromycin; ERY, erythromycin; +, positive; −, negative; MIC50/MIC90, the MIC values for 50% or 90% of bacterial growth inhibition.
Relationship Between Telithromycin MICs Distribution and ST Clonality
Subsequently, MLST was performed to determine the ST distribution of E. faecalis and E. faecium isolates. As shown in Supplementary Tables 1, 2, ST16, ST30, and ST179 were the predominant STs in 44 STs detected from E. faecalis isolates. In total, 25 STs were identified in E. faecium isolates, the main STs were ST18, ST78, and ST80. In addition, the relationships between telithromycin and ERY MICs distributions in the predominant ST isolates are shown in Tables 5, 6. The data indicated that E. faecalis with telithromycin MIC ≤ 1, 2, 4, ≥8 μg/mL accounted for 26.6% (42/158), 9.5% (15/158), 47.5% (75/158), 16.4% (26/158) and that the rates of E. faecium isolates with telithromycin MIC ≤ 1, 2, 4, ≥8 μg/mL were 15.7% (8/51), 2% (1/51), 23.5% (12/51), 58.8% (30/51), respectively. However, the ERY-resistant rates of E. faecalis and E. faecium isolates with the top three STs reached 98.7% (156/158) and 94.1% (48/51), respectively. Moreover, among the predominant STs isolates, 87.1% (135/158) of E. faecalis and 80.4% (41/51) of E. faecium were shown alongside the erm genes carriage (Tables 5, 6), demonstrating clonal clustering toward these predominant STs.
TABLE 5.
TEL MIC values of predominant STs in E. faecalis isolates with the ermB gene.
| ST | No. (%) | TEL MIC (μg/mL) |
ERY MIC (μg/mL) |
ermB | ||||||||
| ≤0.5 | 1 | 2 | 4 | ≥8 | MIC50/MIC90 | ≤0.5 | 1–4 | ≥8 | MIC50/MIC90 | |||
| ST16 | 78 (27.9) | 18 | 3 | 9 | 36 | 12 | 4/8 | 0 | 6 | 72 | >256/>256 | 67 |
| ST30 | 8 (2.9) | 7 | 0 | 1 | 0 | 0 | 0.125/2 | 0 | 2 | 6 | 128/>256 | 6 |
| ST179 | 72 (25.7) | 14 | 0 | 5 | 39 | 14 | 4/8 | 2 | 7 | 63 | >256/>256 | 62 |
MIC, minimum inhibitory concentration; TEL, telithromycin; ERY, erythromycin; MIC50/MIC90, the MIC values for 50% or 90% of bacterial growth inhibition.
TABLE 6.
TEL MIC values of predominant STs in E. faecium isolates carrying the ermB and ermC genes.
| ST | No. (%) | TEL MIC (μg/mL) |
ERY MIC (μg/mL) |
ermB | ermC | ||||||||
| ≤0.5 | 1 | 2 | 4 | ≥8 | MIC50/MIC90 | ≤0.5 | 1–4 | ≥8 | MIC50/MIC90 | ||||
| ST18 | 18 (12.3) | 3 | 0 | 0 | 0 | 15 | 8/8 | 0 | 1 | 17 | >256/>256 | 1 | 17 |
| ST78 | 26 (21.3) | 4 | 1 | 0 | 9 | 12 | 4/8 | 3 | 1 | 22 | 128/>256 | 16 | 3 |
| ST80 | 7 (5.7) | 0 | 0 | 1 | 3 | 3 | 4/8 | 0 | 0 | 7 | >128/>128 | 4 | 0 |
MIC, minimum inhibitory concentration; TEL, telithromycin; ERY, erythromycin; MIC50/MIC90, the MIC values for 50% or 90% of bacterial growth inhibition.
