Abstract
Heparan sulfate proteoglycans consist of a small family of proteins decorated with one or more covalently attached heparan sulfate glycosaminoglycan chains. These chains have intricate structural patterns based on the position of sulfate groups and uronic acid epimers, which dictate their ability to engage a large repertoire of heparan sulfate–binding proteins, including extracellular matrix proteins, growth factors and morphogens, cytokines and chemokines, apolipoproteins and lipases, adhesion and growth factor receptors, and components of the complement and coagulation system. This review highlights recent progress in the characterization of the so-called “heparan sulfate interactome,” with a major focus on systems-wide strategies as a tool for discovery and characterization of this subproteome. In addition, we compiled all heparan sulfate–binding proteins reported in the literature to date and grouped them into a few major functional classes by applying a networking approach.
Keywords: extracellular matrix, glycosaminoglycan, heparan sulfate, heparin, proteoglycan, proteomics, systems biology
Introduction
Heparan sulfate (HS) proteoglycans (HSPGs) are integral components of the glycocalyx, a carbohydrate-rich layer surrounding virtually every cell in vertebrates and invertebrates. HS is a linear polysaccharide assembled from N-acetyl-d-glucosamine (GlcNAc) and d-glucuronic acid (GlcA). The chains undergo a series of modifications during their assembly, including the N-deacetylation and N-sulfation of clusters of GlcNAc residues, epimerization of GlcA to l-iduronic acid (IdoA), and installation of sulfate moieties at C6 and occasionally C3 of glucosamine residues and at C2 in IdoA and more rarely in GlcA units. The sulfated IdoA-containing domains vary in length, position, and spacing within a chain and in the extent of modification, giving rise to enormous structural heterogeneity. These modified segments of the chains orient negative charges in different spatial patterns, which can be complementary to the arrangement of positively charged residues in the binding sites in proteins that recognize HS and the related polysaccharide, heparin. Electrostatic interactions coupled with hydrogen bonding and van der Waals contacts impart additional specificity. Binding of HS to proteins results in protein sequestration and protection, presentation at the cell surface, oligomerization, allosteric activation and internalization, as HSPGs can also act as endocytic receptors.
Given their location in the glycocalyx, it is not surprising that HSPGs orchestrate a myriad of biological processes (Fig. 1). Spatiotemporal regulation of the chemical structure of the HS chains modulates their ability to engage a broad spectrum of HS-binding proteins (HSBPs). The complete network of HS–HSBP interactions, or the “HS interactome,” constitutes a dynamic system that is regulated in a tissue-specific and developmentally restricted manner.1 HSBPs are diverse and include structural proteins, membrane receptors, growth factors, enzymes, and enzyme inhibitors. In contrast to lectins, which can be sorted into evolutionarily related protein families by their folds and carbohydrate-binding domains, most HSBPs are structurally unrelated, and their capacity to bind HS is believed to have arisen through convergent evolution. Some HSBPs are highly specific for HS, whereas others are promiscuous and can engage related glycosaminoglycan (GAG) classes, such as chondroitin/dermatan sulfate and keratan sulfates. As a consequence, HSBPs are functionally diverse, but nevertheless play key roles in orchestrating coagulation, lipid metabolism, extracellular matrix (ECM) assembly, growth factor signaling, cell adhesion, and many other biological processes.
Figure 1.
Overview of the major glycosaminoglycan types and their interaction with proteins at the cell membrane and in the extracellular matrix.
Growing evidence indicates that a large number of HSBPs can physically (or functionally) interact with each other, suggesting that the regulation of the HS interactome, at the systems level, may have far-reaching implications for global orchestration of cellular events. This review deals with the HS interactome from a systems biology perspective and focuses on insights from recent studies where systems-wide methods have been used to determine the scope of this subproteome and to define its global structural and functional features. We also performed comprehensive literature and database searches to summarize all the HSBPs reported to date and took a systematic approach to their analysis by applying a network biology computational approach. Our results reveal that most HSBPs can be grouped into a limited number of protein–protein interaction networks, with remarkable functional commonalities. From an evolutionary point of view, we speculate that the acquisition of the HS-binding capacity in such a diverse group of proteins might be linked to the emergence of multicellularity, as a way of dealing with increased biological complexity.
Systems-wide Strategies to Map the HS Interactome
Since the advent of the “Omics-revolution,” systems-wide approaches have been deployed to determine the scope of the HS interactome. One important insight that emerged from these studies is that several hundred soluble and membrane proteins can interact with HS chains, sometimes with remarkable selectivity. One common approach to large-scale analysis of HSBPs relies on glycan affinity chromatography and mass spectrometry (MS). Large quantities of HS have only recently become available, and instead, heparin has been used as a proxy to fractionate and identify HSBPs from different fluids and tissues. Heparin is a highly sulfated subtype of HS that is mainly produced by connective tissue mast cells, where it plays a key role in the storage of proteases and biogenic monoamines.2,3 Heparin is typically purified from porcine and bovine sources and is routinely used in the clinics as an anticoagulant because it binds to antithrombin and accelerates the inactivation of the procoagulation enzymes thrombin and factor Xa.4 Heparin-affinity matrices are inexpensive and therefore an economical material for HSBP screening. However, its fine structure differs significantly from cellular HS by having a higher degree of sulfation and charge density, and thus exhibits promiscuous binding characteristics similar to cation exchange resins. With the assumption that proteins that bind to heparin also bind HS, heparin-affinity chromatography in conjunction with proteomics strategies is a simple approach to address the identity and scope of the HS interactome in various cells and tissues, with the caveat that some proteins may bind to heparin but have weak affinity for HS (or vice versa).
