Abstract
Erythromycin is a commonly used broad-spectrum antibiotic, but resistance to this antibiotic makes its use less effective. Considerable efforts, beside finding alternatives, are needed to enhance its antimicrobial effect and stability against bacteria. Tetrahedral framework nucleic acids (tFNAs), a novel delivery vehicle with a three-dimensional nanostructure, have been studied as a carrying platform of antineoplastic drugs. In this study, the use of tFNAs in delivering erythromycin into Escherichia coli (E. coli) was investigated for the first time. The tFNAs vehicle increased the bacterial uptake of erythromycin and promoted membrane destabilization. Moreover, it increased the permeability of the bacterial cell wall, and reduced drug resistance by improving the movement of the drug across the membrane. The tFNAs-based delivery system enhanced the effects of erythromycin against E. coli. It may therefore provide an effective delivery vehicle for erythromycin in targeting antibiotic-resistant bacteria with thick cell wall.
Keywords: Tetrahedral framework nucleic acids, Drug delivery system, Escherichia coli, Drug penetration disorder, Erythromycin
Graphical abstract
Highlights
-
•
The tFNAs-based delivery system enhanced the effects of erythromycin against E. coli.
1. Introduction
Erythromycin is a macrolide antibiotic that can irreversibly bind to the 50S subunit of bacterial ribosomes, resulting in the ribosomal inability to polymerize specific amino acid sequences during protein synthesis, by blocking the process of peptide transfer and mRNA transfer, and blocking the growth of the peptide chain, ultimately playing a bacteriostatic role [1,2]. The development of bacterial resistance is one of the greatest challenges to human health. Antibiotic resistance refers to the enhanced survival ability of bacteria in the presence of antibiotics and the reduced effectiveness of antibiotics against bacteria. Due to the incorrect use of drugs, drug-resistant strains have developed rapidly in the recent decades. According to clinical statistics, the drug resistance rate of pathogenic bacteria has reached 30–50%, and it increases by 5% every year [3,4]. The causes of erythromycin resistance mainly include [[5], [6], [7], [8]]: 1. The N6 position in nucleotide A2058 is monomethylated or dimethylated. Methylation can interfere with the formation of hydrogen bonds, leading to a significant decrease in the affinity between macrolides and ribosomal 50s subunits, resulting in the production of resistant strains; 2. the decrease in membrane permeability may lead to the decrease of drug uptake and accumulation, which may lead to drug resistance; 3. the induction of efflux pump may lead to a reduce intracellular concentration. (see Scheme 1)
Scheme 1.
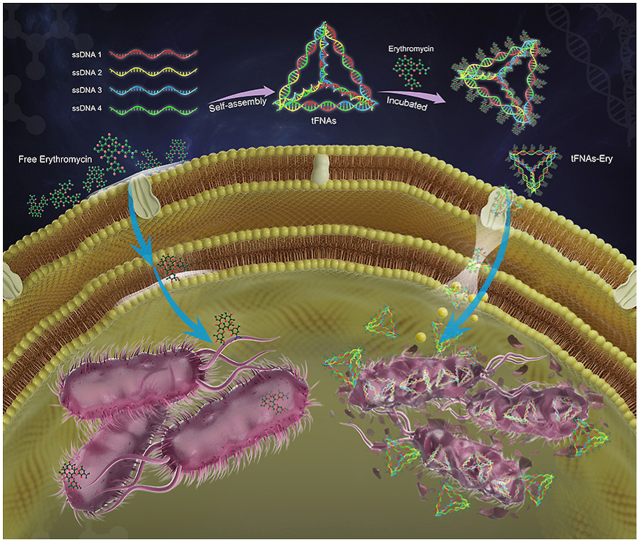
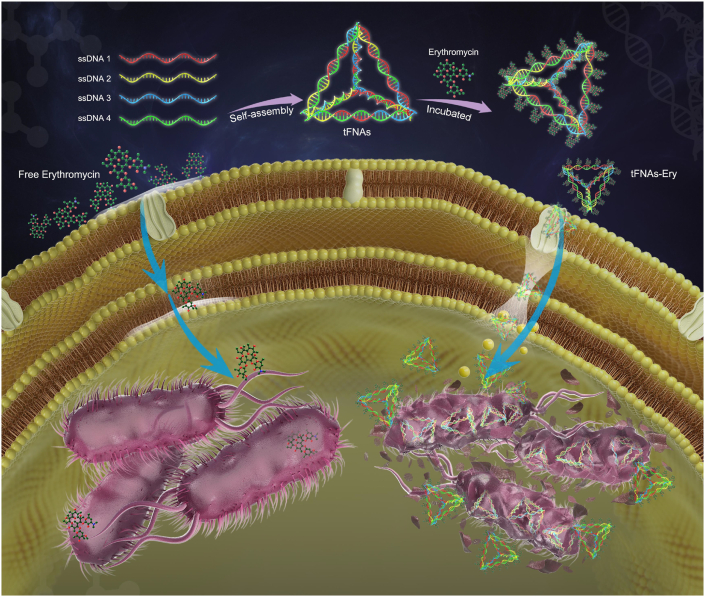
Schematic illustration of the inhibition process of tFNAs-Ery in E. coli. tFNAs increase the erythromycin efficiency by delivering it more inside the cells.
Nucleic acids are important genetic materials in pathogenic microorganisms, which play a definite role in a series of important life phenomena, such as growth, heredity, and variation. Bacterial nucleic acids can be used to detect the pathogenicity and existence of drug-resistant genes in microorganisms [9,10]. Four single-stranded DNA (ssDNA) molecules self-assemble into tetrahedral configurations, forming Tetrahedral Framework Nucleic Acids (tFNAs), based on the highly specific complementary base pairing principle [[11], [12], [13]]. Previous studies showed that tFNAs have an excellent biocompatibility and a favorable ability to permeate bacterial and mammalian cells, that have been recognized as a promising delivery vehicle used in delivering drug molecules [[14], [15], [16], [17], [18], [19]].
In recent years, the use of nanoparticles and other antibacterial materials to achieve bacterial control has attracted the attention of researchers all over the world [20,21]. The application of nucleic acid materials in antibiotic research is mainly focused on target detection, and not on the improvement of antibacterial activity. To reduce the outflow of antibiotics from bacteria that are not sensitive to antibiotics, strengthening antibiotic delivery via endocytosis using tFNAs - could be an option [[22], [23], [24], [25]]. In this study, we used tFNAs to deliver erythromycin (tFNAs-Ery), a commonly used broad-spectrum antibiotic, and evaluated its antibacterial properties against Escherichia coli (E. coli). Due to the strong transmembrane penetrative ability of tFNAs, tFNAs-Ery enable more erythromycin to enter the cell efficiently, and exert antibacterial properties more effectively compared with erythromycin alone. In addition, morphological observation revealed that even under the same erythromycin concentration, erythromycin transported using tFNAs did more damage to the bacterial membrane structure than did free erythromycin. tFNAs-Ery was more lethal than free erythromycin, possibly due to the changes in the cell membrane permeability made by tFNAs which allowed for an increased drug penetration.