The Effects of Telithromycin Against Biofilm Formation and Eradication of E. faecalis Clinical Isolates
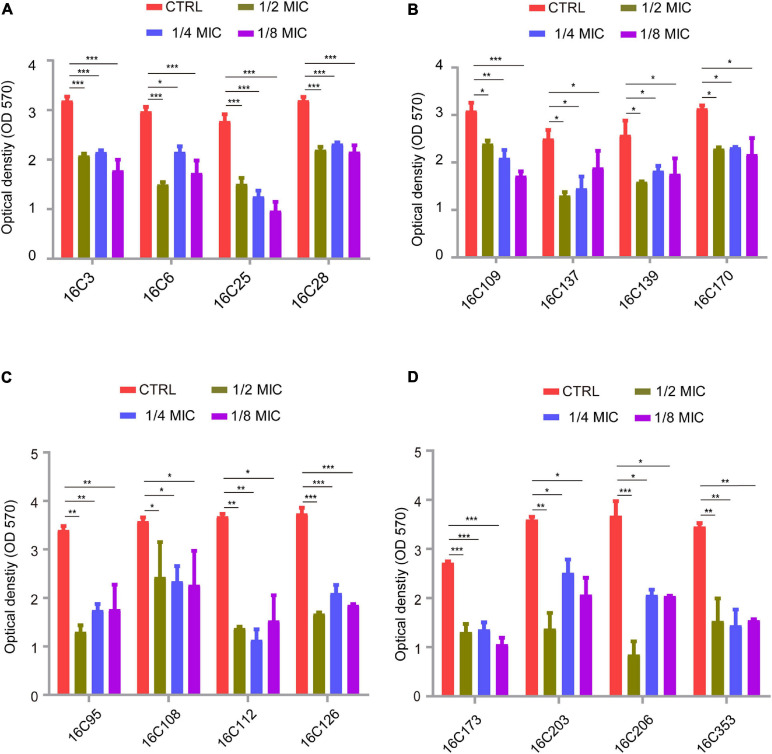
Many studies have demonstrated that E. faecalis isolates have a higher capacity of producing biofilms than E. faecium (Duprè et al., 2003; Sandoe et al., 2003; Zheng et al., 2017), thus E. faecalis isolates are usually chosen for biofilm analysis. Among our 280 E. faecalis isolates, 16 E. faecalis isolates showed a higher biofilm-forming ability, these specific 16 strains were thus tested for biofilm formation. The isolation sites of the 16 E. faecalis strains are listed in Supplementary Table 3. The inhibitory effect of telithromycin on the biofilm formation of these 16 E. faecalis isolates was determined by crystal violet staining. The MIC values of AMP, VAN, LZD, and telithromycin against these isolates are listed in Supplementary Table 4. As shown in Figure 1, 1/2 × MIC, 1/4 × MIC, or 1/8 × MIC of telithromycin could inhibit the biofilm formation of the 16 E. faecalis isolates.
FIGURE 1.
Telithromycin inhibiting biofilm formation of 16 E. faecalis isolates. (A,B) The four isolates per group were treated with TEL at 1/2 ×, 1/4 ×, and 1/8 × MICs, respectively. All of the eight isolates with the TEL MIC at 8 μg/mL. (C,D) The eight isolates of the two groups were treated with TEL at 1/2 ×, 1/4 ×, and 1/8 × MICs. The MICs were 0.25, 0.5, 0.125, 0.125, 0.5, 0.125, 0.25, and 0.25 μg/mL, respectively. TSBG without antimicrobials was used as an untreated control. The data were presented as the mean ± SEM (n = 3 experiments), one-way analysis of variance (ANOVA), and a post hoc-Dunnett test, *p < 0.05, **p < 0.01, ***p < 0.001, compared with the CTRL group.
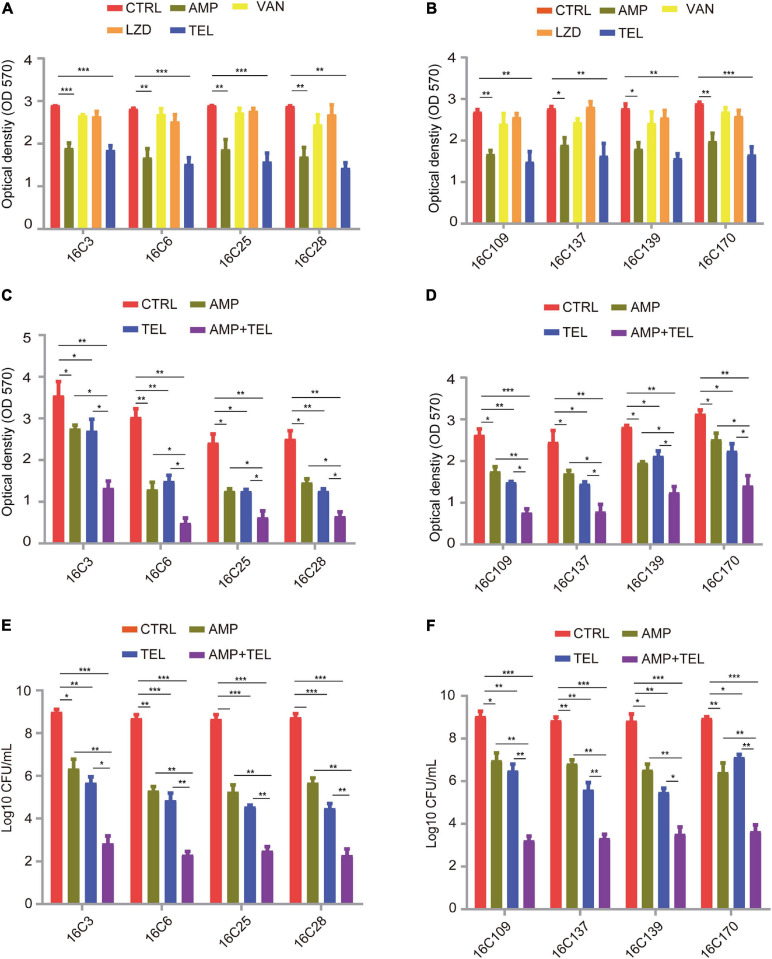
Finally, to dissect the effect of telithromycin on the established biofilms of E. faecalis isolates. Here, eight specific E. faecalis isolates were chosen for analysis by crystal violet staining. As shown in Figures 2A,B, the established biofilms of E. faecalis reduced by almost 40% after treatment with 8 × MIC of telithromycin or ampicillin, whereas 8 × MIC of vancomycin or linezolid only showed slight effects. Due to the high susceptibility rate of the E. faecalis isolates toward ampicillin (Table 1), the effects of telithromycin combined with ampicillin on the established biofilms of these eight E. faecalis isolates were further evaluated. The data showed that the combination of telithromycin and ampicillin resulted in an approximate 70% reduction in the established biofilms than with telithromycin or ampicillin alone (Figures 2C,D). Additionally, a colony-forming unit (CFU) assay was performed to quantify the viable cells of the established biofilms. Consistently, the data further confirmed that telithromycin combined with ampicillin could kill more than 70% of adherent cells in the established biofilms than telithromycin or ampicillin alone (Figures 2E,F). Therefore, these results suggest that the combination of telithromycin and ampicillin is an effective way to reduce the established biofilms in the E. faecalis isolates.