A large array of classical growth factors, including members of the fibroblast growth factor (FGF), bone morphogenic protein (BMP), vascular endothelial cell growth factor (VEGF), and the wingless (WNT) family of growth factors and morphogens, were initially isolated and identified through heparin-affinity chromatography.5–10 Later, their interaction with cellular HS was also established, giving support to a model in which ternary signaling complexes involving HSPGs, growth factor ligands, and high-affinity cellular receptors make up the functional signaling complex.11,12 The HS interactome in human body fluids has also been interrogated in a similar fashion.13–17 Plasma, in particular, is easy to collect and is a valuable source of clinical information. In addition to classical plasma HSBPs such as antithrombin and platelet factor 4 (PF-4), multiple components of the coagulation and the complement system can interact with HS. In fact, the enrichment of HSBPs within these cascades of interconnected protein–protein interaction networks is highly suggestive of a role for HS as a potential global regulator of these complex processes.
Similar strategies have been applied for large-scale analysis of HSBPs from microbes and parasites. Most pathogens do not synthetize HS or heparin but have nonetheless evolved HS-binding or degrading capacity to exploit the glycocalyx and to facilitate key host–pathogen interactions. For example, proteomics screening of merozoite proteins from Plasmodium falciparum has revealed that multiple members of the PfRhopH complex display high affinity for heparin.18 These proteins are secreted by rhoptries, specialized apical organelles that are critical for parasite invasion and nutrient acquisition.19 Similar attempts to define the HS interactome of Toxoplasma gondii identified novel HSBPs involved in parasite development, suggesting that host HS not only regulates invasion but also modulates certain aspects of parasite maturation.20 Studies of bacteria have applied similar strategies to identify novel HS-binding virulence factors.21 Toxins from viperid snake venoms and antimicrobial compounds from egg white have been identified as well.22,23
Although many HSBPs have been reported over the years, membrane-bound and membrane-associated HSBPs are often difficult to characterize due to low abundance, solubility issues, protease resistance, and contamination by intracellular proteins (e.g., histones) that strongly bind to heparin-affinity matrices and outcompete less abundant membrane proteins. To circumvent these problems, cell-surface biotinylation,24 membrane fractionation,25 and limited proteolysis26 strategies have been applied before heparin-affinity chromatography. These approaches have revealed a large number of cell-surface HSBPs, including novel cellular receptors and adhesion molecules. However, these refined strategies have so far only been applied to a few cell lines and tissues, so the true scope of the membrane HS interactome is still largely unknown.
Novel strategies are needed to identify HSBPs that specifically bind to discrete HS oligosaccharide motifs. Many HSBPs depend on interactions with specific HS structural motifs defined by sulfation patterns and GlcA/IdoA content to exert their biological functions. One classical example of clinical relevance is the highly specific interaction of antithrombin with heparin, which is largely mediated by binding a discrete pentasaccharide sequence in heparin to a domain in antithrombin containing key positively charged residue brought into proximity in the folded protein.27,28 Low-molecular weight heparins such as enoxaparin (Lovenox), contain this pentasaccharide-binding site and are widely used clinically to treat different medical conditions, including venous thromboembolism, stroke, and myocardial infarction.29 The identification of similarly specific binary interactions has been hampered by difficulties in isolation and preparation of reference material that can be used for rational structure-and-function studies. Accelerated progress in chemical and chemoenzymatic GAG synthesis is therefore desperately needed.30–33 Challenges and opportunities in this field have been reviewed elsewhere.34 Finally, HS–protein interactions are also dependent on specific molecular features of the HS-binding sites in HSBPs. Unfortunately, the identification of protein determinants that facilitate HS recognition turned out to be less straightforward than initially predicted.
HS-binding Sites: One Story, Many Tales
In 1989, Cardin and Weintraub35 published a key paper recognizing the importance of the primary amino acid sequence of HSBPs for HS recognition. By applying sequence analysis and molecular modeling to a series of well-characterized HSBPs, semiconserved linear sequences of basic amino acids (lysine, arginine and histidine) interspersed by other, often hydrophobic, residues were identified. On this basis, two consensus binding sequences were initially proposed: [-X-B-B-X-B-X-] and [-X-B-B-B-X-X-B-X-], where B denotes a basic amino acid and X a hydrophobic amino acid residue. Modeling of these motifs predicted their presence in alpha-helixes, with the basic residues aligned on the same side of the helix toward the solvent and the hydrophobic residues pointing toward other structural features in the protein. Given that alpha-helices have 3.4 residues per turn, several of the basic residues were expected to align along one face of the helix, creating patches of positively charged amino acids that could interact with the negatively charged sulfate and carboxylic acid groups of the HS chains. Following a similar approach, Margalit et al.36 proposed that a 20-Å distance between the basic amino acids (most frequently arginine) is crucial for the interaction of several HSBPs with heparin.
