Abstract
Background
Brain lipid metabolism appears critical for cognitive aging, but whether alterations in the lipidome relate to cognitive decline remains unclear at the system level.
Methods
We studied participants from the Three-City study, a multicentric cohort of older persons, free of dementia at time of blood sampling, and who provided repeated measures of cognition over 12 subsequent years. We measured 189 serum lipids from 13 lipid classes using shotgun lipidomics in a case-control sample on cognitive decline (matched on age, sex and level of education) nested within the Bordeaux study center (discovery, n = 418). Associations with cognitive decline were investigated using bootstrapped penalized regression, and tested for validation in the Dijon study center (validation, n = 314).
Findings
Among 17 lipids identified in the discovery stage, lower levels of the triglyceride TAG50:5, and of four membrane lipids (sphingomyelin SM40:2,2, phosphatidylethanolamine PE38:5(18:1/20:4), ether-phosphatidylethanolamine PEO34:3(16:1/18:2), and ether-phosphatidylcholine PCO34:1(16:1/18:0)), and higher levels of PCO32:0(16:0/16:0), were associated with greater odds of cognitive decline, and replicated in our validation sample.
Interpretation
These findings indicate that in the blood lipidome of non-demented older persons, a specific profile of lipids involved in membrane fluidity, myelination, and lipid rafts, is associated with subsequent cognitive decline.
Funding
The complete list of funders is available at the end of the manuscript, in the Acknowledgement section.
Keywords: Cognitive dysfunction, Dementia, Membrane lipids, Metabolomics, Lipidomics, Serum biomarker
Research in Context.
Evidence before this study
While cognitive decline and dementia evolve slowly over years, a long-term prospective approach is critical to identify early biomarkers and etiological risk pathways. Prior lipidomics studies were often cross-sectional or with short follow-up, and were thus as likely to reveal lipid changes due to deteriorating cognitive health as to reflect causal pathways.
Added value of this study
Our prospective approach, where investigation of the blood lipidome precedes ascertainment of cognitive decline over 12 years, enhanced by a validation effort, provides a methodologically robust early signature of cognitive aging in the blood lipidome.
Implications of all the available evidence
Our findings extend preliminary evidence suggesting a dysregulation of the blood lipidome toward decreasing membrane lipids (involved in neurone membrane fluidity, myelination and lipid raft microdomains), as a very early biomarker in the course of cognitive decline.
Alt-text: Unlabelled box
1. Introduction
Lipids are major constituents of neuronal membranes and play a central role in brain functioning. Converging evidence has recently implicated lipid metabolism in brain aging, including in neurodegenerative diseases such as late-onset Alzheimer's disease (AD) [1], [2], [3]. Epidemiological studies have shown that elevated blood cholesterol and triglycerides in midlife, and lower blood levels of omega-3 long-chain polyunsaturated fatty acids (LC-PUFA) are associated with higher risks of cognitive impairment and dementia [4,5]. Neuropathological studies further reported AD brains to contain an abnormal composition of two major classes of brain phospholipids, i.e. phosphatidylcholines (PC) and phosphatidylethanolamines (PE) [6,7]. Although this suggests a role for specific lipids in brain aging and cognitive function, a more complete picture of the lipid profiles and changes in lipid metabolism that could help understand cognitive changes with aging is lacking at the systematic level.
Lipidomics is a technology that may help better understand the overall pattern of lipid (dys)regulation in relation to brain aging as it allows to investigate hundreds of lipids and their metabolites in biological fluids and tissues. For example, the blood lipidomics studies conducted thus far in AD and cognitive impairment have revealed promising associations with (i) apolar lipids, including triglycerides [8], [9], [10] and cholesterol esters (CE) [8,11] and with (ii) polar, structural lipids, which are incorporated in cell membranes (e.g., PC [8,[10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22]], sphingomyelins [SM] [9,10,[18], [19], [20], [21],23,24], and ceramides [Cer] [10,21,23,24]). However, most of these studies failed to measure PE, i.e. a main class of lipids that is strongly altered in AD brain [7,25,26]. In addition, previous studies were often cross-sectional or with short follow-up and their data have thus most likely been influenced by behavioral and metabolic changes due to the deteriorating cognitive health itself (i.e., reverse causation).
Brain aging is a heterogeneous process driven by both normal aging and pathology (i.e., dementia and AD); both are closely related (e.g., aging is the primary risk factor for dementia), share many risk factors and underlying mechanisms, and together determine an individual's course of cognitive decline. In dementia and AD science, there is therefore a strong biological rationale to investigate the continuum of brain aging globally, through cognitive decline [27]. As brain aging evolves slowly over the years, a long-term prospective approach is critical if one wants to study metabolic profiles in relation to early biomarkers and etiological risk pathways. So far, few groups have taken such a prospective approach. In addition, while reproducibility is critical in -omics studies, validation in an external sample was rarely performed, and lipidomics studies in relation to cognitive impairment often had inconsistent results [13,[17], [18], [19],23,28].
The large Three-City (3C) cohort study provides unique prospective data to decipher molecular signatures of accelerated cognitive decline. Our objective was to identify, in the 3C study, a robust and valid serum lipid signature associated with subsequent cognitive decline 12 years later.
2. Methods
2.1. Study population
The 3C study is an ongoing multicentric population-based cohort on dementia that was initiated in 1999–2000. It included 9 294 non-institutionalized community dwellers aged 65 years and older from three study centers in France: Bordeaux (South-West, n = 2 104), Dijon (Middle-East, n = 4 931), and Montpellier (South, n = 2 259) [29]. Data collected at baseline included sociodemographic and lifestyle characteristics, medical information, neuropsychological testing, blood pressure, and anthropometric measurements. Fasting serum was sampled for the formation of a biobank. Follow-up visits were scheduled every two to three years after baseline examination. At each visit, participants underwent a battery of neuropsychological tests administered by a trained psychologist during face-to-face interviews. Diagnosis of dementia was established by a committee of neurologists after review of all existing information (including MRI when available) using the Diagnostic and Statistical Manual of Mental Disorders, (4th Edition) [30,31].
2.2. Ethics
The Consultative Committee for the Protection of Persons participating in Biomedical Research at Kremlin-Bicêtre University Hospital (Paris, France) approved the 3C study protocol (CPP n°99–28, June 10, 1999) and all participants provided informed consent. Personal data were anonymized prior to analysis.