2. Material and methods
2.1. Synthesis of tFNAs and drug loading
The specific synthesis details of tFNAs can be seen in previous research reports [[26], [27], [28]]. S1–S4 single DNA strands -Table 1 showed the sequence of bases for each strand of DNA. To form tFNAs-Ery, erythromycin solution (different content: 4000, 2000, 1000, 500 and 250 μg) was incubated with tFNAs (100 nM) at room temperature for 24 h, and then centrifuged at 10 000 rpm for 10 min with a 15KD ultrafiltration centrifuge tube to remove unloaded drug molecules and single strand of DNA. The loading efficiency of erythromycin into tFNAs was analyzed with UV–vis spectroscopy (U–3900H, HITACHI, JAPAN) to detect erythromycin absorbance peak at 236 nm. According to the content reflected by the peak area, the loaded drug content was calculated (Table 2). The loading efficiency of erythromycin into tFNAs can be calculated as follows:
Table 1.
Base Sequences of Each Specific ssDNA.
| ss DNA | Base sequence (5’→3′) |
|---|---|
| S1 | ATTTATCACCCGCCATAGTAGACGTATCACCAGGCAGTTGAGACGAACATTCCTAAGTCTGAA |
| S2 | ACATGCGAGGGTCCAATACCGACGATTACAGCTTGCTACACGATTCAGACTTAGGAATGTTCG |
| S3 | ACTACTATGGCGGGTGATAAAACGTGTAGCAAGCTGTAATCGACGGGAAGAGCATGCCCATCC |
| S4 | ACGGTATTGGACCCTCGCATGACTCAACTGCCTGGTGATACGAGGATGGGCATGCTCTTCCCG |
Table 2.
Loading efficiency of Erythromycin into tFNAs.
| Sample | Initial Erythromycin (μg) | Abs peak area/total peak area (%) | Loading Erythromycin (μg) | Loading Efficiency (%) |
|---|---|---|---|---|
| 1 | 4000 | 47.34 | 1893.6 | 47.34 |
| 2 | 2000 | 61.85 | 1237 | 61.85 |
| 3 | 1000 | 81.34 | 813.4 | 81.34 |
| 4 | 500 | 83.65 | 418.25 | 83.65 |
| 5 | 250 | 79.32 | 198.3 | 79.32 |
2.2. Identification of the successfully synthesized tFNAs and tFNAs-Ery
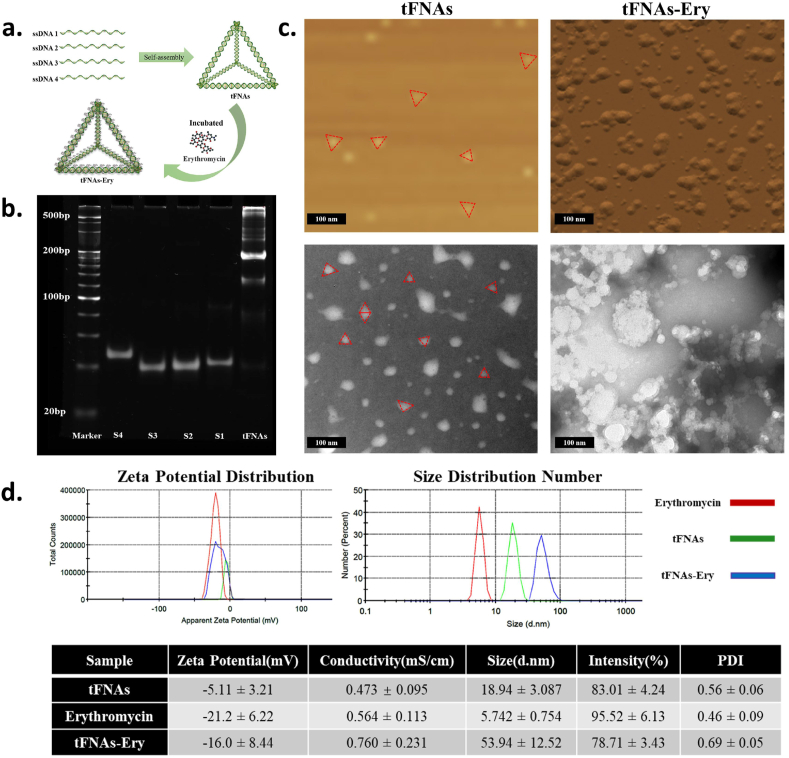
For verification of tFNAs, 8% polyacrylamide gel electrophoresis (PAGE) was used to measure the molecular weights of four ssDNAs and tFNAs, Transmission electron microscopy (TEM, Libra200, Zeiss, German) and Atomic force microscopy (AFM, SPM-9700 instrument, Shimadzu, Kyoto, Japan) was performed for morphological structural observation of tFNAs and tFNAs-Ery. The hydrodynamic sizes and zeta potentials of tFNAs, erythromycin and tFNAs-Ery were measured using a Zeta sizer Nano-ZS (Malvern Instruments, England).
2.3. Cytotoxicity assay
HUVECs and L929 were purchased from the American Type Culture Collection (ATCC®CRL1730™). HUVECs and L929 cells was cultured in high-glucose DMEM (0.1 mM non-essential amino acids, 4 mM l-glutamine, 10% FBS and 1% penicillin-streptomycin antibiotics) and RPMI-1640 culture medium (10% FBS and 1% penicillin-streptomycin antibiotics), respectively. HUVECs and L929 cells were seeded in a 96-well plate (5000 per well) and incubated with tFNAs (100, 200, 250, 500 and 750 nM) and (tFNAs-Ery 5, 50, 500 μg/mL), respectively, for 24 h and 48 h. The changes in proliferation of HUVECs and L929 cells was detected with the cell counting kit-8 (CCK-8; Dojindo, Japan). Specifically, 100 μL medium was mixed with 10 μL CCK-8 reagent per well and measured by a microplate reader (Thermo Scientific, USA). HUVECs and L929 were cultured in 12-well plates (3000 cells per well) for immunofluorescent staining. Please refer to supplementary document to check the specific procedure.
2.4. Cultivation of the microorganism
The Escherichia coli (E. coli) strain (BNCC133264) was obtained from BeNa Culture Collection (Beijing, China). A bacteria colony was picked up using gunpoint and was dipped in 3 mL Luria-Bertani (LB, pH 7.4) medium, cultivated in no carbon dioxide incubator at 37 °C. A turbidimeter was used to measure the bacterial concentration after the E. coli cells were isolated by centrifugation (4000 rpm, 3 min) and rinsed with PBS buffer. Measuring the optical density at 600 nm (OD600nm) to determine the bacterial content. All tools and utensils were sterilized by autoclaving at 120 °C for 30 min and ultraviolet disinfection for 60 min before experiment.
2.5. Antibacterial assay
In order to observe the effect of tFNAs on E. coli growth, the OD600nm of bacteria was observed after 24 h of co-culture with different concentrations of tFNAs (100–500 nM). The control group was the E. coli with no treatment. The E. coli were adjusted with an initial density of 5 × 105 CFU/mL to set up different experimental groups in the 96-well plates for the minimum inhibitory concentration (MIC) test. For detailed experimental procedures, please refer to supplementary document. The OD600nm values of each group was recorded from 0 to 24 h through an automated spectrophotometer (BioTek Instruments, USA) and was drawn as the growth curve. The OD600nm values of the erythromycin, ssDNA-Ery and tFNAs-Ery group were recorded at 24 h to compare the bacterial growth between groups. Sterile straws were used to absorb the diluted E. coli liquid from the 96 wells plate corresponding to drug concentration of 0.25 × MICEry and inoculate it on MHB agar plate into a 37 °C incubator for 24 h, for the enumeration by plate count.