Following these initial findings, the patterns (and spacing) of basic amino acids within HS-binding sites have been further refined.37 Although the presence of these motifs suggests that a protein might bind to HS, at least half of the known HSBPs do not contain Cardin–Weintraub sequences. In fact, HS-binding sites can also be located on beta strands and sheets, in which case the positive residues would need to alternate along the strand to generate a positively charged surface for docking. In other cases, binding sites are generated by folding of different protein domains, generating a common positively charged surface that is favorable for interaction, for example, in antithrombin. Although a certain number of binding principles have been established, the idea of well-conserved motifs within HS-binding sites is still difficult to reconciliate with the low degree of sequence conservation across the large number of HSBPs currently known.
As the structures of new HSBPs are determined, new insights have also emerged. Recent studies by Torrens et al. demonstrated that minimal structural motifs denoted as “CPC clip motifs” (C: cationic and P: polar residues) are conserved among most heparin-binding domains deposited in the Protein Data Bank (PDB) database.38,39 These motifs are not necessarily part of continuous linear structural elements and are thought to act as “staples” to pin the GAG chains onto the protein-binding sites. Similarly, an exhaustive collection of experimentally determined HSBPs was recently reported.25 This data set was subjected to sophisticated network analysis, to elucidate so far unnoticed structural commonalities. In total, 437 non-redundant HSBPs were analyzed using novel sequence similarity metrics and graph analysis. Again, the linear amino acid sequences across all HSBPs were found to be highly variable, ruling out a universal HS-binding sequence. However, several shorter (mostly tripeptides) and widely spaced motifs were found to be conserved. Based on these findings, the authors proposed a model in which the three-dimensional arrangement of these motifs on the protein surface, and not the primary sequence per se, is what determines the structural basis for HS/heparin–protein interactions. In addition, it is also possible that the oligomerization state of a given HSBP might influence the properties of a particular HS-binding site, for example, by including structural determinants originating from separate protein subunits. In summary, the data so far are consistent with the idea that HSBPs use different structural elements (and binding modes) to engage the HS motifs, which probably translates into different affinities, specificities, and biological responses.
Systems-wide Approaches to Characterize HS/Heparin-binding Sites
Chemical Methods
As the number of reported HSBPs grows with time, there is also need for general methods to accurately identify key residues involved in HS recognition. Sequence analysis in conjunction with site-specific mutagenesis and biophysical methods has been successful in establishing structure–function relationships for specific HS–HSBP interactions.40 However, faster approaches are needed to replace these labor-intensive methods. In that line, Vivès et al.41 reported a procedure based on chemical crosslinking of individual HSBPs to 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC)/N-hydroxysuccinimide (NHS)-activated heparin beads. Protein–carbohydrate conjugates were subjected to proteolytic digestion using thermolysin, an endopeptidase with broad specificity, after which the remaining crosslinked peptide fragments, covering the heparin-binding site, were identified by Edman degradation. The workflow was applied to three known HSBPs: the pseudorabies virus (PRV) envelope glycoprotein gC, the CC chemokine RANTES, and the C-terminal fragment of the laminin-5 α3 chain, resulting in successful mapping of their HS-binding sites. Application of this method to mixtures of proteins has not yet been reported.
An alternative MS-based method was also reported by Ori et al.42 Their “Protect-and-Label” strategy is based on the treatment of HSBPs with sulfo-NHS-acetate to chemically modify the exposed primary amines. As the labeling is performed while the protein is bound to heparin beads, lysine residues involved in binding are generally protected from derivatization. Labeled proteins are then displaced from the beads by salt elution and subjected to NHS-biotin treatment to specifically tag the protected lysines in the binding site. Proteins are then trypsinized, and biotinylated peptides are isolated and characterized through MS. Three known HSBPs—FGF-2, PF-4, and pleiotrophin—were analyzed through this protocol, rendering detailed information on their HS/heparin-binding sites. Although very promising, both of these approaches heavily rely on NHS chemistry, which mainly targets exposed lysine residues. Therefore, very little information can be obtained for other amino acids such as asparagine, serine, or tyrosine that are also commonly found in HS/heparin-binding sites. Notwithstanding, these protocols have the potential to be easily adapted to medium/high-throughput formats to complement the information derived from HSBP global discovery screenings.
Computational Methods
New insights into the nature of HS–HSBP interactions have been uncovered through the power of computational algorithms modeled on available structural data. Biomolecular docking techniques followed by molecular dynamics (MD) simulations are useful strategies to interrogate protein–glycan interactions at atomic resolution. The computational treatment of HS–protein interactions is, however, very challenging. Some of the difficulties relate to the high conformational flexibility of HS oligosaccharides, the indispensability of solvent and electrolytes to understand the interaction under physiologic conditions, and the poor complementarity between HSBPs and the ligand in their interfaces. Moreover, only a handful HSBPs in complex with HS/heparin oligosaccharides have been solved and deposited in public repositories (~100). Nonetheless, computational strategies can facilitate the identification of putative HS-binding sites and might also give clues as to the specificity and selectivity toward different HS sequences or motifs.