FIGURE 2.
Telithromycin alone or combined with ampicillin eradicating the established biofilms of eight E. faecalis isolates. (A,B) A total of eight isolates formed mature biofilms for 24 h, then treated with AMP, VAN, LZD, or TEL at 8 × MICs for 48 h. The MICs of AMP, VAN, and TEL for these isolates were 2, 1, and 8 μg/mL, respectively. The MICs of LZD for 16C3, 16C6, 16C25, and 16C28 isolates were 4 μg/mL, the MICs for 16C109, 16C137, 16C139, and 16C170 were 2 μg/mL. (C,D) A total of eight isolates formed mature biofilms for 24 h, then were treated with AMP and TEL alone or TEL combined with AMP at 8 × MICs for 48 h. The MICs of AMP and TEL for these isolates were 2 and 8 μg/mL, respectively. (E,F) A total of eight isolates formed mature biofilms for 24 h, then were treated with AMP and TEL alone or TEL combined with AMP at 8 × MICs for 48 h. Then the adherent cells in these established biofilms were detected by the CFU numbers. The MICs of AMP and TEL for these isolates were 2 and 8 μg/mL, respectively. TSBG without antimicrobials was used as an untreated control throughout. The data were presented as the mean ± SEM (n = 3 experiments), one-way analysis of variance (ANOVA), and post hoc-Dunnett test, *p < 0.05, **p < 0.01, ***p < 0.001, compared with the CTRL group.
Discussion
Enterococci species are commensal bacteria in the human gastrointestinal tract with the ability to cause various nosocomial infections. They not only have multiple inherent antimicrobial resistances, but are also able to acquire mutations and/or new resistance genes (Cetinkaya et al., 2000; Raza et al., 2018). Telithromycin, a novel ketolide antimicrobial agent, can be utilized for the treatment of respiratory infections. It retains its activity against most macrolide-resistant strains of Streptococcus pneumoniae and Streptococcus pyogenes (Spiers and Zervos, 2004; Wolter et al., 2008; Togami et al., 2009; Uzun et al., 2014). In addition, the efficacy of telithromycin against Enterococcus has been studied worldwide (Baltch et al., 2001; Bonnefoy et al., 2001; Mensa et al., 2003; Min et al., 2011), and it has been proven to be more effective against enterococci than some of first- and second-generation macrolides. For instance, telithromycin was found to be more potent against enterococci than erythromycin in a mouse peritonitis model (Singh et al., 2000). The MIC50/MIC90 of telithromycin were 8/32 μg/mL against vancomycin-resistance enterococci strains, whereas erythromycin and clarithromycin were completely inactive (Hamilton-Miller and Shah, 1998). In this study, the MIC50/MIC90 of ERY were >256/>256 μg/mL, and the resistance rate of ERY reached more than 75% in both E. faecalis and E. faecium isolates (Tables 1, 2). However, the MIC50/MIC90 of telithromycin in E. faecalis and E. faecium isolates were found to be as low as 2/8 and 4/8 μg/mL, respectively (Tables 1, 2). This study thus delineated additional evidence to support the fact that telithromycin could be a promising antimicrobial drug for use against enterococci. Of note, the majority of clinical E. faecalis isolates remain susceptible to β-lactams which are inhibitors of cell wall synthesis, and time-kill curve studies demonstrated that telithromycin (4 × MIC) combined with ampicillin (4 × MIC) could kill more than 100 times more planktonic cells in comparison to telithromycin or ampicillin alone in two E. faecalis isolates (Supplementary Figures 2A,B), suggesting that the combination of telithromycin and ampicillin could improve bactericidal activities against E. faecalis.
