Abstract
Aims
Epidermal growth factor receptor (EGFR) T790M mutations can be detected in the circulating tumour DNA from plasma of patients with non-small cell lung cancer (NSCLC) to triage patients for osimertinib eligibility and monitor patients longitudinally for development of T790M-mediated resistance.
Methods
Using droplet digital PCR (ddPCR), we examined the EGFR T790M status of 343 sequential patients with NSCLC and correlated mutational status with demographic and clinical features. Where available, serial T790M blood test results were assessed to identify clinical triggers and timing of repeat testing.
Results
Of the 343 patients with liquid biopsy test results, 24% were T790M positive. No clear clinical correlation with a T790M positive test result was identified in this study, although the number of metastatic sites did correlate significantly with the presence of EGFR sensitising mutations (L858R or exon 19 deletion) in patient plasma, as a measure of tumour DNA shedding. Of the 59 serial blood tests from patients that initially tested negative, 14% were positive on sequential testing, at a time interval up to 6 months after an initially negative blood test.
Conclusions
The ddPCR test for EGFR T790M mutations effectively triaged 24% of patients for treatment with osimertinib, avoiding the need for invasive tissue biopsy in these patients. Our findings suggest that initial and repeat ctDNA testing can be used to monitor for acquired EGFR T790M resistance for NSCLC.
Keywords: biomarkers, tumor; lung neoplasms; carcinoma
Introduction
Patients with non-small cell lung cancer (NSCLC) with sensitising mutations in the epidermal growth factor receptor (EGFR) gene are eligible for treatment with EGFR tyrosine kinase inhibitors (TKI) as first-line systemic therapy. The most common EGFR sensitising mutations are the L858R or exon 19 deletion mutations, which cause constitutive activation of the EGFR tyrosine kinase.1 2 On diagnosis, guidelines recommend testing for EGFR mutations in patients with NSCLC to identify those eligible for TKIs.3 Although response to first-generation TKIs is rapid and durable, nearly all patients treated with first-line TKIs experience acquired resistance, resulting in progression or relapse. The primary mechanism of drug resistance leading to progression is the development of the EGFR T790M resistance mutation, occurring in 60% of cases.4 In 2015, the third-generation TKI osimertinib was shown to be effective for the treatment of EGFR T790M-positive NSCLCs.5 Furthermore, the AURA3 phase 3 trial demonstrated superior response rate and progression-free survival in those treated with first-line osimertinib as compared with first-generation TKIs,6 although mechanisms that confer resistance to osimertinib have been identified.7 8
EGFR T790M can be detected in tumour tissue (surgical biopsy or cytology specimens) or from cell-free circulating tumour DNA (ctDNA) extracted from peripheral blood by liquid biopsy. Current testing recommendations for EGFR T790M detection in lung cancer indicate that identification of the mutation in cell-free DNA of patients with progression or secondary clinical resistance to EGFR TKIs is appropriate, with subsequent testing of the tumour specimen if the plasma result is negative.9 Testing of ctDNA from liquid biopsies offers distinct advantages over tumour tissue testing, including reduced cost, non-invasive nature of the procedure, rapid turnaround time and potential for longitudinal monitoring by serial biopsy.10 A negative plasma test for T790M may reflect either the absence of the T790M mutation (true negative) or a false negative due to the lack of or unquantifiable levels of ctDNA available in that patient’s blood. To distinguish between a true negative and false negative result, ctDNA may also be tested for the EGFR sensitising mutation known to be present in that patient from prior diagnostic testing. A negative plasma EGFR T790M test should result in reflex tissue testing, where feasible, to confirm the mutational status of that patient and ensure better identification of those eligible for osimertinib treatment.9 11 Alternately, patients that test negative for T790M by ctDNA assessment may benefit from repeat blood specimen testing in situations where more invasive procedures are not possible. However, clinical characteristics that indicate increased likelihood of detecting ctDNA in peripheral blood (eg, clinical status, extent of disease progression) and the optimal timing of repeat blood collection are currently unknown.
Identification of an EGFR mutation, either sensitising or resistance, in patient circulating cell-free DNA may be used as an indication of tumour DNA ‘shedding’.12 A positive plasma test result for EGFR sensitising mutations has been shown to correlate with the number and size of metastatic lesions in patients with lung cancer.13 However, correlation between the number or size of metastatic sites and the presence of the EGFR T790M resistance mutation in cell-free DNA has not yet been reported.