As basic residues such as lysines and arginines play pivotal roles in HS recognition, one popular way of predicting HS/heparin-binding sites is through calculation of the protein surface electrostatic potential. The localization of electropositive patches on a protein surface by computational tools, such as APBS and DeepView, is often a good indicator of the presence of a putative binding site. Such popular approaches are built on the assumption that HS–protein interactions are mainly driven by electrostatic forces, which might not be entirely correct in all cases. In fact, the cumulative data suggest a substantial contribution of non-Coulombic forces as well.40 More sophisticated approaches rely on sulfated probes to identify areas of solvent displacement and to initially locate regions that are energetically favorable for HS binding.43,44 New online servers for prediction of binding sites have also been developed during the last few years, making computational approaches easier to access by the biological community.45
Once a binding site has been located, HS oligosaccharide libraries can be probed by molecular docking techniques.46,47 These approaches aim to achieve a close fit of a ligand into a targeted site, giving insights into the affinity of binding and preferable binding poses. As discussed above, chemical and enzymatic synthesis of HS oligosaccharides is very challenging. One advantage of virtual library screenings is that thousands of different structures and conformations can be quickly tested “in silico.” Promising candidate ligands can be identified in this way and experimentally verified later through other biochemical or biophysical methods. Recently, sophisticated protocols have been reported, where combinatorial virtual library screening algorithms were applied to a large set of HS oligosaccharides, ranging from disaccharide to hexasaccharides.48 All potential hits were subsequently parsed through rigorous logical filters capable of segregating the structures into “high-affinity” and “high-specificity” sequences. Compared with other biomolecules such as polypeptides or nucleic acids, HS oligosaccharides contain a high number of rotatable bonds, which essentially limits computational docking to disaccharide, tetrasaccharide, and hexasaccharide, which can then be stitched together to generate longer potential ligands. One caveat is that it is sometimes difficult to distinguish between 180° pairs of GAG oligosaccharide orientations, which might look very similar in either direction.49
Docking information alone cannot render accurate information on the free energy of binding. Typically, when ligands have been fit into putative binding sites, the complexes are subjected to MD simulation workflows and energy minimization routines. MD simulations facilitate the analysis of the trajectories of structural coordinates as a function of time under the influence of a force field. These force fields entail a collection of equations designed to reproduce molecular geometry and selected properties of the analyzed structures. Several glycan-specific force fields have been developed during the last years, including GLYCAM and CHARMM.50,51 The total free energy of binding and the contribution of individual residues can be deduced by averaging the mechanical contributions of the interacting atoms. Application of these approaches to annexin A2 and PECAM-1 has revealed high- and low-affinity binding sites, as well as a dependency on the length of the GAG chains for proper interaction.52
A final detail to keep in mind is that HS–protein interactions are affected by solvent interactions, but taking this into consideration is computationally expensive. A few studies have shown that docking and MD simulations with or without solvent give different results from an energetics perspective.50,52,53 In fact, the analysis of GAG–protein interfaces in structures deposited in the PDB database reveals that they are generally more hydrated than protein–protein interfaces. Indeed, half of the interactions at GAG–protein interfaces appear to involve water. Although many challenges still remain, the development of computational tools has the potential to accelerate the rational design of oligosaccharide mimetics capable of fine tuning GAG–protein interactions in different biological systems. Progress in this area is of critical importance.
Systems-wide Functional Analysis of the HS Interactome
From a functional perspective, very few studies have considered the HS interactome as a coherent subproteome. In a previous study, 435 non-redundant HSBPs were compiled from the literature and subjected to network analysis to identify functional and structural patterns associated with HS binding.25 Interestingly, it was noted that HSBPs form interactome networks with high average clustering coefficients and densely interconnected functional modules. As more recent studies have expanded this subproteome, we decided to revisit the mammalian HS interactome by taking a slightly different approach that emphasizes the characteristic modularity of HSBP interaction networks. We used the 435 proteins previously reported by Ori et al. and supplemented them with new HSBPs identified in other high-throughput studies.14–17,25 A final list of 530 non-redundant proteins was compiled (Supplemental Table 1). This final HSBP list was used to generate a protein network based on physical and functional protein–protein associations (PPAs). High-confidence PPAs (association score >0.7) were extracted from the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database. STRING is an integrated collection of protein–protein interactions based on direct binding data and/or inferred associations from the literature, public databases, and other repositories.54 At the time of this study, the whole STRING-DB high-confidence PPA network contained a total of 15,131 proteins and 359,776 associations, with an average of 47.6 associations per protein. Of the 530 HSBPs in our compiled list, 488 mapped to the high-confidence PPA network. This new STRING subnetwork was then selected for further analysis, reflecting 3136 associations with an average of 12.8 associations per protein. Functional enrichment analysis of the network was performed through the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Supplemental Table 2).55 DAVID was run using default settings with thresholds: count ≥2 and EASE ≤0.1. The EASE score is a modified Fisher exact p value which was further adjusted using a Benjamini correction. Notably, only ~25% of the input HSBPs were automatically ascribed with HS/heparin-binding functions, indicating that the current level of annotation in public repositories is less than satisfactory.
