Abstract
Background & Aims
Nonalcoholic fatty liver disease (NAFLD) is becoming a severe liver disorder worldwide. Autophagy plays a critical role in liver steatosis. However, the role of autophagy in NAFLD remains exclusive and under debate. In this study, we investigated the role of S100 calcium binding protein A11 (S100A11) in the pathogenesis of hepatic steatosis.
Methods
We performed liver proteomics in a well-established tree shrew model of NAFLD. The expression of S100A11 in different models of NAFLD was detected by Western blot and/or quantitative polymerase chain reaction. Liver S100A11 overexpression mice were generated by injecting a recombinant adenovirus gene transfer vector through the tail vein and then induced by a high-fat and high-cholesterol diet. Cell lines with S100a11 stable overexpression were established with a recombinant lentiviral vector. The lipid content was measured with either Bodipy staining, Oil Red O staining, gas chromatography, or a triglyceride kit. The autophagy and lipogenesis were detected in vitro and in vivo by Western blot and quantitative polymerase chain reaction. The functions of Sirtuin 1, histone deacetylase 6 (HDAC6), and FOXO1 were inhibited by specific inhibitors. The interactions between related proteins were analyzed by a co-immunoprecipitation assay and immunofluorescence analysis.
Results
The expression of S100A11 was up-regulated significantly in a time-dependent manner in the tree shrew model of NAFLD. S100A11 expression was induced consistently in oleic acid–treated liver cells as well as the livers of mice fed a high-fat diet and NAFLD patients. Both in vitro and in vivo overexpression of S100A11 could induce hepatic lipid accumulation. Mechanistically, overexpression of S100A11 activated an autophagy and lipogenesis process through up-regulation and acetylation of the transcriptional factor FOXO1, consequently promoting lipogenesis and lipid accumulation in vitro and in vivo. Inhibition of HDAC6, a deacetylase of FOXO1, showed similar phenotypes to S100A11 overexpression in Hepa 1–6 cells. S100A11 interacted with HDAC6 to inhibit its activity, leading to the release and activation of FOXO1. Under S100A11 overexpression, the inhibition of FOXO1 and autophagy could alleviate the activated autophagy as well as up-regulated lipogenic genes. Both FOXO1 and autophagy inhibition and Dgat2 deletion could reduce liver cell lipid accumulation significantly.
Conclusions
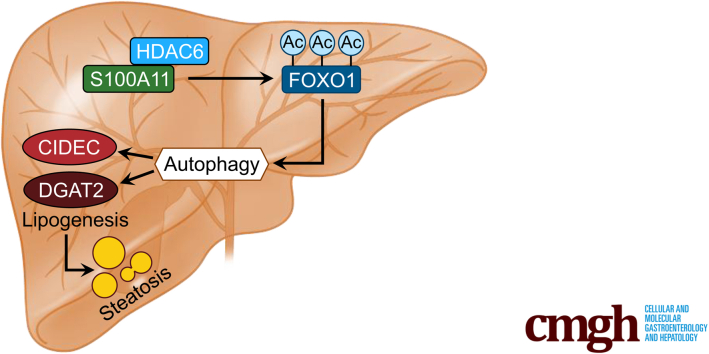
A high-fat diet promotes liver S100A11 expression, which may interact with HDAC6 to block its binding to FOXO1, releasing or increasing the acetylation of FOXO1, thus activating autophagy and lipogenesis, and accelerating lipid accumulation and liver steatosis. These findings indicate a completely novel S100A11-HDAC6-FOXO1 axis in the regulation of autophagy and liver steatosis, providing potential possibilities for the treatment of NAFLD.
Keywords: NAFLD, S100A11, FOXO1, Autophagy, Lipid Metabolism
Abbreviations used in this paper: Ac-FOXO1, acetylated forkhead box O1; ATG7, autophagy related 7; CE, cholesterol ester; CIDEC, cell death-inducing DFFA-like effector c; co-IP, co-immunoprecipitation; CON, control; CRISPR/Cas9, clustered regularly interspaced short palindromic repeats/cas9; DGAT2, diacylglycerol O-acyltransferase 2; FOXO1, forkhead box O1; GRB2, growth factor receptor-bound protein 2; HDAC6, histone deacetylase 6; HFD, high-fat diet; HFHC, high-fat and high-cholesterol diet; HFLC, high-fat low-cholesterol; LC3-II, LC3-phosphatidylethanolamine conjugate; LD, lipid droplets; LDL-c, low-density lipoprotein c; mRNA, messenger RNA; MS, mass spectrometry; NAFLD, nonalcoholic fatty liver disease; OA, oleic acid; PBS, phosphate-buffered saline; qPCR, quantitative polymerase chain reaction; RFP, red fluorescent protein; S100A11, S100 calcium binding protein A11; SDS, sodium dodecyl sulfate; SILAC, stable isotope labeling technology of amino acids in cell culture; SIRT1, Sirtuin 1; TBA, tubastatin A; TG, triglyceride; WT, wild-type
Graphical abstract
Summary.
The calcium binding protein S100A11 is highly conserved in vertebrates and expressed in many tissues. Autophagy is a critical player in nonalcoholic fatty liver disease development. Here, we show that dietary lipids up-regulate S100A11 expression, which accelerates FOXO1-mediated autophagy and lipogenesis via the deacetylase histone deacetylase 6, leading to hepatic steatosis.
Nonalcoholic fatty liver disease (NAFLD) is a chronic and progressive liver disorder afflicting approximately 25% of the world’s population.1, 2, 3 It begins with the presence of simple liver steatosis with little or even no alcohol consumption4 and may develop further into nonalcoholic steatohepatitis, fibrosis, and cirrhosis.5 Generally, liver lipids are stored in lipid droplets (LDs) and maintain homeostasis via a balanced flux of lipid uptake, synthesis, lipolysis, and excretion. Abnormal LD accumulation occurs when this balance is disturbed, which then ensures liver steatosis. Although numerous studies have uncovered distinct factors contributing to the pathogenesis of NAFLD, the precise mechanisms underlying the progression of NAFLD remain largely unclear owing to its highly heterogeneous clinical manifestations.
Autophagy has been shown to play a critical role in the development of NAFLD. Autophagy is an adaptive response to both extracellular and intracellular stress, such as nutrient deprivation or damaged cytoplasmic components including organelles and proteins.6,7 Recent studies have linked autophagy to hepatocyte lipid metabolism. Specifically, impaired hepatic autophagy causes increased LD formation8 and liver steatosis9 via the promotion of triglyceride storage in LD. Moreover, pharmacologic modulation of autophagy improves nonalcoholic and alcoholic hepatic steatosis in mouse models.10, 11, 12 In contrast, other studies have shown that LD numbers were increased rather than decreased under an activated autophagy condition induced by starvation in cell models.13, 14, 15 In addition, the inhibition of autophagy using a pharmacologic approach or the deletion of autophagy genes decreased LD numbers14 and protected the liver from steatosis.16, 17, 18, 19 Despite its controversial and arguable role in NAFLD, autophagy still is thought to facilitate LD formation and lipid accumulation in hepatocytes, which plays a significant part in hepatic steatosis development.
FOXO1, a transcription factor belonging to the forkhead box O (FOXO) family, has been underscored as a key mediator of autophagy in different cell types and disease conditions.20, 21, 22, 23, 24, 25, 26 Under nutrient restriction, FOXO1 is activated to promote lipid catabolism through up-regulation of the expression of lysosomal lipase in mammalian adipocytes27 and D melanogaster,28 and the expression of adipose triglyceride lipase in murine adipocytes.29 In contrast, FOXO1 recently was shown to enhance autophagy to regulate LD growth and size via LD protein fat-specific protein 27, also known as cell death–inducing DFFA-like effector C (CIDEC), in adipocytes.30 Overall, these studies raise the possibility that the role of FOXO1 in regulating lipid metabolism may be context-dependent. However, it is completely unknown how FOXO1 is activated to regulate lipid metabolism via autophagy, and the upstream regulation of FOXO1 during autophagy remains incompletely understood.
S100 calcium-binding protein A11 (S100A11; also known as S100C) is highly conserved in vertebrates and expressed in many tissues.31,32 S100A11 is involved in many biological processes such as cell apoptosis33 and inflammation,34 and a number of human cancers including bladder cancer35 and prostate cancer.36 Regardless of this involvement, the specific roles of S100A11 in lipid metabolism, especially in NAFLD, have not been reported to date. We previously successfully established a tree shrew model of NAFLD that could mimic the progression from simple hepatic steatosis to fibrosis within 10 weeks by feeding a high-fat and high-cholesterol diet (HFHC).37 Based on this animal model, we used proteomics to explore the underlying mechanisms of NAFLD and found that S100A11 was up-regulated progressively in a time-dependent manner. In vivo and in vitro overexpression of S100A11 promoted lipid accumulation and liver steatosis via FOXO1-induced autophagy, suggesting that S100A11 is a novel pathogenic factor of NAFLD.
Results
Up-regulation of the Calcium-Binding Protein S100A11 in In Vitro and In Vivo Models of NAFLD
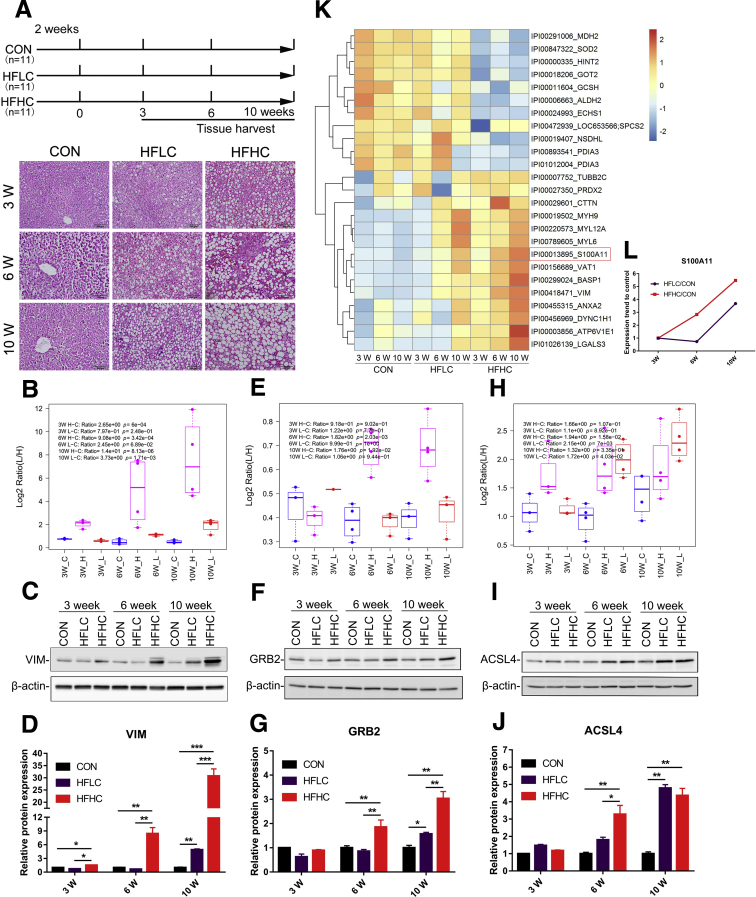
We previously used high-fat low-cholesterol (HFLC) and HFHC diets to establish a tree shrew model of NAFLD.37,38 Compared with the control group (CON), the experimental group showed progressive steatosis with obvious lipid accumulation in a time-dependent manner (Figure 1A). To explore proteins contributing to the progression of NAFLD, a comparative proteomic analysis of tree shrew livers at 3 time points was conducted using stable isotope labeling technology of amino acids in cell culture (SILAC). In total, 3589 proteins were identified and quantified, of which some proteins showed differential abundance in the HFLC and HFHC groups compared with the matched CON in a time-dependent manner (Table 1, Table 2, Table 3). To further validate the proteome results, a subset of proteins with differential abundance and commercially available antibodies were used to perform Western blot. Vimentin is a type III intermediate filament protein involved in liver steatosis.39 Growth factor receptor-bound protein 2 (GRB2) is an adaptor protein, and the suppression of GRB2 expression was implicated in improved hepatic steatosis.40 Long-chain acyl-CoA synthetase 4 is involved in lipid metabolism and overexpressed in African American NAFLD patients.41 In line with the proteomic data, the expression of all 3 proteins was up-regulated consistently in a time-dependent manner, particularly in the HFHC group (Figure 1B–J).
Figure 1.
Liver proteomics analysis and its validation in a tree shrew model of NAFLD and the screening of S100A11 from liver proteomics. (A) Experimental design (top panel), H&E staining of tree shrew liver after 10 weeks of a HFLC and HFHC diet (bottom panel). Original magnification: 20×. Scale bars: 100 μm. (B–D) Validation of vimentin (VIM). (E–G) Validation of GRB2. (H–J) Validation of long-chain acyl-CoA synthetase 4 (ACSL4). Top panel: proteomics results, C, CON; L, HFLC; and H, HFHC groups. Middle panel: Western blot results of tree shrew liver proteins. Bottom panel: quantification of Western blot results from the middle panel. The target proteins were normalized to β-actin. n = 3–4 individual animals. (K) Heatmap generated by the proteins with continuously decreased or increased expression in the HFLC and HFHC groups from 3 to 10 weeks. (L) The expression tendency of S100A11 protein. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
Table 1.
Top 10 Proteins Significantly (Fold > 1.5; P < .05) Increased in the HFHC Group at All 3 Time Points Compared With Matched Controls
| Protein name | Symbol | Fold change |
||
|---|---|---|---|---|
| 3 weeks | 6 weeks | 10 weeks | ||
| S100 calcium-binding protein A11 | S100A11 | 1.77 | 8.57 | 15.70 |
| Vimentin | VIM | 2.65 | 9.08 | 14.01 |
| Brain acid soluble protein 1 | BASP1 | 2.34 | 3.15 | 5.44 |
| Synaptic vesicle membrane protein VAT-1 homolog | VAT1 | 1.65 | 2.93 | 3.86 |
| Myosin-9 | MYH9 | 1.81 | 2.29 | 2.88 |
| Cytoplasmic dynein 1 heavy chain 1 | DYNC1H1 | 1.75 | 2.13 | 2.62 |
| Myosin light chain 6 | MYL6 | 1.61 | 1.92 | 2.61 |
| Myosin light chain 12A | MYL12A | 1.62 | 1.73 | 2.34 |
| V-type proton adenosine triphosphatase subunit E1 | ATP6V1E1 | 1.53 | 1.52 | 2.21 |
| Src substrate cortactin | CTTN | 1.64 | 3.01 | 2.16 |
VIM, vimentin.
Table 2.
Top 10 Proteins Significantly (Fold <1/1.5, P < .05) Decreased in HFHC Group at All 3 Time Points Compared With Matched Control
| Protein name | Symbol | Fold change |
||
|---|---|---|---|---|
| 3 weeks | 6 weeks | 10 weeks | ||
| Glycine cleavage system H protein | GCSH | 0.35 | 0.18 | 0.38 |
| NAD(P) dependent steroid dehydrogenase-like | NSDHL | 0.58 | 0.35 | 0.39 |
| Superoxide dismutase | SOD2 | 0.46 | 0.52 | 0.42 |
| Malate dehydrogenase | MDH2 | 0.50 | 0.64 | 0.49 |
| Aspartate aminotransferase | GOT2 | 0.42 | 0.62 | 0.49 |
| Protein disulfide-isomerase A3 | PDIA3 | 0.59 | 0.57 | 0.51 |
| Enoyl-CoA hydratase | ECHS1 | 0.51 | 0.53 | 0.51 |
| Histidine triad nucleotide-binding protein 2 | HINT2 | 0.47 | 0.59 | 0.55 |
| Aldehyde dehydrogenase | ALDH2 | 0.49 | 0.47 | 0.56 |
| Signal peptidase complex subunit 2 | SPCS2 | 0.06 | 0.59 | 0.57 |
NOTE. The list of the other proteins significantly changed in the HFHC group is available upon request. NAD(P), nicotinamide adenine dinucleotide (phosphate).