2.3. Nested case-control sampling
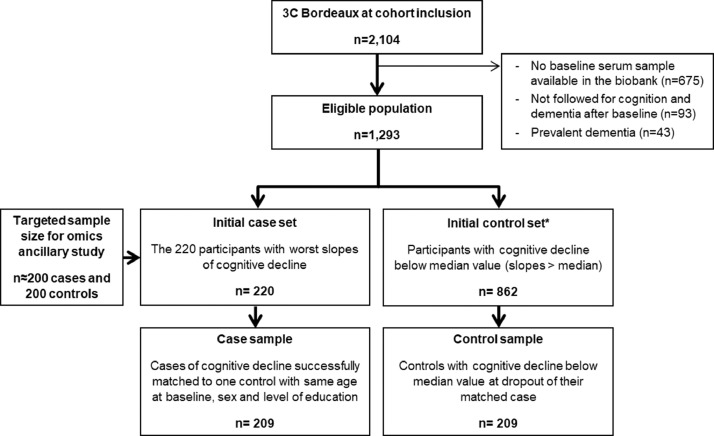
In 2016, an ancillary study was initiated in 3C to investigate metabolomics and lipidomics profiles in relation to subsequent trajectories of cognitive decline [32]. This omics sub-study was based on a case-control study on cognitive decline, nested within the Bordeaux study center. Eligible participants had serum samples available in the biobank, no dementia diagnosis at the time of blood draw at baseline, and at least one repeated cognitive examination over the subsequent 12 years (with a median of three and a maximum of five repeated examination at visit V = 2, 4, 7, 10 and 12 years) (Fig. 1).
Fig. 1.
Flowchart of case-control sampling in the discovery sample
* For each case (with last cognitive measure in visit Vmax), potential controls included participants: (i) followed at least up to Vmax, (ii) not defined as a case in Vmax, (iii) with a slower decline up to Vmax (i.e., with a slope of cognitive change better than the median when only using repeated cognitive data up to Vmax).
To build the case-control study on cognitive decline, we used as primary outcome the change in a composite score of global cognition, including the multiple cognitive domains impaired in dementia. The composite score was defined at each follow-up as the average of Z-scores of five neuropsychological tests: (i) the Mini-Mental State Examination (assessing global cognitive performance) [33], (ii) the Benton Visual Retention Test (assessing visual working memory and attention) [34], (iii) the Isaac's Set Test (assessing verbal fluency) [35], (iv) the Trail-Making Test part A (assessing processing speed) [36], and (v) the Trail-Making Test part B (assessing executive functioning) [36].
We estimated individual slopes of cognitive change using linear mixed models robust to informative dropout (i.e., a mixed model specific to each pattern of follow-up) [37]. The repeated composite cognitive scores were normalized using a latent process mixed model (to ensure Gaussian assumption) [38] before being entered as a dependent variable in the linear mixed model. The model included an intercept (that represented the level of composite cognitive score at baseline), a slope (that represented the annual change in scores over time), both a random intercept and a random slope to account for inter-individual variability (as well as a binary indicator for the first cognitive assessment). To account for informative dropout, we used pattern mixture modeling that reduces potential bias due to informative attrition in longitudinal studies [37]. Participants were combined into four groups on the basis of their last available measurement at V = 4, 7, 10 or 12 years of follow-up: Group 1 with pattern of follow-up {1,0,0,0}; Group 2 with patterns {1,1,0,0} or {0,1,0,0}; Group 3 with patterns {1,1,1,0}, {0,1,1,0}, {1,0,1,0} or {0,0,1,0}; and Group 4 with patterns {1,1,1,1}, {0,1,1,1}, {1,0,1,1}, {1,1,0,1}, {0,0,1,1}, {0,1,0,1}, {1,0,0,1} or {0,0,0,1}, where 0 indicates missing and 1 indicates observed for each of the four time points of observation. This resulted in four groups representing a monotone missing-data pattern (assuming that intermittent missing observations are missing at random). We ran a mixed model specific to each of these four pattern groups. From these models, we extracted individual slopes (as the sum of estimated fixed effect + predicted individual random effect).
Cases were identified as the 220 participants with the worst slopes of cognitive decline (a number slightly higher than the 200 cases initially planned in the ancillary study, to allow for loss of a few cases for which no matched control would be found during the subsequent matching procedure). Among the individuals still in the cohort at the last visit of the case (noted Vmax), we randomly selected a control according to the following criteria: (i) same age (±3 years), sex and educational level (<vs ≥ secondary school), and (ii) a slope of cognitive change up to Vmax better than the median. Overall, 209 cases were successfully individually matched to one control, leading to a total sample size of n = 418 subjects. Mean follow-up of cases was 8.9 years (sd=2.6 years), with 29, 72, 54 and 54 cases sampled at V = 4, 7, 10 and 12 years respectively. Average slopes in cases and controls were −0.26 (95% CI −0.36; −0.16) and −0.07 (95% CI −0.11; −0.03) standard units/years respectively (refer to Supplementary Table S1 for average neuropsychological test scores at each visit).
For validation purpose, a second case-control sample was built within the Dijon study center (with sample size limited to 157 case-control pairs due to budget restrictions). Mean follow-up of cases was 4.3 years (sd=1.6), with 131, 26 and 3 cases sampled at V = 4, 7 and 10 years, respectively. Average slopes in cases and controls were −0.38 (95%CI −0.44; −0.32) and −0.14 (−0.20; −0.08) standard units/years, respectively.
2.4. Assessment of serum lipids
Fasting serum samples collected at baseline were stored at −80 °C in a biobank until use. Lipid extraction and shotgun mass spectrometry analyses were conducted by Lipotype GmbH (Dresden, Germany) [39]. The Lipotype Shotgun Lipidomics platform consists of the automated extraction of samples, an automated direct sample infusion and high-resolution Orbitrap mass spectrometry including lipid class-specific internal standards to assure absolute quantification of lipids. An in-house developed software – LipotypeXplorer – is used for identification of lipids in the mass spectra [40,41]. Details are provided in Supplementary Method 1.
We assumed missing values to be mostly due to concentrations under the limit of quantification. Missing values affect the correlation between variables and deteriorate the performance of multivariate analyses. As previously suggested [42], to reduce the influence of missing values in the analysis, we studied only lipid species with ≥80% non-missing values in either the cases or the controls (discovery and validation samples were pooled at this stage, to ensure the selection of a common set of lipids). The remaining missing values were imputed to zero. Lipids were expressed as percentages of total lipids (to correct for variability in total lipid) and standardized. A description of lipids in the control reference samples is provided in Supplementary Table S2.
2.5. Statistics
2.5.1. Discovery analysis
In descriptive analyses, lipidomics data were investigated at the lipid class level. The correlation structure of the lipidome was described using a Spearman correlation network.
In multivariable exploratory analyses, we used Least Absolute Shrinkage and Selection Operator (LASSO)-penalized conditional logistic regression [43] – a variable selection regression method based on a penalization in the estimation of regression parameters, adapted to the analysis of matched case-control data. As LASSO may lead to unstable solutions (in part due to the influence of extreme values), we enhanced the robustness of the modeling strategy by repeating the model with a thousand bootstrapped samples (i.e., randomly resampling cases and matched controls) [44]. We thus applied a bootstrapped version of LASSO-penalized conditional logistic regression to identify lipids robustly associated with the odds of cognitive decline in the case-control study (our general methodology is detailed in Supplementary Figure 1). Models were conditioned on matching variables and were adjusted for BMI, total number of medications regularly consumed, and use of lipid-lowering treatment (statins and fibrates). For each sample, the optimal LASSO-penalty was chosen as the one providing maximum likelihood by leave-pair-out cross validation, allowing the selection of the most predictive set of lipids. We calculated the frequency of selection of each lipid across the 1000 bootstrapped samples.