At present, optimal timing of EGFR T790M testing in patients with NSCLC with markers of progression or relapse is unclear. In an effort to clarify which clinical features may be indicative of the likelihood of a positive T790M plasma test result, we examined corresponding clinical metrics with T790M positive cell-free DNA. Our clinical laboratory service initiated clinical testing for the EGFR T790M mutation in blood by droplet digital (ddPCR) in 2017 (University Health Network (UHN), Toronto, Canada). Using data from a sequential cohort of patients seen at UHN for NSCLC, we evaluated the positivity rate for the EGFR TKI resistance mutation T790M and the presence of the EGFR exon 19 deletion and L858R TKI sensitising mutations, as indicators of tumour DNA shedding. Clinical metrics assessed included age, sex and smoking status demographics, as well as duration of TKI treatment, blood protein levels of lactate dehydrogenase (LDH) and number and location of metastatic lesions. Furthermore, we investigated what clinical triggers may be present to indicate the need for repeat testing by liquid biopsy and evaluated the time interval between subsequent retesting of plasma from the same patient.
Methods
Patient cohort
The study cohort included 343 sequential patients with NSCLC tested for EGFR T790M by ddPCR between July 2017 and October 2018. Of the patient cohort, 107 were seen by Medical Oncology at UHN for care, while 236 were specimens sent from other hospital sites within Ontario. Referral criterial were based on testing algorithm recommendations for EGFR T790M for lung cancer.11 These criteria included EGFR T790M testing for ctDNA from plasma of patients with NSCLC with a known EGFR sensitising mutation treated with an EGFR TKI that display clinical or radiological progression. Additional follow-up testing for EGFR T790M was performed on 84 patients who had undetectable levels of ctDNA on initial testing (as shown by the failure to detect the known EGFR sensitising mutation in the sample). This included 51 patients with serial blood specimens, 34 patients with subsequent core-needle biopsy (CNB) tissue specimens and 16 patients with fine-needle aspiration (FNA) cytology specimens. Surgical biopsies were obtained from various sites within the lungs, including the right upper, right lower and left lower lung, as well as axillary and internal mammary lymph nodes and the chest wall. For those patients with sequential blood specimens for EGFR T790M, the average time to follow-up testing was 78 days (range 6–340 days).
Specimens and sample processing
Liquid biopsy specimens for ctDNA analysis were provided in cell-free DNA blood collection tubes, as per testing recommendations.11 A total of 471 specimens were received, from 343 patients. Plasma (8–12 mL) was isolated from two tubes of whole blood (approximately 20 mL) by centrifugation, followed by cell-free DNA isolation using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Germantown, Maryland, USA). DNA was isolated from formalin-fixed paraffin-embedded (FFPE) tissue of CNB surgical and FNA cytology specimens using the Maxwell 16 FFPE Plus LEV DNA Purification Kit (Promega, Madison, Wisconsin, USA). Quantification of cell-free DNA and tissue DNA was performed using the QuantiFluor ONE dsDNA System on the Quantus Fluorometer (Promega).
Droplet digital PCR assay
EGFR mutation detection was performed using the QX200 AutoDG Droplet Digital PCR System and the Qx200 Droplet Reader (Bio-Rad Laboratories, Hercules, California, USA) according to the manufacturer’s instructions, and analysed using QuantaSoft Analysis Software V.1.7.4.0917 (Bio-Rad Laboratories). A total of ≥6000 droplets with ≥1200 signal producing events for blood specimens or ≥600 events for FFPE specimens were required for a sample to meet the minimum acceptable quality criteria for reporting. Samples were reported as inconclusive if <6000 droplets, or fewer than 1200 signal producing events for blood specimens or 600 events for FFPE specimens were detected. Samples were reported as positive for EGFR T790M at a variant allele frequency (VAF) of ≥0.5% or ≥6 positive events and were reported as positive for EGFR L858R or exon 19 deletion mutations if a VAF of ≥1% was observed.
Interpretation of ddPCR assay
The lower limit of detection of ddPCR for EGFR T790M, L858R and exon 19 deletion mutations was validated to 0.5% VAF. Three result interpretations were possible on the ddPCR assay: positive, negative or inconclusive. A positive result for EGFR T790M was interpreted as detection of the T790M mutation in the patient plasma at a VAF of ≥0.5%, and a low-level positive was interpreted as detection of the mutation at a VAF of 0.3%–0.5%. A negative result was interpreted as undetectable for the T790M mutation and positive for the EGFR sensitising mutation (either the EGFR L858R variant or exon 19 deletion) that occurred within the tumour tissue of that patient, known from prior clinical testing. A negative result was also interpreted when testing for the sensitising mutation was not possible due to the sensitising mutation being unknown or not reported, lack of availability of a ddPCR assay for the particular sensitising mutation that occurred in that patient (ie, a sensitising mutation other than L858R or exon 19 deletion) or due to insufficient DNA obtained from the plasma specimen to offer sensitising mutation testing. An inconclusive result was interpreted when both EGFR T790M and the known sensitising mutation were undetectable in the patient plasma (ie, inability to confirm the presence of ctDNA in the sample), or when minimal acceptable quality metrics for EGFR T790M testing were not met.