Ever since erythromycin has been clinically applied, the MLSB resistance phenotype has been widely found in erythromycin-resistant isolates of many bacteria species. For instance, Weisblum B et al. demonstrated that erythromycin-resistant S. aureus was related to the MLSB resistance phenotype after the clinical application of erythromycin (Weisblum and Demohn, 1969). This MLSB phenotype was determined by the erm genes producing erythromycin-resistant methylases, which induced methylation at specific adenosine residues on the 23S rRNA, thus leading to the resistance of the newly synthesized ribosome to MLSB antibiotics (Westh et al., 1995; Ackermann and Rodloff, 2003). Previous studies have described that the emergence of high-level erythromycin resistance narrowed the clinical application of macrolides and their application might result in a poor prognosis, more severe recurrence, and higher mortality for the treatment of bacterial infections (Min et al., 2011; Celik et al., 2014). While the chemical structure of telithromycin is derived from erythromycin, telithromycin has a low potential for drug-drug interaction, and is less likely to induce MLSB resistance than macrolides with a 14- or 15-member ring (Benes, 2004). In this study, the ermA and ermB genes were found in ERY-resistant isolates of E. faecalis, and ermA, ermB, and ermC genes were detected in E. faecium isolates (Tables 3, 4). However, telithromycin remained active against enterococci with erm genes. Recombination is involved in the genetic variation of resistance and virulence determinants, which might promote the hospital adaptation of Enterococcus bacteria such as E. faecalis and E. faecium (Homan et al., 2002; Ruiz-Garbajosa et al., 2006). In this study, E. faecalis isolates were grouped into 44 distinct STs, with the predominant STs being ST16 and ST179, which belonged to the clonal complex (CC16) (Bai et al., 2018). In addition, 25 STs were determined in E. faecium isolates, among which ST18 and ST78 were the main positive STs (Tables 5, 6). These results further confirmed that the presence of erm genes had minimal effects on the sensitivity of the predominant ST strains to telithromycin, suggesting its potential application for the treatment of some multi-resistant enterococci infections.
Evidence continues to accrue documenting the crucial role of biofilm formation in enterococcal infection (Ch’ng et al., 2019). The majority of clinical E. faecalis isolates are capable of forming biofilms on inanimate and living surfaces, which may promote antibiotic tolerance and reduce susceptibility to environmental influences or antimicrobial pressures (Paganelli et al., 2012; Holmberg and Rasmussen, 2016). Thus, targeting the biofilm formation of E. faecalis may potentially contribute to the treatment of enterococcal infections. As previously described, telithromycin could be considered as a novel inhibitor of S. aureus biofilms, which may be a result of the decreased expression of biofilm formation-related genes (Woo et al., 2017; Zheng et al., 2020), but there are no available data on the contribution of telithromycin susceptibility to biofilm phenotype in E. faecalis. The present study firstly showed that subinhibitory concentrations of telithromycin could inhibit the biofilm formation of 16 E. faecalis isolates (Figure 1). However, the data regarding biofilm formation inhibition does not show a dose-dependent effect. Therefore, further investigation is needed to characterize the inhibitory effect of telithromycin on the biofilm formation of E. faecalis.
Because biofilm prevention is not always possible, the removal of pre-existing enterococcal biofilms remains a necessity. Although the antibiofilm activity of antibiotics is likely hampered by poor penetration or slowed cell division in metabolically dormant biofilms, the use of antibiotics as first-line treatment for biofilm-associated infections is commonplace (Stewart, 2002). For example, the most recommended antibiotic treatment of endocarditis caused by E. faecalis involves ampicillin combined with gentamicin for 4–6 weeks (Habib et al., 2015). In this study, both telithromycin and ampicillin exhibited promising biofilm-inhibiting activity (40% reduction) in E. faecalis (Figures 2A,B), these results are consistent with those previously reported in the in vitro activities of telithromycin or ampicillin against S. aureus or E. faecalis biofilms (Di Domenico et al., 2019; Zheng et al., 2020). Notably, treatment with an 8 × MIC telithromycin/ampicillin combination exhibited a visible reduction (∼70%) in mature E. faecalis biofilms (Figures 2C,D) and killed 70% of adherent cells in the established biofilms (Figures 2E,F). Therefore, the combination of telithromycin with ampicillin as an anti-adherence or anti-biofilm strategy appears to be much more promising for the treatment of biofilm-associated enterococcal infections.
Accumulating evidence has revealed that there is a lower likelihood of resistance developing through the clinical use of telithromycin. For example, spontaneous resistance to telithromycin was at a low frequency in vitro. Telithromycin does not induce MLSB resistance and it shows low potential in selecting resistance or cross-resistance (Felmingham, 2001). In addition, pharmacodynamic studies suggested that telithromycin was generally well tolerated and a once-daily 800 mg oral dose of telithromycin maintains an effective concentration in plasma for the treatment of respiratory tract infections involving the key respiratory pathogens (Drusano, 2001; Namour et al., 2001). However, information regarding kill kinetics and post-antibiotic effects for telithromycin against enterococci is limited. Thereafter, there is inadequate clinical evidence to suggest an optimal dosage regimen for telithromycin against enterococci. The present study demonstrated the antimicrobial susceptibility and antibiofilm activity of telithromycin against Enterococcus isolates in vitro, suggesting that this compound might be useful, alone or in combination, in some difficult to treat enterococcal infections. The further results of in vivo studies should provide some evidence to support such a possibility.
