Abstract
Background
Liquid biopsy based on 5-hydroxymethylcytosine (5hmC) signatures of plasma cell-free DNA (cfDNA) originating from tumor cells provides a novel approach for early diagnosis in hepatocellular carcinoma (HCC). Here, we sought to develop a reliable model using cfDNA 5hmC signatures and protein biomarkers for diagnosis and prognosis of HCC.
Patients and methods
We carried out genome-wide 5hmC sequencing of cfDNA samples collected from 165 healthy volunteers, 62 liver cirrhosis (LC) patients and 135 HCC patients. A sensitive 5hmC diagnostic model was developed based on 5hmC signatures selected by sparse Partial Least Squares Discriminant Analysis and cross-validation to define the weighted diagnostic score (wd-score). Then, we combined protein biomarkers with the wd-score to build a more robust score (HCC score) by logistic regression.
Results
The distribution pattern of differential 5hmC regions could clearly distinguish HCC patients, LC patients and healthy volunteers. The wd-score based on 64 5hmC signatures in cfDNA achieves 93.24% of area under the curve (AUC) to distinguish HCC patients from non-HCC patients, and the HCC score by combing protein biomarkers achieves 92.75% of AUC to distinguish HCC patients from LC patients. Meanwhile, the HCC score showed high capacity for screening high recurrence risk patients after receiving surgical resection, and appeared to be an independent indicator for both relapse-free survival (P = 0.00865) and overall survival (P = 0.000739). Furthermore, the values of the HCC score in patients' longitudinal plasma samples were positively associated with tumor burden dynamics during follow-up.
Conclusion
We have developed and validated a novel non-invasive liquid biopsy strategy for HCC diagnosis, prognosis and surveillance during HCC progression.
Key words: liquid biopsy, 5-hydroxymethylcytosine signatures, plasma cell-free DNA, protein biomarkers, hepatocellular carcinoma
Highlights
-
•
A novel liquid biopsy approach was developed for HCC diagnosis, prognosis, and surveillance.
-
•
Novel HCC score defined here showed excellent capacity in distinguishing HCC patients from liver cirrhosis patients.
-
•
HCC score could predict the early recurrence risk for HCC patients after receiving surgical operation.
-
•
HCC score could sensitively monitor tumor burden and progression of HCC in longitudinal.
Introduction
Hepatocellular carcinoma (HCC) is the third most common cause of cancer-related death in China.1 Hepatitis B virus (HBV) infection and liver cirrhosis (LC) account for the most relevant HCC risk factors.2 Patients with HCC diagnosed at early stage could receive curative therapeutic treatments, such as hepatectomy and liver transplantation, and thus have a better clinical outcome than those diagnosed at advanced stages.3 However, even so, the prognosis of HCC patients is still poor, with 5-year overall survival (OS) rates less than 50%. Thus, early diagnosis and accurate prognosis prediction are essential to guide HCC patients' personalized management, which could significantly improve the long-term survival. Serum protein biomarkers [alpha-fetoprotein (AFP) and des-gamma-carboxy prothrombin (DCP)] and imaging methods [ultrasonography/computed tomography (CT)/magnetic resonance imaging (MRI)] are the main clinical examinations for HCC diagnosis and surveillance after treatment,4,5 but their usage has been severely limited in the early detection of HCC due to the relatively low sensitivity and specificity.
Circulating cell-free DNA (cfDNA) is short-fragmented DNA (∼160 bp) in the blood, which is mainly derived from necrotic/apoptotic normal cells and tumor cells.6 The usefulness of cfDNA as a non-invasive diagnostic and prognostic biomarker, which could significantly overcome tumor heterogeneity, highlighted in HCC and other cancers by quantitating tumor-specific single-nucleotide variants, copy number variants and epigenetic aberrations.7, 8, 9 5-Hydroxymethylcytosine (5hmC) signature, the oxidation product of 5-methylcytosines (5mC) by the ten-eleven translocation enzymes, is widely reflected in the activation state of gene expression involved in early tumorigenesis.10,11 Recent studies have shown that cfDNA 5hmC signatures of HCC patients could provide higher clinical sensitivity for early HCC diagnosis than the protein biomarker AFP.11, 12, 13 However, considering the heterogeneity among HCC patients and the inherent disadvantages in conventional liquid biopsy, the highly sensitive cfDNA 5hmC detection still could not fully meet the clinical diagnostic and prognostic needs of HCC patients. Therefore, it is a feasible approach to integrate protein biomarkers with cfDNA detection to further improve the sensitivity, specificity and accuracy of existing clinical diagnostic methods. Previously, our and other studies have proved that the combination of mutation-based circulating tumor DNA (ctDNA, a small part of cfDNA coming from tumor cells) detection and protein biomarkers could further improve their sensitivity to early diagnosis and prognosis of HCC.8,14 However, due to the mutation heterogeneity among HCC patients, it is very difficult to combine mutation-based ctDNA detection in clinical use before knowing the pre-existing mutation sites in tumor tissue. Based on the cfDNA 5hmC signatures, we could integrate cfDNA detection with protein biomarkers without knowing the pre-existing tumor information, which might be a feasible strategy for improving the diagnosis and prognosis efficiency of HCC patients in clinic.
In this study, by utilizing the sensitive and reliable 5hmC-Seal method and machine learning modeling, we obtained the genome-wide distribution of cfDNA 5hmC profiles in healthy volunteers, LC and HCC patients to build a diagnostic model for distinguishing HCC patients from LC patients and healthy volunteers. Then, we further combined the weighted diagnostic score (wd-score), which was calculated based on the cfDNA 5hmC diagnostic model, with traditional protein biomarkers (AFP and DCP) to construct a novel HCC diagnostic score (HCC score). Finally, we verified its sensitivity and specificity in HCC diagnosis, recurrence risk assessment after hepatectomy and tracking the dynamics of tumor burden in long-term follow-up patients during HCC progression.