Given the previously reported modularity of HSBP networks, we applied the Louvain method for community detection to better identify and group HSBP clusters displaying higher density of interconnected nodes than expected by random chance.56 This method is based on a heuristic algorithm that aims to detect communities (or cluster of nodes) by optimizing modularity. Modularity is a measure of the tendency of nodes in a large network to display higher local interconnectivity (or edge density) compared with their connections with the rest of the network. This community clustering is an iterative process that is repeated until maximum modularity is achieved, and a hierarchy of communities is generated. By applying this method, we managed to isolate 15 communities (Supplemental Table 3). The identified clusters were further segregated via force-directed visualization algorithms and subjected to functional enrichment analysis. What follows is a description of several of these clusters.
Complement and Chemokines
Interestingly, nearly 75% of all input proteins grouped into five major Louvain clusters (Fig. 2A–E). As shown in Fig. 2A, one of this clusters was largely composed of two major classes of HSBPs, including complement factors and chemokines. These proteins were heavily interconnected through two main nodes, corresponding to complement factor C3 and factor C5. C3 is a key component of the complement system and a point of convergence for the classic, alternative, and lectin activation pathways. Assembly of the C3 convertase through any of these pathways results in proteolytic cleavage of C3 into C3a and C3b, two important factors that promote leukocyte recruitment, pathogen opsonization, and phagocytosis. Similarly, C5 is cleaved into C5a and C5b by the C5 convertase, promoting phagocyte chemotaxis and assembly of the membrane attack complex, respectively. Notably, both C3a and C5a can directly regulate chemokine expression through engagement of G-protein-coupled seven transmembrane domain receptors (C3aR and C5aR).57,58
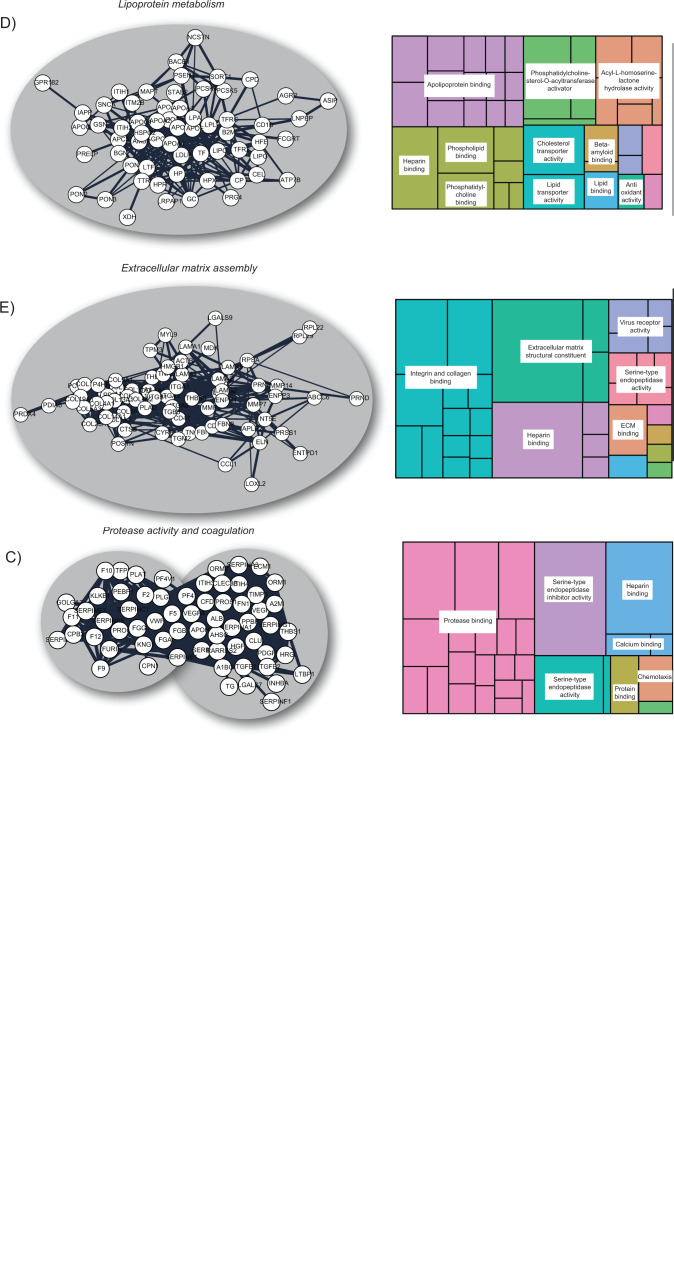
Figure 2.
Most heparan sulfate–binding proteins can be clustered into a few distinct protein groups based on physical and/or functional associations. Almost 75% of the mapped heparan sulfate interactome was related to (A) chemokine and complement proteins, (B) growth factors and ion sensing, (C) protease activity and coagulation, (D) lipoprotein metabolism and (E) ECM assembly. Abbreviation: ECM, extracellular matrix.