Table 3.
Proteins Significantly Changed in HFLC Group at Different Time Points
| Protein name | Symbol | Fold change |
||
|---|---|---|---|---|
| 3 weeks | 6 weeks | 10 weeks | ||
| Proteins significantly (fold > 1.5; P > .05) increased in the 6-week HFLC and 10-week HFLC compared with matched control | ||||
| High-mobility group AT-hook 1 | HMGA1 | 1.34 | 2.24 | 5.37 |
| Histone H3.2 | HIST2H3A | 0.67 | 3.38 | 3.53 |
| Myosin-9 | MYH9 | 1.16 | 2.10 | 3.26 |
| Myosin light chain 6 | MYL6 | 1.07 | 1.82 | 2.85 |
| Tropomyosin alpha-4 | TPM4 | 1.03 | 1.51 | 2.77 |
| Ras-related protein Rab-18 | RAB18 | 1.62 | 2.97 | 2.68 |
| Myosin light chain 12A | MYL12A | 1.08 | 1.64 | 2.63 |
| Myosin light chain 1 | MYL1 | 1.02 | 1.77 | 2.58 |
| Carnitine O-palmitoyltransferase 2 | CPT2 | 0.65 | 1.97 | 2.45 |
| Charged multivesicular body protein 4b | CHMP4B | 1.88 | 1.98 | 2.34 |
| Keratin 8 | KRT8 | 0.70 | 1.69 | 2.19 |
| Keratin 18 | KRT18 | 0.79 | 1.73 | 2.11 |
| FLII actin remodeling protein | FLII | 1.11 | 1.93 | 2.09 |
| Charged multivesicular body protein 1A | CHMP1A | 0.72 | 1.78 | 1.97 |
| Lamin A/C | LMNA | 0.77 | 1.68 | 1.97 |
| Acyl-CoA synthetase long chain family member 4 | ACSL4 | 1.10 | 2.15 | 1.72 |
| V-type proton adenosine triphosphatase subunit d 1 | ATP6V0D1 | 0.78 | 1.80 | 1.67 |
| Ras-related protein Rap-1b | RAP1B | 0.75 | 1.51 | 1.58 |
| G-protein subunit γ5 | GNG5 | 0.90 | 1.51 | 1.55 |
| Vesicle-associated membrane protein-associated protein A | VAPA | 0.79 | 1.79 | 1.52 |
| Mitochondrial calcium uniporter | MCU | 0.54 | 1.28 | 3.74 |
| Protein significantly (fold > 1.5; P < .05) increased in the HFLC groups at all 3 time points compared with matched control | ||||
| Acyl-CoA dehydrogenase very long chain | ACADVL | 1.52 | 1.75 | 1.93 |
NOTE. The list of the other proteins significantly changed in the HFLC group is available upon request.
ACSL4, long-chain acyl-CoA synthetase 4.
Remarkably, the expression of 25 proteins showed a similar tendency of either increasing or decreasing in a time- and cholesterol-dependent manner (Figure 1K) (Table 1, Table 2, Table 3). We focused on these up-regulated proteins and hypothesized that they might be involved in the progression of NAFLD because their expression was correlated positively to the severity of liver steatosis, similar to vimentin, GRB2, and long-chain acyl-CoA synthetase 4 (Figure 1K). Therefore, we ranked the top 10 proteins with increased expression levels in the HFHC group at all 3 time points. Among them, the expression level of S100A11 was increased most prominently (Table 1). Similarly, the proteome analysis showed that the expression of S100A11 gradually increased not only in a time-dependent manner, but probably also was affected by cholesterol levels (Figure 1L).
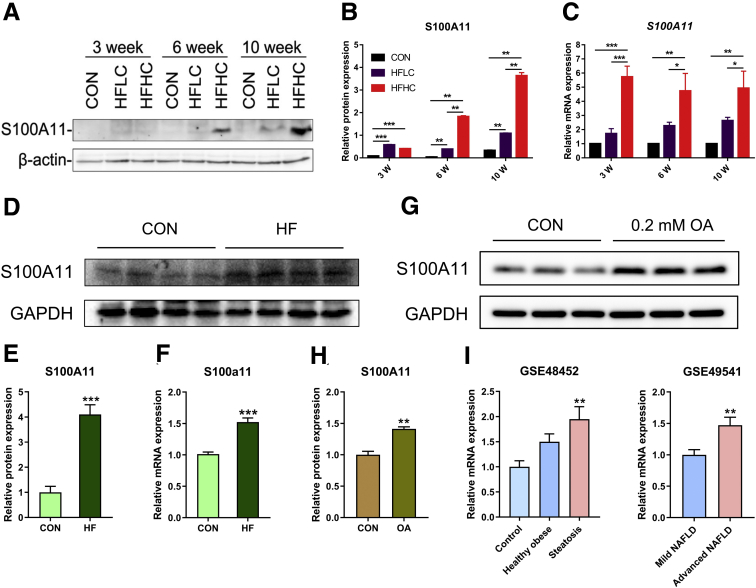
S100A11 has been reported to participate in various cellular responses, however its role in NAFLD remains completely unknown. Using Western blot (Figure 2A and B) and quantitative polymerase chain reaction (qPCR) (Figure 2C), respectively, we found that the S100A11 protein and transcripts indeed increased progressively in both the HFLC and HFHC groups from weeks 3 to 10. To confirm these observations, we measured the S100A11 expression in multiple in vitro and in vivo models of NAFLD. Both the messenger RNA (mRNA) and protein levels of S100A11 consistently and significantly increased in the liver of the mouse model of NAFLD induced by a high-fat diet (35% fat by weight) (Figure 2D–F). The treatment of Hepa 1–6 cells with oleic acid (OA, 0.2 mmol/L) significantly promoted the protein expression of S100A11 (Figure 2G and H). Furthermore, a literature review showed that the mRNA expression of S100A11 also was up-regulated in mouse NAFLD as well as in baboon and human alcoholic fatty liver disease42, 43, 44, 45, 46 (Table 4). More importantly, by searching the published human NAFLD transcriptome data (GEO data sets, https://www.ncbi.nlm.nih.gov/gds), we found that the liver S100A11 level was gradually up-regulated from controls to healthy obese (without hepatic steatosis) patients and hepatic steatosis patients (GSE48452 in Figure 2I), and the liver S100A11 level in advanced NAFLD patients also was significantly higher than that in mild NAFLD patients (GSE49541 in Figure 2I). Collectively, the lines of results consistently showed that S100A11 may play an uncharacterized role in NAFLD from mice, tree shrews, and human beings.
Figure 2.
Up-regulation of the calcium-binding protein S100A11 in in vitro and in vivo models of NAFLD. (A) S100A11 expression detected by Western blot in tree shrew liver. (B) Quantification of Western blot results from panel A. The target protein was normalized to β-actin. n = 4 individual animals for each group. (C) Relative mRNA expression of S100A11 in tree shrew liver detected by qPCR. β-actin was used as an internal control. n = 4 individual animals for each group. (D) Protein expression of S100A11 detected by Western blot in mouse liver induced by the HFD. (E) Quantification of Western blot results from panel D. The target protein was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). n = 4 individual animals for each group. (F) mRNA expression of S100A11 in mouse liver detected by qPCR. GAPDH was used as an internal control. n = 4 individual animals for each group. (G) S100A11 expression detected by Western blot in Hepa 1–6 cells induced for 24 hours by 0.2 mmol/L OA. (H) Quantification of Western blot results from panel G. The target protein was normalized to GAPDH. (I) The GSE data showed the expression of liver S100A11 in human NAFLD. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. GSE, series records of Gene Expression Omnibus database.
Table 4.
Significant Up-regulation (P < .05) of S100A11 mRNA in Different Fatty Liver Models
| Species | Disease | Fold change | References |
|---|---|---|---|
| Mice | NAFLD | 2.89 | 42 |
| Mice | NAFLD | 3.98 | 43 |
| Mice | NAFLD | 2.53 | 44 |
| Mice | NAFLD | 2.59 | 45 |
| Baboon | ALD | 1.70 | 46 |
| Human | ALD | 2.70 | 46 |
ALD, alcoholic liver disease.
Hepatic Overexpression of S100A11 Exacerbated Liver Steatosis in Mice Fed an HFHC Diet
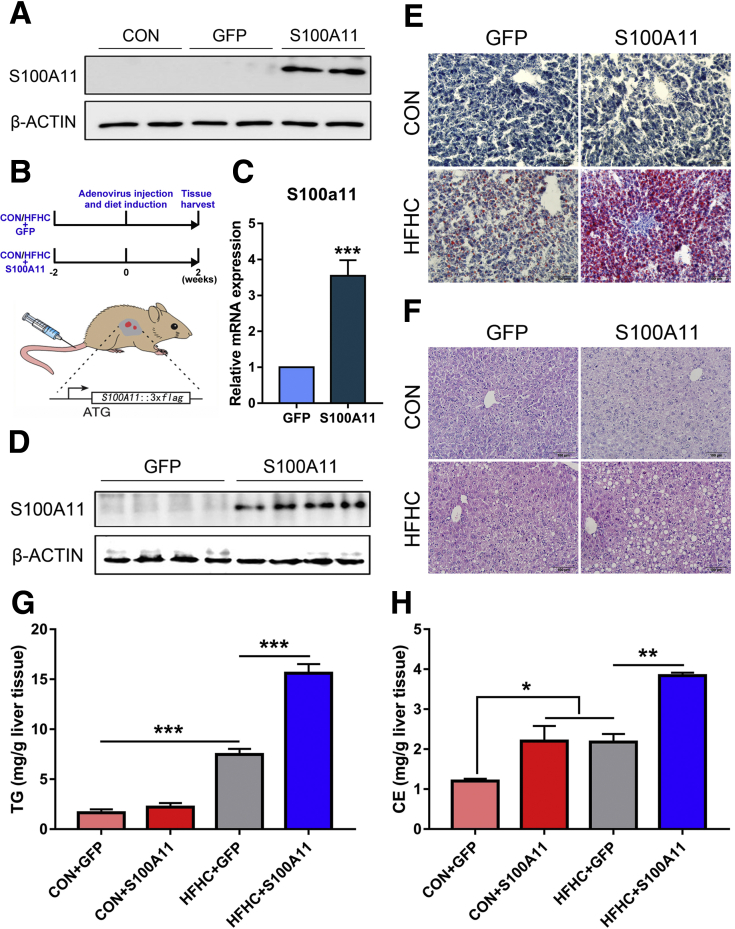
Given the tremendous limitation of research methods and reagents using tree shrews, we opted to use the well-established mouse model to determine whether S100A11 is a positive causal factor in liver steatosis. A mouse S100A11 (S100A11-flag octapeptide-green fluorescent protein [FLAG-GFP]) expression system was generated and subsequently packaged with adenovirus and confirmed via Western blot (Figure 3A). The packaged adenovirus with either S100A11-FLAG-GFP (S100A11) or FLAG-GFP (GFP) then was injected into tail veins of the mice (Figure 3B). The infection efficiency and specificity were tested in vivo (Figure 3C and D). These 2 groups of mice were fed a chow diet (CON) or HFHC diet for 2 weeks before being killed and liver tissue was harvested (Figure 3B).
Figure 3.
Overexpression of S100A11 in the liver exacerbated liver lipid deposition in mice fed the HFHC diet. (A) Infection efficiency of packaged adenovirus was tested by Western blot in vitro. (B) Schematic diagram of the experimental timeline and adenovirus injection into the veins of the mice tails. (C) qPCR and (D) Western blot detection of S100A11 adenovirus expression efficiency in mice liver. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control to normalize the S100a11 gene expression. n = 4 individual animals for each group. (E and F) Representative figures of Oil Red O and H&E staining of mice liver. Original magnification: 20×. Scale bars: 100 μm. (G and H) Quantification of liver TG and CE contents through gas chromatography. n = 3–4 individual animals for each group. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
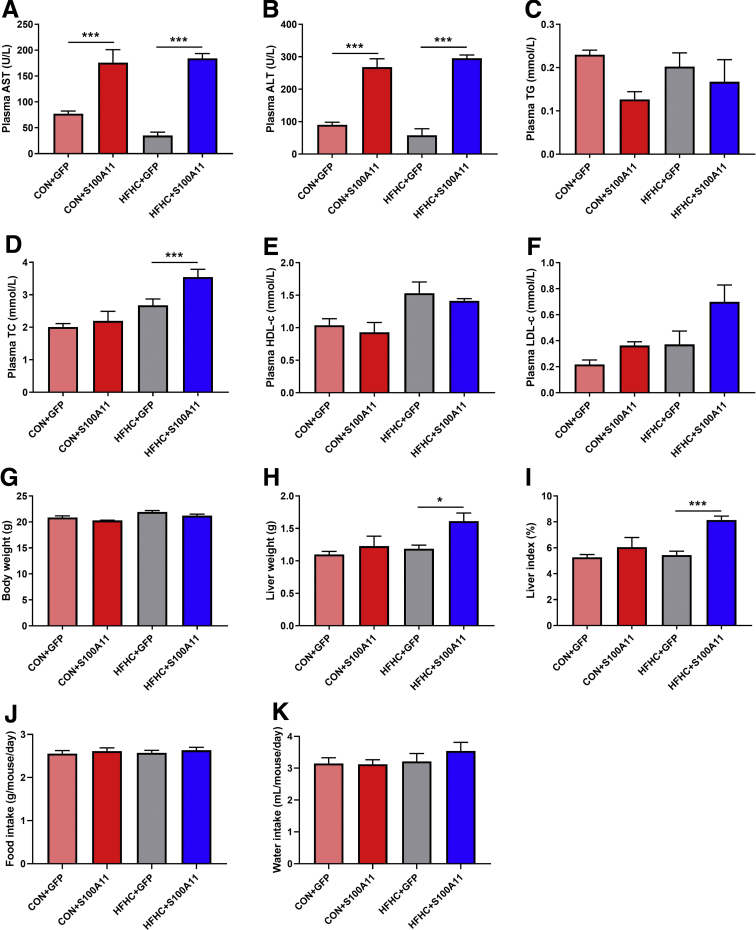
Compared with the GFP control, overexpression of S100A11 significantly increased the plasma levels of aspartate transaminase and alanine transaminase in both the CON and HFHC diet groups (Figure 4A and B), suggesting an injured liver. However, no obvious effects on body weight, food and water intake, or plasma levels of triglycerides (TGs) and low-density lipoprotein c (LDL-c) were observed, except a slightly increased level of plasma total cholesterol in the HFHC group (Figure 4C–K). Although liver histology showed that GFP and S100A11 treatments did not lead to apparent liver lipid accumulation, lipid analysis through gas chromatography showed that the level of liver cholesterol esters (CEs) but not TGs was somewhat increased in the mice fed the CON diet (Figure 3E–H).
Figure 4.
Plasma biochemical indicators, body weight, liver weight, and food and water intake of the mice treated with S100A11 overexpression and CON or HFHC diets. (A) The plasma level of aspartate aminotransferase (AST). (B) The plasma level of alanine aminotransferase (ALT). (C) The plasma level of TGs. (D) The plasma level of total cholesterol (TC). (E) The plasma level of high-density lipoprotein cholesterol (HDL-c). (F) The plasma level of LDL-c. (G) Body weight. (H) Liver weight. (I) Liver index (liver weight/body weight × 100%). (J and K) Food and water intake. n = 4–6 individual animals. ∗P < .05 and ∗∗∗P < .001.