Clinical assessment
Assessment of the clinical features at the time of relapse and EGFR T790M plasma testing was conducted for all patients seen by Medical Oncology at UHN for care. Clinical metrics selected for evaluation included: age, sex, smoking status, tumour size, number and location of metastases, radiological status of progression, date and type of TKI treatment, as well as blood protein measurements for LDH, aspartate aminotransferase (AST), alanine aminotransferase (ALT) and alkaline phosphatase (ALP).
Statistics
Associations between the EGFR T790M test result and clinical features were assessed using the Fisher exact test for the categorical variables and the Mann-Whitney for the continuous variables for the 107 patients treated at UHN to determine statistical significance. For each patient, the first positive T790M blood test result was used in associations; if there were no positive blood tests, the most recent blood test was used.
Results
Evaluation of EGFR T790M in circulating cell-free DNA samples
Circulating tumour DNA specimens from a cohort of patients with NSCLC that experienced relapse or progression on first generation TKIs were assessed for the presence of the EGFR T790M mutation in circulating cell-free DNA. A total of 471 specimens from 343 patients were assessed. Following extraction, an average quantity of 1.69 ng/µL of DNA was obtained (range 0.01–72.00 ng/µL). Of the 343 patients assessed, 24% (82/343) tested positive for the resistance mutation on the first test, 50% (171/343) tested negative and 26% (90/343) were inconclusive. In addition to the analysis of the EGFR T790M resistance mutation, the most common EGFR sensitising mutations (L858R and exon 19 deletions) were also assessed in the circulating cell-free DNA of those patients analysed. The EGFR sensitising mutation status was provided by the referring centre for 55% (130/236) of patients referred from sites other than UHN and was unavailable or not reported for the remaining 45% (106/236) of patients.
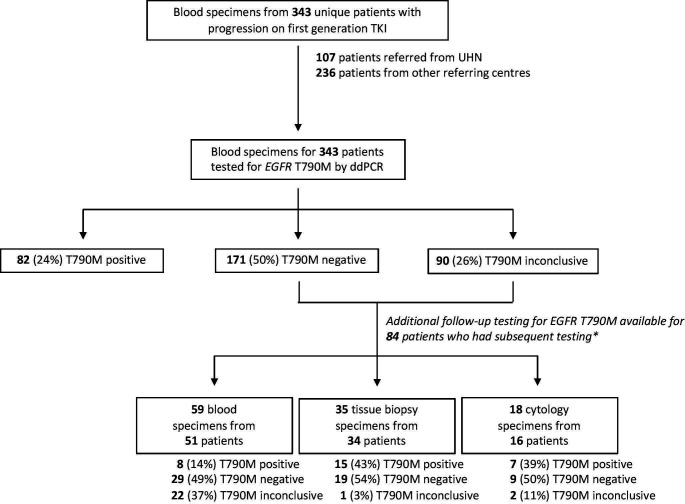
Of those 261 patients with a negative or inconclusive test result for EGFR T790M on initial testing that were tested for the EGFR sensitising mutation known to occur in that patient from prior clinical testing, 13% (33/261) were positive and 33% (86/261) were negative or inconclusive. A further 54% (142/261) of samples were not tested for the EGFR sensitising mutation due to the following reasons: lack of reporting of the sensitising mutation type by the referring centre (122/142); an alternate sensitising mutation occurred that was not captured by the ddPCR tests used in this study, including G719S/C/A, L861Q, N771delinsHV (12/142); or insufficient DNA material was available (8/142). A summary of the EGFR T790M mutational status of patients assessed in this study, including additional testing of patient plasma for ctDNA or reflex tissue biopsy and cytology testing, is shown in figure 1.
Figure 1.
Cohort of patients with lung cancer referred for epidermal growth factor receptor (EGFR) T790M ctDNA mutational status assessment. *Some patients received multiple subsequent tests, including serial blood and/or tissue biopsies from the same patient. TKI, tyrosine kinase inhibitor; UHN, University Health Network.