The role of HS in the regulation of chemokine activity (e.g., the establishment of chemokine gradients and modulation of signal transduction) has been extensively reviewed.59 Less is known about the impact of HS on complement-mediated activities. One notable exception is the complement factor H (CFH), a key fluid-phase regulator of the alternative complement pathway.60–62 During infections, CFH protects host cells from complement-mediated cytotoxicity by interacting with cell-surface HS chains (and sialic acids). Specifically, CFH modulates the inactivation of C3b by acting as a cofactor of complement factor I (CFI) and by accelerating the decay of C3 convertase. As HS is normally not expressed by microbes, this is an effective strategy to distinguish between self and non-self entities during the activation of the complement cascade. Familial mutations in the HS-binding domain of CFH are associated with impaired HS binding, resulting in a severe form of thrombotic microangiopathy known as atypical hemolytic uremic syndrome.63,64 Similarly, allotypic variants of the CFH gene are strongly associated with the development of age-related macular degeneration (AMD), one of the leading causes of adult blindness in the Western world.65 Progression of AMD has been found to correlate with changes in the HS composition of the human Bruch’s membrane/choroid, leading to focal deposits of lipids and proteins.66
As shown in Fig. 2A, most complement factors are de facto HSBPs, raising the possibility of HS serving as a global regulator of the complement system. In fact, kinetic studies of many complement proteins reveal a relevant range of affinities (from 2 to 320 nM) toward heparin, which could be pharmacologically exploited to fine tune the degree of activation of the complement system in clinical settings.67 This observation, together with the aforementioned functional interconnections between complement factors and chemokines, indicate that the impact of heparin and heparin-like compounds on complement activation is an area that warrants further investigation.
Growth Factor and Growth Factor Receptors
A second cluster was characterized by a large variety of growth factors and growth factor receptors (Fig. 2B), including members of the hepatocyte growth factor, platelet-derived growth factor, Ephrin, and the FGF protein family of receptor tyrosine kinases (RTKs). RTKs modulate mitogenic, angiogenic, and metastatic mechanisms and constitute major pharmacologic targets for the treatment of cancer.68 Upon ligand binding, RTKs undergo conformational changes, homo/hetero-oligomerization, autophosphorylation of the intracellular tyrosine kinase domain, and transcriptional activation of important factors that regulate cell behavior. In many cases, as it has been extensively reviewed for the FGF/FGF receptor axis, HS serves as a coreceptor of RTK signaling by assisting in ternary complex formation and by fine tuning the activation threshold of the signaling complexes.69 The development and use of HS mimetic compounds to regulate the activity of HS-responsive RTKs is an area of intense research.70 Other growth factors captured in the cluster included members of the BMP signaling pathway such as BMP2 and BMP4, as well as known BMP antagonists such as Fstl1. Genetic alteration of the fine structure of HS affects downstream BMP signaling, leading to a wide range of morphogenetic defects in bone and lung development.71
A few proteins involved in sodium and calcium sensing and transport were also clustered together with the growth factors and growth factor receptors. There is some data pointing to a role for the syndecan family of HSPGs in the regulation of stretch-activated ion channels. Indeed, recent studies have provided evidence for the presence of ion channels within focal adhesions that surprisingly operate in concert with transmembrane proteoglycans, such as syndecan 4.72 These ion gates and transporters seem to impact cell migration and adhesion by regulating the cytosolic ion environment, which in turn modulates both cytoskeleton rearrangement and the formation of cell adhesion molecule complexes. Accordingly, the extracellular and pericellular pH is known to impact the stability of focal adhesions. A recent review on this topic can be found elsewhere.73 The scope of this phenomenon and the exact mechanisms that operate through this novel syndecan-ion channel axis still need to be fully elucidated, including the role of the GAG chains (if any) vs the HSPG core protein. Interestingly, our association of ion sensing and transport with growth factor signaling raises the question whether regulation of some ion channels by specific growth factor signaling pathways could perhaps be modulated by HSPGs, in a glycan-dependent fashion. This intriguing possibility could be relevant for diseases such as fibrosis and certain types of cancer, where a perturbation of the cell’s ionic balance and changes in HSPG expression are known to be contributing factors.
Protease Activity and Coagulation
Regulation of the coagulation system is another striking example of how HSPGs coordinate complex protein–protein interaction networks. As shown in Fig. 2C, many HSPGs involved in the regulation of the coagulation cascade were captured by the Louvain clustering. As expected, functional enrichment reflected the impact of HS chains on protease activity and included both serine proteases and serine protease inhibitors (serpins). All serpins exert their function through a common mechanism, which entails dramatic conformational changes and covalent inactivation of the targeted proteases. Interestingly, some of the best studied HS-binding serpins such as antithrombin (serpin C1) and heparin cofactor II (serpin D1) can inhibit procoagulant factors (e.g., thrombin and factor Xa), whereas others such as serpin A5 target anticoagulant factors (e.g., protein C). In general, the rate of these interactions is largely accelerated by binding to GAGs. As a side note, it has been shown that in jawless fish such as lampreys, the peptide hormone precursor angiotensinogen can moonlight as a potent heparin-dependent thrombin inhibitor in addition to regulating vascular tone.74,75 These two ancestral functions were uncoupled in gnathostomes during evolution, but perhaps some of the mammalian coagulation serpins might still display undiscovered vasomodulatory properties. The evolutionary history of the HS interactome and its coevolution with the HS biosynthetic machinery are areas that have not been explored and that warrant further investigation. Finally, a few growth factors and cytokines such as VEGF and PF4 were also found within this cluster. The tight modularity of this interactome and its molecular diversity highlight the multiple levels of regulation that complex biological processes such as the coagulation cascade can exhibit and raises the question of the potential role of cellular HS as a global modulator.