In contrast, the HFHC diet markedly induced liver lipid deposition, and overexpression of S100A11 further exaggerated this effect (Figure 3E and F). Likewise, gas chromatography quantification of TG (Figure 3G) and CE (Figure 3H) further validated the presence of steatosis in mice bearing S100A11 overexpression and fed the HFHC diet, suggesting that S100A11 is indeed a pathogenic factor contributing to liver steatosis.
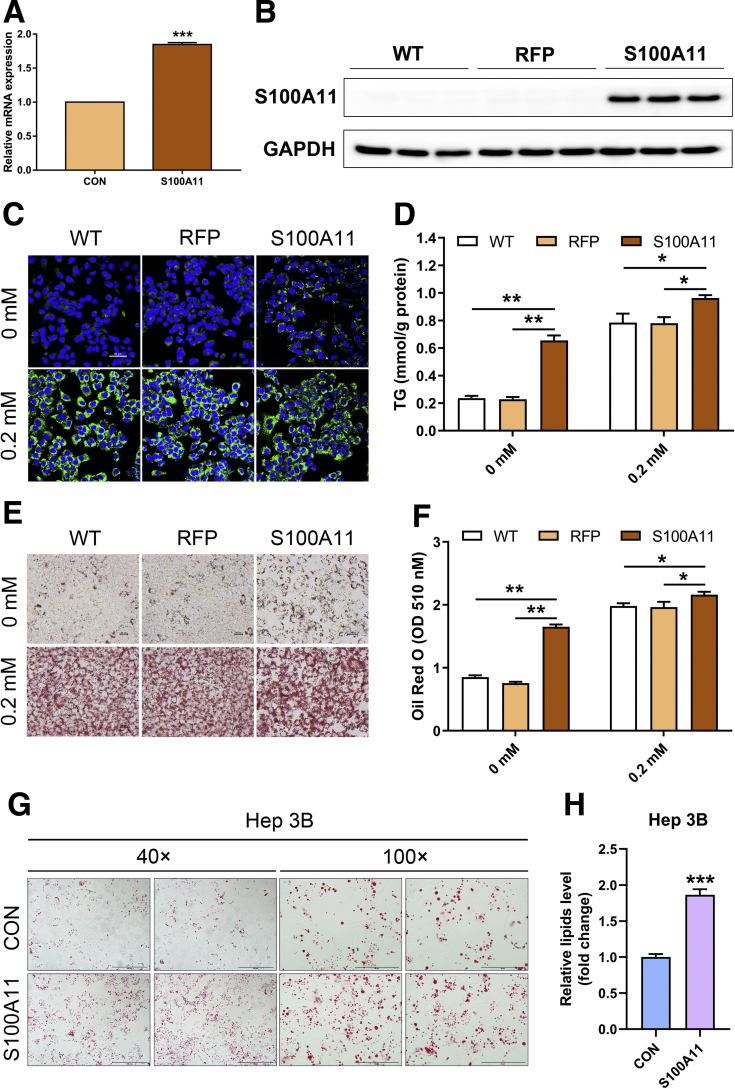
In Vitro Overexpression of S100A11-Induced Lipid Accumulation
To reinforce the hypothesis in vitro, S100A11 was fused with FLAG and red fluorescent protein (RFP) to generate a Hepa 1–6 cell line stably expressing S100A11 (S100A11-FLAG-RFP). Compared with the control (RFP), overexpression of S100A11 increased the expression of S100a11 mRNA (Figure 5A) and S100A11 protein (Figure 5B). Using Bodipy staining to visualize neutral lipids, we found that the S100A11-overexpressing cells accumulated much more lipids than the wild-type cells or RFP cells under both treatments with or without OA (0.2 mmol/L) (Figure 5C). Furthermore, quantification of the TG levels of those cells consistently confirmed the significant consequence of S100A11 overexpression in TG deposits, regardless of OA administration (Figure 5D), as with Oil Red O staining (Figure 5E and F). Similarly, we also established a human Hep 3B cell line with S100A11 stable overexpression and observed similar results as found in the Hepa 1–6 cells (Figure 5G and H). Thus, these results convincingly support that the positive role of S100A11 to enhance hepatic lipid accumulation was mirrored both in vivo and in vitro.
Figure 5.
S100A11 overexpression induced lipid accumulation in Hepa 1–6 and Hep 3B cells. (A and B) qPCR and Western blot detection of the S100A11 expression in Hepa 1–6 stable cell lines by puromycin selection for 2 weeks. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. (C) Bodipy staining of the S100A11 overexpression Hepa 1–6 cells treated with or without 0.2 mmol/L OA for 24 hours. Original magnification: 20×. (D) TG measurement in the S100A11 overexpression Hepa 1–6 cells treated with or without 0.2 mmol/L OA for 24 hours. (E) Oil Red O staining of the S100A11 overexpression Hepa 1–6 cells treated with or without 0.2 mmol/L OA for 24 hours. Original magnification: 20×. (F) Quantification of Oil Red O staining from panel E. (G) Oil Red O staining of the S100A11 overexpression Hep 3B cells treated with 0.1 mmol/L OA for 12 hours. Original magnification: 40× (left two panels) and 100× (right two panels). (H) Quantification of Oil Red O staining from panel G. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
Overexpression of S100A11 Induced Lipid Biosynthesis In Vivo and In Vitro
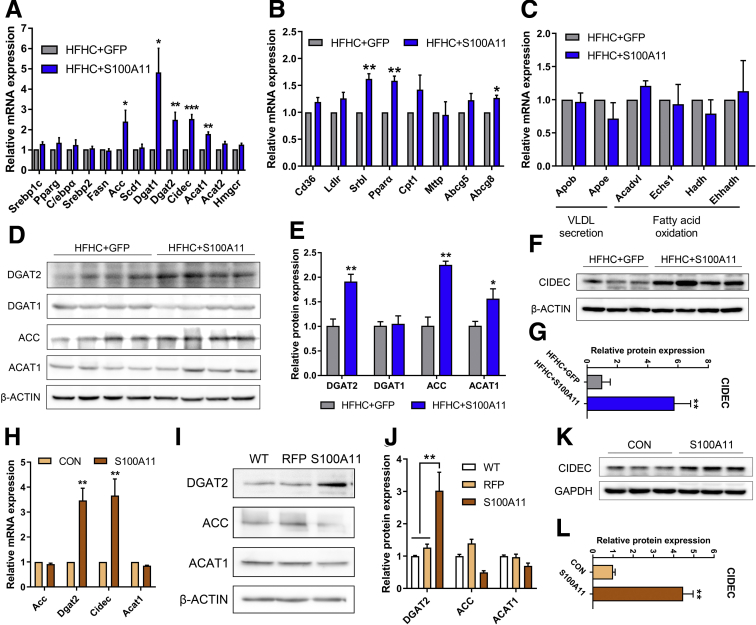
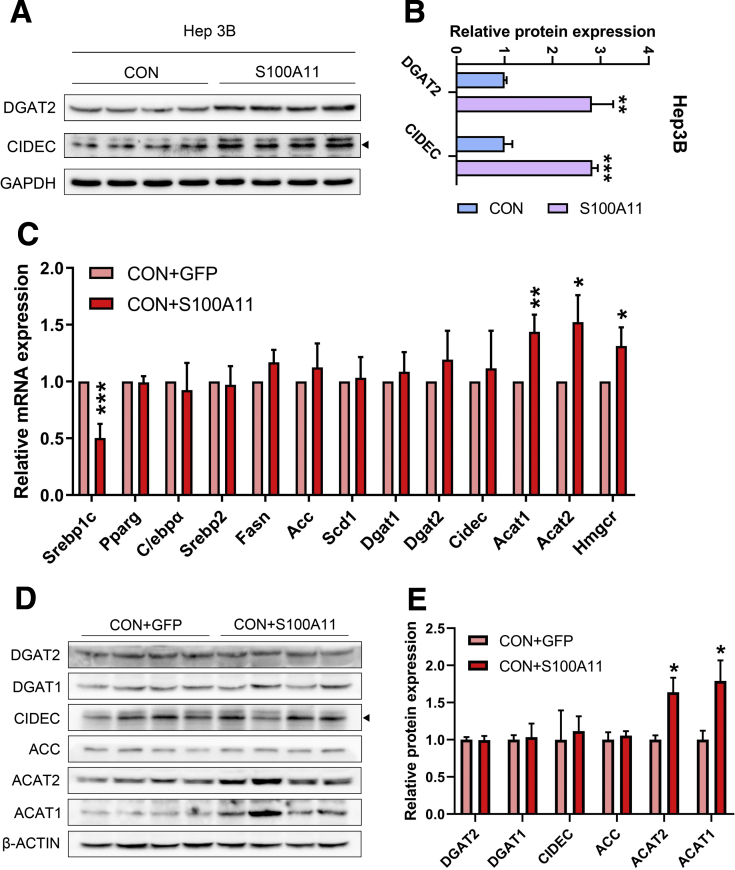
Lipid accumulation often is derived from an imbalance of lipid biosynthesis, lipolysis, uptake, and efflux. Because overexpression of S100A11 promotes hepatic lipid accumulation in vivo and in vitro, the expression of an array of well-known genes involved in the biosynthesis of TG and CE, such as acetyl-CoA carboxylase (Acc), diglyceride acyltransferase 2 (Dgat2), and acetyl-CoA acetyltransferase 1 (Acat1), was examined. CIDEC plays a critical role in lipid droplet growth.47 The qPCR results revealed that the mRNA levels of acetyl-CoA carboxylase, Dgat2, Cidec, and acetyl-CoA acetyltransferase 1 (Figure 6A), as well as Srb1, Ppara, and Abcg8 (Figure 6B), were up-regulated significantly in the livers of the HFHC-fed, S100A11-overexpressing mice compared with those of the control group. In contrast, the transcriptional expression of other genes involved in fatty acid oxidation and very-low-density lipoprotein secretion showed no significant changes in the livers of the HFHC-fed, S100A11-overexpressing mice (Figure 6C). The protein levels of DGAT2, ACC, ACAT1, and CIDEC also consistently were increased in the S100A11-overexpressing mice (Figure 6D–G). Likewise, both the mRNA and protein levels of DGAT2 and CIDEC were up-regulated significantly in the S100A11-overexpressing Hepa 1–6 cells (Figure 6H–L) and Hep 3B cells (Figure 7A and B). Interestingly, the transcriptional and translational expression of genes involved in CE synthesis but not TG synthesis were increased significantly in the livers of the S100A11-overexpression mice group compared with that of the GFP mice group fed the CON diet (Figure 7C–E), which might account for the increased CE level but not the TG level in their livers (Figure 3G and H). Taken together, these results imply that the overexpression of S100A11 promotes lipid accumulation, possibly through the up-regulation of lipogenic genes Dgat2 and Cidec in vitro and in vivo.
Figure 6.
In vivo and in vitro S100A11 overexpression increased the expression of neutral lipid synthesis genes and LD fusion factors. (A–C) mRNA level of genes involved in lipid synthesis, LD fusion, VLDL secretion, and fatty acid oxidation in mice liver detected by qPCR. β-actin was used as an internal control. n = 4 individual animals for each group. (D and E) Protein level and quantification in mice liver by Western blot. The target proteins were normalized to β-actin. n = 4 individual animals for each group. (F and G) Protein level and quantification of CIDEC in mice liver by Western blot. The CIDEC protein was normalized to β-actin. n = 3–4 individual animals for each group. (H) mRNA level of genes in Hepa 1–6 cells detected by qPCR. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. (I and J) Protein level and quantification in the Hepa 1–6 cells detected by Western blot. The target proteins were normalized to β-actin. (K and L) Protein level and quantification of the CIDEC in the Hepa 1–6 cells detected by Western blot. The CIDEC protein was normalized to GAPDH. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. VLDL, very-low-density lipoprotein.
Figure 7.
In vitro Western blot detection of proteins related to lipogenesis in S100A11 overexpression Hep 3B cells and in vivo qPCR and Western blot detection of lipogenic genes and proteins in the liver of the CON + GFP and CON + S100A11 mice groups. (A) Protein level of the lipogenic proteins in the S100A11 stable overexpression Hep 3B cells treated with 0.1 mmol/L OA for 12 hours. (B) Quantification of the related proteins from panel A. The target proteins were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (C) mRNA expression of lipogenic genes detected by qPCR. n = 4 individual animals. β-actin was used as an internal control. (D and E) Western blot detection and quantification of related proteins. The target proteins were normalized to β-actin. n = 4 individual animals. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
Overexpression of S100A11 Activated Autophagy Process via FOXO1
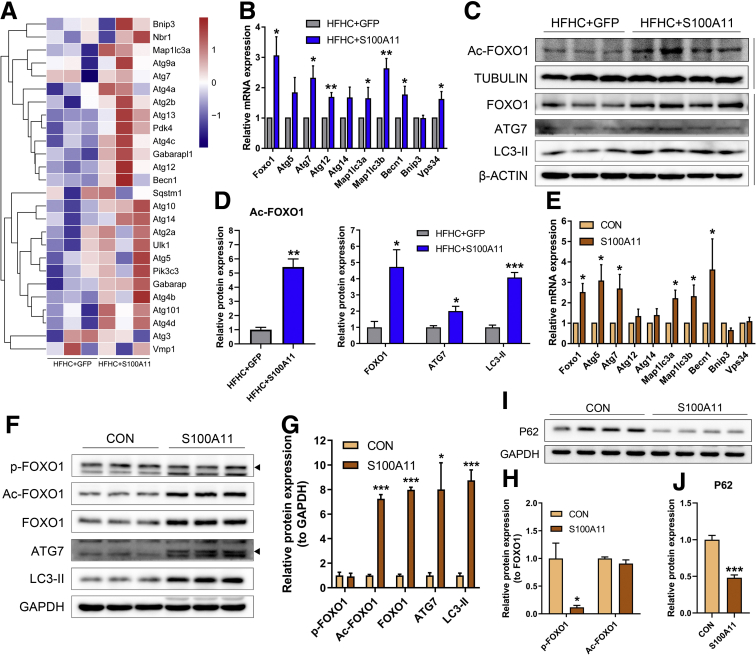
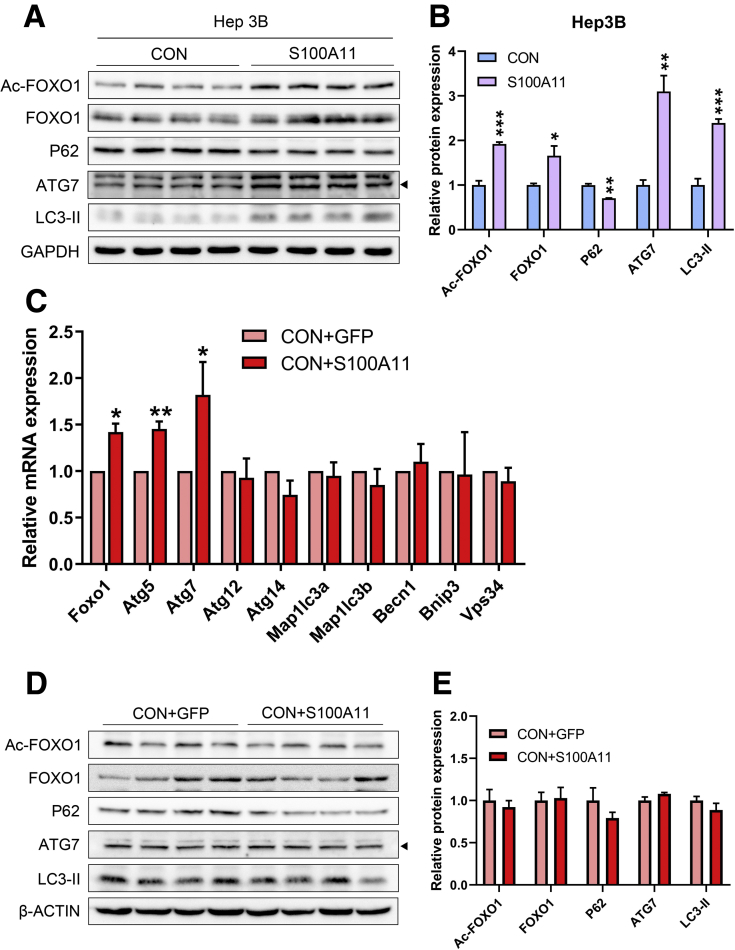
To explore the underlying mechanisms of S100A11 in lipid accumulation via activation of lipogenesis in both mouse liver and Hepa 1–6 cells, liver RNA sequencing initially was performed between the HFHC + GFP and HFHC + S100A11 mouse groups. Interestingly, RNA sequencing data analysis showed that the majority of genes involved in autophagy showed a tendency of up-regulation in the HFHC + S100A11 group compared with the HFHC + GFP group (Figure 8A). Previous studies showed that activation of autophagy could increase liver lipid accumulation.16, 17, 18, 19 We found that the mRNA expression of several autophagy marker genes including Atg7, Becn1, and Map1lc3a/b significantly increased in both the liver of the HFHC-fed mice (Figure 8B) and Hepa 1–6 cells (Figure 8E) under the condition of S100A11 overexpression. The protein levels of autophagy related 7 (ATG7) and LC3-phosphatidylethanolamine conjugate (LC3-II), 2 widely used autophagy markers, were consistently and notably up-regulated under S100A11 overexpression both in vivo (Figure 8C and D) and in vitro (Figure 8F and G). Another autophagic protein, the widely used P62,48 was decreased significantly in the S100A11 overexpression Hepa 1–6 cells (Figure 8I and J) and Hep 3B cells (Figure 9A and B), indicating an activated autophagic status. Similarly, the mRNA levels of some of the autophagic genes, such as Atg5 and Atg7, were slightly but significantly increased in the CON + S100A11 group compared with the CON + GFP group (Figure 9C–E).