2.6. The ability of tFNAs-Ery to penetrate the bacterial cell wall membrane into the E. coli
To observe the inside-bacterial localization of tFNAs and tFNAs-Ery, the confocal laser microscope (TCS SP8; Leica, Wetzlar, Germany) was performed to catch bacterial images. ssDNA, tFNAs and tFNAs-Ery modifying by Cyanine-5 (Cy5) were incubated with E. coli (5 × 105 CFU/mL) in LB liquid medium at 37 °C for 3 h. The strain was centrifuged and collected and stained with SYTO-9 dye for 15 min. It was resuspended in PBS after washing three times by PBS. To measure bacterial uptake rate of ssDNA, tFNAs and tFNAs-Ery, the E. coli strain (10 000 cells for each sample) was measured using flow cytometer (FC500 Beckman, IL USA) after centrifuged and collected and suspended in 500 μL of PBS.
2.7. Assessment of the leakage of cytoplasm
100 nM tFNAs, erythromycin and tFNAs-Ery were separately incubated with 5 × 105 CFU/mL E. coli. The control group was untreated E. coli. All the samples were centrifuged (4000 rpm, 5 min) and filtrated to obtain the supernatants after incubation at 37 °C for 3 h. Atomic absorption spectrometer (AAS; SpectrAA 220FS, VARIAN, America) was used to analyzed the concentration of [Na+] and [K+] inside the obtained supernatants. Subsequently, the supernatants were additionally subjected to the o-Nitrophenyl-β-D-Glucopyranosides (ONPG) test, which involved incubation with 4 mM ONPG (20 h, 37 °C) followed by measurement of the optical density at 420 nm (OD420nm) in a 96-well plate through a microplate reader.
2.8. The morphological observation of E. coli
The E. coli treated by 100 nM tFNAs, erythromycin and tFNAs-Ery respectively were collected and isolated. AFM, Scanning Electron Microscopy (FEI, INSPECT F, USA) and TEM were used to observe the morphological changes of the E. coli. The AFM sample (a drop of bacteria solution) suspended in PBS was dripped on cleaved mica dried for 20 min before putting into the instrument. For SEM observation, the E. coli cells were fixed overnight in 4% paraformaldehyde at 4 °C, dehydrated with anhydrous ethanol concentration gradient and then dried in vacuum and were coated with gilded film with their surface. TEM samples (E. coli cells fixation dehydration) were prepared with firmware copper carrier mesh for observation.
2.9. Stability analysis of tFNAs and tFNAs-Ery
tFNAs (250 nM) and tFNAs -Ery was co-incubated cultivated with 10% FBS, HUVECs cell lysate and E. coli lysate, respectively. Gel electrophoresis was used to observe the change of band for the nanostructure of tFNAs at 0–24 h.
2.10. Data analysis
SPSS 19.0 (IBM, Armonk, NY) was used for data analysis. ANOVA and t-test were conducted to obtain variance between-group. All quantitative results were presented as mean ± standard deviation (SD). If the two-tailed P value was <0.05(*), <0.01(**), and <0.001(***), it can be considered that the data were significantly different.
3. Results
3.1. Characterization of tFNAs and tFNAs-Ery
8% polyacrylamide gel electrophoresis (PAGE), which was used to measure the molecular weight of tFNAs, showed that tFNAs is a self-assembled structure of four ssDNAs, each having a molecular weight of about 50 bp, while the tFNAs having a molecular weight of about 200 bp (Fig. 1b). Furthermore, atomic force microscopy (AFM) and transmission electron microscopy (TEM) demonstrated that tFNAs marked with dotted red line had a triangular shape, which is in accordance with previous studies, while tFNAs-Ery had small particles around and inside the tFNAs (Fig. 1c). The sizes and zeta potentials of the tFNAs, erythromycin alone and tFNAs-Ery were determined using dynamic light scattering (DLS). The average zeta potential of tFNAs-Ery changed from −5.11 ± 3.21 mV to −16.0 ± 8.44 mV as the tFNAs was loaded with increasing amounts of erythromycin. The average size of tFNAs was 18.94 ± 3.087 nm, while that of tFNAs-Ery was 53.94 ± 12.52 nm (Fig. 1d). The results above highlight the convenience and efficiency of fabricating the tFNAs-Ery complex.
Fig. 1.
Characterization of tFNAs and tFNAs-Ery. a. Schematic illustration synthesis of tFNAs-Ery. b. Confirmation of the successful synthesis of tFNAs by polyacrylamide gel electrophoresis (PAGE). The relative size of single-stranded DNA, partial assembly and tFNAs. c. Atomic Force Microscope Images and Transmission Electron Microscope Images of tFNAs (red dotted line) and tFNAs-Ery. d. Zeta potentials and Size of tFNAs and tFNAs-Ery. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. The biocompatibility of tFNAs and tFNAs-Ery
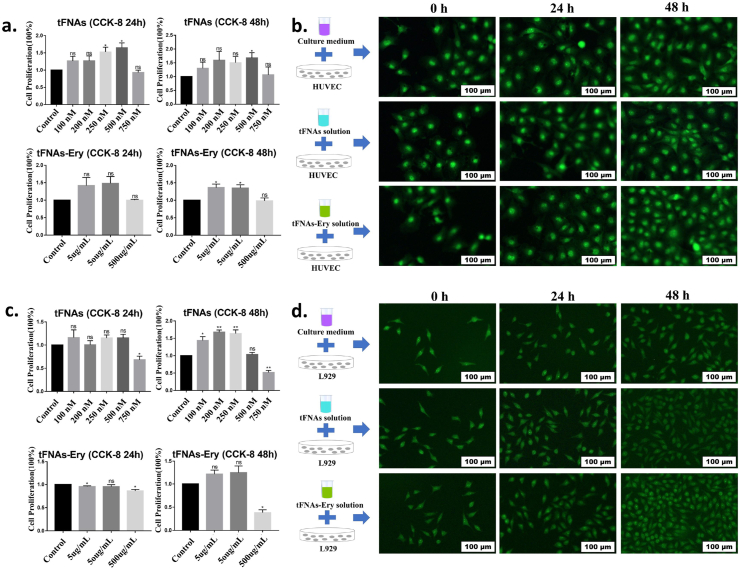
The cytotoxicity of tFNAs and tFNAs-Ery is an important consideration for further biological applications. Fig. 2a shows that even at a high dose (750 nM tFNAs; 500 μg/mL tFNAs-Ery), tFNAs and tFNAs-Ery showed no obvious HUVEC cytotoxicity after incubation for 24 h and 48 h. On the other hand, growth inhibition in L929 cells were observed at 750 nM and 500 μg/mL of tFNAs and tFNAs-Ery, respectively (Fig. 2c). To investigate the effect of tFNAs and tFNAs-Ery on the maintenance of HUVEC and L929 integrities, cell morphologies were investigated via immunofluorescent staining. Fig. 2b and d shows that the HUVEC and L929 cells exposed to tFNAs and tFNAs-Ery maintained normal cellular morphologies and showed normal cell proliferation. Notably, these results suggested that 0–500 nM of tFNAs and 0–50 μg/mL of tFNAs- Ery are safe for further in vivo use.