Lipoprotein Metabolism
Another Louvain cluster was highly enriched in proteins involved in lipoprotein metabolism and cholesterol transport (Fig. 2D). There is abundant evidence of the critical role of HSPGs in hepatic clearance of triglyceride-rich lipoproteins (TRLs) in both mice and humans.76–78 Genetic studies have highlighted the role of HSPG syndecan 1 as an important endocytic receptor of TRL remnants, together with other classical receptors, such as the low-density lipoprotein receptor (LDLR) and the LDLR-related protein 1 (LRP1). Sulfation of the HS chains of syndecan 1 is crucial for mediating interactions with multiple apolipoproteins, such as apolipoprotein B (ApoB), apolipoprotein E, and apolipoprotein A-V located on TRL-remnant populations of defined sizes.79
Accumulation of LDL particles in the subendothelial matrix is an early event during the development of atherosclerosis that triggers subsequent pathologic events such as LDL oxidation, vascular inflammation, monocyte recruitment, smooth muscle cell proliferation, and atherosclerotic plaque formation. Interestingly, HSPGs such as perlecan can contribute to vascular inflammation and atherosclerosis through its binding to ApoB-rich lipoproteins.80,81 On the contrary, there is some evidence of its anti-inflammatory role through regulation of migration and proliferation of vascular smooth muscle cells, revealing the complex and sometimes paradoxical roles played by HSPGs, depending on the tissue context and the fine structure of the HS chains.82
One of the proteins within this cluster of HSBPs, lipoprotein lipase (LPL), is an important factor normally anchored to the luminal side of the endothelium, where it degrades very-low-density lipoproteins and chylomicrons, and releases free fatty acids. LPL displays high affinity toward heparin because its polypeptide sequence contains multiple heparin-binding sites. Moreover, intravenous injection of heparin rapidly increases the LPL levels in blood, suggesting a displacement mechanism.83 Although endothelial HS was largely assumed to be the physiologic ligand of LPL in the vasculature, more detailed studies have demonstrated that another protein called glycosylphosphatidylinositol-anchored high-density lipoprotein binding protein 1 (GPIHBP1) is the actual receptor for LPL.84 GPIHBP1 binds to LPL via negatively charged protein surfaces, and this interaction can be outcompeted by exogenous addition of heparin. This is a remarkable example of the critical difference between heparin and HS in biological systems, and a lesson to not draw quick conclusions about physiologic HS–HSBP interactions, when only heparin is used as a surrogate. Nevertheless, other studies have suggested that vascular HSPGs can modulate the LPL transport.85,86
ECM Assembly
Finally, the largest cluster of HSBPs was enriched in proteins involved in ECM assembly (Fig. 2E). HSPGs are major components of the ECM, where they act as flexible molecular scaffolds for structural proteins involved in tissue organization, as well as key modulators of cell-surface signaling complexes that regulate cell adhesion, migration, and cytoskeleton reorganization. Specifically, the majority of HSBPs in this cluster are integral components of the basement membrane such as laminins, and cell surface integrins. The ECM is also a key element of the tumor microenvironment, and HS–HSBP interactions therein have a profound impact on oncogenic processes affecting tumor progression, including the regulation of growth factor availability and the degree of immune infiltration and tumor angiogenesis.87,88
Evolutionary Perspective
Despite the large structural and molecular diversity of the HSBPs, if the collection is treated as a coherent subproteome, they tend to group into protein–protein interaction networks with common functional themes. One appealing explanation for this phenomenon may find its basis within an evolutionary framework. We suggest there is a potential link between the evolutionary pressure driving the acquisition of HS-binding capacity, and the concomitant emergence of multicellularity and the increased biological complexity associated with it. The fact that most unicellular organisms lack the biosynthetic tools to assemble complex GAG polysaccharides is a strong indication that this might be the case. During the evolution of unicellular life forms toward multicellularity, the flux of chemical information is dramatically increased because cells are not only responsive to cell-autonomous and environmental changes, but they are also expected to integrate and respond to communication flows originating from adjacent cells. These relationships become more imperative as we move from cells, to tissues, to organs, and to organ systems. As discussed before, most HSBPs are either present at the cell membrane or embedded in the ECM, which constitute the point of first contact in intercellular communication. Many of the biological processes in which HSBPs are involved, such as cellular signaling, directly adhere to the notion of chemical communication flows that need to be carefully integrated to facilitate coordinated responses. Perhaps, the evolutionary acquisition of HS-binding ability constituted an additional global checkpoint that was advantageous to multicellular biological systems, to quickly adapt and more effectively respond to changing environmental cues. If that is the case, the large chemical space that can be coded into HS structures would constitute a flexible arsenal that can be rapidly deployed to fine tune the effect and timing of these responses.