Correlation of clinical metrics with EGFR T790M mutation status
Clinical characteristics at the time of relapse were investigated for the cohort of patient treated at UHN and correlated with the occurrence of a positive ctDNA test result for EGFR T790M (table 1). Evaluation of patient demographics, including age, sex and smoking status demonstrated that none of these features correlated significantly with EGFR T790M mutational status. Clinical features related to tumour progression, including tumour size, number of metastases and location of metastases to the central nervous system, bone, liver, adrenals and/or lymph nodes were also evaluated, and no clinically significant associations were identified for these features. However, a trend towards a negative EGFR T790M ctDNA test result was observed in the small subset of patients with metastatic spread only within the lungs (table 1). Prior treatment with TKIs, including duration and type of TKI therapy, as well as plasma LDH, AST, ALT and ALP levels was also evaluated in association with EGFR T790M status and none of these metrics reached significance in our analyses.
Table 1.
Correlation of epidermal growth factor receptor (EGFR) T790M mutation status in plasma with demographics and clinical features of patients treated at the University Health Network (UHN)
| Characteristic | All patients (n=107) |
EGFR T790M positive (n=34) |
EGFR T790M negative or inconclusive (n=73) |
P value |
| Age, N | 103 | 32 | 71 | 0.36 |
| Mean, years (SD) | 65.6 (12.9) | 67.5 (12) | 64.8 (13.3) | |
| Median, years (min, max) | 64 (31, 99) | 71 (43, 86) | 63 (31, 99) | |
| Sex, N | 106 | 33 | 73 | 0.83 |
| Male, N (%) | 66 (62) | 20 (61) | 46 (63) | |
| Female, N (%) | 40 (38) | 13 (39) | 27 (37) | |
| Smoking status, N | 104 | 33 | 71 | 0.82 |
| Never, N (%) | 27 (26) | 9 (27) | 18 (25) | |
| Former, N (%) | 77 (74) | 24 (73) | 53 (75) | |
| Number of metastatic sites, N | 105 | 32 | 73 | 0.16 |
| Mean, N (SD) | 3.4 (1.6) | 3.7 (1.1) | 3.3 (1.7) | |
| Median, N (min, max) | 3 (0, 9) | 3.5 (2, 7) | 3 (0, 9) | |
| Metastases, N | 104 | 32 | 72 | |
| CNS, N (%) | 51 (49) | 14 (44) | 37 (51) | 0.53 |
| Bone, N (%) | 57 (55) | 21 (66) | 36 (50) | 0.2 |
| Liver, N (%) | 19 (18) | 7 (22) | 12 (17) | 0.59 |
| Lymph node, N (%) | 62 (60) | 21 (66) | 41 (57) | 0.52 |
| Adrenal, N (%) | 12 (12) | 3 (9) | 9 (13) | 0.75 |
| None (disease in lung only), N (%) | 9 (9) | 0 (0) | 9 (13) | 0.05 |
| Size of large tumour, N | 80 | 28 | 52 | 0.4 |
| Mean, mm (SD) | 2.9 (2.7) | 3.4 (3.6) | 2.7 (2) | |
| Median, mm (range) | 2.2 (0.3, 19) | 2.7 (0.3, 19) | 2.2 (0.4, 10.8) | |
| Prior TKI treatment, N | 104 | 33 | 71 | 0.84 |
| First-generation (gefitinib, erlotinib), N (%) | 95 (91) | 31 (94) | 64 (90) | |
| Second-generation (afatinib), N (%) | 9 (9) | 2 (6) | 7 (10) | |
| TKI treatment duration, N | 93 | 33 | 70 | 0.74 |
| Mean, days (SD) | 527.2 (418.4) | 471.7 (220.4) | 553.4 (483.9) | |
| Median, days (min, max) | 422 (28, 2149) | 458 (162, 1205) | 406 (28, 2149) | |
| LDH levels, N | 93 | 29 | 64 | 0.83 |
| ≤220 U/L, N (%) | 47 (51) | 14 (48) | 33 (52) | |
| >220 U/L, N (%) | 46 (49) | 15 (52) | 31 (48) | |
| EGFR sensitising mutation, N | 83 | 29 | 55 | 0.32 |
| L858R, N (%) | 35 (42) | 11 (38) | 24 (44) | |
| Exon 19 deletion, N (%) | 44 (53) | 18 (62) | 26 (48) | |
| Other (G719S/C/A, L861Q, N771delinsHV), N (%) | 4 (5) | 0 (0) | 4 (7) |
CNS, central nervous system; LDH, lactate dehydrogenase; TKI, tyrosine kinase inhibitor.