Fig. 2.
Cytotoxicity test of tFNAs and tFNAs-Ery. a. HUVEC cells proliferation analyzed by CCK-8 assay after being cocultured with tFNAs and tFNAs-Ery at 24 h and 48 h. Statistical analysis (n = 4): *, p < 0.05; **, p < 0.01. b. Observation of HUVEC morphological changes after exposure to tFNAs and tFNAs-Ery by fluorescent microscopy. c. L929 cells proliferation analyzed by CCK-8 assay after being cocultured with tFNAs and tFNAs-Ery at 24 h and 48 h. Statistical analysis (n = 4): *, p < 0.05; **, p < 0.01. d. Observation of L929 morphological changes after exposure to tFNAs and tFNAs-Ery by fluorescent microscopy.
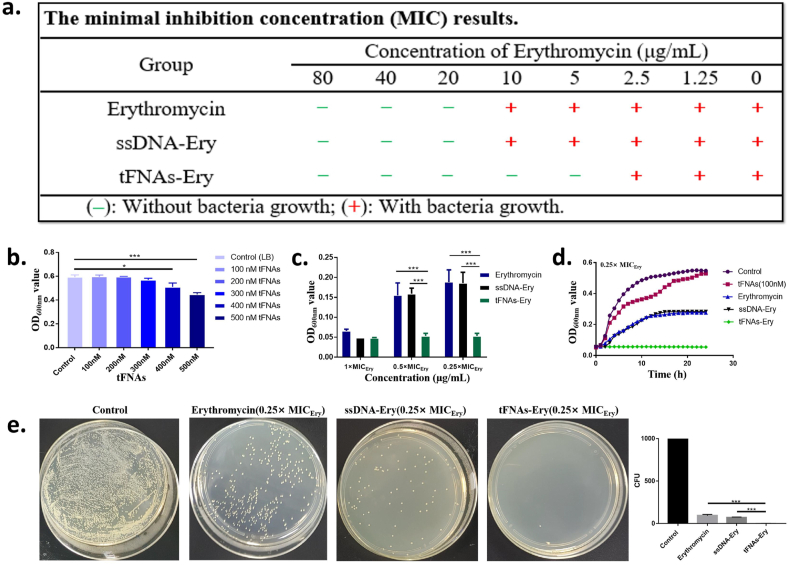
3.3. Determination of antibacterial activity in vitro
The optical density of E. coli at 600 nm (representing the density of bacteria) was measured after 24 h of incubation with erythromycin, ssDNA-Ery, and tFNAs-Ery, respectively. As shown in Fig. 3a, tFNAs-Ery showed a better antibacterial effect compared to erythromycin and ssDNA-Ery, based on MIC results. 0–300 nM of tFNAs had no significant effect on the growth of E. coli (Fig. 3b). The MIC of the erythromycin on E. coli is denoted as MICEry and the MIC of tFNAs-Ery was a quarter of MICEry. The OD600nm value of different groups as the concentration of the erythromycin changes from 1 × MICEry to 0.25 × MICEry as showed in Fig. 3c. The density of bacteria of tFNAs-Ery group was significantly less than that of erythromycin group and ssDNA-Ery group at 0.5 × MICEry and 0.25 × MICEry. In groups treated with the same erythromycin treatment concentration, 24-h E. coli growth curves revealed that tFNAs-Ery had suppressed E. coli growth more than ssDNA-Ery and erythromycin (Fig. 3d). The CFU counts also demonstrated the above results (Fig. 3e). The plate colony count of the erythromycin and ssDNA-Ery treatment group had more colonies than those of tFNAs-Ery group through intuitive observation. The above results indicated that erythromycin has a better effect with the aid of tFNAs but ssDNA didn't work.
Fig. 3.
Antibacterial activity of tFNAs-Ery on E. coli. a. The minimal inhibition concentration (MIC) results of erythromycin, ssDNA-Ery and tFNAs-Ery. b. The effect of different concentration of tFNAs on E. coli proliferation. The control group was the E. coli with no treatment. Statistical analysis (n = 3): *, p < 0.05; **, p < 0.01; ***, p < 0.001. c. Comparison of the antibacterial effects of erythromycin, ssDNA-Ery and tFNAs-Ery at 1 × MICEry, 0.5 × MICEry, 0.25 × MICEry by OD600nm values. Statistical analysis (n = 3): *, p < 0.05; **, p < 0.01; ***, p < 0.001. d. The 24-h growth curves of E. coli after treating by100nM tFNAs, 0.25 × MICEry of erythromycin, ssDNA-Ery and tFNAs-Ery. The control group was the E. coli with no treatment. e. Comparison of the plate colony count of E. coli in erythromycin, ssDNA-Ery and tFNAs-Ery groups at 0.25 × MICEry.
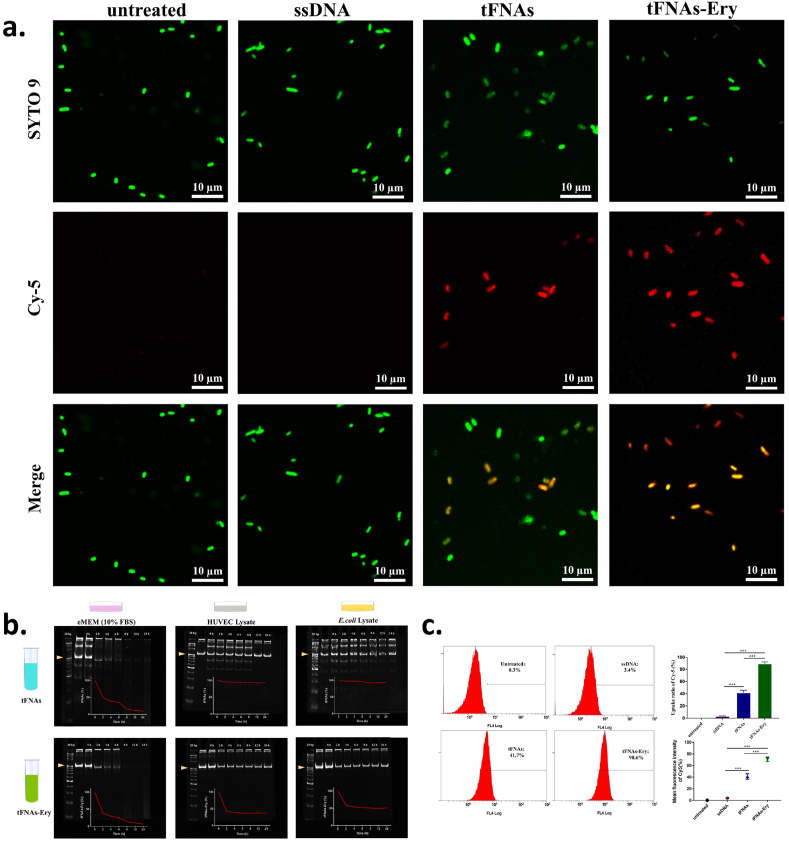
3.4. Bacterial uptake of tFNAs and tFNAs-Ery
Since erythromycin needs to enter the bacterial cell to manifest its bactericidal effect, the bacterial uptake of erythromycin is a mandatory first step for its mechanism of action. To determine whether tFNAs could successfully transport erythromycin inside E. coli cells, we tested the bacterial uptake of tFNAs and tFNAs-Ery via flow cytometry and confocal laser scanning microscopy (CLSM). As shown in Fig. 4a, untreated bacteria showed green fluorescence after incubation with SYTO-9, whereas Cy-5- labeled tFNAs and tFNAs-Ery showed red fluorescence. Therefore, obvious bacterial uptake of the fluorescent tFNAs or tFNAs-Ery appeared yellow in merged images. The CLSM images, which were consistent with the flow cytometry results, showed that the uptake ratio of tFNAs-Ery was more than that of tFNAs alone. Flow cytometry showed that the E. coli cellular uptake of Cy5-labeled tFNAs reached about 41.7% in 90 min, while Cy5-labeled tFNAs-Ery reached up to 90.6%. However, Cy-5-labeled ssDNA had difficulty in entering E. coli cells (Fig. 4c). Under the same drug concentration, tFNAs itself had no antibacterial effect, while tFNAs-Ery had a stronger antibacterial effect than erythromycin alone. These indicate that the tFNAs helped increase the concentration of erythromycin inside the target cells.