Methods
Patients and sample collection
In total, 362 adult participants were retrospectively enrolled in this study, including 165 healthy volunteers, 62 LC patients and 135 HCC patients from Mengchao Hepatobiliary Hospital of Fujian Medical University (Fujian, China). The corresponding inclusion and exclusion criteria are provided in Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2020.100021. Informed consent was obtained from all participants before the study. All HCC patients firstly underwent hepatectomy when diagnosed with HCC and received follow-up according to the standard HCC management. Peripheral blood samples were collected from 135 patients at preoperative and post-operative (1-3 months) time points. In addition, 127 peripheral blood samples were collected from 10 HCC patients with long-term follow-up at preoperative, post-operative and multiple follow-up time points. Available CT/MRI scans were acquired and interpreted by two certified radiologists, and corresponding protein biomarkers (AFP and DCP) were also detected (the cut-off levels of AFP and DCP were 20 ng/ml and 40 mAU/ml, respectively).
All collection and usage of human samples was in accordance with the principles of the Declaration of Helsinki and approved by the Institution Review Board of Mengchao Hepatobiliary Hospital of Fujian Medical University (Fujian, China).
5hmC sequencing profiling
5hmC Library construction was carried out as previously described11 and detailed in Supplementary Methods, available at https://doi.org/10.1016/j.esmoop.2020.100021. Trimmomatic (version 0.38) was used to trim low-quality bases and adaptor sequences of reads.15 Three types of spike-in DNA sequences (5C, 5mC, 5hmC) were used to evaluate the capture efficiency of the final sequencing libraries (Supplementary Figure S1A, available at https://doi.org/10.1016/j.esmoop.2020.100021). The percentage that reads aligned to 5hmC spike-in DNA divided by reads aligned to three spike-in DNA sequences was calculated as the capture efficiency. For each sample, we firstly used MACS2 (version 2.1.2) callpeak command with parameters ‘-f BED -t {pebed} --nolambda --nomodel --extsize 170 -g hs’ to identify candidate 5hmC modified regions.16 All 5hmC modified regions were filtered by the ENCODE hg19 blacklist, and filtered to remove peaks that extend beyond the ends of chromosomes or on chromosomes X, Y, or on the mitochondrial genome. We obtained a non-redundant set of fixed-width peaks for each sample in the iterative removal procedure as previously described.17 Bedtools (version 2.25.0) was used to obtain the fragment count of each peak. Fragments per kilobase per million was calculated for each peak as its 5hmC modification level.18
Comparison between liver disease patients and healthy volunteers
Samples from HCC patients and LC patients were grouped as the liver disease group. We used the Wilcoxon–Mann–Whitney test to identify differential 5hmC modified regions between the liver disease group and healthy controls. The differentially 5hmC modified regions (DhMPs) were defined as regions with a P value <0.01 and |log2FC| > 0.7.
Development of the diagnostic model and wd-scores
We applied sparse Partial Least Squares Discriminant Analysis (sPLS-DA) using the mixOmics Package in R to develop the diagnostic model.19 Our samples were randomly split into a training set and a validation set; 75% of samples were used for the original training set [liver disease patients (n = 147) and healthy volunteers (n = 123)] and 25% of samples were used as the original validation set [liver disease patients (n = 50) and healthy volunteers (n = 42)]. To avoid model overfitting, fivefold cross-validation was carried out to select the best candidate 5hmC marker regions, in which the original training set was randomly divided into five parts. In each iteration, we randomly chose four parts of the samples for the training set and one part for the validation set. Then, we selected the DhMPs (Wilcoxon–Mann–Whitney test, P value < 0.01 and |log2FC| > 0.7) between liver disease patients and healthy volunteers in the training set. We then used these DhMPs to train a classifier by sPLS-DA function, and evaluated the classifier with the validation set. A panel of 5hmC marker regions cross-validated in all iterations was retained for the final diagnostic model. Finally, the original validation set was used to evaluate the final diagnostic model.
Wd-score and HCC score
We based our wd-score calculation on the coefficients of 5hmC marker regions and 5hmC level for each individual.
The coefficient of 5hmC marker regions was calculated as:
(n represents the number of principal components that the diagnostic model remained).
The wd-score for each sample was calculated as:
(k represents the markers).
The HCC score, a combined score of AFP, DCP and wd-score, was evaluated to distinguish HCC patients and LC patients by the logistic regression as:
Based on having the highest value of the Youden index (sensitivity + specificity − 1), we determined the optimal thresholds of wd-score and HCC score for prediction in the training set.
Statistical analysis
Visualization of the statistical results was carried out using R (version 3.6.1). The Wilcoxon–Mann–Whitney test was applied to do the comparison between the two groups, and Benjamini and Hochberg correction was used to correct the raw P value. The R packages Rtsne and pheatmap were used for dimension reduction and clustering analysis. AUROC function of R packages mixOmics was used to generate ROC curves to evaluate the performance of prediction models. Survival analysis and visualization were carried out with the R package ‘survminer’.