Patient demographics and clinical metrics were also correlated with the presence of ctDNA, measured via assessment of the presence of EGFR sensitising or resistance mutations in patient plasma (table 2), as an indication of patients who shed tumour DNA into peripheral circulation (ie, ctDNA ‘shedders’). Age, sex and smoking status did not correlate with the presence of ctDNA. However, a greater number of metastatic sites and metastases to the bone and liver did correlate significantly with the presence of tumour DNA shedding in patient plasma.
Table 2.
Correlation of circulating tumour DNA shedding in patient plasma with demographics and clinical features of patients treated at University Health Network
| Characteristic | All patients (n=107) |
Shedders* of circulating tumour DNA (n=48) |
Non-shedders of circulating tumour DNA (n=59) |
P value |
| Age, N | 103 | 46 | 57 | 0.66 |
| Mean, years (SD) | 65.6 (12.9) | 65.2 (12.6) | 66 (13.3) | |
| Median, years (min, max) | 64 (31, 99) | 63 (39, 88) | 65 (31, 99) | |
| Sex, N | 106 | 47 | 59 | 0.69 |
| Male, N (%) | 66 (62) | 28 (60) | 38 (64) | |
| Female, N (%) | 40 (38) | 19 (40) | 21 (36) | |
| Smoking status, N | 104 | 47 | 57 | 1 |
| Never, N (%) | 27 (26) | 12 (26) | 15 (26) | |
| Former, N (%) | 77 (74) | 35 (74) | 42 (74) | |
| Number of metastatic sites, N | 105 | 46 | 59 | 0.0012 |
| Mean, N (SD) | 3.4 (1.6) | 3.9 (1.3) | 2.9 (1.7) | |
| Median, N (min, max) | 3 (0, 9) | 4 (2, 7) | 3 (0, 9) | |
| Metastases, N | 104 | 46 | 58 | |
| CNS, N (%) | 51 (49) | 22 (48) | 29 (50) | 0.85 |
| Bone, N (%) | 56 (54) | 31 (67) | 25 (43) | 0.018 |
| Liver, N (%) | 18 (17) | 13 (28) | 5 (9) | 0.017 |
| Lymph node, N (%) | 61 (59) | 30 (65) | 31 (53) | 0.24 |
| Adrenal, N (%) | 12 (12) | 6 (13) | 6 (11) | 0.76 |
| None (disease in lung only), N (%) | 9 (9) | 0 (0) | 9 (16) | 0.004 |
| Size of large tumour, N | 79 | 40 | 39 | 0.31 |
| Mean, mm (SD) | 3 (2.7) | 3.3 (3.2) | 2.7 (2.1) | |
| Median, mm (range) | 2.2 (0.3, 19) | 2.5 (0.3, 19) | 2.1 (0.4, 10.8) | |
| Prior TKI treatment, N | 104 | 46 | 58 | 0.67 |
| First-generation (gefitinib, erlotinib), N (%) | 95 (91) | 42 (91) | 53 (91) | |
| Second-generation (afatinib), N (%) | 9 (9) | 4 (9) | 5 (9) | |
| TKI treatment duration, N | 103 | 47 | 56 | 0.96 |
| Mean, days (SD) | 523.2 (419.1) | 502.2 (367.6) | 540.9 (460.4) | |
| Median, days (min, max) | 419 (28, 2149) | 423 (28, 2015) | 406 (30, 2149) | |
| LDH levels, N | 93 | 43 | 50 | 0.54 |
| ≤220 U/L, N (%) | 47 (51) | 20 (47) | 27 (54) | |
| >220 U/L, N (%) | 46 (49) | 23 (53) | 23 (46) | |
| EGFR sensitising mutation, N | 83 | 43 | 40 | 0.11 |
| L858R, N (%) | 35 (42) | 18 (42) | 17 (42) | |
| Exon 19 deletion, N (%) | 44 (53) | 25 (58) | 19 (48) | |
| Other (G719S/C/A, L861Q, N771delinsHV), N (%) | 4 (5) | 0 (0) | 4 (10) |
*Shedders of tumour DNA are defined as patients with a positive plasma test result for either the epidermal growth factor receptor (EGFR) sensitising mutation (L858R or exon 19 deletion) or resistance mutation (T790M), as a measure of the presence of tumour DNA within the plasma specimen.