Fig. 4.
The bacterial uptake and stability of tFNAs and tFNAs-Ery. a. Confocal laser scanning microscopy images of bacterial uptake of tFNAs and tFNAs-Ery in E. coli at 90 min. Green fluorescence shows the total number of live bacteria; Red fluorescence come from Cy5-ssDNA, Cy5-tFNAs or Cy5-tFNAs-Ery; Yellow fluorescence in the merge images represent Cy5-ssDNA, Cy5-tFNAs or Cy5-tFNAs-Ery co-located with the E. coli; Statistical analysis (n = 3): ***, p < 0.001. b. polyacrylamide gel electrophoresis (PAGE) analysis of the stability of tFNAs and tFNAs-Ery nanostructure after incubation with eMEM medium (with 10% FBS), HUVEC lysate (Protein concentration:500 μg/mL) and E. coli lysate (Protein concentration:500 μg/mL) at 37 °C for 0–24 h, respectively. c. Flow cytometry analysis of the uptake rates of E. coli incubated with ssDNA, tFNAs and tFNAs-Ery. The control group was the E. coli with no treatment; Statistical analysis (n = 3): ***, p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. Stability analysis of tFNAs and tFNAs-Ery
According to the above results, we found that the tetrahedral spatial structure of tFNAs is the key to its ability to efficiently carry erythromycin into the E. coli cells. Although the effective concentration of free erythromycin against E. coli is 20 μg/mL, our results demonstrated that even 5 μg/mL of tFNAs-Ery was evidently bactericidally effective for 24 h. It is expected that tFNAs will remain structurally stable in the environment where bacteria grow and metabolize, and that their transport efficiency will not be reduced. To analyze the stability of tFNAs and tFNAs-Ery, we incubated tFNAs (250 nM) with 10% FBS, cell (HUVECs) lysate and E. coli lysate, respectively. Gel electrophoresis showed that the band for the tFNAs nanostructure remained almost unchanged after 12 h of incubation in 10% FBS, cell lysate and E. coli lysate, respectively (Fig. 4b). Furthermore, the band for the tFNAs could still be observed with a slightly attenuated intensity even after 24 h of incubation. These results indicate that tFNAs may be stable against nuclease degradation in biological media.
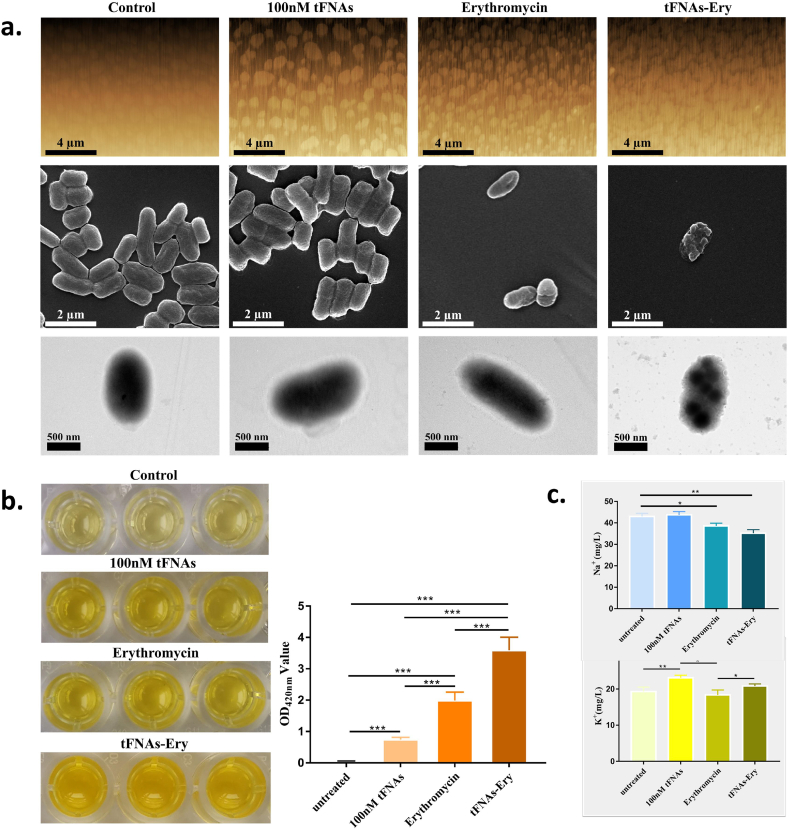
3.6. Morphological observation of E. coli cells
After the confirmation of the increased uptake of tFNAs-Ery by E. coli cells, individual cellular morphology was observed. The AFM images demonstrated that tFNAs alone caused little membrane changes, and erythromycin alone caused membrane deformation in E. coli cells. On the other hand, tFNAs-Ery caused the most serious membrane deformation, as demonstrated by the presence of debris, and break-off fragments. The SEM images revealed that the E. coli cells shrunk and formed more pores after receiving tFNAs-Ery, as compared with cells treated with erythromycin alone. Moreover, there was no difference between the tFNAs group and the control group. The TEM micrographs demonstrated more details of the cytoplasm of E. coli cells treated with tFNAs-Ery (Fig. 5a).
Fig. 5.
Morphological changes and the leakage of cytoplasm. a. AFM, SEM and TEM images of E. coli after treatment with 100 nM tFNAs, erythromycin and tFNAs-Ery. The control group was the E. coli with no treatment. b. Leakage of β-galactosidase manifested by OD420nm values of the supernatants of E. coli treated by 100 nM tFNAs, erythromycin and tFNAs-Ery. Statistical analysis (n = 3): *, p < 0.05; **, p < 0.01. c. Relative values of [K+] and [Na+] concentration of the supernatants of E. coli treated by 100 nM tFNAs, erythromycin and tFNAs-Ery, measured by AAS. Statistical analysis (n = 3): *, p < 0.05; **, p < 0.01.