Results
Study design and patient characteristics
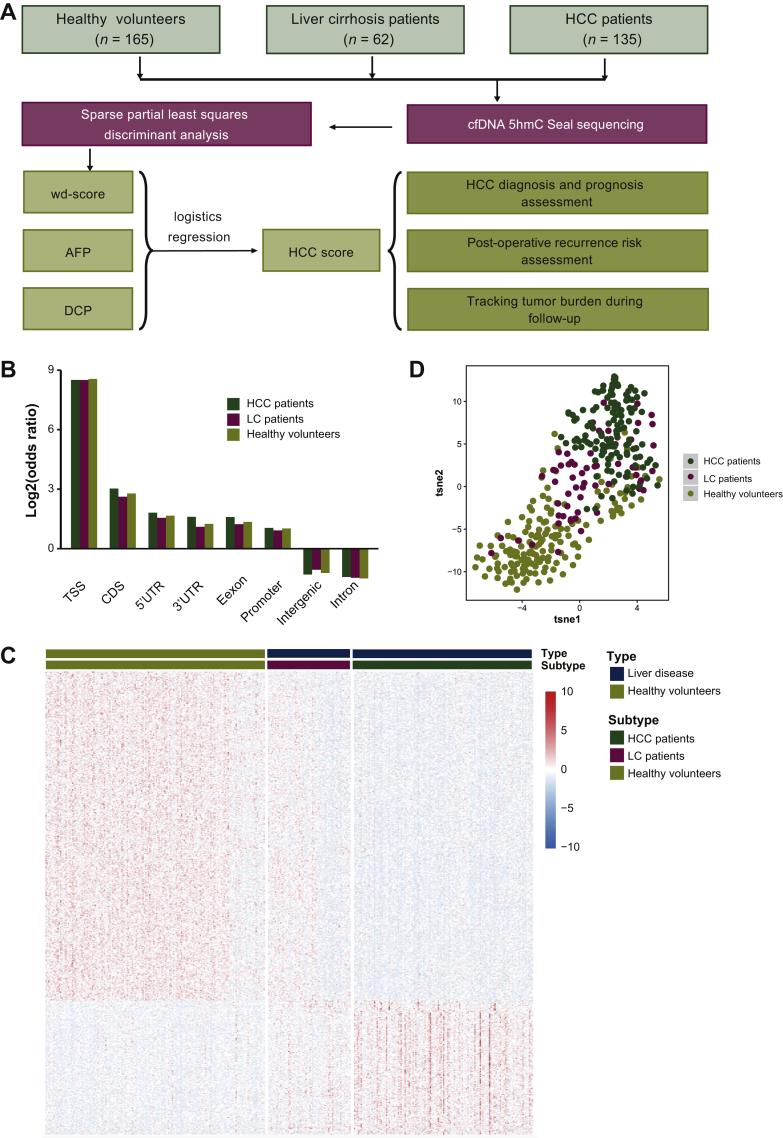
To study the potential value of cfDNA 5hmC profiling as a diagnostic and prognostic biomarker for HCC, a total of 362 participants, including 165 healthy volunteers, 62 LC patients and 135 HCC patients were enrolled. The study designs are summarized in Figure 1A. In total, 165 plasma samples from healthy volunteers, 62 from LC patients and 347 from HCC patients were collected. Among the 347 plasma samples collected from HCC patients, 135 were preoperative samples from 135 HCC patients, 105 of them were post-operative samples from 105 HCC patients and the remaining 107 were multiple follow-up samples collected from 10 HCC patients with long-term follow-up. Detailed clinical information for 135 HCC enrolled patients before surgical operation is presented in Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2020.100021. Some 83.7% (113/135) of the enrolled HCC patients had concomitant LC. All patients were firstly diagnosed as HCC and did not receive any treatment before surgery. In these enrolled patients, 71.11% showed positive AFP (>20 ng/ml) in preoperative plasma and only 38.52% of them had AFP level exceeding 400 ng/ml, which is the confident detection threshold for HCC.
Figure 1.
The overview of study design and 5hmC modified regions distribution in enrolled participants.
(A) The workflow chart of the study design, a total of 362 subjects, including 165 healthy volunteers, 62 LC patients and 135 HCC patients were enrolled in analysis, and a novel HCC diagnostic score (HCC score) was finally construct by combining a cfDNA 5hmC diagnostic model with traditional protein biomarkers (AFP and DCP) for HCC diagnosis, recurrence risk assessment after hepatectomy and tracking the dynamics of tumor burden in long-term follow-up patients during HCC progression. (B) The genomic element distribution of 5hmC modified regions. (C) The unsupervised hierarchical clustering of 620 fixed-length differentially 5hmC modified regions between liver disease patients and healthy volunteers. (D) The distribution among enrolled participants based on the differential 5hmC profile.
5hmC, 5-hydroxymethylcytosine; AFP, alpha-fetoprotein; CDS, coding DNA sequences; cfDNA, cell-free DNA; DCP, des-gamma-carboxy prothrombin; HCC, hepatocellular carcinoma; LC, liver cirrhosis; Tsne, t-distributed stochastic neighbor embedding; TSS, transcription start site; UTR, untranslated region; wd-score, weighted diagnostic score.
HCC diagnostic model based on cfDNA 5hmC profiling
For building an accurate and convenient diagnostic model, cfDNA 5hmC libraries of HCC patients, LC patients and healthy controls were sequenced to generate a median number of 36.7 million reads in each sample (Methods). Genomic element distribution showed that the 5hmC modified regions enriched in transcription-related elements (promoters, untranslated regions, transcriptional start sites) and coding regions (exons, coding DNA sequences) were consistent with a previous study12 (Figure 1B). Since LC and HCC are both serious liver diseases, their disease status is significantly different from healthy controls. Therefore, the DhMPs in the whole genome from HCC patients, LC patients and healthy volunteers were firstly used for testing the feasibility to distinguish liver disease patients (HCC and LC) from healthy volunteers. In total, 620 fixed-length DhMPs between liver diseases and healthy controls (Wilcoxon signed-rank test, false discovery rate < 0.01, |log fold change| > 0.7) were found to have a capacity to distinguish liver disease patients from healthy volunteers, with the potential for HCC diagnosis (Figure 1C). As shown in Figure 1D, the differential 5hmC profile in LC patients was distributed among HCC patients and healthy volunteers, which is consistent with the history of HCC progression, since most HCC patients were developed from HBV associated LC background in China.