Conclusion
In summary, this study presents the effective antimicrobial activity of telithromycin against clinical enterococci isolates from China in comparison to that of ERY. Importantly, this study further demonstrated that telithromycin could inhibit the biofilm formation of E. faecalis and that telithromycin combined with ampicillin resulted in enhanced antimicrobial and antibiofilm activity. To our knowledge, this is the first study to present new insight into the antibiofilm activity of telithromycin against enterococci and provide further evidence for the potential clinical application of telithromycin/ampicillin combination for the treatment of Enterococcus infections.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
YX conducted the PCR analyses, MLST analysis, biofilms inhibition and eradication of E. faecalis, and drafted the manuscript. JC collected the bacterial isolates, performed the antimicrobials susceptibility tests, and gene manipulation. XS participated in gene manipulation, MLST analysis, and the inhibition of E. faecalis biofilms. GX and PL participated in the collection of bacterial isolates, MLST analysis, and the inhibition and eradication of E. faecalis biofilms. ZY and QD participated in the collection of bacterial isolates, and the inhibition and eradication of E. faecalis biofilms. ZC and JZ designed the study, analyzed the experimental data, and revised the manuscript. All authors have read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Yang Wu (Key Laboratory of Medical Molecular Virology of Ministries of Education and Health, School of Basic Medical Science and Institutes of Biomedical Sciences, Shanghai Medical College, Fudan University) for his excellent technical support and suggestions. The authors would also like to thank Ms. Cynthia Brast (University of Florida, Gainesville, FL, United States) for reviewing the manuscript. This manuscript has been released as a pre-print at ResearchSquare https://www.researchsquare.com/article/rs-24213/v1 (Yanpeng et al., 2020).
Abbreviations
- AMP
ampicillin
- CIP
ciprofloxacin
- DOX
doxycycline
- ERY
erythromycin
- LZD
linezolid
- MICs
minimum inhibitory concentrations
- MIN
minocycline
- MLST
multilocus sequence type
- NIT
nitrofurantoin
- PCR
polymerase chain reaction
- STs
sequence types
- TEC
tetracycline
- TED
tedizolid
- TEL
telithromycin
- TET
tetracycline
- VAN
vancomycin.
Funding. This work was supported by the following grants: the collection of bacterial isolates, antimicrobials susceptibility tests, PCR analyses, and MLST analysis were supported by the Science, Technology and Innovation Commission of Shenzhen Municipality of Key Funds (JCYJ20180508162403996) and basic research funds (JCYJ20180302144721183, JCYJ20180302144340004, JCYJ20180302144345028, and JCYJ20180302144431923). The inhibition and eradication of E. faecalis biofilms were supported by the Sanming Project of Medicine in Shenzhen (No. SMGC201705029) and the Shenzhen Nanshan District Scientific Research Program of the People’s Republic of China (Nos. 2019042, 2019051, 2019046, 2019027, and 2019040); provincial medical fund of Guangdong (2018116164215307), and Shenzhen Key Medical Discipline Construction Fund (No. SZXK06162).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.616797/full#supplementary-material
References
- Ackermann G., Rodloff A. C. (2003). Drugs of the 21st century: telithromycin (HMR 3647)–the first ketolide. J. Antimicrob. Chemother. 51 497–511. 10.1093/jac/dkg123 [DOI] [PubMed] [Google Scholar]
- Arias C. A., Murray B. E. (2012). The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10 266–278. 10.1038/nrmicro2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B., Hu K., Li H., Yao W., Li D., Chen Z., et al. (2018). Effect of tedizolid on clinical Enterococcus isolates: in vitro activity, distribution of virulence factor, resistance genes and multilocus sequence typing. FEMS Microbiol. Lett. 3:365 10.1093/femsle/fnx284 [DOI] [PubMed] [Google Scholar]
- Baldassarri L., Cecchini R., Bertuccini L., Ammendolia M. G., Iosi F., Arciola C. R., et al. (2001). Enterococcus spp. produces slime and survives in rat peritoneal macrophages. Med. Microbiol. Immunol. 190 113–120. 10.1007/s00430-001-009698 [DOI] [PubMed] [Google Scholar]
- Baltch A. L., Smith R. P., Ritz W. J., Bopp L. H. (2001). Inhibitory and bactericidal effects of telithromycin (HMR 3647, RU 56647) and five comparative antibiotics, used singly and in combination, against vancomycin-resistant and vancomycin-susceptible enterococci. Chemotherapy 47 250–260. 10.1159/000048531 [DOI] [PubMed] [Google Scholar]
- Benes J. (2004). Telithromycin. Klin. Mikrobiol. Infekc. Lek. 10 16–21. [PubMed] [Google Scholar]