3.7. Assessment of the leakage of cytoplasm
Based on the pore formation and cell shrinkage observed above, cytoplasm leakage and cell permeability were further assessed. β-galactosidase, an enzyme which can turn o-Nitrophenyl-β-D-Glucopyranosides (ONPG) into o-nitrophenol, a yellow product, is found in the normally impermeable E. coli cytoplasm. An increase in leakage indicates an increased membrane permeability, which can be quantitatively measured via the ONPG test. It was demonstrated that tFNAs-Ery led to a higher galactosidase leakage than erythromycin alone (Fig. 5b). Measuring the changes in intracellular [K+] and [Na+] is another strategy for quantitatively assessing the leakage and evaluating permeability, since a suitable content of [K+] concentration is necessary for E. coli survival. Consistent with the ONPG test, the tFNAs-Ery treatment led to a higher leakage of [K+] from E. coli cells than treatment with erythromycin alone. In addition, compared with the control group, the tFNAs-Ery treatment led to more changes in the [Na+] concentration than erythromycin alone (Fig. 5c).
4. Discussion
With the rapid development of DNA nanotechnology, a variety of self-assembled DNA nanomaterial with different sizes and spatial structures have been developed. The list of applications of DNA nanostructure research is endless in the field of molecular biology, including the use of these nanomaterials as -biosensors, vehicles, for drug delivery, and so on [[29], [30], [31], [32], [33], [34], [35], [36], [37]]. At present, the antibacterial application of DNA nanostructures mainly focused on optimizing bactericidal performance and cytotoxicity by combining these nanostructures with other antibacterial substances [[38], [39], [40], [41]]. However, few studies have used DNA nanomaterials as transport carriers of commonly used antibiotics to address a certain degree of bacterial resistance. The most significant characteristic of gram-negative bacteria, such as E. coli, is their cell membrane composition, which includes an inner membrane, a watery periplasm containing a peptidoglycan layer, and an outer membrane. It is precisely because of the particularity of these structures that most antibiotics have difficulty crossing the bacterial cell membrane to produce an antibacterial effect. In addition, gram-negative bacteria also have efflux pumps, and drug sensors that can promote drug exit and monitor drug entry into the cell, respectively. Thus, it is very difficult to develop effective drugs against gram-negative bacteria [[42], [43], [44], [45], [46], [47]]. Nanomaterials are gradually playing more important roles in antimicrobial studies, and many researchers have obtained valuable results. F. Nassar et al. developed a new uracil derivative and tested its antibacterial, antioxidant and anticancer activities. They found that the material was more effective against gram-positive bacteria than the control drug, cefoperazone. It also has high antibacterial activity against gram-negative bacteria [48]. Jin Hyunyeom and other researchers [49] used gold nanoparticles and DNA aptamers that bind to antimicrobial peptides and efficiently transfer them into mammalian cells. The use of these nanoparticles improves not only the stability of antimicrobial peptides, but also their effectiveness. Xiangyuan Ouyang et al. [50] had constructed a DNA nanoribbon with a width of 16 nm that was found to be a novel clinically relevant metallo-β-lactamase. Their discoveries provided a new platform for designing macromolecular inhibitors combined with β-lactam antibiotics against multidrug-resistant bacteria. In general, the combination of DNA nanostructures and antibiotics may be another promising research direction in the future. In our study, tFNAs-Ery were active against E. coli that are less sensitive to erythromycin alone, they displayed a better in vitro antibacterial effectivity compared to erythromycin alone, a strong stability in the humoral environment, high bacteria-permeating ability and low resistance development. tFNAs-Ery is not a new antibiotic, it is a revised version of erythromycin, which restored the potency of the antibiotic. This makes the use of tFNAs an interesting and a useful strategy for future studies on antibiotic delivery.
5. Conclusion
In conclusion, we utilized tFNAs as a delivery platform for erythromycin. Our results demonstrated that tFNAs can enhance the antibacterial effects of erythromycin against E. coli as a result of an increased bacterial uptake. The reason for the increased bacterial uptake may be the augmentation of the permeability of microbial membranes due to interactions with tFNAs. Furthermore, tFNAs-Ery was stable in the simulated bacterial internal environment in the first 8 h. Studies on the application of tFNAs delivery on other antibiotics, as well as on its effectivity against other types of bacteria, should be pursued in the future. In the development of tFNAs, the existing analysis focuses on its drug carrying capacity to improve the local concentration of antibiotics. Future studies should look into directly eradicating drug-resistant bacteria and reducing the MIC of antibiotics at the genetic level by designing special aptamers. The use of nucleic acid technology can inhibit or up-regulate the expression of specific genes, which may lead to alterations in bacterial growth and metabolism upon exposure to antibiotics.
Funding
This study was funded by the National Key R&D Program of China [2019YFA0110600] and National Natural Science Foundation of China [81970916, 81671031].
Data statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Yue Sun: Conceptualization, Investigation, Methodology, Project administration, Writing - original draft. Yuhao Liu: Data curation, Formal analysis, Investigation, Writing - review & editing. Bowen Zhang: Methodology, Investigation, Software. Shirong Shi: Data curation, Methodology, Resources. Tao Zhang: Data curation, Investigation, Writing - review & editing. Dan Zhao: Data curation, Methodology, Software. Taoran Tian: Resources, Methodology, Writing - review & editing. Qirong Li: Writing - review & editing. Yunfeng Lin: Project administration, Funding acquisition, Supervision, Writing - review & editing.