CNS, central nervous system; LDH, lactate dehydrogenase; TKI, tyrosine kinase inhibitor.
Assessment of reflex tissue and sequential blood testing for EGFR T790M
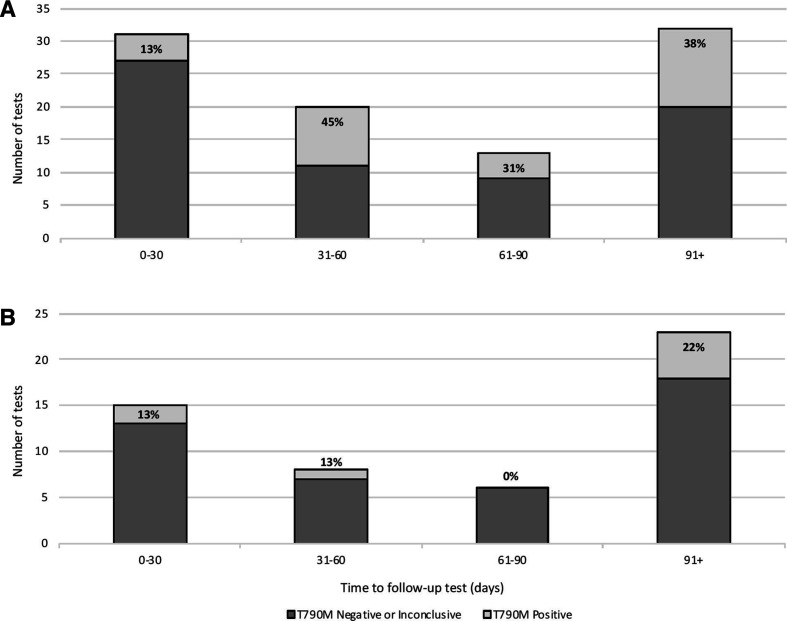
When feasible, patients with an initially negative or inconclusive ddPCR test underwent follow-up ddPCR testing after some period of time. Overall, 59 repeat serial blood specimen results were available for 51 patients, including 43 patients with 1 additional blood specimen and 8 patients with 2 follow-up blood specimens. Of those 59 specimens where repeat blood draws were available, 86% (51/59) remained negative or inconclusive, while 14% (8/59) became positive on further testing. The positivity rate of blood, tissue biopsy and FNA cytology specimens tested for EGFR T790M by ddPCR is summarised in figure 1. For the subset of patients that had follow-up testing by tissue biopsy or cytology, a positive EGFR T790M test result on sequential testing occurred in 43% (15/35) of reflex tissue biopsy specimens and 39% (7/18) of reflex cytology specimens. A summary of test results for those patients with an initial negative or inconclusive blood test for EGFR T790M that received reflex tissue or serial blood testing is included in figure 2, and the number of tests positive for EGFR T790M at each time interval, including test results for reflex tissue testing and sequential blood testing, is summarised in figure 3. Of those patients with sequential blood tests, approximately 13% (2/15) of patients received a positive EGFR T790M test result within 30 days of retesting (figure 3B). Over 20% (5/18) of patients with a repeat blood specimen tested at 90 days or longer received a positive result for EGFR T790M (figure 3B), with 2 patients developing a positive test result 6 months following an initially inconclusive test result (figure 2B).
Figure 2.
Reflex tissue or serial liquid biopsy testing of patients with a negative or inconclusive blood test for epidermal growth factor receptor (EGFR) T790M on initial testing, including (A) all patients with multiple measurements, and (B) only patients with multiple sequential tests where a subsequent test was positive for EGFR T790M.
Figure 3.
Number of sequential tests positive for epidermal growth factor receptor (EGFR) T790M by ddPCR and time interval following initial blood test. (A) The percentage of EGFR T790M positive or negative/inconclusive follow-up tests performed on any sequential blood specimens, reflex tissue biopsy and reflex tissue cytology specimens at 30-day intervals following an initially negative or inconclusive blood test. (B) The percentage of EGFR T790M positive or negative/inconclusive tests performed on only the subset of patients with available sequential blood specimens at 30-day intervals following an initially negative or inconclusive blood test.