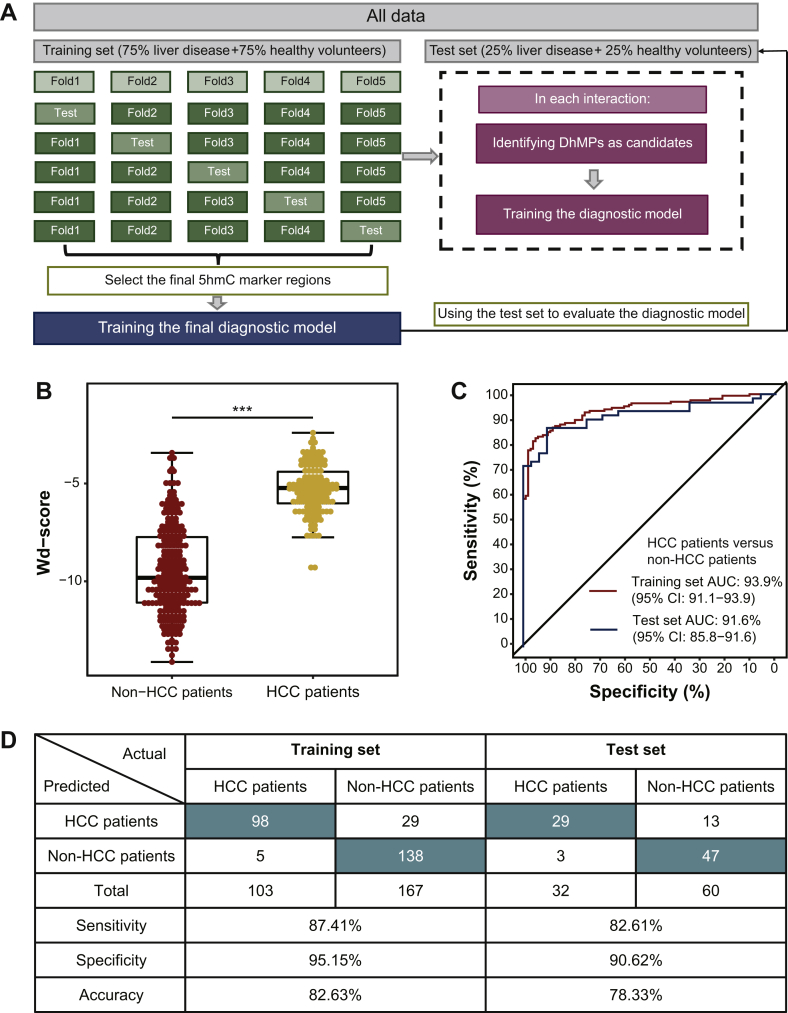
Next, we further carried out sPLS-DA to develop a diagnostic model for distinguishing liver disease patients (HCC and LC ) from healthy volunteers with 64 DhMPs, which includes genes implicated in HCC progression (Figure 2A and Supplementary Table S3, available at https://doi.org/10.1016/j.esmoop.2020.100021). Our diagnostic model exhibited an excellent distinguishing capacity between liver disease patients and healthy volunteers in both the training set (AUC = 98.8%) and the test set (AUC = 87.2%, Supplementary Figure S1B, available at https://doi.org/10.1016/j.esmoop.2020.100021). The wd-scores were computed based on the 5hmC levels of those 5hmC marker regions in cfDNA, and showed that the wd-score in liver disease patients was significantly higher than those in healthy volunteers (Wilcoxon signed-rank test, P = 1.25e−48, Supplementary Figure S1C, available at https://doi.org/10.1016/j.esmoop.2020.100021). At a wd-score cut-off of −7.8931, the wd-score achieved 91.87% and 80.95% of accuracy to distinguish liver disease patients from healthy volunteers in the training set and test set, respectively (Supplementary Figure S1D, available at https://doi.org/10.1016/j.esmoop.2020.100021). Moreover, early diagnosis could significantly improve the prognosis of HCC patients. Significantly, by analyzing cfDNA in high-risk LC patients, our wd-score could also distinguish HCC patients from non-HCC patients (Wilcoxon signed-rank test, P = 4.33e−33, Figure 2B), showing excellent capacity in both the training set (AUC = 93.9%) and test set (AUC = 91.6%) (Figure 2C). At a wd-score cut-off of −7.187891, the wd-score achieved 82.63% and 78.33% of accuracy to distinguish HCC patients from non-HCC patients in the training set and test set, respectively (Figure 2D). In addition, to explore the changed dynamics of wd-score in the HCC patients after receiving surgical resection, we further calculated the wd-score of the matched post-operative plasma samples from 105 HCC patients. The results showed that the post-operative wd-score was significantly decreased when compared with the matched preoperative wd-score (Wilcoxon signed-rank test, P = 5.63e−18, Supplementary Figure S1E, available at https://doi.org/10.1016/j.esmoop.2020.100021). Finally, we further validated the performance of our diagnostic model in 26 pairs of HCC samples and matched para-tumor tissue from published research.12 As expected, the wd-scores in HCC tissues were significantly higher than those in matched para-tumor tissues (Wilcoxon signed rank-test, P = 3.44e−7, Supplementary Figure S1F, available at https://doi.org/10.1016/j.esmoop.2020.100021), which is consistent with the results in liquid biopsy. Taken together, these results revealed that our weighted 5hmC diagnostic score could distinguish well between HCC patients, LC patients and healthy volunteers, and also indicate the patient's disease status after receiving surgical resection.
Figure 2.
Building an HCC diagnostic model based on cfDNA 5hmC signatures.
(A) The modeling method of the cfDNA 5hmC diagnostic model for distinguishing HCC patients from non-HCC patients. (B) The level of wd-score in HCC patients and non-HCC patients. (C) Receiver operating characteristic (ROC) curves of the cfDNA 5hmC diagnostic prediction model with DhMPs in the training set and test set. (D) Confusion matrices built from the cfDNA 5hmC diagnostic prediction model for distinguishing HCC patients from non-HCC patients in the training set and test set.
5hmC, 5-hydroxymethylcytosine; AUC, area under curve; cfDNA, cell-free DNA; CI, confidence interval; DhMPs, differentially 5hmC modified regions; HCC, hepatocellular carcinoma; wd-score, weighted diagnostic score.