Declaration of competing interest
There is no conflict to declare.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2020.12.027.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Seiple I.B., Zhang Z., Jakubec P., Langlois-Mercier A., Wright P.M., Hog D.T., Yabu K., Allu S.R., Fukuzaki T., Carlsen P.N., Kitamura Y., Zhou X., Condakes M.L., Szczypiński F.T., Green W.D., Myers A.G. A platform for the discovery of new macrolide antibiotics. Nature. 2016;533(7603):338–345. doi: 10.1038/nature17967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui W., Ma S. Recent advances in the field of 16-membered macrolide antibiotics. Mini Rev. Med. Chem. 2011;11(12):1009–1018. doi: 10.2174/138955711797247734. [DOI] [PubMed] [Google Scholar]
- 3.Aminov R.I. A brief history of the antibiotic era: lessons learned and challenges for the future. Front. Microbiol. 2010;1:134. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Veerapandian M., Yun K. Functionalization of biomolecules on nanoparticles: specialized for antibacterial applications. Appl. Microbiol. Biotechnol. 2011;90(5):1655–1667. doi: 10.1007/s00253-011-3291-6. [DOI] [PubMed] [Google Scholar]
- 5.Dinos G.P. The macrolide antibiotic renaissance. Br. J. Pharmacol. 2017;174(18) doi: 10.1111/bph.13936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biljana A., Jill B., Ana Č., Milan M., Nevena S., Predrag N. 16-membered macrolide antibiotics: a review. Int. J. Antimicrob. Agents. 2018;51(3) doi: 10.1016/j.ijantimicag.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 7.Laurent P., Aurélie J., Polymyxins N. Patrice. Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 2017;30(2) doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohanam L., Priya L., Selvam E.M., Shivekar S.S., Menon T. Molecular mechanisms of efflux pump mediated resistance in clinical isolates of multidrug resistant pseudomonas aeruginosa. Int. J. Infect. Dis. 2016;45 [Google Scholar]
- 9.Lim A., Naidenov B., Bates H., Willyerd K., Snider T., Couger M.B., Chen C., Ramachandran A. Nanopore ultra-long read sequencing technology for antimicrobial resistance detection in mannheimia haemolytica. J. Microbiol. Methods. 2019;159 doi: 10.1016/j.mimet.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Lepoitevin M., Ma T., Bechelany M., Janot J.-M., Balme S. Functionalization of single solid state nanopores to mimic biological ion channels: a review. Adv. Colloid Interface Sci. 2017:250. doi: 10.1016/j.cis.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Zhang T., Tian T., Zhou R., Li S., Ma W., Zhang Y., Liu N., Shi S., Li Q., Xie X., Ge Y., Liu M., Zhang Q., Lin S., Cai X., Lin Y. Design, fabrication and applications of tetrahedral DNA nanostructure-based multifunctional complexes in drug delivery and biomedical treatment. Nat. Protoc. 2020;15(8):2728–2757. doi: 10.1038/s41596-020-0355-z. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J., Zhang M., Gao Y., Qin X., Zhang T., Cui W., Mao C., Xiao D., Lin Y. Tetrahedral framework nucleic acids promote scarless healing of cutaneous wounds via the akt-signaling pathway. Signal Transduct Targeted Thera. 2020;5(1):120. doi: 10.1038/s41392-020-0173-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S., Tian T., Zhang T., Cai X., Lin Y. Advances in biological applications of self-assembled DNA tetrahedral nanostructures. Mater. Today. 2019;24:57–68. doi: 10.1016/j.mattod.2018.08.002. [DOI] [Google Scholar]
- 14.Liu Y., Sun Y., Li S., Liu M., Qin X., Chen X., Lin Y. Tetrahedral framework nucleic acids deliver antimicrobial peptides with improved effects and less susceptibility to bacterial degradation. Nano Lett. 2020;20(5):3602–3610. doi: 10.1021/acs.nanolett.0c00529. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y., Xie X., Ma W., Zhan Y., Mao C., Shao X., Lin Y. Multi-targeted antisense oligonucleotide delivery by a framework nucleic acid for inhibiting biofilm formation and virulence. Nano-Micro Lett. 2020;12(6):117–129. doi: 10.1007/s40820-020-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sirong S., Yang C., Taoran T., Songhang L., Shiyu L., Yuxin Z., Xiaoru S., Tao Z., Yunfeng L., Xiaoxiao C. Effects of tetrahedral framework nucleic acid/wogonin complexes on osteoarthritis. Bone Res. 2020;8:6. doi: 10.1038/s41413-019-0077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhan Y., Ma W., Zhang Y., Mao C., Shao X., Xie X., Wang F., Liu X., Li Q., Lin Y. DNA-based nanomedicine with targeting and enhancement of therapeutic efficacy of breast cancer cells. ACS Appl. Mater. Interfaces. 2019;11(17):15354–15365. doi: 10.1021/acsami.9b03449. [DOI] [PubMed] [Google Scholar]
- 18.Liu M., Ma W., Li Q., Zhao D., Shao X., Huang Q., Hao L., Lin Y. Aptamer-targeted DNA nanostructures with doxorubicin to treat protein tyrosine kinase 7-positive tumours. Cell Prolif. 2019;52(1):e12511. doi: 10.1111/cpr.12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma W., Zhan Y., Zhang Y., Xie X., Mao C., Lin Y. Enhanced neural regeneration with a concomitant treatment of framework nucleic acid and stem cells in spinal cord injury. ACS Appl. Mater. Interfaces. 2020;12(2):2095–2106. doi: 10.1021/acsami.9b19079. [DOI] [PubMed] [Google Scholar]
- 20.Panchal P., Ogunsona E., Mekonnen T. Trends in advanced functional material applications of nanocellulose. Processes. 2018;7(1) [Google Scholar]
- 21.Emma W., Antoin L., Aine W., Fiona R. The use of nanoparticles in anti-microbial materials and their characterization. Analyst. 2008;133(7) doi: 10.1039/b715532h. [DOI] [PubMed] [Google Scholar]
- 22.Hadiya S., Liu X., Abd El-Hammed W., Elsabahy M., Aly S.A. Levofloxacin-loaded nanoparticles decrease emergence of fluoroquinolone resistance in escherichia coli. Microb. Drug Resist. 2018;24(8):1098–1107. doi: 10.1089/mdr.2017.0304. [DOI] [PubMed] [Google Scholar]
- 23.Bush K., Courvalin P., Dantas G., Davies J., Eisenstein B., Huovinen P., Jacoby G.A., Kishony R., Kreiswirth B.N., Kutter E., Lerner S.A., Levy S., Lewis K., Lomovskaya O., Miller J.H., Mobashery S., Piddock L.J., Projan S., Thomas C.M., Tomasz A., Tulkens P.M., Walsh T.R., Watson J.D., Witkowski J., Witte W., Wright G., Yeh P., Zgurskaya H.I. Tackling antibiotic resistance. Nat. Rev. Microbiol. 2011;9(12):894–896. doi: 10.1038/nrmicro2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013;12(5):371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 25.Hemeg H.A. Nanomaterials for alternative antibacterial therapy. Int. J. Nanomed. 2017;12:8211–8225. doi: 10.2147/IJN.S132163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao C., Pan W., Shao X., Ma W., Zhang Y., Zhan Y., Gao Y., Lin Y. The clearance effect of tetrahedral DNA nanostructures on senescent human dermal fibroblasts. ACS Appl. Mater. Interfaces. 2019;11(2):1942–1950. doi: 10.1021/acsami.8b20530. [DOI] [PubMed] [Google Scholar]