The serum lipid signature associated with cognitive decline was defined from the lipids selected in at least 60% of the bootstrapped samples. There is no gold-standard for the choice of such threshold. This cut-off was chosen post hoc, as a reasonable tradeoff to optimize the likelihood of the signature to represent a true signal (i.e., including lipids selected in many bootstraps), while favoring findings obtained in the original sample (i.e., avoiding too many lipids selected in bootstrapped samples but not in the original sample).
In a subsequent step, we defined a lipid signature score based on unbiased regression coefficients by entering the lipids selected in the signature in an unpenalized conditional logistic regression (confidence intervals were not estimated at this step as they are known to be biased due to post-selection inference) [45]. To limit overfitting, the association of the lipid signature score with cognitive decline was evaluated using leave-pair-out cross-validation (i.e., computing the lipid signature score of each pair using regression coefficients estimated in the discovery case-control sample deprived of this pair) [46].
Finally, in sensitivity analyses on the discovery stage, we used lipid intensity values expressed as absolute concentrations (pmol/µL) instead of proportions of total lipid concentrations (used in the main analysis).
2.5.2. Validation analysis
We used two different approaches for validation of our signature. As the discovery was based on a multivariate approach, we first focused on the overall lipid signature and evaluated whether the signature score as defined in the discovery stage was also associated with cognitive decline in the validation sample. Second, we examined multivariate associations of each individual lipid included in the signature with cognitive decline in the validation sample. Odds ratios were estimated using a conditional logistic regression including all lipids simultaneously, conditioned on matching variables and adjusted for BMI, the number of drugs consumed and the use of lipid-lowering treatment. Lipids were considered replicated if they were associated with cognitive decline in the same direction as in the discovery sample and with a statistical threshold at p<0.10.
2.5.3. Supplementary analyses
We evaluated the impact of APOE-ɛ4 or diabetes in our findings in several ways. First, we evaluated possible confounding by APOE-ɛ4 or diabetes in the relation of each selected lipid and of the signature score with cognitive decline. Second, we investigated effect modification by testing for an interaction between the lipid signature score and both APOE-ɛ4 and diabetes on cognitive decline.
Statistical analyses were performed using R software version 3.6.1 (igraph 1.2.4.1, penalized 0.9–51, and survival 2.44–1.1 packages).
2.6. Role of funders
Funders had no role in study design, data collection, data analyses, interpretation or writing of report.
3. Results
3.1. Discovery stage
Participants were 76 years-old on average at blood draw, 66% were female and approximately 29% reached secondary school (Table 1; left column). Compared with controls, individuals with cognitive decline were twice as often carriers of the APOE-ε4 allele, were almost three-times more often diabetics, consumed more medications, and had slightly lower global cognitive performances (as indicated by the Mini Mental State Examination) at baseline.
Table 1.
Baseline characteristics of cases of cognitive decline and matched controls in the discovery (n = 209 case-control pairs) and validation (n = 157 case-control pairs) samples.
| Discovery sample |
Validation sample |
|||
|---|---|---|---|---|
| Baseline characteristics | Cases | Controls | Cases | Controls |
| Age (years) | 75.9 (4.5) | 75.7 (4.2) | 76.5 (5.2) | 76.1 (4.8) |
| Sex, female | 66.0 | 66.0 | 62.4 | 62.4 |
| Education, ≥ secondary school | 28.7 | 28.7 | 31.8 | 31.8 |
| APOE-ε4 carrier | 26.2 | 12.0 | 26.8 | 19.1 |
| BMI (kg/m²) | 26.8 (4.4) | 26.1 (3.6) | 25.5 (4.5) | 25.0 (3.7) |
| Number of drugs consumed | 5 [3–7] | 4 [3–6] | 5 [3–7] | 4 [3–5] |
| Lipid-lowering treatment | 33.5 | 36.4 | 31.2 | 29.3 |
| Diabetes1 | 13.2 | 5.7 | 12.1 | 6.4 |
| Hypertension2 | 78.5 | 76.1 | 80.9 | 83.4 |
| Current smoker | 4.8 | 4.3 | 7.0 | 2.6 |
| History of cardiovascular diseases | 33.5 | 27.8 | 42.0 | 29.9 |
| Plasma triglycerides (mmol/L) | 1.4 (0.8) | 1.3 (0.6) | 1.3 (0.6) | 1.2 (0.5) |
| Plasma total cholesterol (mmol/L) | 5.8 (1.0) | 5.8 (0.9) | 5.8 (1.0) | 5.8 (0.9) |
| MMSE score (range 0–30) | 26.9 (2.2) | 28.0 (1.6) | 25.7 (2.4) | 28.7 (1.0) |
Values are mean (SD), median [IQR] or percentages.
Fasting glucose ≥7.2 mmol/L or specific medication.
Blood pressure ≥140/90 mmHg or specific medication.
Abbreviations: APOE-ε4, allele ε4 for the apolipoprotein E gene; BMI, Body Mass Index; MMSE, Mini-Mental State Examination.
3.1.1. Description of the lipidome
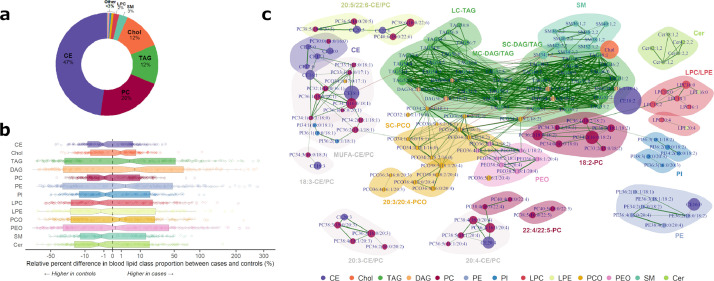
The analytical pool comprised 189 lipids belonging to 13 classes: (i) apolar species, including diacylglycerols (DAG, n = 6), triacylglycerols (TAG, n = 37), free cholesterol (Chol), and cholesterol esters (CE, n = 16), and (ii) polar (structural) species, including phosphatidylcholines (PC, n = 48), phosphatidylethanolamines (PE, n = 7), phosphatidylinositols (PI, n = 13), lysophosphatidylcholines (LPC, n = 6), lysophosphatidyl-ethanolamines (LPE, n = 2), ether-phospholipids (possibly plasmalogens) ether-PC (PCO, n = 29) and ether-PE (PEO, n = 8), sphingomyelins (SM, n = 11), and ceramides (Cer, n = 5). The apolar lipids TAG, Chol and CE accounted for >70% of total lipid concentration. Among polar lipids, PC was the most frequent class, accounting for 21% of total lipids, while the other polar species were less concentrated (<5%) in the lipidome (Fig. 2A). Relative percentage differences between cases and controls were generally small (around 1%; Fig. 2B), and not statistically significant at the lipid class level (all p>0.05 for univariate conditional logistic regressions). The correlation network of the lipidome revealed strong Spearman correlations between: (i) lipids belonging to the same class (e.g., SM class, at the top-right of Fig. 2C) and (ii) lipids with a similar fatty acid composition (e.g., long chain CE/PC with polyunsaturated fatty-acids 20:5/22:6, at the top-left of Fig. 2C).
Fig. 2.