Improve HCC diagnostic accuracy by combining 5hmC of cfDNA and protein biomarkers
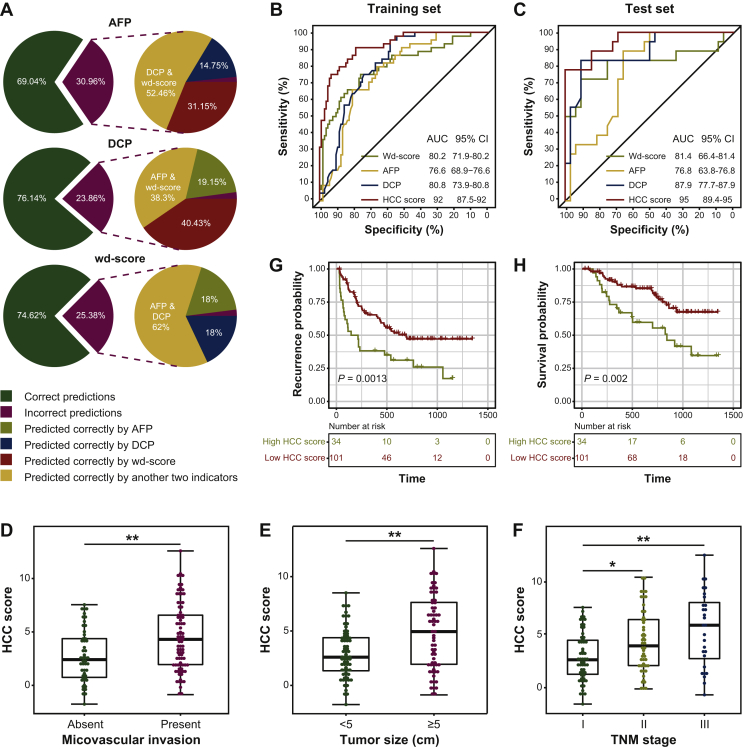
As is well known, LC patients are at high risk to develop HCC and need frequent examinations for HCC surveillance. Therefore, we next focus on the diagnostic ability to distinguish between LC and HCC. The wd-score in HCC patients was significantly higher than those in LC patients (Wilcoxon signed-rank test, P = 7.4e−12, Supplementary Figure S2A, available at https://doi.org/10.1016/j.esmoop.2020.100021), showing excellent capacity in distinguishing HCC patients from LC patients (Supplementary Figure S2B and C, available at https://doi.org/10.1016/j.esmoop.2020.100021). Compared with the translational protein biomarkers, the diagnostic capacity of the wd-score (accuracy = 74.62%) is better than AFP (accuracy = 69.04%) and comparable to DCP (accuracy = 76.14%) in our enrolled patients (Supplementary Table S4, available at https://doi.org/10.1016/j.esmoop.2020.100021). Surprisingly, we found that our wd-score had a complementary diagnostic capacity with AFP and DCP in distinguishing HCC patients from LC patients. Some 30.96% and 23.86% patients were predicted the wrong disease status by AFP and DCP, and 83.60% and 78.73% of those patients could be correctly predicted by wd-score, respectively; while 25.38% samples were incorrect in the 5hmC-based model, 80% and 80% of them could be predicted correctly by AFP and DCP, respectively (Figure 3A). Therefore, we further combined the wd-score, AFP and DCP to develop a novel HCC diagnostic score (HCC score) using logistics regression: HCC score = 1.5530 + 0.2319 ∗ log2(AFP) + 0.6690 ∗ log2(DCP) + 0.9644 ∗ wd-score. As expected, our HCC score exhibited an excellent diagnostic performance on HCC patients and LC patients. As shown in Figure 3B and C, comparing with our wd-score, AFP or DCP alone, the time-dependent ROC analysis showed that the HCC score could further improve HCC diagnostic ability in the training set and test set (training set: AUC = 92%, accuracy = 82.31%, sensitivity = 78.64% and specificity = 90.91%; test set: AUC = 95%, accuracy = 88%, sensitivity = 93.75% and specificity = 77.78%). Moreover, further study on the clinicopathologic characteristics of our enrolled HCC patients revealed that HCC scores were positively associated with microvascular invasion (MVI), tumor size, TNM (tumor–node–metastasis) stage (A review of HCC staging systems; https://cco.amegroups.com/article/view/2528/3943) (Figure 3D-F). Multivariate Cox regression analysis indicated that the HCC score could serve as an independent factor of recurrence (P = 0.025) and survival (P = 0.09892, Supplementary Table S5, available at https://doi.org/10.1016/j.esmoop.2020.100021). The Kaplan–Meier analysis showed that patients with a higher HCC score (cut-off value as 6.216063 in preoperative plasma) at diagnosis had a shorter relapse-free survival (log-rank, P = 0.0013, Figure 3G) and a shorter OS time (log-rank, P = 0.002, Figure 3H). Taken together, these results suggested that the HCC score had a better diagnostic and prognostic potential in HCC for clinical evaluation.
Figure 3.
The development of a novel HCC diagnostic score for HCC diagnosis.
(A) The complementary diagnostic ability among the wd-score, AFP and DCP in distinguishing HCC patients from LC patients. (B) The receiver operating characteristic (ROC) curves of HCC score, AFP, DCP and wd-score in the training set. (C) The ROC curves of HCC score, AFP, DCP and wd-score in the test set. (D) The preoperative HCC score in HCC patients with and without microvascular invasion. (E) The preoperative HCC score in HCC patients with large tumor size (≥5 cm) and small tumor size (<5 cm). (F) The preoperative HCC score in HCC patients with different TNM stage. The relapse-free survival (G) and overall survival (H) curves of HCC patients with low or high HCC score at preoperative time point in the enrolled cohort.
AFP, alpha-fetoprotein; AUC, area under curve; CI, confidence interval; DCP, des-gamma-carboxy prothrombin; HCC, hepatocellular carcinoma; TNM, tumor–node–metastasis; wd-score, weighted diagnostic score.