Figure 8.
Overexpression of S100A11 promoted FOXO1 and autophagy marker expression in vivo and in vitro. (A) Heatmap generated by RNA sequencing data indicated that autophagy was up-regulated in the mice liver treated by S100A11 overexpression and the HFHC diet for 2 weeks. n = 3 individual animals for each group. (B) mRNA expression of FOXO1 and autophagy marker genes in vivo detected by qPCR. β-actin was used as an internal control. n = 4 individual animals for each group. (C and D) Western blot and quantification of Ac-FOXO1, FOXO1, ATG7, and LC3-II in the liver of the hepatic S100A11 overexpression and HFHC diet–treated mice. The target proteins were normalized to tubulin or β-actin as indicated. n = 3–4 individual animals. (E) mRNA expression of FOXO1 and autophagy marker genes in vitro detected by qPCR. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. (F) Western blot detection of phosphorylated, acetylated, and unphosphorylated FOXO1, ATG7, and LC3-II in the S100A11 overexpression Hepa 1–6 cells. (G) Quantification of Western blot results from panel F. The target proteins were normalized to GAPDH. (H) Quantification of Western blot results from panel F. p-FOXO1 and Ac-FOXO1 were normalized to FOXO1. (I and J) Protein level and quantification of P62 in the S100A11 overexpression Hepa 1–6 cells detected by Western blot. The target protein was normalized to GAPDH. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. p-FOXO1, phosphorylated forkhead box O1.
Figure 9.
In vitro Western blot detection of proteins related to autophagy in S100A11 overexpression Hep 3B cells and in vivo qPCR and Western blot detection of autophagic genes and proteins in the liver of the CON + GFP and CON + S100A11 mice groups. (A) Protein level of the autophagic proteins in the S100A11 stable overexpression Hep 3B cells treated with 0.1 mmol/L OA for 12 hours. (B) Quantification of the related proteins from panel A. The target proteins were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (C) mRNA expression of autophagic genes detected by qPCR. n = 4 individual animals. β-actin was used as an internal control. (D and E) Western blot detection and quantification of related proteins. The target proteins were normalized to β-actin. n = 4 individual animals. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
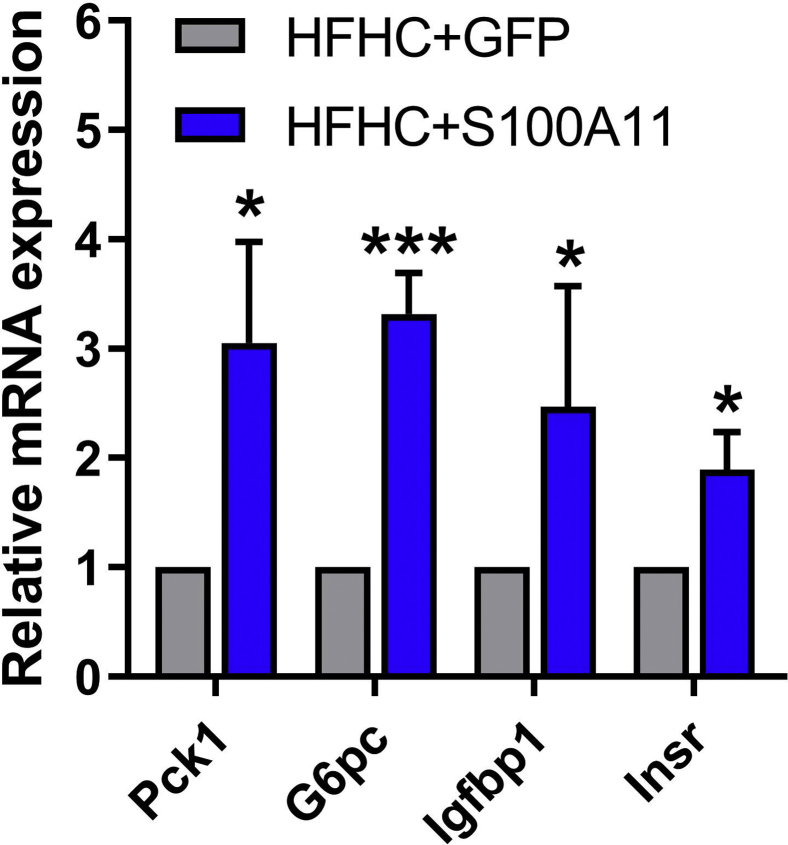
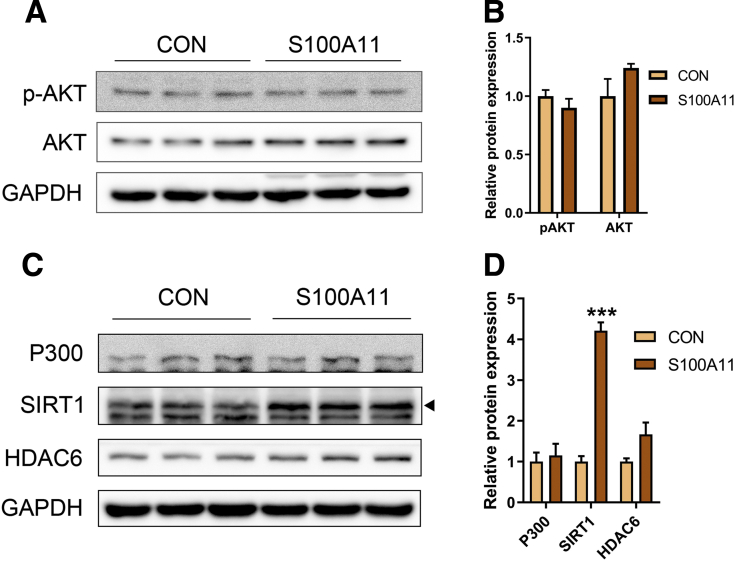
The transcription factor FOXO1 is a direct mediator of autophagy.21 FOXO1 can be phosphorylated by activated Protein Kinase B to promote its degradation in proteasome49 or be acetylated by P300.50 Moreover, it recently was reported that FOXO1 regulates CIDEC under autophagy conditions.30 In this study, the mRNA level of FOXO1 as well as the nonphosphorylated (but not the phosphorylated) level of FOXO1 protein increased significantly in both the Hepa 1–6 cells and liver of the HFHC-fed mice with S100A11 overexpression (Figure 8B–G). To find evidence of the activation of FOXO1, we further assessed the changes in FOXO1-targeted genes, such as Pck1 and G6pc,51 Igfbp1,52 and Insr,53 in the HFHC-induced S100A11 overexpression mouse livers and found that the expression of these genes increased significantly (Figure 10), showing the activation of FOXO1. High levels of acetylated FOXO1 (Ac-FOXO1) protein also were detected in the HFHC + S100A11 group (Figure 8C and D) and S100A11-overexpressing Hepa 1–6 cells (Figure 8F–H), as well as in Hep 3B cells (Figure 9A and B). However, overexpression of S100A11 affected neither the phosphorylated Protein Kinase B protein level (Figure 11A and B) nor the P300 protein level (Figure 11C and D). This suggests that S100A11 overexpression could up-regulate autophagy through activated FOXO1 expression and acetylation by factors other than P300.
Figure 10.
qPCR analysis of target genes of FOXO1. mRNA expression of FOXO1 target genes in the S100A11 overexpression mouse livers treated by the HFHC diet. n = 4 individual animals. β-actin was used as an internal control. ∗P < .05 and ∗∗∗P < .001.
Figure 11.
Western blot analysis of phosphorylation and acetylation modification-related protein expression in the S100A11 overexpression Hepa 1–6 cells. (A) Western blot results of phosphorylated protein kinase B and AKT. (B) Quantification of Western blot results from panel A. The target proteins were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (C) Western blot results of P300, SIRT1, and HDAC6 in the S100A11 overexpression Hepa 1–6 cells. (D) Quantification of Western blot results from panel C. The target proteins were normalized to GAPDH. ∗∗∗P < .001.
Inhibition of Histone Deacetylase 6 Increased Lipid Accumulation Through Increased Expression and Acetylation of FOXO1 in Hepatocytes
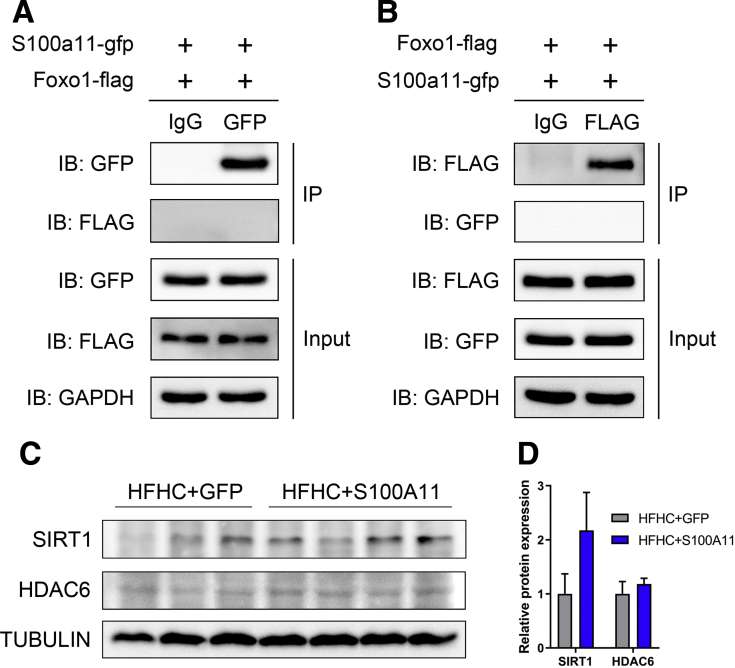
To analyze whether S100A11 could interact directly with FOXO1 to facilitate the acetylation of FOXO1, we constructed recombinant plasmids of S100a11-gfp and Foxo1-flag and co-expressed them into 293T cells. However, the co-immunoprecipitation (co-IP) experiment did not show a direct interaction between S100A11 and FOXO1 (Figure 12A and B). Therefore, other factors mediating S100A11 and FOXO1 were hunted by an IP experiment using S100A11-FLAG-RFP as a bait protein in stable S100A11 overexpression Hepa 1–6 cells, and proteins that interacted with S100A11 were detected by mass spectrometry (MS). Among those proteins (the entire list of proteins is available by request), 2 deacetylases, Sirtuin 1 and histone deacetylase 6 (HDAC6), were identified.
Figure 12.
Co-IP assays of S100A11 and FOXO1 proteins in 293T cells and Western blot analysis of acetylation modification-related protein expression in mice liver. The indicated recombinant plasmids were co-expressed in 293T cells for 48 hours and then the cell lysates were analyzed by Co-IP assays. (A) S100A11 did not interact with FOXO1. (B) FOXO1 did not interact with S100A11. (C) Western blot results of SIRT1 and HDAC6 in the HFHC-fed S100A11 overexpression mice liver. (D) Quantification of Western blot results from panel C. The target proteins were normalized to tubulin. n = 3–4 individual animals. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
In further examination, the protein level of SIRT1 increased in the S100A11 overexpression Hepa 1–6 cells (Figure 11C and D) and in the liver of the HFHC-fed mice with S100A11 overexpression (Figure 12C and D). A previous study showed that SIRT1 could bind directly to FOXO1 and mediate the deacetylation of FOXO1.20 To validate its responsibility in FOXO1 deacetylation, SIRT1 activity in the wild-type Hepa 1–6 cells was inhibited by treatment with a specific inhibitor (EX-527, 10 μmol/L). However, the lipid content of the cells and acetylation of FOXO1 did not change (Figures 13A–C and 14A and B), suggesting that SIRT1 may not be the factor mediating acetylation of FOXO1 in this study.
Figure 13.