Description of the blood lipidome (class contributions [panel a], boxplots of case-control differences in class proportions [panel b] and Spearman correlation network [panel c]) in the discovery sample (n = 418).
Only Spearman correlations with absolute value >0.6 are drawn. Green links indicates positive correlations, and red links indicate negative correlations. Link width is proportional to the strength of correlation. Node size is proportional to average lipid proportion to total lipid concentration. Groups of lipids were automatically detected based on nodes betweenness and labels were attributed post-hoc.
Abbreviations: CE, cholesterol esters; Cer, ceramides; Chol, cholesterol; DAG, diacylglycerols; LC, long-chain; LPC, lyso-phosphatidylcholines; LPE, lyso-phosphatidylethanolamines; MC, medium-chain, MUFA, monounsaturated fatty acid; PC, phosphatidylcholines; PCO, ether-phosphatidylcholines; PE, phosphatidylethanolamines; PEO, ether-phosphatidylethanolamines; PI, phosphatidylinositol; SC, short-chain; SM, sphingomyelins; TAG, triglycerides.
3.1.2. Identification of a serum lipid signature of subsequent cognitive decline
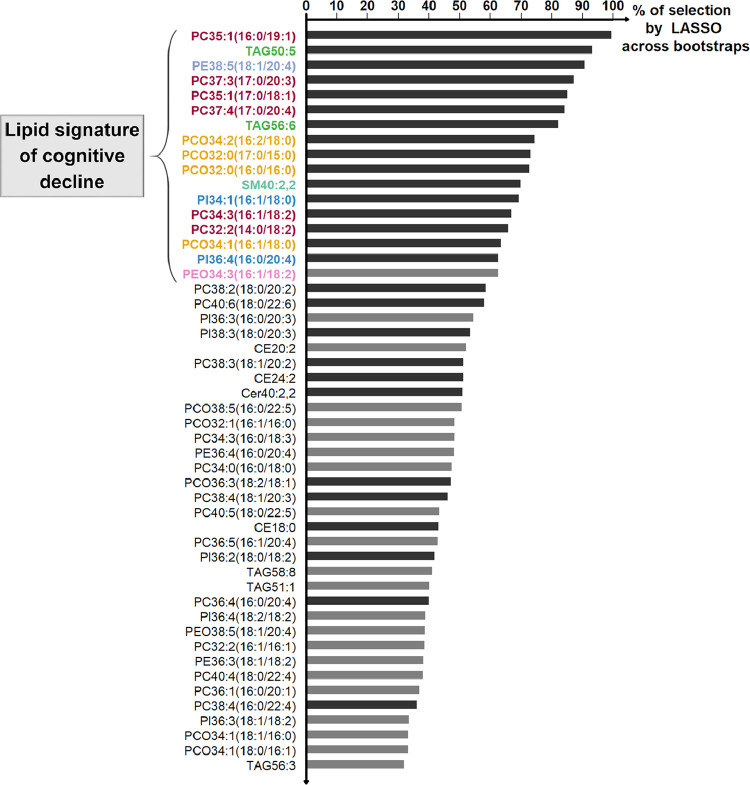
The bootstrap-LASSO discovery analysis identified a signature of 17 lipids associated with cognitive decline (Fig. 3). This signature was largely dominated by polar lipids, including: PC and short-chain PCO (representing half of the 17 lipids); a few PI and PE/PEO species; and the sphingomyelin SM40:2,2. Apolar lipids were less dominant in the signature and were represented by a few unsaturated TAG species (see Fig. 3 for specific names of each of the 17 lipids).
Fig. 3.
The serum lipid signature selected in the discovery stage
Lipids were ranked by decreasing frequency of selection across bootstraps. Only top 50 lipids are displayed. Dark gray bars indicate lipids selected on the initial LASSO regression model, and light gray bars indicate those selected across bootstrapped samples only. We retained the 17 lipids selected in >60% of bootstraps (with names highlighted in color).
Sensitivity analyses using lipids expressed in pmol/µL yielded comparable results.
3.2. Validation stage
Characteristics of the validation sample were generally similar to those of the discovery sample, except that cases of the validation sample were slightly older in age (76.3 versus 75.8 years-old) and had more often history of vascular conditions (42.0 vs 33.5%); moreover, compared to the discovery sample, cases of the validation sample had slightly lower global cognitive performances at baseline (MMSE=25.7 vs 26.9) (Table 1; right column).
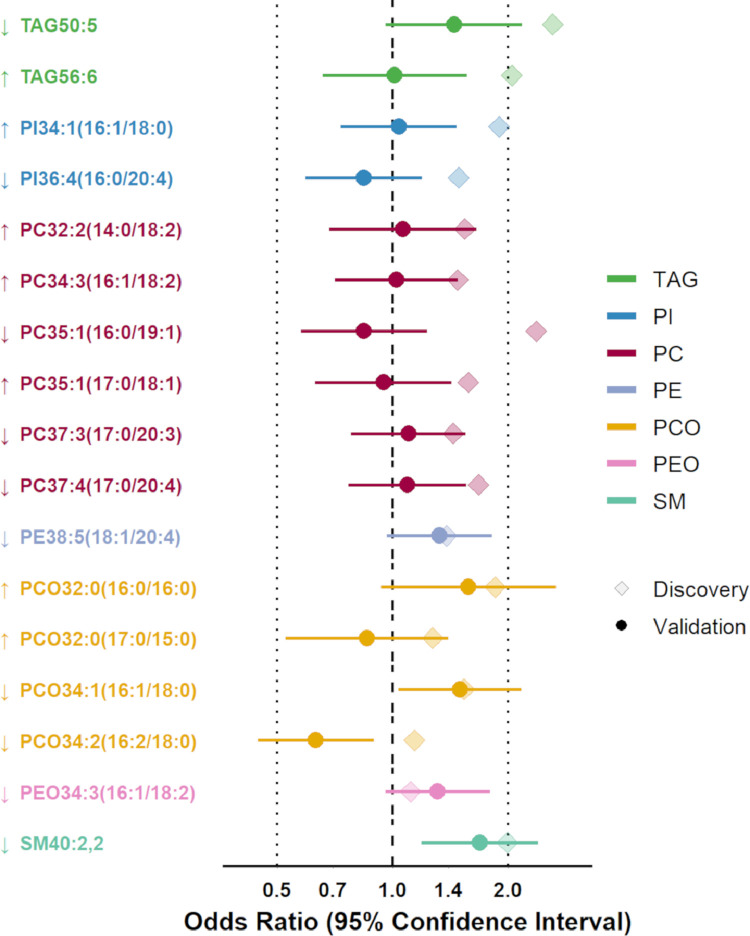
The lipid signature score was associated with greater cognitive decline in both discovery and validation stages (OR for 1SD-increase in score=3.39 and 1.39 [95% CI 1.08–1.78], respectively). In secondary analyses on individual lipids, we replicated 6 out of the 17 lipids (Fig. 4). Lower levels of five lipids, including TAG50:5 (ORs for 1SD-decrease=2.61 and 1.44 [0.96–2.17] in discovery and validation, respectively), PE38:5(18:1/20:4) (1.38 and 1.32 [0.97–1.81]), PCO34:1(16:1/18:0) (1.53 and 1.50 [1.04–2.16]), PEO34:3(16:1/18:2) (1.11 and 1.31 [0.96–1.79]), and SM40:2,2 (1.99 and 1.69 [1.19–2.40]), and higher level of PCO32:0(16:0/16:0) (OR for 1SD-increase=1.85 and 1.58 [0.93–2.66] in discovery and validation) were associated with greater cognitive decline.