- Bonnefoy A., Guitton M., Delachaume C., Le Priol P., Girard A. M. (2001). In vivo efficacy of the new ketolide telithromycin (HMR 3647) in murine infection models. Antimicrob. Agents Chemother. 45 1688–1692. 10.1128/aac.45.6.1688-1692.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik S., Cakirlar F. K., Torun M. M. (2014). Presence of vancomycin, aminoglycosides, and erythromycin resistance genes in enterococci isolated from clinical samples in Turkey. Clin. Lab. 60 1801–1806. 10.7754/clin.lab.2014.140211 [DOI] [PubMed] [Google Scholar]
- Cetinkaya Y., Falk P., Mayhall C. G. (2000). Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13 686–707. 10.1128/cmr.13.4.686-707.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch’ng J. H., Chong K. K. L., Lam L. N., Wong J. J., Kline K. A. (2019). Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 17 82–94. 10.1038/s41579-018-0107-z [DOI] [PubMed] [Google Scholar]
- Costerton J. W., Stewart P. S., Greenberg E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284 1318–1322. 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- Darouiche R. O. (2004). Treatment of infections associated with surgical implants. N. Engl. J. Med. 350 1422–1429. 10.1056/NEJMra035415 [DOI] [PubMed] [Google Scholar]
- Di Domenico E. G., Rimoldi S. G., Cavallo I., D’Agosto G., Trento E., Cagnoni G., et al. (2019). Microbial biofilm correlates with an increased antibiotic tolerance and poor therapeutic outcome in infective endocarditis. BMC Microbiol. 19:228 10.1186/s12866-019-15961592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rosa R., Creti R., Venditti M., D’Amelio R., Arciola C. R., Montanaro L., et al. (2006). Relationship between biofilm formation, the enterococcal surface protein (Esp) and gelatinase in clinical isolates of Enterococcus faecalis and Enterococcus faecium. FEMS Microbiol. Lett. 256 145–150. 10.1111/j.1574-6968.2006.00112.x [DOI] [PubMed] [Google Scholar]
- Douthwaite S., Champney W. S. (2001). Structures of ketolides and macrolides determine their mode of interaction with the ribosomal target site. J. Antimicrob. Chemother. 48 1–8. 10.1093/jac/48.suppl_2.1 [DOI] [PubMed] [Google Scholar]
- Drusano G. (2001). Pharmacodynamic and pharmacokinetic considerations in antimicrobial selection: focus on telithromycin. Clin. Microbiol. Infect. 7 24–29. [PubMed] [Google Scholar]
- Duprè I., Zanetti S., Schito A. M., Fadda G., Sechi L. A. (2003). Incidence of virulence determinants in clinical Enterococcus faecium and Enterococcus faecalis isolates collected in Sardinia (Italy). J. Med. Microbiol. 52 491–498. 10.1099/jmm.0.050385030 [DOI] [PubMed] [Google Scholar]
- Eliopoulos G. M. (1997). Vancomycin-resistant enterococci. Mechanism and clinical relevance. Infect. Dis. Clin. North Am. 11 851–865. 10.1016/s0891-5520(05)7039370397 [DOI] [PubMed] [Google Scholar]
- Felmingham D. (2001). Microbiological profile of telithromycin, the first ketolide antimicrobial. Clin. Microbiol. Infect. 7 2–10. [PubMed] [Google Scholar]
- Fiore E., Van Tyne D., Gilmore M. S. (2019). Pathogenicity of Enterococci. Microbiol. Spectr. 4:7 10.1128/microbiolspec.GPP3-00532018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib G., Lancellotti P., Antunes M. J., Bongiorni M. G., Casalta J. P., Del Zotti F., et al. (2015). 2015 ESC Guidelines for the management of infective endocarditis: The Task Force for the Management of Infective Endocarditis of the European Society of Cardiology (ESC). Eur. Heart J. 36 3075–3128. 10.1093/eurheartj/ehv319 [DOI] [PubMed] [Google Scholar]
- Hamilton-Miller J. M., Shah S. (1998). Comparative in-vitro activity of ketolide HMR 3647 and four macrolides against gram-positive cocci of known erythromycin susceptibility status. J. Antimicrob. Chemother. 41 649–653. 10.1093/jac/41.6.649 [DOI] [PubMed] [Google Scholar]
- Hidron A. I., Edwards J. R., Patel J., Horan T. C., Sievert D. M., Pollock D. A., et al. (2008). NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect. Control. Hosp. Epidemiol. 29 996–1011. 10.1086/591861 [DOI] [PubMed] [Google Scholar]
- Holmberg A., Rasmussen M. (2016). Mature biofilms of Enterococcus faecalis and Enterococcus faecium are highly resistant to antibiotics. Diagn. Microbiol. Infect. Dis. 84 19–21. 10.1016/j.diagmicrobio.2015.09.012 [DOI] [PubMed] [Google Scholar]
- Homan W. L., Tribe D., Poznanski S., Li M., Hogg G., Spalburg E., et al. (2002). Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40 1963–1971. 10.1128/jcm.40.6.1963-1971.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics N. S., Burgess J. G. (2015). Extracellular DNA in oral microbial biofilms. Microbes Infect. 17 531–537. 10.1016/j.micinf.2015.03.015 [DOI] [PubMed] [Google Scholar]