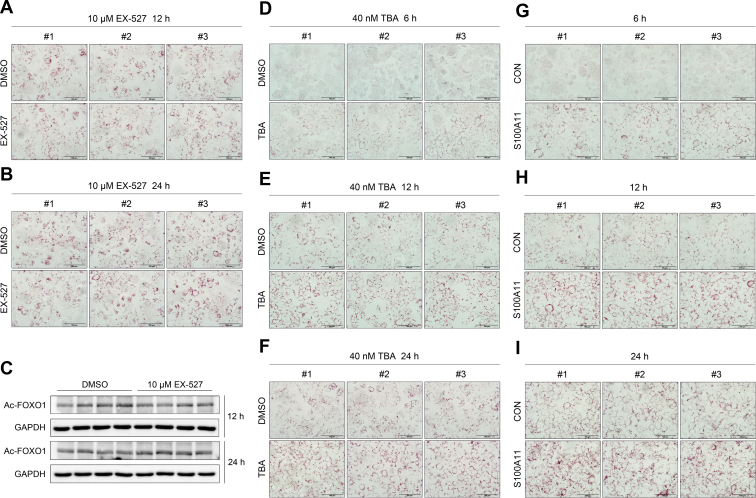
Oil Red O staining of wild-type and S100A11 overexpression Hepa 1–6 cells, and Western blot analysis of wild-type Hepa 1–6 cells treated with EX-527. (A) Results of wild-type Hepa 1–6 cells treated with 0.1 mmol/L OA and 10 μmol/L EX-527 for 12 hours. (B) Results of wild-type Hepa 1–6 cells treated with 0.1 mmol/L OA and 10 μmol/L EX-527 for 24 hours. Original magnification: 40×. Scale bars: 100 μm. (C) Western blot detection of Ac-FOXO1 in wild-type Hepa 1–6 cells treated with 0.1 mmol/L OA and 10 μmol/L EX-527 for 12 and 24 hours. (D-F) Results of wild-type Hepa 1–6 cells treated with 0.1 mmol/L OA and 40 nmol/L TBA for 6, 12, and 24 hours, respectively. Original magnification: 40×. Scale bars: 100 μm. (G–I) Results of S100A11 overexpression Hepa 1–6 cells treated with 0.1 mmol/L OA for 6, 12, and 24 hours, respectively. Original magnification: 40×. Scale bars: 100 μm. DMSO, dimethyl sulfoxide; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
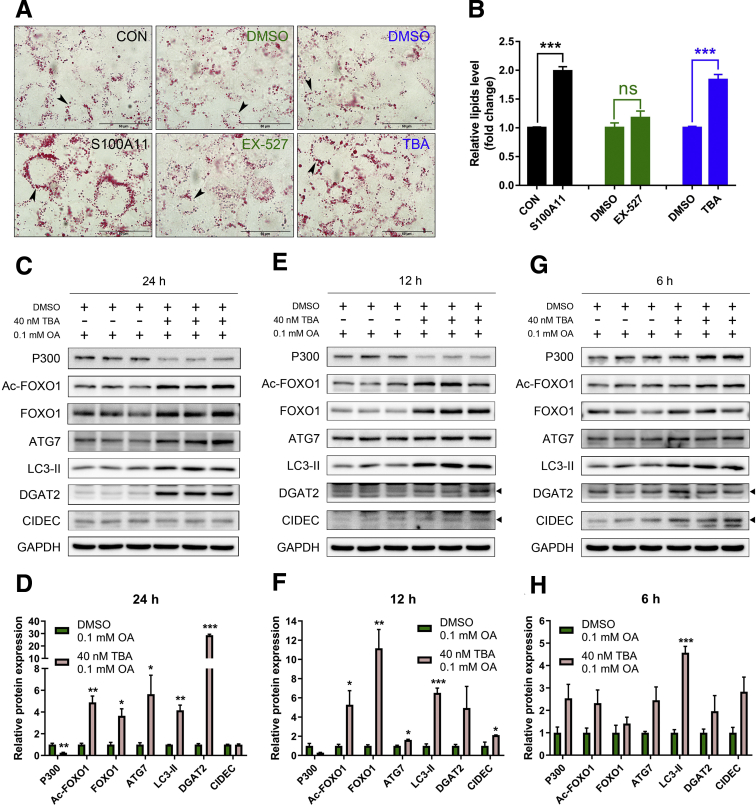
Similarly, a specific inhibitor (tubastatin A [TBA], 40 nmol/L) of HDAC6 was used to treat the wild-type Hepa 1–6 cells and showed significant lipid accumulation in a time-dependent manner (Figures 13D–F and 14A and B). This mimicked the phenomena induced by S100A11 overexpression (Figures 13G–I and 14A and B). Subsequently, after treatment with 40 nmol/L TBA for 24 hours, the protein levels of FOXO1 and Ac-FOXO1 increased significantly (Figure 14C and D), as did ATG7 and LC3-II (Figure 14C and D). The lipogenic protein DGAT2 also consistently showed a markedly high expression in the 40 nmol/L TBA-treated group (Figure 14C and D). Likewise, the expression levels of the proteins treated with TBA for 6 and 12 hours were similar but to a lesser extent (Figure 14E–H). Collectively, these data indicated that both HDAC6 inhibition and S100A11 overexpression in hepatocytes showed similar phenotypes, suggesting that HDAC6 and S100A11 may function in the same pathway.
Figure 14.
Overexpression of S100A11 or inhibition of HDAC6 increased lipid accumulation in hepatocytes. (A) Oil Red O staining of stable S100A11 overexpression Hepa 1–6 cells treated with 0.1 mmol/L OA (left panel), wild-type Hepa 1–6 cells treated with 10 μmol/L EX-527 and 0.1 mmol/L OA (middle panel), and wild-type Hepa 1–6 cells treated with 40 nmol/L TBA and 0.1 mmol/L OA (right panel). All cells were treated for 24 hours. Original magnification: 100×. Scale bars: 50 μm. (B) Quantification of the results from panel A by ImageJ. Western blot detection and quantification of P300, Ac-FOXO1, FOXO1, ATG7, LC3-II, DGAT2, and CIDEC in the wild-type Hepa 1–6 cells treated with 0.1 mmol/L OA and 40 nmol/L TBA for (C and D) 24 hours, (E and F) 12 hours, and (G and H) 6 hours. The target proteins were normalized to GAPDH. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. DMSO, dimethyl sulfoxide; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
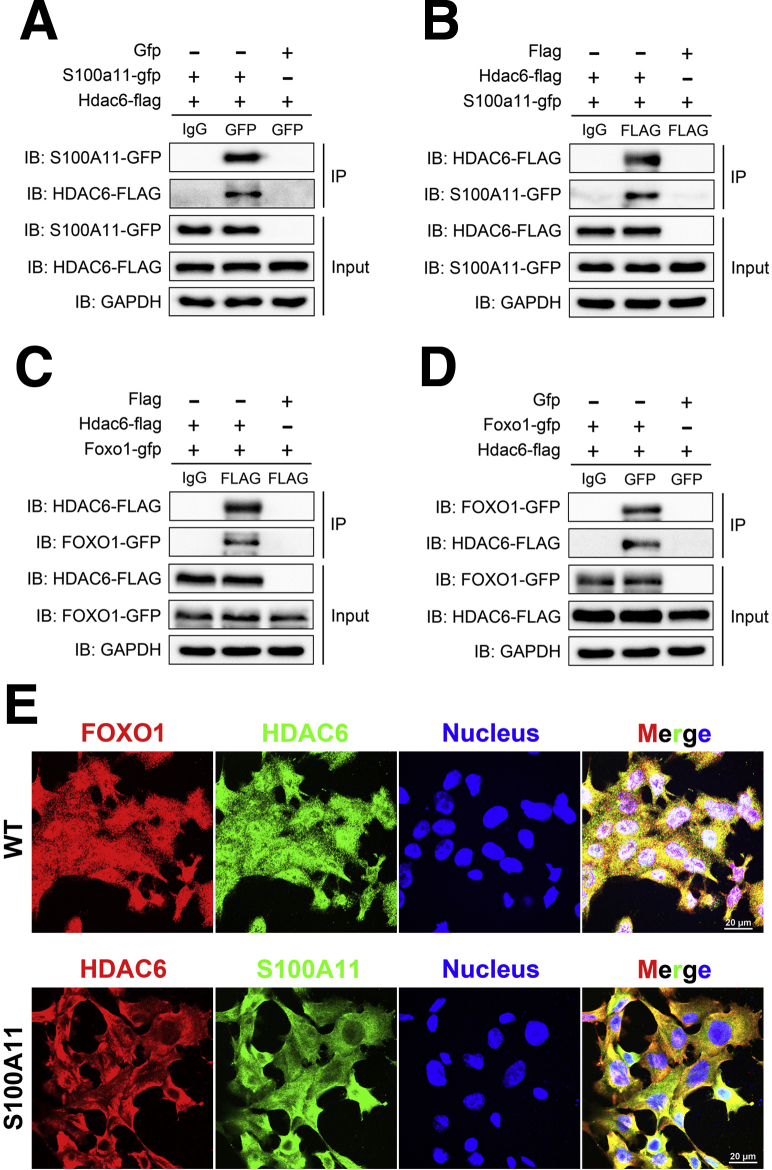
According to these results, we hypothesized that HDAC6 may be a deacetylase of FOXO1. Because our co-IP results showed a direct physical interaction between S100A11 and HDAC6, we wondered whether such an interaction also happens between HDAC6 and FOXO1. We co-expressed the recombinant plasmid of S100a11-gfp and Hdac6-flag, and Hdac6-flag and Foxo1-gfp in 293T cells, respectively. The co-IP assay showed that S100A11 and HDAC6 could be precipitated by each other (Figure 15A and B), as could HDAC6 and FOXO1 (Figure 15C and D). Moreover, co-staining of FOXO1 and HDAC6 in the wild-type Hepa 1–6 cells (Figure 15E, upper panel) and co-staining of S100A11 and HDAC6 in the S100A11 overexpression Hepa 1–6 cells (Figure 15E, lower panel) produced their strong co-localization, further supporting a direct interaction between HDAC6 and FOXO1 and/or S100A11. Collectively, this evidence consistently showed an S100A11–HDAC6–FOXO1 axis coordinately regulating autophagy and lipid metabolism in liver cells.
Figure 15.
Co-IP and immunofluorescence analysis. (A and B) Co-IP assays of S100A11 and HDAC6 interactions as well as (C and D) HDAC6 and FOXO1 interactions in 293T cells. In all of the co-IP experiments, the cells were transfected with the indicated plasmids for 48 hours and the cell lysates were immunoprecipitated and detected with the indicated antibodies. (E) Immunofluorescent staining of FOXO1 and HDAC6 in the wild-type Hepa 1–6 cells (upper panel) and HDAC6 and S100A11 in the S100A11 overexpression Hepa 1–6 cells (lower panel). Original magnification: 100×. Scale bars: 20 μm. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IB, immunoblotting; WT, wild-type.
Inhibition of FOXO1 and Autophagy or Deletion of Dgat2 Alleviated S100A11-Induced Lipid Accumulation
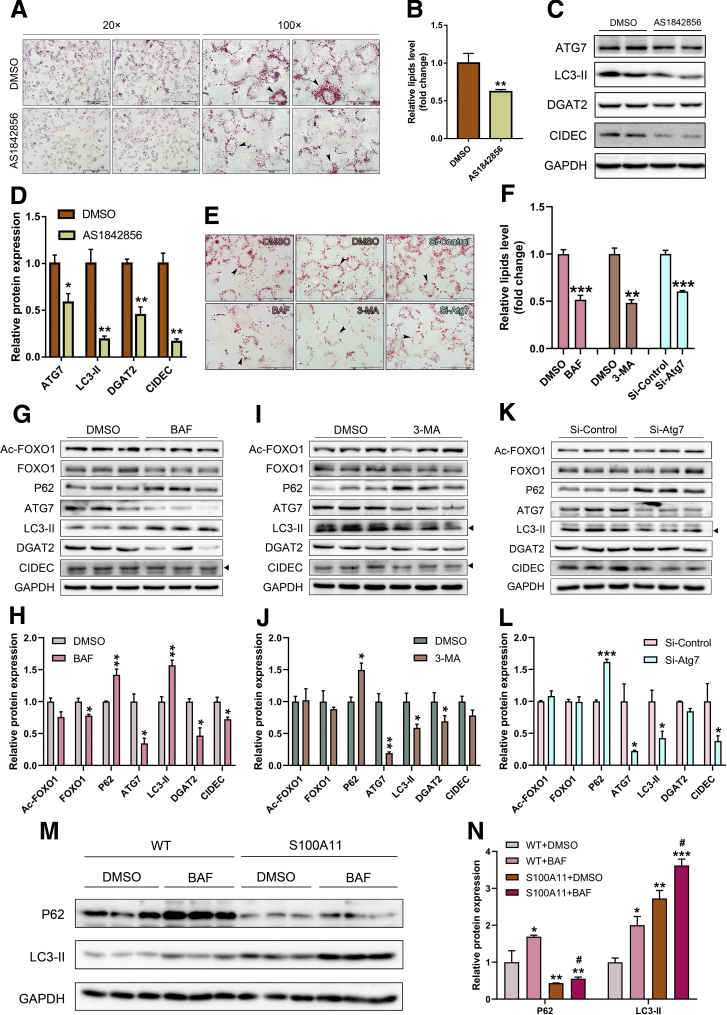
Because overexpression of S100A11 activated autophagy and FOXO1 expression, we hypothesized that FOXO1-mediated autophagy might promote Dgat2 and CIDEC expression and eventually lead to lipid accumulation. The S100A11-overexpressing Hepa 1–6 cells were treated with AS1842856, a specific inhibitor of FOXO1 and autophagy.30 Oil Red O staining results showed that AS1842856 treatment led to a dramatically attenuated lipid accumulation (Figure 16A and B). The protein levels of autophagy markers ATG7 and LC3-II as well as lipogenic genes DGAT2 and CIDEC significantly decreased accordingly (Figure 16C and D), suggesting that FOXO1 is required/necessary for S100A11-activated autophagy and lipogenesis. To further confirm the roles of autophagy in this study, more specific autophagy inhibitors bafilomycin A1 and 3-methyladenine and small interfering RNA of Atg754 were used to treat the S100A11 overexpression of Hepa 1–6 cells. We found that the lipid accumulation in the S100A11 overexpression Hepa 1–6 cells decreased significantly (Figure 16E and F). The autophagy process and lipogenesis also were down-regulated significantly (Figure 16G–N). Taken together, these results consistently showed that activated autophagy is required for S100A11 overexpression–induced lipid accumulation.
Figure 16.
Inhibition of FOXO1 and autophagy decreased lipid content and the level of proteins related to the autophagy process in the S100A11 overexpression Hepa 1–6 cells. (A) Oil Red O staining of the S100A11 overexpression Hepa 1–6 cells after treatment with FOXO1 inhibitor (0.25 μmol/L AS1842856) for 6 hours. Left four images: Original magnification, 20×; scale bars: 200 μm. Right four images: Original magnification: 100×; scale bars: 50 μm. (B) Quantification of the results from panel A by ImageJ. (C and D) Results of Western blot detection and the quantification of related proteins in the Hepa 1–6 cells after treatment with AS1842856 (0.25 μmol/L) for 6 hours. The target proteins were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (E) Oil Red O staining of stable S100A11 overexpression Hepa 1–6 cells treated with 10 nmol/L bafilomycin A1 (BAF) (left panel), 2 mmol/L 3-methyladenine (3-MA) (middle panel), and RNA interference of Atg7 (right panel). Original magnification: 100×; scale bars: 50 μm. (F) Quantification of the results from panel E by ImageJ. Western blot detection and quantification of related proteins in the S100A11 overexpression Hepa 1–6 cells treated with (G and H) 10 nmol/L BAF, (I and J) 2 mmol/L 3-MA, and (K and L) RNA interference of Atg7, respectively. The target proteins were normalized to GAPDH. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. For the autophagy-inhibitor treatment, all cells were cultured with a medium containing 0.1 mmol/L OA and the related inhibitor for 12 hours. For the RNA interference of Atg7, the cells were first treated with a small interfering (si) RNA of Atg7 for 48 hours and then incubated with 0.1 mmol/L OA for 12 hours. (M) Autophagy flux data. All cells were treated with dimethyl sulfoxide (DMSO) or 10 nmol/L BAF for 12 hours. (N) Quantification of the related proteins in panel M. The target proteins were normalized to GAPDH. Significant difference between the control (WT + DMSO) and a specific treatment (WT + BAF, S100A11 + DMSO, and S100A11 + BAF): ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. Significant difference between the S100A11 + DMSO and S100A11 + BAF: #P < .05.
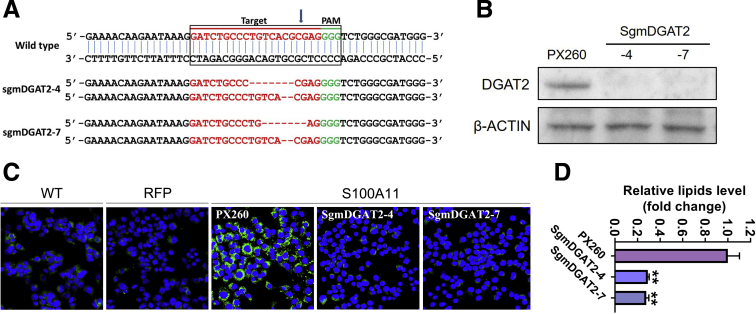
Inversely, knocking out the Dgat2 gene in the S100A11 overexpressing Hepa 1–6 cells (Figure 17A and B) by clustered regularly interspaced short palindromic repeats/cas9 technology decreased lipid accumulation as indicated by Bodipy staining (Figure 17C and D), showing that the aberrant hepatic lipid deposition caused by S100A11 overexpression could be reversed efficiently by Dgat2 deficiency.