Fig. 4.
Multivariate associations of the 17 lipids from the serum lipid signature with the odds of subsequent cognitive decline in the discovery (n = 418) and validation (n = 314) case-control samples.
Odds Ratios were estimated using a conditional logistic regression including all lipids simultaneously, conditioned on matching variables (age at blood draw, sex and level of education) and adjusted for Body Mass Index, the number of drugs consumed and the use of lipid-lowering treatment. ORs are for a 1 SD-increase (↑) or 1 SD decrease (↓) of proportion to total lipids in serum (note that we imposed all discovery ORs to be greater than 1, to reflect the magnitude of the beta coefficients included in the prediction risk score; the sign of regression coefficients is reflected by the direction of arrows [increasing arrow = positive coefficient, i.e., increasing lipid level increases risk of cognitive decline; decreasing arrow = negative coefficient, i.e., decreasing lipid level increases risk). Confidence intervals are not valid in post-selection inference and were thus not estimated in the discovery sample.
In supplementary analyses further controlling for APOE-ε4 or diabetes, associations of either individual lipids or the signature score with cognitive decline were virtually unchanged (results not shown). Furthermore, we found evidence of effect modification by APOE-ε4, yet limited to the validation sample, where the lipid signature score was more strongly associated with cognitive decline in APOE-ε4 carriers (OR for 1SD-increase in score=2.51 [95% CI 1.32–4.78] and 1.26 [0.95–1.67] in APOE-ε4 carriers vs non-carriers respectively; P for interaction=0.04). However, when examining individual lipids, we found no evidence of effect modification by APOE-ε4 for any of the lipid identified in the signature (P for interaction >0.10 in validation sample). No evidence of interaction with APOE-ε4 was found in the discovery sample, and there was no significant interaction with diabetes in any sample.
4. Discussion
In this prospective population-based study, we discovered and validated a robust lipid signature within the serum lipidome of non-demented participants that was strongly associated with their subsequent cognitive decline over 12 years. Among the six lipids successfully validated, lower levels of triglyceride TAG50:5 and four membrane lipids (a phosphatidylethanolamine [PE38:5(18:1/20:4)], two ether-phospholipids [PCO34:1(16:1/18:0), PEO34:3(16:1/18:2)], and a sphingomyelin [SM40:2,2]), and higher levels of the ether-phospholipid PCO32:0(16:0/16:0) were associated with greater odds of cognitive decline. Moreover, association of the overall lipid signature with cognitive decline seemed stronger among APOE-ɛ4 carriers, although evidence was limited to the validation sample.
So far, very few prospective studies have examined associations between the blood lipidome and subsequent cognitive impairment or incident dementia or AD, and most of them had a small to moderate follow-up [13,[15], [16], [17], [18], [19],28,47,48,22]. Moreover, lipidomics is a rapidly evolving field, and our shotgun approach enabled the inclusion of rare species such as PE/PEO, that have seldom been investigated in relation to cognitive outcomes in earlier studies (e.g., many studies used the Biocrates platform [Absolute IDQ p180 kit] that does not include PE/PEO) [13,15,[17], [18], [19],22]. In addition, not only does the general structure (e.g., head group [e.g., phosphocholine], total length and saturation) matters, but also the fatty acid composition of lipids influences their metabolism and biological effects, and the level of detail for these molecular characterizations differs greatly between analytical platforms [19,28,47]. As a result, the heterogeneity between lipidomics studies in terms of epidemiological design and analytical platform limits their comparability [49].
Overall, previous lipidomics studies have suggested a potential dysregulation of PCO species early in the development of cognitive impairment/AD [13,[15], [16], [17], [18], [19]]. In an untargeted metabolomics study, the only statistically significant difference in the entire metabolome of AD participants compared to controls was lower blood levels of one cluster of PCO/plasmalogens [16]. These findings are generally in line with our results, where PCO emerged as a key class of lipids in our signature. Interestingly, one PCO from our signature, i.e. PCO34:2, was associated with a greater risk of AD in the Baltimore Longitudinal Study of Aging [18], and consistently, with a greater cognitive decline in our validation stage. Surprisingly, however, the association of this lipid with cognitive decline was in the opposite direction in the discovery stage. Overall, when looking into the details of the species, there has been very few overlap across studies in the literature, and conflicting directions of association were reported in relation to cognitive impairment/AD (see reviews) [50,51].
Furthermore, while the total pool of blood triglycerides has been associated with higher cognitive decline [4], some lipidomics studies, that allow molecular characterization of specific species, have yielded opposite results. Such studies for instance reported reduced plasma levels of specific triglycerides in AD patients [8,9], and an inverse association between the proportion of triglycerides in Very Low Density Lipoprotein and the risk of dementia [28]. The biological reason for such inverse associations remains unclear and deserves further research.
As with the global lipidome, previous studies focusing on specific lipid classes have also been inconsistent. Our findings of a higher cognitive decline with lower blood SM40:2,2 levels are consistent with two cross-sectional studies that also linked lower blood SM levels to AD status and to lower total brain volume [21,52]. However, prospective studies have yielded heterogeneous results [18,19,23,24]. For example, in one of the largest studies conducted to date on the blood sphingo-lipidome and dementia risk, higher blood SM levels were associated with an increased risk of AD in men, but a reduced risk in women (i.e., an interaction with sex, that we did not found in our study) [24]. Finally, our prospective study supports and extends previous results on reduced levels of PEO plasmalogens in both the brain and blood of AD patients, even at the earliest stages of the disease [26,53].
Validation has been rarely performed in previous lipidomics studies, which certainly contributes to the divergence of results in the literature. Yet, validation is critical in research on blood biomarkers of cognitive aging and dementia, as there is an inherent and large heterogeneity across studies due to the complexity of labile exposures, such as the lipidome, and of the outcomes with long-term evolutions, like cognitive aging and dementia. For instance, although derived from the same cohort study, the characteristics of our validation sample were quite different from the discovery sample (e.g., participants were older, had slightly lower cognitive performances and more vascular risk factors at baseline). Despite these differences, we could still replicate both our global signature and some specific individual lipids as well, which suggests our findings captured a biologically relevant signature.
The lipid signature we identified may indeed play an important and biologically relevant role in brain aging. For example, SM and ether phospholipids like PCO/PEO, with their most abundant species being plasmalogens, are enriched in the myelin sheath and are key components of “lipid rafts” [1,54]. They are relevant for these specific membrane microdomains, which act as dynamic platforms for several cellular processes, including signaling pathways, molecular trafficking and protein interactions [1]. Lipid rafts further play a critical role in neurotransmission and synaptic plasticity [55], all essential for proper cognitive functioning and in addition, their structure and composition have recently been shown to influence amyloid beta production in AD [56]. Furthermore, phospholipids with unsaturated acyl chains are enriched in neuronal membranes and contribute to their fluidity [57]. It has been hypothesized that AD could be closely related to imbalances in the proportion of unsaturated fatty acids (decreasing) versus saturated fatty acids (increasing) in membrane phospholipids [58]. Accordingly, we found lower levels of one PE with two unsaturated fatty acids (PE38:5(18:1/20:4)) and higher levels of one saturated PCO (PCO32:0(16:0/16:0)) associated with greater cognitive decline.