Recurrence risk assessment of HCC score in HCC patients after receiving surgical resection
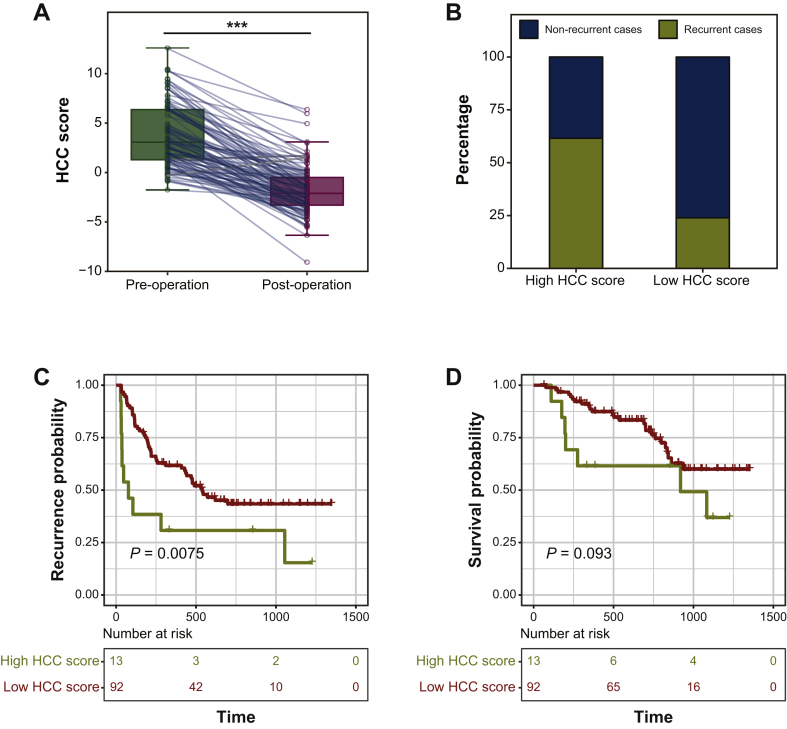
HCC patients' tumor burden would be significantly decreased or completely eliminated after receiving surgical resection, making their overall state more closely related to that of LC patients or healthy volunteers. Therefore, assessing the potential recurrence risk of HCC patients after surgical resection could decide which patients need to receive additional adjuvant therapies (such as transarterial chemoembolization). We then examined the usefulness of the HCC score in recurrence risk assessment after hepatectomy. As expected, the post-operative HCC score was significantly decreased when compared with the matched preoperative HCC score (Wilcoxon signed-rank test, P = 6.09e−27, Figure 4A). When the cut-off value of the HCC score in post-operative plasma was 1.058031, which is the predicting threshold for distinguishing between HCC and LC, the recurrence rate of the patient group with a high HCC score was significantly higher than that of the patient group with a low HCC score within 6 months (61.54% versus 23.91%; Figure 4B) or within 1 year (69.23% versus 38.04%; Supplementary Figure S3A, available at https://doi.org/10.1016/j.esmoop.2020.100021). Further analysis revealed that almost all of these patients with a high score at the post-operative time point had MVI (12/13) or higher TNM stage (TNM II and TNM III, 11/13), which related to higher risk of recurrence after surgical operation. Moreover, the HCC score in post-operative plasma could also serve as an independent factor for recurrence (P = 0.00865) in multivariate Cox regression analysis and the only indicator for survival in univariate Cox regression analysis (P = 0.000739, Supplementary Table S6, available at https://doi.org/10.1016/j.esmoop.2020.100021). The Kaplan–Meier analysis showed that patients with high HCC scores in post-operative plasma had higher recurrence risk (log-rank, P = 0.0075) and a relative shorter OS time (log-rank, P = 0.093) (Figure 4F and G). Taken together, these data also suggested that our HCC score could indeed indicate the risk of early recurrence for HCC patients after receiving a surgical operation.
Figure 4.
Assessment recurrence risk in HCC patients after receiving surgical resection.
(A) The level of HCC score in the preoperative plasma and in the matched post-operative plasma collected from HCC patients. (B) The recurrence rate within 6 months in different patient groups with high or low HCC score in post-operative plasma based on a cut-off value of 1.058031. The relapse-free survival (C) and overall survival (D) curves of HCC patients with low or high HCC score at post-operative time point in the enrolled cohort.
HCC, hepatocellular carcinoma.
Real-time tracking tumor burden during HCC progression
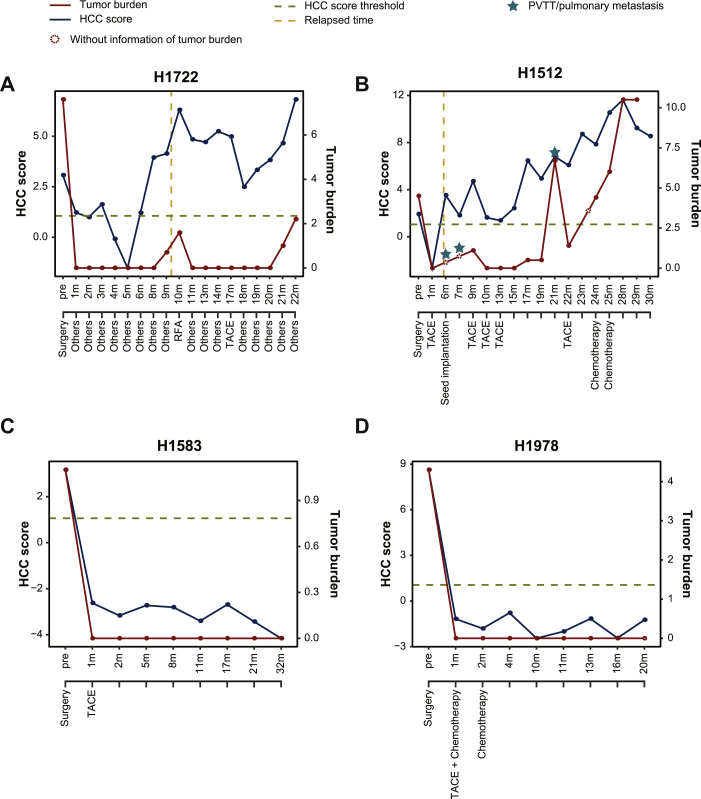
To further explore whether HCC score could dynamically track tumor burden during HCC progression, we retrospectively analyzed 127 sequential follow-up plasma samples collected from 10 HCC patients with HBV-associated LC history. For patients with recurrence, the HCC score in patient H1722 could be clearly detected at a high level before receiving a surgical operation and decreased significantly after surgery within 2 months, but still remained above the predictive threshold, suggesting a higher risk of recurrence (Figure 5A). The observed HCC score were increased at the 3rd month and kept a continuous declination at the 4th and 5th periods during medical treatment. However, the HCC score increased to a high value from the 6th to 10th months, suggesting that tumor recurrence occurred in this patient. According to MRI imaging, a tumor lesion (0.7 × 0.7 cm) could be clearly observed at the 9th month and continued to grow until the 10th month (1.59 × 1.51 cm). Then, he received a second surgical resection and the HCC score were correspondingly dropped; oscillation remained above the predictive threshold at the 11th-20th month. Significantly, the HCC score may have the ability to successfully predict the second recurrence of patient H1722, as it increased before a tumor lesion (1.0 × 0.8 cm) screened by MRI imaging in the 18th month to the 20th month (Figure 5A). The consistent trend of tumor burden and HCC scores were also observed in patients H1512 (Figure 5B), H1526 (Supplementary Figure S4A, available at https://doi.org/10.1016/j.esmoop.2020.100021) and H1570 (Supplementary Figure S4B, available at https://doi.org/10.1016/j.esmoop.2020.100021). However, due to the heterogeneity among patients, we still found that the HCC score could not completely detect the small recurrence or metastasis in two HCC patients (H1940 and H1786) at some follow-up points (Supplementary Figure S4C and D, available at https://doi.org/10.1016/j.esmoop.2020.100021). For other patients without recurrence after undergoing surgery and adjuvant transarterial chemoembolization treatment, patients H1583, H1978, H1550 and H1925 all showed no abnormal levels of HCC score in follow-up time points (Figure 5C and D and Supplementary Figure S4E and F, available at https://doi.org/10.1016/j.esmoop.2020.100021). Taken together, after cross-comparison with MRI imaging results, these results suggest that tracking the level of HCC score in patients' longitudinal plasma samples during follow-up may provide a sensitive strategy for real-time monitoring of tumor burden and evaluating prognosis outcomes.