Figure 17.
Deletion of Dgat2 decreased lipid content in the S100A11 overexpression Hepa 1–6 cells. (A) Sequencing of Dgat2 knockout using the CRISPR/Cas9 system. Dgat2 was amplified by genomic PCR and then sequenced to verify the changes in gene sequences. (B) Western blot detection of Dgat2 deletion in the S100A11 overexpression Hepa 1–6 cells using the CRISPR/Cas9 method. (C and D) Bodipy staining and quantification by ImageJ of the Hepa 1–6 cells with S100A11 overexpression and Dgat2 deletion. WT, wild-type. ∗∗P < .01.
Discussion
NAFLD is a severe liver disease, and numerous factors contribute to its development. Although some studies have reported that S100A11 is an important factor in diseases such as cancer35,36 and rheumatoid arthritis,55 its role in lipid metabolism has not yet been addressed. Several lines of evidence, including ours, convincingly support that S100A11 is a pathogenic factor of NAFLD. First, our liver proteomics of the tree shrew model of NAFLD as well as a time-course microarray analysis of mice liver steatosis56 consistently showed that S100A11 was up-regulated significantly in a time-dependent manner. Second, the up-regulation of S100A11 was confirmed further in the OA-treated mice Hepa 1–6 cells, human Hep 3B cells, liver of the HF-induced NAFLD mouse model, and especially the human liver transcriptome data of NAFLD (Figure 2 and Table 4), as well as baboon and human alcoholic fatty liver disease (Table 4). Third, overexpression of S100A11 indeed promoted lipid accumulation in the Hepa 1–6 cells and liver of the HFHC-treated mice. Thus, up-regulation of S100A11 definitely leads to liver steatosis, showing an uncharacterized role of S100A11 in NAFLD.
Autophagy has been linked to NAFLD because it can reduce intracellular LD.8, 9, 10, 11 Increased lipid accumulation has been observed in autophagy-deficient hepatocytes and Atg5 knocked-out mouse embryonic fibroblasts). Hence, autophagy can protect the liver from steatosis.12 However, we found that activated autophagy by the in vivo and in vitro overexpression of S100A11 actually promoted lipid accumulation in both cell and animal models. In addition, inhibition of autophagy could relieve S100A11 overexpression–induced lipid metabolism. Therefore, this study showed that S100A11 acts as a regulator of both autophagy and lipid metabolism. Other research also showed that starvation-induced autophagy significantly increased the abundance of LD in mouse embryonic fibroblasts, which then could be blocked by the deletion of the critical autophagy gene Atg5.15 Similarly, mice lacking another core gene of autophagy fak-family Interacting protein of 200 kDa in hepatocytes were protected from starvation-induced or high-fat diet (HFD)-induced lipid deposition in the liver.18 In addition, mice lacking Atg7 in the liver showed a marked decrease in hepatic lipids and were protected from HFD-induced obesity and insulin resistance.16 Moreover, knockdown of Atg5 or Atg7 in 3T3-L1 preadipocytes lowered lipid content,57 and knockout of Atg7 in adipose tissue also protected the mice from obesity and insulin resistance caused by HFD.57,58 Collectively, these studies, including ours, indicated that under certain circumstances, activated autophagy probably accelerated metabolic diseases such as NAFLD, suggesting that its beneficial or detrimental role may be context-dependent.
It has been widely reported that FOXO1 plays a crucial role in the autophagic process.21, 22, 23, 24, 25,59 In this study, we showed that overexpression of S100A11 significantly increased the expression levels of FOXO1 as well as autophagy markers ATG7 and LC3-II in vivo and in vitro and also activated the expression of CIDEC and DGAT2 to promote hepatic lipid accumulation. In support of that, a recent report also showed that FOXO1 regulates adipogenesis and LD growth via a FOXO1–autophagy–CIDEC axis in adipocytes.30 Furthermore, inhibition of FOXO1 reduced not only the expression of autophagy markers ATG7 and LC3-II, as well as lipogenic proteins CIDEC and DGAT2, but also hepatic lipid accumulation. Therefore, our study consistently showed that FOXO1 plays synergic roles in coordinately regulating autophagy and lipid metabolism. More importantly, we showed that S100A11 is an upstream regulator of FOXO1, and S100A11 overexpression induces lipid deposition in a FOXO1 and autophagy-dependent manner.
Although overexpression of S100A11 could stimulate the accumulation of Ac-FOXO1, the co-IP results failed to show a direct interaction between S100A11 and FOXO1 (Figure 12A and B). Furthermore, our IP-MS results showed that deacetylase HDAC6 could interact physically with S100A11. More importantly, inhibition of HDAC6 in the Hepa 1–6 cells showed similar results to S100A11 overexpression, including the accumulation of lipids, up-regulation of FOXO1 and Ac-FOXO1, activation of autophagy, and increased expression of lipogenic pathways (Figures 13 and 14). Coincidentally, a recent study also showed that HDAC6 can bind and deacetylate FOXO1 in T helper 17 cells.60 Hence, HDAC6 may act as a deacetylase to mediate the deacetylation of FOXO1. Our co-IP assay and immunofluorescence results showed the physical interactions not only between S100A11 and HDAC6, but also between HDAC6 and FOXO1 (Figure 15). Therefore, we speculate that S100A11 may competitively bind with HDAC6 to modulate the accumulation of Ac-FOXO1, uncovering that S100A11 acts as an upstream regulator of FOXO1 via HDAC6 in regulating autophagy and lipogenesis.
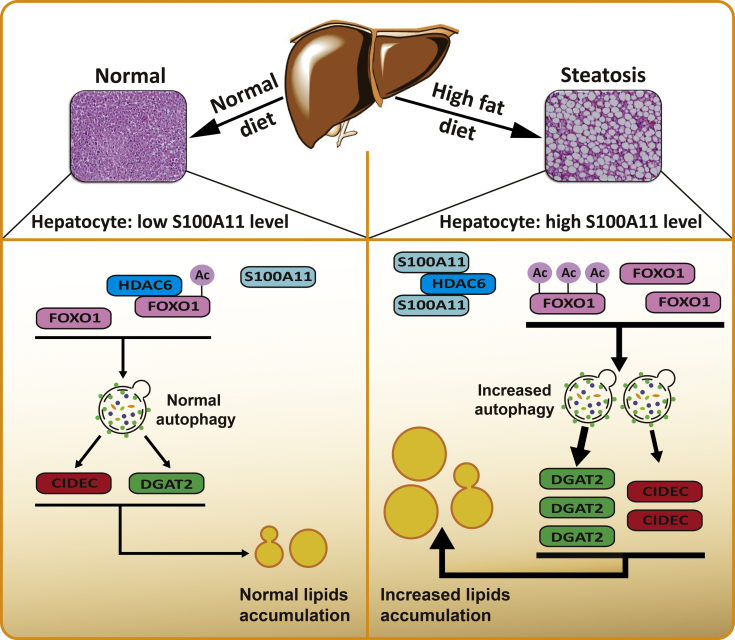
Collectively, we propose that dietary lipids drive S100A11 expression, which may interact with HDAC6 to block its binding to FOXO1, thus releasing or increasing the acetylation of FOXO1, which consequently activates autophagy and lipogenesis, leading to lipid accumulation and liver steatosis (Figure 18). In summary, our study showed a S100A11–HDAC6–FOXO1 axis that synergistically regulates autophagy and lipid metabolism, providing a unique insight into the pathogenies of NAFLD.
Figure 18.
A working model of S100A11 for promoting autophagy and lipid accumulation in hepatocytes.
Materials and Methods
Animal Model
The tree shrew model of NAFLD was established as previously described.37 The mouse model of HFD-induced NAFLD was generated as described in our previous study.61 For S100a11 overexpression in the liver of mice, the recombinant adenovirus gene transfer vector bearing a mouse S100a11 coding sequence sequence combination with 3 × FLAG and GFP was injected through the tail vein of the mice. After injection, the mice were divided into 2 groups and fed either a control diet (D12102C, 4% fat by weight; Research Diets, Inc, New Brunswick, NJ) or a HFHC (D12109C, 20% fat, 1.25% cholesterol, and 0.5% sodium cholate by weight; Research Diets, Inc). The mice were treated with S100a11 overexpression and an HFHC diet for 2 weeks and then killed to collect samples.
All of the animal experiments were conducted according to the guidelines approved by the Animal Ethics Committee of the Kunming Institute of Zoology, Chinese Academy of Science (approval number: SYDW20120105001). The animals were housed 1 animal per cage at room temperature, maintained at 21°C ± 2°C and 50%–70% humidity, with natural lighting and free access to food and water.
Harvest of Blood and Liver Tissues
After the animals were killed, blood and liver samples were harvested. Blood was harvested in an anticoagulant tube containing heparin sodium and then centrifuged at 3000 rpm at room temperature. The plasma levels of aspartate aminotransferase, alanine aminotransferase, TGs, total cholesterol, high-density lipoprotein cholesterol, and LDL-c were measured as previously described.38 Liver samples were fixed in 10% formalin or snap-frozen by liquid nitrogen and then stored at -80°C for later analysis.
Liver Proteomics
SILAC-based proteomic analysis of the livers of tree shrews was performed as previously described.62 Briefly, the liver tissue of each animal was homogenized and mixed with a SILAC-labeled cell mixture consisting of HEK 293, Huh7, and human colorectal carcinoma cell line cells to create a super-SILAC mix. The super-SILAC mix was digested into peptides, and the peptides were analyzed by high-performance liquid chromatography MS/MS. The MS data were first processed using MaxQuant software (version 1.5.1.0; http://maxquant.org), and then analyzed using bioinformatics methods.
Histologic Analysis
Liver specimens were fixed in 10% neutral-buffered formalin, embedded in paraffin, sectioned at 5 μm, and stained with H&E. For Oil Red O staining, liver samples were first embedded in an optimal cutting temperature compound, sectioned at 30 μm, and then stained with Oil Red O to identify neutral lipids (red) and counterstained with hematoxylin for nuclei (blue).
Lipid Measurement
The measurement method for TGs and CEs in the liver was the same as previously described.37 A TG kit (A110-1-1; Nanjing Jiancheng, Jiangsu, China) was used to measure the TGs in the Hepa 1–6 cells according to the manufacturer’s instructions. To quantify the Oil Red O staining, in some cases, the cells were first stained with Oil Red O and washed with phosphate-buffered saline (PBS), and then incubated for 5 minutes with isopropanol. After incubation, the isopropanol absorbance was measured at 510 nm. In other cases, microscopic photographs of the Oil Red O staining were analyzed by ImageJ software (National Institutes of Health, Bethesda, MD).
Cell Culture
Hepatocyte cell lines Hepa 1–6 and Hep 3B and 293T cells were cultured in Dulbecco's modified Eagle medium (C11995500CP; Gibco, Beijing, China) supplemented with 10% fetal bovine serum (10099-141; Gibco) as well as 100 U/mL penicillin and streptomycin in 5% CO2 at 37°C. The culture medium was changed every 2 days unless otherwise stated.
For OA (O7501; Sigma-Aldrich, Shanghai, China) treatment, 10 mmol/L OA stock solution was added to the culture medium to a final concentration of 0.1 or 0.2 mmol/L, and then the cells were treated for either 6, 12, or 24 hours according to the specific experimental purpose.
Bodipy and Oil Red O Staining
The cells were first seeded on coverslips in a 12-well plate and then treated with a specific compound. After treatment, the cells were fixed with 4% paraformaldehyde in PBS for 1 hour at room temperature or overnight at 4°C. The coverslips then were washed for 5 minutes × 3 times with PBS. For Bodipy staining, the cells were stained with 20 μg/mL Bodipy (D3922; Invitrogen, Carlsbad, CA) for 20 minutes, washed 3 times with PBS, counterstained with 4′,6-diamidino-2-phenylindole (D8417; Sigma-Aldrich) for 2 minutes, and washed 3 times with PBS. For Oil Red O staining, the cells were washed for 2 minutes with 60% isopropanol, stained with 0.18% Oil Red O (O1391; Sigma-Aldrich) working solution for 20 minutes, and washed 3 times with PBS. The coverslips then were mounted on glass slides with glycerol jelly mounting medium (C0187; Beyotime, Jiangsu, China) to visualize lipid droplets under a microscope (BX53; Olympus, Tokyo, Japan).
Construction of S100a11 Stable Overexpression Cell Line
A lentiviral vector was used to generate S100a11-FLAG-RFP and control RFP lentiviral constructs, which then were used to package lentivirus in a well-established 293T cell line system. Next, Hepa 1–6 cells or Hep 3B cells were infected with the lentivirus and screened with puromycin for 2 weeks. After screening, the positive monoclonal cells were selected from the enriched cell mix and verified by Western blot.
Plasmid Construction and Co-IP
Full-length mouse S100a11, Hdac6, and Foxo1 coding sequences were amplified by PCR according to sequences in the NCBI (Bethesda, MD) consensus coding sequence database (S100a11, CCDS38525.1; Hdac6, CCDS40845.1; and Foxo1, CCDS17343.1) and then cloned into pCS2+-C-Flag or pEGFP-N1 vectors.
The recombinant plasmids were transfected into 293T cells using Lipofectamine 3000 (L3000015; Thermo Fisher Scientific, Shanghai, China) according to the manufacturer’s instructions. After 48 hours, the cells were harvested and lysed in an IP buffer (50 mmol/L Tris-HCl, 150 mmol/L NaCl, 1 mmol/L EDTA, 1% Nonidet P 40, and 0.25% sodium deoxycholate) containing protease inhibitor cocktail (P8340; Sigma-Aldrich) and phenylmethylsulfonyl fluoride. The cell lysates were centrifuged at 13,000 × g for 10 minutes at 4°C. Immunoprecipitation was performed by adding primary anti-FLAG antibody (F3165; Sigma-Aldrich) or anti-GFP antibody (ab290; Abcam, Shangai, China) to the cell lysates and then rocking overnight at 4°C. This then was paired with the species-matched mouse IgG (12-371; Sigma-Aldrich) or rabbit IgG (ab172730; Abcam) as a negative control. The cell lysates then were incubated with protein G agarose (20398; Thermo Fisher Scientific) or protein A agarose (ab193255; Abcam) according to the binding capacity of the primary antibody to protein G or A agarose and rocked for 4 hours at 4°C. After washing 4 times with the IP buffer, the immunoprecipitate was resuspended in a 2 × sodium dodecyl sulfate (SDS) sample buffer and boiled at 95°C for 5 minutes for later Western blot analysis.
Immunofluorescence Analysis
Cells were washed with PBS and fixed with 4% paraformaldehyde for 10 minutes at room temperature and washed with PBS 3 times for 5 minutes each, then permeabilized with 0.2% Triton X-100 (order number: A600198; Sangon Biotech, Shanghai, China) in PBS at room temperature for 5 minutes. The cells then were washed 3 times and incubated in a blocking buffer (5% bovine serum albumin in PBS) at room temperature for 1 hour. After blocking, the cells were washed once and incubated with either primary anti-FOXO1 (14952; CST, Shanghai, China) or anti-HDAC6 (12834-1-AP; Proteintech, Wuhan, China) antibodies in wild-type Hepa 1–6 cells or anti-S100A11 (10237-1-AP; Proteintech) and anti-HDAC6 (GTX84377; GeneTex, Irvine, TX) in the S100A11 overexpression Hepa 1–6 cells. All of the primary antibodies were diluted in an antibody dilution buffer (1% bovine serum albumin in PBS) at 1:100 and the cells were incubated with related primary antibodies overnight at 4°C. The cells then were washed 3 times and incubated with related secondary anti-mouse IgG (H+L) and F(ab′)2 fragment (Alexa Fluor 555 conjugate) antibodies at 1:1000 (4409; CST) and anti-rabbit IgG (H+L) and F(ab')2 fragment (Alexa Fluor 488 conjugate) at 1:1000 (4412; CST) for 1 hour at room temperature in a dark and moist environment. The cells then were washed 3 times and stained with Hoechst 33342 (1:1000 in PBS) for 3 minutes at room temperature in a dark and moist environment. Images were acquired using a Nikon A1MP+ confocal microscope system (Nikon, Tokyo, Japan).