Discussion
This retrospective analysis of EGFR T790M mutation detection by ddPCR in patients with NSCLC demonstrated the effective clinical implementation of published testing recommendations.9 11 Appropriate collection of blood using cell-free DNA blood collection tubes and accurate reporting of known EGFR sensitising mutation status from prior clinical testing were noted as important preanalytical criteria. Evaluation of the presence of the EGFR sensitising mutation known to occur in each patient from prior testing effectively enabled determination of those patients considered to be a ‘true negative’ for EGFR T790M versus those patients with results reported as inconclusive due to the inability to determine whether circulating tumour DNA was present in that sample. Of those patients evaluated in this study, 45% did not have a reported EGFR sensitising mutation provided by the referring laboratory. In patients where the EGFR T790M mutation was not detected in the blood specimen, a positive result for the known EGFR sensitising mutation would indicate that tumour DNA was shed into the patient plasma. Conversely, a negative plasma test result for the sensitising mutation would indicate that the tumour tissue in that patient was not shedding DNA into the patient plasma, and thus ctDNA was not detected. Knowledge of the known sensitising mutation identified through prior clinical testing therefore offers utility in the interpretation of ddPCR testing results for EGFR T790M, and it is recommended that these data be provided by laboratories at the time of referral.
Of the 343 patients with blood specimens received for EGFR T790M testing, 24% tested positive for the mutation, which is comparable to the positivity rate of approximately 27% reported in the literature.14 Although no clinically significant associations were identified between EGFR T790M positivity and tumour size, and number or location of metastases, all patients with local metastases that occurred only within the lungs tested negative for EGFR T790M, indicating that a negative ctDNA test result is more likely to occur in patients that do not have extrathoracic disease spread. Of the 82 patients that tested positive for EGFR T790M, 49% of patients carried the exon 19 deletion sensitising mutation and 27% were known to carry the L858R sensitising variant, while the sensitising mutation status of the remaining 24% of patients was not reported. This disproportionate enrichment of patients with the exon 19 deletion genotype has been reported to occur15 and suggests an underlying difference in the development of TKI resistance in these patient cohorts.
The presence of tumour DNA shedding, as measured by the presence of EGFR sensitising or resistance mutations in liquid biopsy specimens, was observed in our data to correlate with the number of metastatic sites and metastases to the bone and liver in particular, a finding which has similarly been reported in the literature.13 However, this correlation has not been previously identified for the EGFR T790M mutation in particular, and indeed our data do not suggest a correlation between plasma T790M status and number or size of metastases. Therefore, although EGFR T790M status does not correlate with number or location of metastases, this feature does in fact correlate with tumour DNA shedding.
Of the 261 patients with a negative or inconclusive blood test for EGFR T790M, reflex tissue CNB or FNA cytology specimens were available for 13% and 6%, respectively. Tumour tissue testing is known to yield a higher rate of T790M detection as compared with blood testing,14 and tumour heterogeneity can result in variation in T790M status at different tumour sites.16 In the present study, the positivity rate of EGFR T790M obtained from tumour tissue and cytology testing is therefore unsurprisingly higher than that of sequential blood analysis, with a positivity rate of 14% for serial blood specimens, with 43% of tissue biopsies and 39% of cytology specimens positive for T790M.
For the patient population where reflex tissue testing is not possible, the optimal timing of retesting these patients by plasma testing for ctDNA remains unclear. We evaluated those patients who received serial blood tests to investigate what clinical indicators might be present to trigger a repeat liquid biopsy. Although no clear clinical trigger was identified, our findings demonstrate that a positive test result was identified in 13% of patients retested within 60 days following an initially negative or inconclusive test, and two patients developed a positive test 6 months after a negative blood test. Of note, the majority of those who become positive for EGFR T790M on sequential blood tests initially received an inconclusive blood test. This suggests that it is particularly important to retest those patients who received an inconclusive result on initial testing. Our data demonstrate that repeat ctDNA testing may be clinically indicated when tissue testing is not feasible and that a substantial percentage of patients with an initially negative test will become positive for EGFR T790M ctDNA at a later date, if offered sequential testing. This finding highlights the importance of further studies to investigate appropriate timing of repeat blood specimen testing for monitoring for EGFR T790M status and repeat ctDNA testing should be offered if tissue reflex testing is not possible, particularly when clinically indicated due to signs of progression.
Take home messages.
The droplet digital PCR test for epidermal growth factor receptor (EGFR) T790M mutations effectively triaged 24% of patients for treatment with third-generation tyrosine kinase inhibitors, avoiding the need for invasive tissue biopsy in these patients.
A positive test result was identified in 13% of patients retested within 60 days following an initially negative or inconclusive test, and positive test results correlated with increased tumour shedding.
Repeat ctDNA testing may be clinically indicated when tissue testing is not feasible and a substantial percentage of patients with an initially negative test will become positive for EGFR T790M ctDNA at a later date, if offered sequential testing.