Figure 5.
The dynamic change of HCC score in serial plasma samples during HCC progression.
The time-course demonstration of quantified HCC score levels and tumor burden in patients H1722 (A), 1522 (B), H1583 (C) and H1978 (D), respectively. Red lines indicate the level of HCC score and green lines indicate the level of tumor burden. Clinical treatments of these patients are also shown as del operator with different color.
HCC, hepatocellular carcinoma; PVTT, portal vein tumor thrombus; RFA, radiofrequency ablation; TACE, transarterial chemoembolization.
Discussion
Early diagnosis and accurate prognosis prediction are crucial for HCC management, including HCC surveillance for the high-risk population and the effective treatment of HCC.5,20 However, this requires long-term continuous monitoring. Compared with tissue biopsy and imaging detection, blood-based liquid biopsy can be used more frequently and is cheaper for routine clinical monitoring, which also could be better accepted by patients.21,22 At present, due to the existence of tumor heterogeneity, AFP and DCP, which are commonly used in clinic, have only reached 41%-70% and 50%-80% of the diagnostic efficacy of HCC, respectively.23 5hmC Signatures in cfDNA have proved to be a reliable epigenetic biomarker for detection and diagnosis of cancer, including HCC, breast cancer and lung cancer.10,24 Technically, compared with detecting single-nucleotide polymorphism in cfDNA, detecting 5hmC signatures in cfDNA only needs to carry out covalent chemical labeling based on low-depth whole genome sequencing, which has the advantages of preventing sequencing biases and cost efficiency, with great potential for clinical application. In this study, unlike identifying differential 5hmC marker genes in other studies,12,25 we developed a diagnostic model based on the differential fixed-length peaks (201 bp) which enriched 5hmC modified bases to improve the accuracy of 5hmC modified region identification. This approach might be more conducive for clinical translation of identified 5hmC marker regions. Moreover, this weighted model has demonstrated a good ability to distinguish among HCC patients, LC patients and healthy volunteers, and had complementary diagnostic capacity with the existing diagnostic protein markers (AFP and DCP). Our 5hmC model could totally achieve an accuracy rate of 93% in distinguishing HCC patients from non-HCC patients, which is equivalent to the previously published 5hmC methylation model,12 although large-scale cases need further verification. Collectively, our results further indicated that cfDNA 5hmC profiling based liquid biopsy may provide a non-invasive and easily accessible approach for early HCC diagnosis, with the potential of becoming a part of HCC management.
Compared with single markers, a diagnostic model which was developed with multiple biomarkers for HCC has proved to be more sensitive and robust to overcome high HCC heterogeneity among patients. Cohen et al.26 developed a CancerSEEK test by combing the mutations in cfDNA and the levels of circulating proteins, achieving a median of 70% sensitivity in eight cancer types, including HCC. Herein, we further build a novel HCC diagnostic score (HCC score) by combining a weighted 5hmC diagnostic model, AFP and DCP to distinguish HCC patients from LC patients, achieving 92% and 95% of AUC in the training set and test set, respectively. In addition to its potential in early detection, HCC scores in preoperative plasma could also serve as indicators of HCC progression with excellent performance in HCC prognosis.
As the radical curative effect of surgery is significantly related to the prognosis of patients, there are no specific guidelines and still a lack of objective evaluation indicators to address which patients should receive intensive treatment after surgery.27 In clinical practice, we generally judge the prognosis according to the patient's tumor pathological characteristics, such as vascular invasion and tumor stage, and monitor HCC recurrence by serial imaging. In this study, we found that assessment of HCC scores in HCC patients after receiving surgical operation could also serve as an independent prognostic indicator for screening out the patients with a high risk of early recurrence and metastasis. Meanwhile, post-operative HCC scores were also found to be significantly correlated with traditional high-risk factors for recurrence of HCC (MVI and TNM, Supplementary Figure S3B and C, available at https://doi.org/10.1016/j.esmoop.2020.100021). In addition, the HCC score was also positively associated with the dynamic of tumor burden during long-term follow-up, although this finding was limited by the relatively small size of enrolled patients. Taken together, our HCC score has an excellent predictive capability, and therefore can be considered as a potential approach for HCC management.