Inhibitor Treatment and RNA Knockdown
For SIRT1 inhibition, Hepa 1–6 cells were treated for 12 and 24 hours with 0.1 mmol/L OA and 10 μmol/L EX-527 (HY-15452; MedChemExpress, Monmouth Junction, NJ). For HDAC6 inhibition, Hepa 1–6 cells were treated for 6, 12, and 24 hours with 0.1 mmol/L OA and 40 nmol/L TBA (SML0044; Sigma-Aldrich). For FOXO1 inhibition, the cells were treated for 6 hours with the specific inhibitor AS1842856 (HY-100596; MedChemExpress) at a final concentration of 0.25 μmol/L. For autophagy inhibition, Hepa 1–6 cells were treated for 12 hours with 0.1 mmol/L OA and 10 nmol/L bafilomycin A1 (HY-100558; MedChemExpress) or with 0.1 mmol/L OA and 2 mmol/L 3-methyladenine (HY-19312; MedChemExpress). For RNA knockdown of Atg7, mouse small interfering RNA of Atg7 was ordered from Ribobio Co (Guangzhou, China) and the related experiment was conducted according to the manufacturer’s instructions.
CRISPR/Cas9-Mediated Gene Knockout
The CRISPR/Cas9 system was used to knockout the Dgat2 gene in the Hepa 1–6 cell. The guide RNA to the Dgat2 gene was designed using an online tool (http://crispr.mit.edu) and inserted into the pX260a plasmid. After construction, the recombinant plasmid was transfected into Hepa 1–6 cells by Lipofectamine 3000 (L3000015, Thermo Fisher Scientific, Shanghai, China), and the transfected cells were enriched by puromycin selection. Afterward, the positive monoclonal cells were selected from the enriched cell mix and verified by Western blot and genomic PCR. The technical details were described in a previous study.63
Protein Extraction and Western blot Analysis
The liver tissue samples and cultured cells were lysed in a lysis buffer (50 mmol/L Tris-HCl, 150 mmol/L NaCl, 1 mmol/L EDTA, 10 mmol/L Sodium fluoride, 1% Triton X-100, 0.1% SDS, and 0.5% sodium deoxycholate) supplemented with protease inhibitor cocktail and phenylmethylsulfonyl fluoride, followed by centrifugation at 13,000 × g for 15 minutes at 4°C. The protein concentrations were determined via the bicinchonininc acid (23227; Thermo Fisher Scientific) method.
Protein samples then were mixed with a 2 × SDS sample buffer and boiled at 95°C for 5 minutes. Proteins were fractionated by SDS polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes. Immunoblots were blocked with 5% bovine serum albumin in Tris-Buffered Saline containing Tween-20 (20 mmol/L Tris-HCl, 140 mmol/L NaCl, 0.05% Tween-20, pH 7.5) and probed with primary antibodies overnight at 4°C. Information on all of the antibodies used in this study is available in Table 5.
Table 5.
Information on Antibodies Used in This Study
| Antibody | Supplier | Product code |
|---|---|---|
| S100A11 | Proteintech (Wuhan, China) | 10237-1-AP |
| DGAT1 | Novus (Littleton, CO) | NB110-41487 |
| DGAT2 | ABclonal (Wuhan, China) | A13891 |
| ACC | CST (Danvers, MA) | 4190 |
| ACAT1 | Proteintech | 16215-1-AP |
| CIDEC | Novus | NB100-430 |
| FOXO1 | CST | 2880 |
| p-FOXO1 | CST | 9461 |
| Ac-FOXO1 | Affinity Biosciences (Cincinnati, OH) | AF2305 |
| ATG7 | ABclonal | A0691 |
| LC3 | Proteintech | 12135-1-AP |
| P300 | Santa Cruz (Dallas, TX) | sc-48343 |
| GFP | Abcam | ab290 |
| FLAG | Sigma-Aldrich (Shanghai, China) | F3165 |
| ACSL4 | Abcam | ab110007 |
| GRB2 | BD (Lexington, KY) | 610111 |
| VIM | Abcam | ab8978 |
| HDAC6 | Proteintech | 12834-1-AP |
| SIRT1 | Proteintech | 13161-1-AP |
| p-AKT | CST | 9271 |
| AKT | CST | 9272 |
| RFP | Abcam | ab62341 |
| GAPDH | Proteintech | 60004-1-Ig |
| β-Actin | Sigma-Aldrich | A5441 |
ACSL4, long-chain acyl-CoA synthetase 4; p-AKT, Protein Kinase B; p-FOXO1, phosphorylated forkhead box O1; VIM, vimentin.
mRNA Extraction and qPCR
Total RNA was extracted from liver tissues using RNAiso Plus (9109; Takara, Beijing, China) and reverse-transcribed to complementary DNA using a PrimeScript RT reagent kit (RR047A; Takara). Real-time qPCR was performed on an ABI 7900HT sequencer (Applied Biosystems, Foster City, CA). β-actin or glyceraldehyde-3-phosphate dehydrogenase was used as a standard control to normalize the relative mRNA expression of a specific gene via the delta-delta cycle threshold method. The sequences of all the primers used in this study are available in Table 6.
Table 6.
Information on Primers Used in This Study
| Primer symbols | Primer sequences (5’-3’) |
|
|---|---|---|
| Forward | Reverse | |
| S100a11 | GCATTGAGTCCCTGATTGCT | ATCTAGCTGCCCGTCACAGT |
| Srebp1c | GTACCTGCGGGACAGCTTAG | CTGCATCTGCCTCAACACAT |
| Pparg | AAGAGCTGACCCAATGGTTG | ACCCTTGCATCCTTCACAAG |
| C/ebpα | GCAGTGTGCACGTCTATGCT | AAGTCTTAGCCGGAGGAAGC |
| Srebp2 | CAGGTGCAGACGGTACAGG | CGTGGTCAACACAAGGGAATC |
| Fasn | ATGCACACTCTGCGATGAAG | CAGTGTTCACAGCCAGGAGA |
| Acc | GAATCTCCTGGTGACAATGCTTATT | GGTCTTGCTGAGTTGGGTTAGCT |
| Scd1 | TTCTTGCGATACACTCTGGTGC | CGGGATTGAATGTTCTTGTCGT |
| Dgat1 | GCTGATCCCAGGTTGTTCAT | GAGACAGCTTTGGCCTTGAC |
| Dgat2 | ACGCAGTCACCCTGAAGAAC | CCCAGGTGTCAGAGGAGAAG |
| Cidec | CGGGTAACCTTCGACCTGTA | GGGTCTTCATTGCAGCATCT |
| Hmgcr | CCGAATTGTATGTGGCACTG | GGTGCACGTTCCTTGAAGAT |
| Acat1 | TTCAGGGAGCCATTGAGAAG | AGTGTTGCTTGCCTGGTAGG |
| Acat2 | CCTGGAGGATGTTGACCTGT | CCTCCATCGATGTTGACCTT |
| Cd36 | TGCTTGCAAATCCAAGAATG | AGAGAGAGCACACACCACCA |
| Ldlr | GCCACATGGTATGAGGTTCC | GCTCGTCCTCTGTGGTCTTC |
| Srb1 | TGGACAAATGGAACGGACTC | GTGAAGCGATACGTGGGAAT |
| Pparα | CCTGAACATCGAGTGTCGAA | CCAGCTTCAGCCGAATAGTT |
| Cpt1 | CTTCCATGACTCGGCTCTTC | AGCTTGAACCTCTGCTCTGC |
| Mttp | AAAGCAGAGCGGAGACAGAG | TGTCTCGAATTGCCTGAGTG |
| Abcg5 | AGGGCCTCACATCAACAGAG | GCTGACGCTGTAGGACACAT |
| Abcg8 | CTGTGGAATGGGACTGTACTTC | GTTGGACTGACCACTGTAGGT |
| Apob | AAGCACCTCCGAAAGTACGTG | CTCCAGCTCTACCTTACAGTTGA |
| Apoe | CTGACAGGATGCCTAGCCG | CGCAGGTAATCCCAGAAGC |
| Acadvl | CTACTGTGCTTCAGGGACAAC | CAAAGGACTTCGATTCTGCCC |
| Echs1 | AGCCTGTAGCTCACTGTTGTC | ATGTACTGAAAGTTAGCACCCG |
| Hadh | GGATTCATCGTGAACCGACT | TCCCAGCTTCATCGCTGTAT |
| Ehhadh | TCTCCTCGGTTGGTGTTCTT | GCTGCTTTGGGTCTGACTCT |
| Foxo1 | TATTGAGCGCTTGGACTGTG | TGGACTGCTCCTCAGTTCCT |
| Pck1 | ATCATCTTTGGTGGCCGTAG | CCTCAGATCTCATGGCTGCT |
| G6pc | CAACAGCTCCGTGCCTATAA | TGGCTTTTTCTTTCCTCGAA |
| Igfbp1 | ATCAGCCCATCCTGTGGAAC | TGCAGCTAATCTCTCTAGCACTT |
| Insr | CCTCAGTGCCAGTGATGTGT | TCGATCCGTTCTCGAAGACT |
| Atg5 | GCGGTTGAGGCTCACTTTAT | ATTCCATGAGTTTCCGGTTG |
| Atg7 | GTTCGCCCCCTTTAATAGTGC | TGAACTCCAACGTCAAGCGG |
| Atg12 | AACAAAGAAATGGGCTGTGG | GAAGGGGCAAAGGACTGATT |
| Atg14 | GCAGCTCGTCAACATTGTGT | TGCGTTCAGTTTCCTCACTG |
| Map1lc3a | CATGAGCGAGTTGGTCAAGA | TTGACTCAGAAGCCGAAGGT |
| Map1lc3b | TTATAGAGCGATACAAGGGGGAG | CGCCGTCTGATTATCTTGATGAG |
| Becn1 | TTTGACCATGCAATGGTAGC | GCTTTTGTCCACTGCTCCTC |
| Bnip3 | TGCTCCGTTTTATCCGTTTC | TCGACTTGACCAATCCCATA |
| Vps34 | GAGACTTCAGGCCTTGCTTG | CAAGTCGTCTCCATGCTTGA |
| Actin | GGCTGTATTCCCCTCCATCG | CCAGTTGGTAACAATGCCATGT |
| Gapdh | CAGCAACTCCCACTCTTCCAC | TGGTCCAGGGTTTCTTACTC |
Mouse Liver mRNA Sequencing
In brief, total RNA of mouse liver was first isolated, quantified, and qualified. Second, sequencing libraries were generated using the NEBNext Ultra RNA Library Prep Kit for Illumina (E7530L; Ipswich, MA) following the manufacturer’s recommendations, and index codes were added to attribute each sample’s sequences. Third, clustering of the index-coded samples was performed on a cBot Cluster Generation System using a HiSeq 4000 PE Cluster Kit (Illumina). After cluster generation, the library preparations were sequenced on an Illumina HiSeq 4000 platform. The whole list of the changed genes is available upon request.
Statistical analyses
Data are presented as means ± SEM. Statistical analysis was performed with a t test or analysis of variance followed by least significance difference multiple comparison using SPSS 20.0 (IBM SPSS Statistics, Armonk, NY). P < .05 was considered statistically significant. All of the figures were produced using GraphPad Prism 7 (GraphPad Software, La Jolla, CA) or Photoshop CS/AI (Adobe, San Jose, CA).
Acknowledgments
The authors thank Metabo-Profile Biotechnology Co, Ltd (Shanghai, China) for lipid analysis.
CRediT Authorship Contributions
Linqiang Zhang, PhD (Conceptualization: Equal; Formal analysis: Equal; Investigation: Equal; Writing nceptualization: Equal;
Zhiguo Zhang, PhD (Conceptualization: Equal; Formal analysis: Equal; Investigation: Equal; Writing nceptualization: Equal;
Chengbin Li, MD (Investigation: Supporting);
Tingting Zhu, MD (Investigation: Supporting);
Jing Gao, PhD (Investigation: Supporting);
Hu Zhou, PhD (Investigation: Supporting);
Yingzhuan Zheng, MD (Investigation: Supporting);
Qing Chang, MD (Investigation: Supporting);
Mingshan Wang, PhD (Investigation: Supporting);
Jieyu Wu, PhD (Investigation: Supporting);
Liyuan Ran, PhD (Investigation: Supporting);
Yingjie Wu, PhD (Resources: Supporting);
Huilai Miao, PhD (Resources: Supporting);
Xiaoju Zou, PhD (Resources: Supporting);
Bin Liang, PhD (Conceptualization: Lead; Funding acquisition: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing ; Projectn:ation: Equal).
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by the Ministry of Science and Technology of the People’s Republic of China grant 2018YFA0800700; the National Natural Science Foundation of China grants 81700520, U1702288, U1702287, 31671230, 91857113, and 31860323; the Yunnan Applied Basic Research Projects 2017FA007, 2018FB117, and 2019FY003021; the Yunnan Oversea High-level Talents Program grants 2015HA039 and 2015HA040 (B.L.); and the Open Program of the Key Laboratory of Animal Models and Human Disease Mechanisms of Chinese Academy of Sciences & Yunnan Province (AMHD-2018-6).
Contributor Information
Yingjie Wu, Email: yyjjwu@yahoo.com.
Huilai Miao, Email: 627225370@qq.com.
Xiaoju Zou, Email: xiaojuzou@163.com.
Bin Liang, Email: liangb73@ynu.edu.cn.