- Jett B. D., Huycke M. M., Gilmore M. S. (1994). Virulence of enterococci. Clin. Microbiol. Rev. 7 462–478. 10.1128/cmr.7.4.462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. E., Draghi D. C., Thornsberry C., Karlowsky J. A., Sahm D. F., Wenzel R. P. (2004). Emerging resistance among bacterial pathogens in the intensive care unit–a European and North American Surveillance study (2000-2002). Ann. Clin. Microbiol. Antimicrob. 3:14 10.1186/1476-0711-314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeaux D., Ghigo J. M., Beloin C. (2014). Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 78 510–543. 10.1128/mmbr.0001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. (2001). Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45 999–1007. 10.1128/aac.45.4.999-1007.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrie T. J., Nelligan J., Costerton J. W. (1982). A scanning and transmission electron microscopic study of an infected endocardial pacemaker lead. Circulation 66 1339–1341. 10.1161/01.cir.66.6.1339 [DOI] [PubMed] [Google Scholar]
- Megran D. W. (1992). Enterococcal endocarditis. Clin. Infect. Dis. 15 63–71. 10.1093/clinids/15.1.63 [DOI] [PubMed] [Google Scholar]
- Mendes R. E., Castanheira M., Farrell D. J., Flamm R. K., Sader H. S., Jones R. N. (2016). Longitudinal (2001-14) analysis of enterococci and VRE causing invasive infections in European and US hospitals, including a contemporary (2010-13) analysis of oritavancin in vitro potency. J. Antimicrob. Chemother. 71 3453–3458. 10.1093/jac/dkw319 [DOI] [PubMed] [Google Scholar]
- Menninger J. R. (1995). Mechanism of inhibition of protein synthesis by macrolide and lincosamide antibiotics. J. Basic Clin. Physiol. Pharmacol. 6 229–250. 10.1515/jbcpp.1995.6.3-4.229 [DOI] [PubMed] [Google Scholar]
- Mensa J., García-Vázquez E., Vila J. (2003). Macrolides, ketolides and streptogramins. Enferm. Infecc. Microbiol. Clin. 21:219. [PubMed] [Google Scholar]
- Min Y. H., Yoon E. J., Kwon A. R., Shim M. J., Choi E. C. (2011). Alterations in regulatory regions of erm(B) genes from clinical isolates of enterococci resistant to telithromycin. Arch. Pharm. Res. 34 2149–2154. 10.1007/s12272-011-12191214 [DOI] [PubMed] [Google Scholar]
- Mohamed J. A., Huang D. B. (2007). Biofilm formation by enterococci. J. Med. Microbiol. 56 1581–1588. 10.1099/jmm.0.4733147330 [DOI] [PubMed] [Google Scholar]
- Murray B. E., Weinstock G. M. (1999). Enterococci: new aspects of an old organism. Proc. Assoc. Am. Physicians 111 328–334. 10.1046/j.1525-1381.1999.99241.x [DOI] [PubMed] [Google Scholar]
- Namour F., Wessels D. H., Pascual M. H., Reynolds D., Sultan E., Lenfant B. (2001). Pharmacokinetics of the new ketolide telithromycin (HMR 3647) administered in ascending single and multiple doses. Antimicrob. Agents Chemother. 45 170–175. 10.1128/aac.45.1.170-175.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M., Chung E. P. (2005). Telithromycin: the first ketolide antimicrobial. Clin. Ther. 27 1144–1163. 10.1016/j.clinthera.2005.08.009 [DOI] [PubMed] [Google Scholar]
- Paganelli F. L., Willems R. J., Leavis H. L. (2012). Optimizing future treatment of enterococcal infections: attacking the biofilm? Trends Microbiol. 20 40–49. 10.1016/j.tim.2011.11.001 [DOI] [PubMed] [Google Scholar]
- Raza T., Ullah S. R., Mehmood K., Andleeb S. (2018). Vancomycin resistant Enterococci: A brief review. J. Pak. Med. Assoc. 68 768–772. [PubMed] [Google Scholar]
- Richards M. J., Edwards J. R., Culver D. H., Gaynes R. P. (2000). Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control. Hosp. Epidemiol. 21 510–515. 10.1086/501795 [DOI] [PubMed] [Google Scholar]
- Ruiz-Garbajosa P., Bonten M. J., Robinson D. A., Top J., Nallapareddy S. R., Torres C., et al. (2006). Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44 2220–2228. 10.1128/jcm.025962595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoe J. A. T., Witherden I. R., Cove J. H., Heritage J., Wilcox M. H. (2003). Correlation between enterococcal biofilm formation in vitro and medical-device-related infection potential in vivo. J. Med. Microbiol. 52 547–550. 10.1099/jmm.0.052015200 [DOI] [PubMed] [Google Scholar]
- Shoji T., Takaya A., Sato Y., Kimura S., Suzuki T., Yamamoto T. (2015). RlmCD-mediated U747 methylation promotes efficient G748 methylation by methyltransferase RlmAII in 23S rRNA in Streptococcus pneumoniae; interplay between two rRNA methylations responsible for telithromycin susceptibility. Nucleic Acids Res. 43 8964–8972. 10.1093/nar/gkv609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert D. M., Ricks P., Edwards J. R., Schneider A., Patel J., Srinivasan A., et al. (2013). Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect. Control. Hosp. Epidemiol. 34 1–14. 10.1086/668770 [DOI] [PubMed] [Google Scholar]
- Singh K. V., Zscheck K. K., Murray B. E. (2000). Efficacy of telithromycin (HMR 3647) against enterococci in a mouse peritonitis model. Antimicrob. Agents Chemother. 44 3434–3437. 10.1128/aac.44.12.3434-3437.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers K. M., Zervos M. J. (2004). Telithromycin. Expert Rev. Anti. Infect. Ther. 2 685–693. 10.1586/14789072.2.5.685 [DOI] [PubMed] [Google Scholar]