Footnotes
Handling editor: Runjan Chetty.
Contributors: TJS and TS contributed to the planning, conduct, data collection and analysis, writing and reporting of all work described in the article. FS contributed to the planning, conduct and analysis of the reported work. SP contributed to the collection and analysis of the clinical data. JW designed, conducted and reported the statistical data. SG and LR contributed to the technical and methodological data collection, and SG contributed to the methodological writing.
Funding: Work in this manuscript was funded in part by AstraZeneca and the Princess Margaret Cancer Foundation
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: This study was approved by the UHN Research Ethics Board (#18-5988).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request. This manuscript contains deidentified participant data.
References
- 1. Sharma SV, Bell DW, Settleman J, et al. . Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169–81. 10.1038/nrc2088 [DOI] [PubMed] [Google Scholar]
- 2. Yun C-H, Boggon TJ, Li Y, et al. . Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 2007;11:217–27. 10.1016/j.ccr.2006.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lindeman NI, Cagle PT, Beasley MB, et al. . Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American pathologists, International association for the study of lung cancer, and association for molecular pathology. J Thorac Oncol 2013;8:823–59. 10.1097/JTO.0b013e318290868f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu HA, Arcila ME, Rekhtman N, et al. . Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240–7. 10.1158/1078-0432.CCR-12-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khozin S, Weinstock C, Blumenthal GM, et al. . Osimertinib for the treatment of metastatic EGFR T790M mutation-positive non-small cell lung cancer. Clin Cancer Res 2017;23:2131–5. 10.1158/1078-0432.CCR-16-1773 [DOI] [PubMed] [Google Scholar]
- 6. Soria J-C, Ohe Y, Vansteenkiste J, et al. . Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 2018;378:113–25. 10.1056/NEJMoa1713137 [DOI] [PubMed] [Google Scholar]
- 7. Kim TM, Song A, Kim D-W, et al. . Mechanisms of acquired resistance to AZD9291: a mutation-selective, irreversible EGFR inhibitor. J Thorac Oncol 2015;10:1736–44. 10.1097/JTO.0000000000000688 [DOI] [PubMed] [Google Scholar]
- 8. Thress KS, Paweletz CP, Felip E, et al. . Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560–2. 10.1038/nm.3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lindeman NI, Cagle PT, Aisner DL, et al. . Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the college of American pathologists, the international association for the study of lung cancer, and the association for molecular pathology. J Mol Diagn 2018;20:129–59. 10.1016/j.jmoldx.2017.11.004 [DOI] [PubMed] [Google Scholar]
- 10. Goldman JW, Noor ZS, Remon J, et al. . Are liquid biopsies a surrogate for tissue EGFR testing? Ann Oncol 2018;29:i38–46. 10.1093/annonc/mdx706 [DOI] [PubMed] [Google Scholar]
- 11. Stockley T, Souza CA, Cheema PK, et al. . Evidence-based best practices for EGFR T790M testing in lung cancer in Canada. Curr Oncol 2018;25:163–9. 10.3747/co.25.4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fan G, Zhang K, Ding J, et al. . Prognostic value of EGFR and KRAS in circulating tumor DNA in patients with advanced non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget 2017;8:33922–32. 10.18632/oncotarget.15412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu Y-J, Zhang H-B, Liu Y-H, et al. . Quantitative cell-free circulating EGFR mutation concentration is correlated with tumor burden in advanced NSCLC patients. Lung Cancer 2017;109:124–7. 10.1016/j.lungcan.2017.05.005 [DOI] [PubMed] [Google Scholar]
- 14. Wan R, Wang Z, Lee JJ, et al. . Comprehensive analysis of the discordance of EGFR mutation status between tumor tissues and matched circulating tumor DNA in advanced non-small cell lung cancer. J Thorac Oncol 2017;12:1376–87. 10.1016/j.jtho.2017.05.011 [DOI] [PubMed] [Google Scholar]
- 15. Ke E-E, Zhou Q, Zhang Q-Y, et al. . A higher proportion of the EGFR T790M mutation may contribute to the better survival of patients with exon 19 deletions compared with those with L858R. J Thorac Oncol 2017;12:1368–75. 10.1016/j.jtho.2017.05.018 [DOI] [PubMed] [Google Scholar]
- 16. Arcila ME, Oxnard GR, Nafa K, et al. . Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res 2011;17:1169–80. 10.1158/1078-0432.CCR-10-2277 [DOI] [PMC free article] [PubMed] [Google Scholar]