In summary, we have developed a novel non-invasive detection system for HCC diagnosis, prognosis and surveillance during HCC progression by combing the sensitive 5hmC signatures and clinical protein biomarkers. Compared with single biomarker detection, the HCC score could effectively provide higher clinical diagnostic accuracy to help screening high-risk individuals, therefore optimizing clinical decisions during HCC management. However, due to the discrepancy between datasets caused by limited sample sizes, the clinical applicability of our model still needs to be validated in large-scale, randomized clinical trials.
Acknowledgments
Funding
This work was supported by National Science and Technology Major Project of China [grant number 2018ZX10302205], regional development project of Fujian Province [grant number 2019Y3001], National Natural Science Foundation of China [grant number 31771426], the Scientific Foundation of the Fuzhou Health Commission [grant number 2019-S-wt3].
Disclosure
The authors have declared no conflicts of interest.
Contributor Information
D. Xie, Email: danxie@scu.edu.cn.
X. Liu, Email: xiaoloong.liu@gmail.com.
J. Liu, Email: drjingfeng@126.com.
Supplementary data
References
- 1.Chen W., Zheng R., Baade P.D. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Wang F.S., Fan J.G., Zhang Z., Gao B., Wang H.Y. The global burden of liver disease: the major impact of China. Hepatology. 2014;60:2099–2108. doi: 10.1002/hep.27406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramesh H. Resection for hepatocellular carcinoma. J Clin Exp Hepatol. 2014;4:S90–S96. doi: 10.1016/j.jceh.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi J., Kim G.A., Han S., Lee W., Chun S., Lim Y.S. Longitudinal assessment of three serum biomarkers to detect very early-sta/ge hepatocellular carcinoma. Hepatology. 2019;69:1983–1994. doi: 10.1002/hep.30233. [DOI] [PubMed] [Google Scholar]
- 5.Bruix J., Reig M., Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150:835–853. doi: 10.1053/j.gastro.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 6.Heitzer E., Auinger L., Speicher M.R. Cell-free DNA and apoptosis: how dead cells inform about the living. Trends Mol Med. 2020;26:519–528. doi: 10.1016/j.molmed.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Xu R.H., Wei W., Krawczyk M. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16:1155–1161. doi: 10.1038/nmat4997. [DOI] [PubMed] [Google Scholar]
- 8.Cai Z., Chen G., Zeng Y. Comprehensive liquid profiling of circulating tumor DNA and protein biomarkers in long-term follow-up patients with hepatocellular carcinoma. Clin Cancer Res. 2019;25:5284–5294. doi: 10.1158/1078-0432.CCR-18-3477. [DOI] [PubMed] [Google Scholar]
- 9.Ye Q., Ling S., Zheng S., Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18:114. doi: 10.1186/s12943-019-1043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W., Zhang X., Lu X. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017;27:1243–1257. doi: 10.1038/cr.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song C.-X., Yin S., Ma L. 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res. 2017;27:1231–1242. doi: 10.1038/cr.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai J., Chen L., Zhang Z. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut. 2019;68:2195–2205. doi: 10.1136/gutjnl-2019-318882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arechederra M. New warning signs on the road: 5-hydroxymethylcytosine-based liquid biopsy for the early detection of hepatocellular carcinoma. Gut. 2019;68:2103–2104. doi: 10.1136/gutjnl-2019-319339. [DOI] [PubMed] [Google Scholar]
- 14.Qu C., Wang Y., Wang P. Detection of early-stage hepatocellular carcinoma in asymptomatic HBsAg-seropositive individuals by liquid biopsy. Proc Natl Acad Sci U S A. 2019;116:6308–6312. doi: 10.1073/pnas.1819799116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Liu T., Meyer C.A. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corces M.R., Granja J.M., Shams S. The chromatin accessibility landscape of primary human cancers. Science. 2018;362:eaav1898. doi: 10.1126/science.aav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohart F., Gautier B., Singh A., Lê Cao K.-A. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13:e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang H.-Y., Chen J., Xia C.-C., Cao L.-K., Duan T., Song B. Noninvasive imaging of hepatocellular carcinoma: from diagnosis to prognosis. World J Gastroenterol. 2018;24:2348. doi: 10.3748/wjg.v24.i22.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marrugo-Ramírez J., Mir M., Samitier J. Blood-based cancer biomarkers in liquid biopsy: a promising non-invasive alternative to tissue biopsy. Int J Mol Sci. 2018;19:2877. doi: 10.3390/ijms19102877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin C.-Q., Yuan C.-H., Qu Z., Guan Q., Chen H., Wang F.-B. Liquid biopsy of hepatocellular carcinoma: circulating tumor-derived biomarkers. Dis Markers. 2016;2016:1427849. doi: 10.1155/2016/1427849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song P., Gao J., Inagaki Y. Biomarkers: evaluation of screening for and early diagnosis of hepatocellular carcinoma in Japan and China. Liver Cancer. 2013;2:31–39. doi: 10.1159/000346220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng C., Stroup E.K., Zhang Z., Chiu B.C.-H., Zhang W. Towards precision medicine: advances in 5-hydroxymethylcytosine cancer biomarker discovery in liquid biopsy. Cancer Commun. 2019;39:12. doi: 10.1186/s40880-019-0356-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiu B.C., Zhang Z., You Q. Prognostic implications of 5-hydroxymethylcytosines from circulating cell-free DNA in diffuse large B-cell lymphoma. Blood Adv. 2019;3:2790–2799. doi: 10.1182/bloodadvances.2019000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen J.D., Li L., Wang Y. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen V.L., Xu D., Wicha M.S., Lok A.S., Parikh N.D. Utility of liquid biopsy analysis in detection of hepatocellular carcinoma, determination of prognosis, and disease monitoring: a systematic review. Clin Gastroenterol Hepatol. 2020;18:2879–2902.e9. doi: 10.1016/j.cgh.2020.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.