- Stewart P. S. (2002). Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 292 107–113. 10.1078/1438-42214196 [DOI] [PubMed] [Google Scholar]
- Sun X., Lin Z. W., Hu X. X., Yao W. M., Bai B., Wang H. Y., et al. (2018). Biofilm formation in erythromycin-resistant Staphylococcus aureus and the relationship with antimicrobial susceptibility and molecular characteristics. Microb. Pathog. 124 47–53. 10.1016/j.micpath.2018.08.021 [DOI] [PubMed] [Google Scholar]
- Togami K., Chono S., Seki T., Morimoto K. (2009). Distribution characteristics of telithromycin, a novel ketolide antimicrobial agent applied for treatment of respiratory infection, in lung epithelial lining fluid and alveolar macrophages. Drug Metab. Pharmacokinet 24 411–417. 10.2133/dmpk.24.411 [DOI] [PubMed] [Google Scholar]
- Uzun B., Güngör S., Pektaş B., Aksoy Gökmen A., Yula E., Koçal F., et al. (2014). [Macrolide-lincosamide-streptogramin B (MLSB) resistance phenotypes in clinical Staphylococcus isolates and investigation of telithromycin activity]. Mikrobiyol. Bul. 48 469–476. 10.5578/mb.7748 [DOI] [PubMed] [Google Scholar]
- van Harten R. M., Willems R. J. L., Martin N. I., Hendrickx A. P. A. (2017). Multidrug-Resistant Enterococcal Infections: New Compounds, Novel Antimicrobial Therapies? Trends Microbiol. 25 467–479. 10.1016/j.tim.2017.01.004 [DOI] [PubMed] [Google Scholar]
- Van Laethem Y., Sternon J. (2003). [Telithromycin, first ketolide]. Rev. Med/. Brux 24 42–46. [PubMed] [Google Scholar]
- Vester B., Douthwaite S. (2001). Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45 1–12. 10.1128/aac.45.1.1-12.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xiong Y., Wang Z., Zheng J., Xu G., Deng Q., et al. (2020). Comparison of solithromycin with erythromycin in Enterococcus faecalis and Enterococcus faecium from China: antibacterial activity, clonality, resistance mechanism, and inhibition of biofilm formation. J. Antibiot. 10.1038/s41429-020-00374372 [Online ahead of print]. [DOI] [PubMed] [Google Scholar]
- Weiner L. M., Webb A. K., Limbago B., Dudeck M. A., Patel J., Kallen A. J., et al. (2016). Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011-2014. Infect. Control. Hosp. Epidemiol. 37 1288–1301. 10.1017/ice.2016.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B. (1995). Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39 577–585. 10.1128/aac.39.3.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B., Demohn V. (1969). Erythromycin-inducible resistance in Staphylococcus aureus: survey of antibiotic classes involved. J. Bacteriol. 98 447–452. 10.1128/jb.98.2.447-452.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westh H., Hougaard D. M., Vuust J., Rosdahl V. T. (1995). Prevalence of erm gene classes in erythromycin-resistant Staphylococcus aureus strains isolated between 1959 and 1988. Antimicrob. Agents Chemother. 39 369–373. 10.1128/aac.39.2.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter N., Smith A. M., Farrell D. J., Northwood J. B., Douthwaite S., Klugman K. P. (2008). Telithromycin resistance in Streptococcus pneumoniae is conferred by a deletion in the leader sequence of erm(B) that increases rRNA methylation. Antimicrob. Agents Chemother. 52 435–440. 10.1128/aac.010741077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S. G., Lee S. Y., Lee S. M., Lim K. H., Ha E. J., Eom Y. B. (2017). Activity of novel inhibitors of Staphylococcus aureus biofilms. Folia Microbiol. 62 157–167. 10.1007/s12223-016-0485484 [DOI] [PubMed] [Google Scholar]
- Yanpeng X., Junwen C., Xiang S., Guangjian X., Peiyu L., Qiwen D., et al. (2020). In vitro antibacterial activity of telithromycin against Enterococcus spp. isolated from China. BMC Microbiol. 2020:24213 10.21203/rs.3.rs-24213/v1 [DOI] [Google Scholar]
- Zheng J. X., Sun X., Lin Z. W., Qi G. B., Tu H. P., Wu Y., et al. (2019). In vitro activities of daptomycin combined with fosfomycin or rifampin on planktonic and adherent linezolid-resistant isolates of Enterococcus faecalis. J. Med. Microbiol. 68 493–502. 10.1099/jmm.0.000945 [DOI] [PubMed] [Google Scholar]
- Zheng J. X., Tu H. P., Sun X., Xu G. J., Chen J. W., Deng Q. W., et al. (2020). In vitro activities of telithromycin against Staphylococcus aureus biofilms compared with azithromycin, clindamycin, vancomycin and daptomycin. J. Med. Microbiol. 69 120–131. 10.1099/jmm.0.001122 [DOI] [PubMed] [Google Scholar]
- Zheng J. X., Wu Y., Lin Z. W., Pu Z. Y., Yao W. M., Chen Z., et al. (2017). Characteristics of and Virulence Factors Associated with Biofilm Formation in Clinical Enterococcus faecalis Isolates in China. Front. Microbiol. 8:2338. 10.3389/fmicb.2017.02338 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.