The main strengths of our study are: (i) a prospective population-based design with a long term follow-up for cognition that allowed us to capture lipid alterations at the very beginning of development of cognitive decline, (ii) the inclusion of a validation sample with a harmonized design and similar analytical platform, and (iii) a robust and novel analytical strategy for both biochemistry (shotgun lipidomics) and statistical analyses (bootstrap LASSO, controlling for a number of risk factors). Such a robust methodology has been seldom used in previous studies and clearly represents an added value of our study.
However, our study has also some limitations. First, lipid measurements were only available at a single time-point and we were thus unable to examine the stability of our results over time. It is likely that signatures within the lipidome vary according to dementia stages, and future studies should examine the dynamics of lipid changes as cognitive aging develops. Second, we did not have information on the fatty acid composition/position/family of all lipids; for example, the fatty acids composition of TAG species was unknown and we were not able to differentiate arachidonic acid (an omega-6 LC-PUFA) from its omega-3 isomer in our phospholipid analysis. Third, although we have addressed many methodological issues, methodological limitations still persist. For example, moderate sample size may have yielded false negative results, and a similar study should therefore be conducted in a larger independent cohort. Moreover, we used an extreme-phenotyping strategy for achieving good statistical power under sample size limitations; despite being limited by use of a matched case-control design, measurement error in estimation of associations induced by such strategy is still possible. In addition, we have attempted to minimize confounding by multiple covariate adjustments, but residual confounding is always an issue in observational studies. Fourth, although dementia and AD are primary causes of accelerated cognitive decline (accordingly, 51% and 52% of cases developed incident dementia during follow-up in the discovery and validation sample respectively, versus <3% of controls), the present case-control study was designed to examine cognitive decline and did not allow a methodologically accurate investigation of incident dementia. Last, results from this study depend on a specific case-control design and may not be generalizable to other populations.
In conclusion, in a longitudinal cohort of older persons, we have identified and replicated a signature within the serum lipidome that was associated with subsequent cognitive decline. Our findings mainly point to an early dysregulation of specific lipids involved in membrane fluidity (PE), myelination and lipid rafts (SM, PCO/PEO) and suggest that a decrease in their unsaturation level is present early in the cognitive aging process. Whether such alterations contribute to the pathophysiology of cognitive aging, or whether they represent only very early biomarkers of neuronal dysfunction and death remains to be elucidated. Anyway, these findings may further help reveal neuroplasticity-related pathways underlying cognitive aging and dementia, and/or allow to study the influence of specific exposures (e.g., nutrition) and the role of ApoE genotype, which deserve further research.
Contributors
CH, CPL and CS designed and conceptualized the study. CK, AK and CS had a major role in the acquisition of data. SLA conducted statistical analysis. BPH and CPL provided significant statistical advice. SLA and CS drafted the manuscript, verified the underlying data, and take full responsibility for data integrity. CM, DYL, MUS, CAL, RGD, LA, BA, PJL, SRR, CDL, ADP, ST and AK provided significant advice. All authors interpreted the data, reviewed and approved the manuscript for intellectual content.
Declaration of Competing Interests
SLA, BPH, CH, CM, DYL, MUS, CAL, RGD, LA, BA, PJL, SRR, CPL, AK and CS report no disclosures. CK is shareholder and employee of Lipotype GmbH. CDL, ADP, and ST report grants from Medical Research Council UK during the conduct of the study.
Acknowledgments
Acknowledgements
The Three-City Study is conducted under a partnership agreement between the Institut National de la Santé et de la Recherche Médicale (INSERM), the Institut de Santé Publique et Développement of the Victor Segalen Bordeaux 2 University and Sanofi-Aventis. The Fondation pour la Recherche Médicale funded the preparation and initiation of the study. The 3C Study is also supported by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, Mutuelle Générale de l'Education Nationale, Institut de la Longévité, Regional Governments of Aquitaine and Bourgogne, Fondation de France, Ministry of Research-INSERM Programme “Cohortes et collections de données biologiques”, French National Research Agency COGINUT ANR-06-PNRA-005, the Fondation Plan Alzheimer (FCS 2009–2012), and the Caisse Nationale pour la Solidarité et l'Autonomie (CNSA). This specific project on metabolomics and cognitive decline is funded by JPI-HDHL (ANR-15-HDHL-0002–05; Medical Research Council UK: MR/N030087/1; PCIN-2015–229-MINECO; CiberFES- FEDER, 2017SGR1546 & ICREA Academia Award from the Generalitat de Catalunya). Raúl González-Domínguez thanks the “Juan de la Cierva” program from MINECO (FJCI-2015–26590). Aniko Korosi is supported by NWO and Alzheimer Nederland, Paul Lucassen by the UvA Urban Mental Health program and Alzheimer Nederland. Sophie Lefèvre-Arbogast was part of the University Research school (Ecole Universitaire de Recherche, EUR) Digital Public Health PhD program, supported within the framework of the French National Research Agency (ANR) “Programme d'Investissement d'Avenir” (Investment for the Future) PIA3 (17-EURE-0019).
Data sharing statement
Anonymized data described in the manuscript will be shared by the 3C scientific committee upon request from any qualified investigator.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103216.