References
- 1.Khan R., Bril F., Cusi K., Newsome P.N. Modulation of insulin resistance in NAFLD. Hepatology. 2019;70:711–724. doi: 10.1002/hep.30429. [DOI] [PubMed] [Google Scholar]
- 2.Younossi Z.M. Non-alcoholic fatty liver disease – a global public health perspective. J Hepatol. 2019;70:531–544. doi: 10.1016/j.jhep.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 3.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 4.Sanyal A.J., Brunt E.M., Kleiner D.E., Kowdley K.V., Chalasani N., Lavine J.E., Ratziu V., McCullough A. Endpoints and clinical trial design for nonalcoholic steatohepatitis. Hepatology. 2011;54:344–353. doi: 10.1002/hep.24376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loomba R., Sanyal A.J. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013;10:686–690. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- 6.Levine B., Klionsky D.J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 7.Lum J.J., DeBerardinis R.J., Thompson C.B. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 8.Rajat S., Susmita K., Yongjun W., Youqing X., Inna N., Masaaki K., Keiji T., Ana Maria C., Czaja M.J. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L., Li P., Fu S., Calay E.S., Hotamisligil G.S. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czaja M.J., Ding W.X., Donohue T.M., Jr., Friedman S.L., Kim J.S., Komatsu M., Lemasters J.J., Lemoine A., Lin J.D., Ou J.H., Perlmutter D.H., Randall G., Ray R.B., Tsung A., Yin X.M. Functions of autophagy in normal and diseased liver. Autophagy. 2013;9:1131–1158. doi: 10.4161/auto.25063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding W.X., Li M., Chen X., Ni H.M., Lin C.W., Gao W., Lu B., Stolz D.B., Clemens D.L., Yin X.M. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin C.W., Zhang H., Li M., Xiong X., Chen X., Chen X., Dong X.C., Yin X.M. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol. 2013;58:993–999. doi: 10.1016/j.jhep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y., Zong W.X., Ding W.X. Recycling the danger via lipid droplet biogenesis after autophagy. Autophagy. 2017;13:1995–1997. doi: 10.1080/15548627.2017.1371394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen T.B., Louie S.M., Daniele J.R., Tran Q., Dillin A., Zoncu R., Nomura D.K., Olzmann J.A. DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev Cell. 2017;42:9–21.e25. doi: 10.1016/j.devcel.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rambold A.S., Cohen S., Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev Cell. 2015;32:678–692. doi: 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim K.H., Jeong Y.T., Oh H., Kim S.H., Cho J.M., Kim Y.N., Kim S.S., Kim D.H., Hur K.Y., Kim H.K., Ko T., Han J., Kim H.L., Kim J., Back S.H., Komatsu M., Chen H., Chan D.C., Konishi M., Itoh N., Choi C.S., Lee M.S. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med. 2013;19:83–92. doi: 10.1038/nm.3014. [DOI] [PubMed] [Google Scholar]
- 17.Li Y., Chao X., Yang L., Lu Q., Li T., Ding W.X., Ni H.M. Impaired fasting-induced adaptive lipid droplet biogenesis in liver-specific Atg5-deficient mouse liver is mediated by persistent nuclear factor-like 2 activation. Am J Pathol. 2018;188:1833–1846. doi: 10.1016/j.ajpath.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma D., Molusky M.M., Song J., Hu C.R., Fang F., Rui C., Mathew A.V., Pennathur S., Liu F., Cheng J.X., Guan J.L., Lin J.D. Autophagy deficiency by hepatic FIP200 deletion uncouples steatosis from liver injury in NAFLD. Mol Endocrinol. 2013;27:1643–1654. doi: 10.1210/me.2013-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata M., Yoshimura K., Furuya N., Koike M., Ueno T., Komatsu M., Arai H., Tanaka K., Kominami E., Uchiyama Y. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun. 2009;382:419–423. doi: 10.1016/j.bbrc.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 20.Hariharan N., Maejima Y., Nakae J., Paik J., Depinho R.A., Sadoshima J. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010;107:1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H.Y., Han J., Cao S.Y., Hong T., Zhuo D., Shi J., Liu Z., Cao W. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuzaki T., Alvarez-Garcia O., Mokuda S., Nagira K., Olmer M., Gamini R., Miyata K., Akasaki Y., Su A.I., Asahara H., Lotz M.K. FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci Transl Med. 2018;10 doi: 10.1126/scitranslmed.aan0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaffner I., Minakaki G., Khan M.A., Balta E.A., Schlotzer-Schrehardt U., Schwarz T.J., Beckervordersandforth R., Winner B., Webb A.E., DePinho R.A., Paik J., Wurst W., Klucken J., Lie D.C. FoxO function is essential for maintenance of autophagic flux and neuronal morphogenesis in adult neurogenesis. Neuron. 2018;99:1188–1203.e1186. doi: 10.1016/j.neuron.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S., Xia P., Huang G., Zhu P., Liu J., Ye B., Du Y., Fan Z. FoxO1-mediated autophagy is required for NK cell development and innate immunity. Nat Commun. 2016;7:11023. doi: 10.1038/ncomms11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong X., Tao R., DePinho R.A., Dong X.C. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem. 2012;287:39107–39114. doi: 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y., Li X., Cai M.Y., Ma K., Yang J., Zhou J., Fu W., Wei F.Z., Wang L., Xie D., Zhu W.G. XBP-1u suppresses autophagy by promoting the degradation of FoxO1 in cancer cells. Cell Res. 2013;23:491–507. doi: 10.1038/cr.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lettieri Barbato D., Tatulli G., Aquilano K., Ciriolo M.R. FoxO1 controls lysosomal acid lipase in adipocytes: implication of lipophagy during nutrient restriction and metformin treatment. Cell Death Dis. 2013;4:e861. doi: 10.1038/cddis.2013.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vihervaara T., Puig O. dFOXO regulates transcription of a drosophila acid lipase. J Mol Biol. 2008;376:1215–1223. doi: 10.1016/j.jmb.2007.12.042. [DOI] [PubMed] [Google Scholar]
- 29.Chakrabarti P., Kandror K.V. FoxO1 controls insulin-dependent adipose triglyceride lipase (ATGL) expression and lipolysis in adipocytes. J Biol Chem. 2009;284:13296–13300. doi: 10.1074/jbc.C800241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L., Zheng L.D., Zou P., Brooke J., Smith C., Long Y.C., Almeida F.A., Liu D., Cheng Z. FoxO1 antagonist suppresses autophagy and lipid droplet growth in adipocytes. Cell Cycle. 2016;15:2033–2041. doi: 10.1080/15384101.2016.1192732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He H., Li J., Weng S., Li M., Yu Y. S100A11: diverse function and pathology corresponding to different target proteins. Cell Biochem Biophys. 2009;55:117. doi: 10.1007/s12013-009-9061-8. [DOI] [PubMed] [Google Scholar]
- 32.Inada H., Naka M., Tanaka T., Davey G.E., Heizmann C.W. Human S100A11 exhibits differential steady-state RNA levels in various tissues and a distinct subcellular localization. Biochem Biophys Res Commun. 1999;263:135–138. doi: 10.1006/bbrc.1999.1319. [DOI] [PubMed] [Google Scholar]
- 33.Makino E., Sakaguchi M., Iwatsuki K., Huh N.H. Introduction of an N-terminal peptide of S100C/A11 into human cells induces apoptotic cell death. J Mol Med (Berl) 2004;82:612–620. doi: 10.1007/s00109-004-0560-1. [DOI] [PubMed] [Google Scholar]
- 34.Cecil D.L., Johnson K., Rediske J., Lotz M., Schmidt A.M., Terkeltaub R. Inflammation-induced chondrocyte hypertrophy is driven by receptor for advanced glycation end products. J Immunol. 2005;175:8296–8302. doi: 10.4049/jimmunol.175.12.8296. [DOI] [PubMed] [Google Scholar]
- 35.Memon A.A., Sorensen B.S., Meldgaard P., Fokdal L., Thykjaer T., Nexo E. Down-regulation of S100C is associated with bladder cancer progression and poor survival. Clin Cancer Res. 2005;11:606–611. [PubMed] [Google Scholar]
- 36.Rehman I., Azzouzi A.R., Cross S.S., Deloulme J.C., Catto J.W., Wylde N., Larre S., Champigneuille J., Hamdy F.C. Dysregulated expression of S100A11 (calgizzarin) in prostate cancer and precursor lesions. Hum Pathol. 2004;35:1385–1391. doi: 10.1016/j.humpath.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 37.Zhang L., Zhang Z., Li Y., Liao S., Wu X., Chang Q., Liang B. Cholesterol induces lipoprotein lipase expression in a tree shrew (Tupaia belangeri chinensis) model of non-alcoholic fatty liver disease. Sci Rep. 2015;5:15970. doi: 10.1038/srep15970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L., Wu X., Liao S., Li Y., Zhang Z., Chang Q., Xiao R., Liang B. Tree shrew (Tupaia belangeri chinensis), a novel non-obese animal model of non-alcoholic fatty liver disease. Biol Open. 2016;5:1545–1552. doi: 10.1242/bio.020875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park J.-E., Kim H.T., Lee S., Lee Y.-S., Choi U.-K., Kang J.H., Choi S.Y., Kang T.-C., Choi M.-S., Kwon O.-S. Differential expression of intermediate filaments in the process of developing hepatic steatosis. Proteomics. 2011;11:2777–2789. doi: 10.1002/pmic.201000544. [DOI] [PubMed] [Google Scholar]
- 40.Shan X., Miao Y., Fan R., Song C., Wu G., Wan Z., Zhu J., Sun G., Zha W., Mu X., Zhou G., Chen Y. Suppression of Grb2 expression improved hepatic steatosis, oxidative stress, and apoptosis induced by palmitic acid in vitro partly through insulin signaling alteration. Vitro Cell Dev Biol Animal. 2013;49:576–582. doi: 10.1007/s11626-013-9646-9. [DOI] [PubMed] [Google Scholar]
- 41.Stepanova M., Hossain N., Afendy A., Perry K., Goodman Z.D., Baranova A., Younossi Z. Hepatic gene expression of Caucasian and African-American patients with obesity-related non-alcoholic fatty liver disease. Obes Surg. 2010;20:640–650. doi: 10.1007/s11695-010-0078-2. [DOI] [PubMed] [Google Scholar]
- 42.Lede V., Meusel A., Garten A., Popkova Y., Penke M., Franke C., Ricken A., Schulz A., Kiess W., Huster D., Schoneberg T., Schiller J. Altered hepatic lipid metabolism in mice lacking both the melanocortin type 4 receptor and low density lipoprotein receptor. PLoS One. 2017;12 doi: 10.1371/journal.pone.0172000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dorn C., Engelmann J.C., Saugspier M., Koch A., Hartmann A., Muller M., Spang R., Bosserhoff A., Hellerbrand C. Increased expression of c-Jun in nonalcoholic fatty liver disease. Lab Invest. 2014;94:394–408. doi: 10.1038/labinvest.2014.3. [DOI] [PubMed] [Google Scholar]
- 44.Do G.M., Oh H.Y., Kwon E.Y., Cho Y.Y., Shin S.K., Park H.J., Jeon S.M., Kim E., Hur C.G., Park T.S., Sung M.K., McGregor R.A., Choi M.S. Long-term adaptation of global transcription and metabolism in the liver of high-fat diet-fed C57BL/6J mice. Mol Nutr Food Res. 2011;55(Suppl 2):S173–S185. doi: 10.1002/mnfr.201100064. [DOI] [PubMed] [Google Scholar]
- 45.Mitsumoto K., Watanabe R., Nakao K., Yonenaka H., Hashimoto T., Kato N., Kumrungsee T., Yanaka N. Time-course microarrays reveal early activation of the immune transcriptome in a choline-deficient mouse model of liver injury. Life Sci. 2017;184:103–111. doi: 10.1016/j.lfs.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 46.Seth D., Leo M.A., McGuinness P.H., Lieber C.S., Brennan Y., Williams R., Wang X.M., McCaughan G.W., Gorrell M.D., Haber P.S. Gene expression profiling of alcoholic liver disease in the baboon (Papio hamadryas) and human liver. Am J Pathol. 2003;163:2303–2317. doi: 10.1016/S0002-9440(10)63587-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puri V., Konda S., Ranjit S., Aouadi M., Chawla A., Chouinard M., Chakladar A., Czech M.P. Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. J Biol Chem. 2007;282:34213–34218. doi: 10.1074/jbc.M707404200. [DOI] [PubMed] [Google Scholar]
- 48.Wang S., Ni H.M., Chao X., Ma X., Kolodecik T., De Lisle R., Ballabio A., Pacher P., Ding W.X. Critical role of TFEB-mediated lysosomal biogenesis in alcohol-induced pancreatitis in mice and humans. Cell Mol Gastroenterol Hepatol. 2020;10:59–81. doi: 10.1016/j.jcmgh.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuzaki H., Daitoku H., Hatta M., Tanaka K., Fukamizu A. Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc Natl Acad Sci U S A. 2003;100:11285–11290. doi: 10.1073/pnas.1934283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perrot V.R., Rechler M.M. The coactivator p300 directly acetylates the forkhead transcription factor foxo1 and stimulates foxo1-induced transcription. Mol Endocrinol. 2005;19:2283–2298. doi: 10.1210/me.2004-0292. [DOI] [PubMed] [Google Scholar]
- 51.Li K., Qiu C., Sun P., Liu D-c, Wu T-j, Wang K., Zhou Y-c, Chang X-a, Yin Y., Chen F., Zhu Y-x, Han X. Ets1-mediated acetylation of FoxO1 is critical for gluconeogenesis regulation during feed-fast cycles. Cell Rep. 2019;26:2998–3010.e2995. doi: 10.1016/j.celrep.2019.02.035. [DOI] [PubMed] [Google Scholar]
- 52.Nakae J., Cao Y., Daitoku H., Fukamizu A., Ogawa W., Yano Y., Hayashi Y. The LXXLL motif of murine forkhead transcription factor FoxO1 mediates Sirt1-dependent transcriptional activity. J Clin Invest. 2006;116:2473–2483. doi: 10.1172/JCI25518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Puig O., Tjian R. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev. 2005;19:2435–2446. doi: 10.1101/gad.1340505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X., Wu R., Liu Y., Zhao Y., Bi Z., Yao Y., Liu Q., Shi H., Wang F., Wang Y. m(6)A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2020;16:1221–1235. doi: 10.1080/15548627.2019.1659617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andres Cerezo L., Sumova B., Prajzlerova K., Veigl D., Damgaard D., Nielsen C.H., Pavelka K., Vencovsky J., Senolt L. Calgizzarin (S100A11): a novel inflammatory mediator associated with disease activity of rheumatoid arthritis. Arthritis Res Ther. 2017;19:79. doi: 10.1186/s13075-017-1288-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oh H.Y., Shin S.K., Heo H.S., Ahn J.S., Kwon E.Y., Park J.H., Cho Y.Y., Park H.J., Lee M.K., Kim E.J., Jung U.J., McGregor R.A., Hur C.G., Choi M.S. Time-dependent network analysis reveals molecular targets underlying the development of diet-induced obesity and non-alcoholic steatohepatitis. Genes Nutr. 2013;8:301–316. doi: 10.1007/s12263-012-0322-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh R., Xiang Y., Wang Y., Baikati K., Cuervo A.M., Luu Y.K., Tang Y., Pessin J.E., Schwartz G.J., Czaja M.J. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y., Goldman S., Baerga R., Zhao Y., Komatsu M., Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sengupta A., Molkentin J.D., Yutzey K.E. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–28331. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiu W., Wang B., Gao Y., Tian Y., Tian M., Chen Y., Xu L., Yao T.P., Li P., Yang P. Targeting histone deacetylase 6 reprograms interleukin-17-producing helper T cell pathogenicity and facilitates immunotherapies for hepatocellular carcinoma. Hepatology. 2020;71:1967–1987. doi: 10.1002/hep.30960. [DOI] [PubMed] [Google Scholar]
- 61.Ran L., Wang X., Mi A., Liu Y., Wu J., Wang H., Guo M., Sun J., Liu B., Li Y., Wang D., Jiang R., Wang N., Gao W., Zeng L., Huang L., Chen X., LeRoith D., Liang B., Li X., Wu Y. Loss of adipose growth hormone receptor in mice enhances local fatty acid trapping and impairs brown adipose tissue thermogenesis. iScience. 2019;16:106–121. doi: 10.1016/j.isci.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J., Xu S., Gao J., Zhang L., Zhang Z., Yang W., Li Y., Liao S., Zhou H., Liu P., Liang B. SILAC-based quantitative proteomic analysis of the livers of spontaneous obese and diabetic rhesus monkeys. Am J Physiol Endocrinol Metab. 2018;315:E294–E306. doi: 10.1152/ajpendo.00016.2018. [DOI] [PubMed] [Google Scholar]
- 63.Xu S., Zou F., Diao Z., Zhang S., Deng Y., Zhu X., Cui L., Yu J., Zhang Z., Bamigbade A.T., Zhang H., Wei X., Zhang X., Liang B., Liu P. Perilipin 2 and lipid droplets provide reciprocal stabilization. Biophys Rep. 2019;5:145–160. [Google Scholar]