- 27.Ma W., Xie X., Shao X., Zhang Y., Mao C., Zhan Y., Zhao D., Liu M., Li Q., Lin Y. Tetrahedral DNA nanostructures facilitate neural stem cell migration via activating rhoa/rock2 signalling pathway. Cell Prolif. 2018;51(6) doi: 10.1111/cpr.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin M., Wang J., Zhou G., Wang J., Wu N., Lu J., Gao J., Chen X., Shi J., Zuo X., Fan C. Programmable engineering of a biosensing interface with tetrahedral DNA nanostructures for ultrasensitive DNA detection. Angew Chem. Int. Ed. Engl. 2015;54(7):2151–2155. doi: 10.1002/anie.201410720. [DOI] [PubMed] [Google Scholar]
- 29.Nuli X., Shiyuan L., Xiaohai Y., Xiaoxiao H., Jin H., Kemin W. DNA tetrahedron nanostructures for biological applications: biosensors and drug delivery. Analyst. 2017;142(18) doi: 10.1039/c7an01154g. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Ma W., Zhan Y., Mao C., Shao X., Xie X., Wei X., Lin Y. Nucleic acids and analogs for bone regeneration. Bone Res. 2018;6:37. doi: 10.1038/s41413-018-0042-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baelo A., Levato R., Julian E., Crespo A., Astola J., Gavalda J., Engel E., Mateos-Timoneda M.A., Torrents E. Disassembling bacterial extracellular matrix with dnase-coated nanoparticles to enhance antibiotic delivery in biofilm infections. J. Contr. Release. 2015;209:150–158. doi: 10.1016/j.jconrel.2015.04.028. h ttps://doi.org/S0168-3659(15)00258-8. [DOI] [PubMed] [Google Scholar]
- 32.Sun H., Chao J., Zuo X., Su S., Liu X., Yuwen L., Fan C., Wang L. Gold nanoparticle-decorated mos2 nanosheets for simultaneous detection of ascorbic acid, dopamine and uric acid. RSC Adv. 2014;4(52):27625–27629. doi: 10.1039/c4ra04046e. [DOI] [Google Scholar]
- 33.Li X., Xie X., Ma Z., Li Q., Liu L., Hu X., Liu C., Li B., Wang H., Chen N., Fan C., Song H. Programming niche accessibility and in vitro stemness with intercellular DNA reactions. Adv. Mater. 2018;30(46) doi: 10.1002/adma.201804861. [DOI] [PubMed] [Google Scholar]
- 34.Li J., Song S., Liu X., Wang L., Pan D., Huang Q., Zhao Y., Fan C. Enzyme-based multi-component optical nanoprobes for sequence-specific detection of DNA hybridization. Adv. Mater. 2008;20(3):497. doi: 10.1002/adma.200701918. [DOI] [Google Scholar]
- 35.Xing K., Chen X.G., Liu C.S., Cha D.S., Park H.J. Oleoyl-chitosan nanoparticles inhibits escherichia coli and staphylococcus aureus by damaging the cell membrane and putative binding to extracellular or intracellular targets. Int. J. Food Microbiol. 2009;132(2–3):127–133. doi: 10.1016/j.ijfoodmicro.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 36.Perche F., Le Gall T., Montier T., Pichon C., Malinge J.M. Cardiolipin-based lipopolyplex platform for the delivery of diverse nucleic acids into gram-negative bacteria. Pharmaceuticals. 2019;12(2) doi: 10.3390/ph12020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Setyawati M.I., Kutty R.V., Tay C.Y., Yuan X., Xie J., Leong D.T. Novel theranostic DNA nanoscaffolds for the simultaneous detection and killing of escherichia coli and staphylococcus aureus. ACS Appl. Mater. Interfaces. 2014;6(24):21822–21831. doi: 10.1021/am502591c. [DOI] [PubMed] [Google Scholar]
- 38.Salamanca C.H., Yarce C.J., Roman Y., Davalos A.F., Rivera G.R. Application of nanoparticle technology to reduce the anti-microbial resistance through beta-lactam antibiotic-polymer inclusion nano-complex. Pharmaceuticals. 2018;11(1) doi: 10.3390/ph11010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge Y., Tian T., Shao X., Lin S., Zhang T., Lin Y., Cai X. Pegylated protamine-based adsorbing improves the biological properties and stability of tetrahedral framework nucleic acids. ACS Appl. Mater. Interfaces. 2019;11(31):27588–27597. doi: 10.1021/acsami.9b09243. [DOI] [PubMed] [Google Scholar]
- 40.Zhang M., Zhu J., Qin X., Zhou M., Zhang X., Gao Y., Zhang T., Xiao D., Cui W., Cai X. Cardioprotection of tetrahedral DNA nanostructures in myocardial ischemia-reperfusion injury. ACS Appl. Mater. Interfaces. 2019;11(34):30631–30639. doi: 10.1021/acsami.9b10645. [DOI] [PubMed] [Google Scholar]
- 41.Meng L., Ma W., Lin S., Shi S., Li Y., Lin Y. Tetrahedral DNA nanostructure-delivered dnazyme for gene silencing to suppress cell growth. ACS Appl. Mater. Interfaces. 2019;11(7):6850–6857. doi: 10.1021/acsami.8b22444. [DOI] [PubMed] [Google Scholar]
- 42.Culp E.J., Waglechner N., Wang W., Fiebig-Comyn A.A., Hsu Y.P., Koteva K., Sychantha D., Coombes B.K., Van Nieuwenhze M.S., Brun Y.V., Wright G.D. Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature. 2020;578(7796):582–587. doi: 10.1038/s41586-020-1990-9. [DOI] [PubMed] [Google Scholar]
- 43.Caruso G. Antibiotic resistance in escherichia coli from farm livestock and related analytical methods: a review. J. AOAC Int. 2018;101(4):916–922. doi: 10.5740/jaoacint.17-0445. [DOI] [PubMed] [Google Scholar]
- 44.Roth N., Käsbohrer A., Mayrhofer S., Zitz U., Hofacre C., Domig K.J. The application of antibiotics in broiler production and the resulting antibiotic resistance in escherichia coli: a global overview. Poultry Sci. 2019;98(4):1791–1804. doi: 10.3382/ps/pey539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sgariglia E., Aconiti Mandolini N., Napoleoni M., Medici L., Fraticelli R., Conquista M., Gianfelici P., Staffolani M., Fisichella S., Capuccella M., Sargenti M., Perugini G. Antibiotic resistance pattern and virulence genesin avian pathogenic escherichia coli (apec) from different breeding systems. Vet. Ital. 2019;55(1):26–33. doi: 10.12834/VetIt.1617.8701.1. [DOI] [PubMed] [Google Scholar]
- 46.Palma N., Pons M.J., Gomes C., Mateu J., Riveros M., García W., Jacobs J., García C., Ochoa T.J., Ruiz J. Resistance to quinolones, cephalosporins and macrolides in escherichia coli causing bacteraemia in peruvian children. J Global Antimicr Res. 2017;11:28–33. doi: 10.1016/j.jgar.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 47.Baron S.A., Rolain J.M. Efflux pump inhibitor cccp to rescue colistin susceptibility in mcr-1 plasmid-mediated colistin-resistant strains and gram-negative bacteria. J. Antimicrob. Chemother. 2018;73(7):1862–1871. doi: 10.1093/jac/dky134. [DOI] [PubMed] [Google Scholar]
- 48.Nassar I.F., Farargy A.F.E., Abdelrazek F.M., Hamza Z. Synthesis of new uracil derivatives and their sugar hydrazones with potent antimicrobial, antioxidant and anticancer activities. Nucleos Nucleot. Nucleic Acids. 2020;1–20 doi: 10.1080/15257770.2020.1736300. [DOI] [PubMed] [Google Scholar]
- 49.Yeom J.H., Lee B., Kim D., Lee J.K., Kim S., Bae J., Park Y., Lee K. Gold nanoparticle-DNA aptamer conjugate-assisted delivery of antimicrobial peptide effectively eliminates intracellular salmonella enterica serovar typhimurium. Biomaterials. 2016;104:43–51. doi: 10.1016/j.biomaterials.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 50.Ouyang X., Chang Y.N., Yang K.W., Wang W.M., Bai J.J., Wang J.W., Zhang Y.J., Wang S.Y., Xie B.B., Wang L.L. A DNA nanoribbon as a potent inhibitor of metallo-beta-lactamases. Chem. Commun. 2017;53(63):8878–8881. doi: 10.1039/c7cc04483f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.