Appendix. Supplementary materials
References
- 1.Di Paolo G., Kim T.-.W. Linking lipids to Alzheimer's disease: cholesterol and beyond. Nat Rev Neurosci. 2011 May;12(5):284–296. doi: 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunkle B.W., Grenier-Boley B., Sims R., Bis J.C., Damotte V., Naj A.C. Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019 Mar;51(3):414. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panza F., D'Introno A., Colacicco A.M., Capurso C., Pichichero G., Capurso S.A. Lipid metabolism in cognitive decline and dementia. Brain Res Rev. 2006 Aug 1;51(2):275–292. doi: 10.1016/j.brainresrev.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Power M.C., Rawlings A., Sharrett A.R., Bandeen-Roche K., Coresh J., Ballantyne C.M. Association of midlife lipids with 20-year cognitive change: a cohort study. Alzheimer’s Dement. 2018 Feb;14(2):167–177. doi: 10.1016/j.jalz.2017.07.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samieri C., Féart C., Letenneur L., Dartigues J.-.F., Pérès K., Auriacombe S. Low plasma eicosapentaenoic acid and depressive symptomatology are independent predictors of dementia risk. Am J Clin Nutr. 2008 Jan 9;88(3):714–721. doi: 10.1093/ajcn/88.3.714. [DOI] [PubMed] [Google Scholar]
- 6.Cunnane S.C., Plourde M., Pifferi F., Bégin M., Féart C., Barberger-Gateau P. Fish, docosahexaenoic acid and Alzheimer's disease. Prog Lipid Res. 2009 Sep 1;48(5):239–256. doi: 10.1016/j.plipres.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Naudí A., Cabré R., Jové M., Ayala V., Gonzalo H., Portero-Otín M. Chapter five - lipidomics of human brain aging and Alzheimer's disease pathology. In: Hurley MJ, editor. International review of neurobiology. 2015. pp. 133–189. editor. Omic Studies of Neurodegenerative Disease: Part B; vol. 122. [DOI] [PubMed] [Google Scholar]
- 8.Proitsi P., Kim M., Whiley L., Simmons A., Sattlecker M., Velayudhan L. Association of blood lipids with Alzheimer’s disease: a comprehensive lipidomics analysis. Alzheimer’s Dement. 2017 Feb;13(2):140–151. doi: 10.1016/j.jalz.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 9.de Leeuw F.A., Peeters C.F.W., Kester M.I., Harms A.C., Struys E.A., Hankemeier T. Blood-based metabolic signatures in Alzheimer’s disease. Alzheimer’s Dement Diagn Assess Dis Monit. 2017;8:196. doi: 10.1016/j.dadm.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olazarán J., Gil-de-Gómez L., Rodríguez-Martín A., Valentí-Soler M., Frades-Payo B., Marín-Muñoz J. A blood-based, 7-metabolite signature for the early diagnosis of Alzheimer's disease. J Alzheimers Dis. 2015 Apr 13;45(4):1157–1173. doi: 10.3233/JAD-142925. [DOI] [PubMed] [Google Scholar]
- 11.Proitsi P., Kim M., Whiley L., Pritchard M., Leung R., Soininen H. Plasma lipidomics analysis finds long chain cholesteryl esters to be associated with Alzheimer's disease. Transl Psychiatry. 2015 Jan 13;5(1):e494. doi: 10.1038/tp.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whiley L., Sen A., Heaton J., Proitsi P., García-Gómez D., Leung R. Evidence of altered phosphatidylcholine metabolism in Alzheimer's disease. Neurobiol Aging. 2014 Feb 1;35(2):271–278. doi: 10.1016/j.neurobiolaging.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mapstone M., Cheema A.K., Fiandaca M.S., Zhong X., Mhyre T.R., MacArthur L.H. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med. 2014 Apr;20(4):415–418. doi: 10.1038/nm.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiandaca M.S., Zhong X., Cheema A.K., Orquiza M.H., Chidambaram S., Tan M.T. Plasma 24-metabolite Panel Predicts Preclinical Transition to Clinical Stages of Alzheimer's Disease. Front Neurol. 2015;6:237. doi: 10.3389/fneur.2015.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casanova R., Varma S., Simpson B., Min K., An Y., Saldana S. Blood metabolite markers of preclinical Alzheimer’s disease in two longitudinally followed cohorts of older individuals. Alzheimer’s Dement. 2016 Jul;12(7):815–822. doi: 10.1016/j.jalz.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orešič M., Hyötyläinen T., Herukka S.-.K., Sysi-Aho M., Mattila I., Seppänan-Laakso T. Metabolome in progression to Alzheimer's disease. Transl Psychiatry. 2011 Dec 13;1(12) doi: 10.1038/tp.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D., Misialek J.R., Boerwinkle E., Gottesman R.F., Sharrett A.R., Mosley T.H. Prospective associations of plasma phospholipids and mild cognitive impairment/dementia among African Americans in the ARIC Neurocognitive Study. Alzheimer’s Dement Diagn Assess Dis Monit. 2017;6:1. doi: 10.1016/j.dadm.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varma V.R., Oommen A.M., Varma S., Casanova R., An Y., Andrews R.M. Brain and blood metabolite signatures of pathology and progression in Alzheimer disease: a targeted metabolomics study. PLoS Med. 2018;15(1) doi: 10.1371/journal.pmed.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toledo J.B., Arnold M., Kastenmüller G., Chang R., Baillie R.A., Han X. Metabolic network failures in Alzheimer’s disease: a biochemical road map. Alzheimer’s Dement. 2017 Sep;13(9):965–984. doi: 10.1016/j.jalz.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li D., Misialek J.R., Boerwinkle E., Gottesman R.F., Sharrett A.R., Mosley T.H. Plasma phospholipids and prevalence of mild cognitive impairment and/or dementia in the ARIC Neurocognitive Study (ARIC-NCS) Alzheimer’s Dement Diagn Assess Dis Monit. 2016;3:73. doi: 10.1016/j.dadm.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han X., Rozen S., Boyle S.H., Hellegers C., Cheng H., Burke J.R. Metabolomics in early alzheimer's disease: identification of altered plasma sphingolipidome using shotgun lipidomics. PLoS ONE. 2011 Jul 11;6(7):e21643. doi: 10.1371/journal.pone.0021643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huo Z., Yu L., Yang J., Zhu Y., Bennett D.A., Zhao J. Brain and blood metabolome for Alzheimer's dementia: findings from a targeted metabolomics analysis. Neurobiol Aging. 2020 Feb 1;86:123–133. doi: 10.1016/j.neurobiolaging.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mielke M.M., Bandaru V.V.R., Haughey N.J., Rabins P.V., Lyketsos C.G., Carlson M.C. Serum sphingomyelins and ceramides are early predictors of memory impairment. Neurobiol Aging. 2010 Jan;31(1):17–24. doi: 10.1016/j.neurobiolaging.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mielke M.M., Haughey N.J., Han D., An Y., Bandaru V.V.R., Lyketsos C.G. The association between plasma ceramides and sphingomyelins and risk of Alzheimer's disease differs by sex and APOE in the Baltimore longitudinal study of aging. J Alzheimers Dis JAD. 2017;60(3):819–828. doi: 10.3233/JAD-160925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farooqui A.A., Rapoport S.I., Horrocks L.A. Membrane phospholipid alterations in Alzheimer's disease: deficiency of ethanolamine plasmalogens. Neurochem Res. 1997 Apr 1;22(4):523–527. doi: 10.1023/a:1027380331807. [DOI] [PubMed] [Google Scholar]
- 26.Han X., Holtzman D.M., McKeel D.W. Plasmalogen deficiency in early Alzheimer's disease subjects and in animal models: molecular characterization using electrospray ionization mass spectrometry. J Neurochem. 2001 May 15;77(4):1168–1180. doi: 10.1046/j.1471-4159.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 27.Morris M.C., Evans D.A., Hebert L.E., Bienias J.L. Methodological issues in the study of cognitive decline. Am J Epidemiol. 1999 May 1;149(9):789–793. doi: 10.1093/oxfordjournals.aje.a009893. [DOI] [PubMed] [Google Scholar]
- 28.Tynkkynen J., Chouraki V., van der Lee S.J., Hernesniemi J., Yang Q., Li S. Association of branched-chain amino acids and other circulating metabolites with risk of incident dementia and Alzheimer’s disease: a prospective study in eight cohorts. Alzheimer’s Dement. 2018;14(6):723–733. doi: 10.1016/j.jalz.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.3C Study Group Vascular factors and risk of dementia: design of the Three-City Study and baseline characteristics of the study population. Neuroepidemiology. 2003 Dec;22(6):316–325. doi: 10.1159/000072920. [DOI] [PubMed] [Google Scholar]
- 30.American Psychiatric Association . 4th edition. American Psychiatric Association; Washington, DC: 1994. DSM-IV: diagnostic and statistical manual of mental disorders; p. 886. [Google Scholar]
- 31.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA work group under the auspices of department of health and human services task force on Alzheimer's disease. Neurology. 1984 Jul;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 32.Low D.Y., Lefèvre-Arbogast S., González-Domínguez R., Urpi-Sarda M., Micheau P., Petera M. Diet-related metabolites associated with cognitive decline revealed by untargeted metabolomics in a prospective cohort. Mol Nutr Food Res. 2019 Jun 20;0(0) doi: 10.1002/mnfr.201900177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Folstein M.F., Folstein S.E., McHugh P.R. Mini-mental state”: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975 Nov;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 34.Benton A.L. Editions du Centre de psychologie appliquée; Paris: 1953. Manuel du test de rétention visuelle: applications cliniques et expérimentales. [Google Scholar]
- 35.Isaacs B., Kennie A.T. The set test as an aid to the detection of dementia in old people. Br J Psychiatry. 1973 Oct;123(575):467–470. doi: 10.1192/bjp.123.4.467. [DOI] [PubMed] [Google Scholar]
- 36.Reitan R.M. Validity of the trail making test as an indicator of organic brain damage. Percept Mot Skills. 1958 Dec 1;8(3):271–276. [Google Scholar]
- 37.Little R.J.A. Modeling the drop-out mechanism in repeated-measures studies. J Am Stat Assoc. 1995;90(431):1112–1121. [Google Scholar]
- 38.Proust C., Jacqmin-Gadda H., Taylor J.M.G., Ganiayre J., Commenges D. A nonlinear model with latent process for cognitive evolution using multivariate longitudinal data. Biometrics. 2006;62(4):1014–1024. doi: 10.1111/j.1541-0420.2006.00573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Surma M.A., Herzog R., Vasilj A., Klose C., Christinat N., Morin-Rivron D. An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur J Lipid Sci Technol. 2015 Oct 1;117(10):1540–1549. doi: 10.1002/ejlt.201500145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herzog R., Schwudke D., Schuhmann K., Sampaio J.L., Bornstein S.R., Schroeder M. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011;12(1):R8. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herzog R., Schuhmann K., Schwudke D., Sampaio J.L., Bornstein S.R., Schroeder M. LipidXplorer: a software for consensual cross-platform lipidomics. PLoS ONE. 2012 Jan 17;7(1):e29851. doi: 10.1371/journal.pone.0029851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J., Zhao X., Lu X., Lin X., Xu G. A data preprocessing strategy for metabolomics to reduce the mask effect in data analysis. Front Mol Biosci. 2015;2:4. doi: 10.3389/fmolb.2015.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Ser B. 1994;58:267–288. [Google Scholar]
- 44.Bach F.R. Proceedings of the 25th international conference on machine learning. 2008. Bolasso: model consistent Lasso estimation through the bootstrap; pp. 33–40. [Google Scholar]
- 45.Belloni A., Chernozhukov V. Least squares after model selection in high-dimensional sparse models. Bernoulli. 2013 May;19(2):521–547. [Google Scholar]
- 46.Airola A., Pahikkala T., Waegeman W., De Baets B., Salakoski T. An experimental comparison of cross-validation techniques for estimating the area under the ROC curve. Comput Stat Data Anal. 2011 Apr 1;55(4):1828–1844. [Google Scholar]
- 47.Bressler J., Yu B., Mosley T.H., Knopman D.S., Gottesman R.F., Alonso A. Metabolomics and cognition in African American adults in midlife: the atherosclerosis risk in communities study. Transl Psychiatry. 2017 Jul;7(7):e1173. doi: 10.1038/tp.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Proitsi P., Kuh D., Wong A., Maddock J., Bendayan R., Wulaningsih W. Lifetime cognition and late midlife blood metabolites: findings from a British birth cohort. Transl Psychiatry. 2018;8(1):203. doi: 10.1038/s41398-018-0253-0. 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lipidomics Standards Initiative Consortium Lipidomics needs more standardization. Nat Metab. 2019 Aug;1(8):745–747. doi: 10.1038/s42255-019-0094-z. [DOI] [PubMed] [Google Scholar]
- 50.Lefèvre-Arbogast S., Wagner M., Proust-Lima C., Samieri C. Nutrition and metabolic profiles in the natural history of dementia: recent insights from systems biology and life course epidemiology. Curr Nutr Rep. 2019 Sep;8(3):256–269. doi: 10.1007/s13668-019-00285-1. [DOI] [PubMed] [Google Scholar]
- 51.Jiang Y., Zhu Z., Shi J., An Y., Zhang K., Wang Y. Metabolomics in the development and progression of dementia: a systematic review. Front Neurosci. 2019 Apr 12;13:343. doi: 10.3389/fnins.2019.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li D., Misialek J.R., Jack C.R., Mielke M.M., Knopman D., Gottesman R. Plasma metabolites associated with brain MRI measures of neurodegeneration in older adults in the atherosclerosis risk in communities–neurocognitive study (ARIC-NCS) Int J Mol Sci. 2019 Apr 9;20(7) doi: 10.3390/ijms20071744. pii: E1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goodenowe D.B., Cook L.L., Liu J., Lu Y., Jayasinghe D.A., Ahiahonu P.W.K. Peripheral ethanolamine plasmalogen deficiency: a logical causative factor in Alzheimer's disease and dementia. J Lipid Res. 2007 Nov;48(11):2485–2498. doi: 10.1194/jlr.P700023-JLR200. [DOI] [PubMed] [Google Scholar]
- 54.Dean J.M., Lodhi I.J. Structural and functional roles of ether lipids. Protein Cell. 2018 Feb;9(2):196–206. doi: 10.1007/s13238-017-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sebastião A.M., Colino-Oliveira M., Assaife-Lopes N., Dias R.B., Ribeiro J.A. Lipid rafts, synaptic transmission and plasticity: impact in age-related neurodegenerative diseases. Neuropharmacology. 2013 Jan 1;64:97–107. doi: 10.1016/j.neuropharm.2012.06.053. [DOI] [PubMed] [Google Scholar]
- 56.Grimm M.O.W., Mett J., Grimm H.S., Hartmann T. APP function and lipids: a bidirectional link. Front Mol Neurosci. 2017;10:63. doi: 10.3389/fnmol.2017.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Farooqui A.A., Horrocks L.A., Farooqui T. Glycerophospholipids in brain: their metabolism, incorporation into membranes, functions, and involvement in neurological disorders. Chem Phys Lipids. 2000 Jun;106(1):1–29. doi: 10.1016/s0009-3084(00)00128-6. [DOI] [PubMed] [Google Scholar]
- 58.González-Domínguez R., García-Barrera T., Gómez-Ariza J.L. Using direct infusion mass spectrometry for serum metabolomics in Alzheimer's disease. Anal Bioanal Chem. 2014 Nov 1;406(28):7137–7148. doi: 10.1007/s00216-014-8102-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.