Future Perspectives
As suggested throughout this review, by coordinated molecular events such as cellular signaling, complement activation, or the coagulation cascade, HSPGs may engage multiple components within the same pathway and across clusters. How these protein–carbohydrate interactions will affect the spatiotemporal dynamics of these processes at a systems level is difficult to predict. As our clustering approach suggests, the functional interconnections between multiple classes of HSBPs indicate that one should take a broader view of how altering a given component in a cluster either pharmacologically or genetically could affect other pathways. As discussed above, dissection of the structural details that facilitate these interactions requires access to well-defined oligosaccharide libraries, as well as novel screening methodologies to measure simultaneous binding of receptors and protein ligands. Given the large number of HSBPs reported so far and the fact that many of them occur in shared pathways, the data clearly indicate that the role of HS in regulating biological processes might be more comprehensive than previously expected.
To the best of our knowledge, it appears that most studies addressing the scope of the HS interactome have mainly focused on qualitative aspects, that is, the complement of HSBPs present in a sample at a given time and set of conditions. However, it is reasonable to assume that quantitative changes might be important as well, especially during pathologic processes. Thus, alterations in the expression of HSPGs or the composition of HS could provide coordinated control over multiple pathways and systems. This possibility may be best exemplified during embryonic development and regeneration, which correlate with large-scale changes in HSPG and HS composition in a temporal and spatially regulated manner.
Realization of the potential of the HS interactome as a molecular window to explore basal cellular physiology and to understand complex diseases might therefore require novel analytical strategies to assess both qualitative and quantitative changes in this subproteome. There is now sufficient evidence showing that HS-mediated functions affect a broad range of physiologic and pathophysiologic processes, making them of general interest to the scientific community and no longer exclusively confined to the realm of glycobiology experts and carbohydrate chemists.
Supplemental Material
Supplemental material, sj-pdf-1-jhc-10.1177_0022155420988661 for A Systems View of the Heparan Sulfate Interactome by Liliana Schaefer, Charles W. Frevert, Alejandro Gómez Toledo, James T. Sorrentino, Daniel R. Sandoval, Johan Malmström, Nathan E. Lewis and Jeffrey D. Esko in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-xlsx-2-jhc-10.1177_0022155420988661 for A Systems View of the Heparan Sulfate Interactome by Liliana Schaefer, Charles W. Frevert, Alejandro Gómez Toledo, James T. Sorrentino, Daniel R. Sandoval, Johan Malmström, Nathan E. Lewis and Jeffrey D. Esko in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-xlsx-3-jhc-10.1177_0022155420988661 for A Systems View of the Heparan Sulfate Interactome by Liliana Schaefer, Charles W. Frevert, Alejandro Gómez Toledo, James T. Sorrentino, Daniel R. Sandoval, Johan Malmström, Nathan E. Lewis and Jeffrey D. Esko in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-xlsx-4-jhc-10.1177_0022155420988661 for A Systems View of the Heparan Sulfate Interactome by Liliana Schaefer, Charles W. Frevert, Alejandro Gómez Toledo, James T. Sorrentino, Daniel R. Sandoval, Johan Malmström, Nathan E. Lewis and Jeffrey D. Esko in Journal of Histochemistry & Cytochemistry
Footnotes
Author Contributions: A.G.T., J.T.S., and J.D.E. wrote the primary draft of the paper. D.R.S., J.M., and N.E.L. provided innumerable insights into the literature and many helpful comments.
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by grants P01 HL131474 (to J.D.E.) and R21 CA199292 (to J.D.E. and N.E.L.), NIGMS R35GM119850 (to N.E.L), and T32GM008806 (to N.E.L).
Contributor Information
Alejandro Gómez Toledo, Department of Cellular and Molecular Medicine, University of California, San Diego, La Jolla, California; Glycobiology Research and Training Center, University of California, San Diego, La Jolla, California; Department of Clinical Sciences, Lund, Division of Infection Medicine, Lund University, Lund, Sweden.
James T. Sorrentino, Bioinformatics and Systems Biology Graduate Program, University of California, San Diego, La Jolla, California Department of Bioengineering, University of California, San Diego, La Jolla, California.
Daniel R. Sandoval, Department of Cellular and Molecular Medicine, University of California, San Diego, La Jolla, California Glycobiology Research and Training Center, University of California, San Diego, La Jolla, California.
Johan Malmström, Department of Clinical Sciences, Lund, Division of Infection Medicine, Lund University, Lund, Sweden.
Nathan E. Lewis, Department of Bioengineering, University of California, San Diego, La Jolla, California Department of Pediatrics, University of California, San Diego, La Jolla, California.
Jeffrey D. Esko, Department of Cellular and Molecular Medicine, University of California, San Diego, La Jolla, California; Glycobiology Research and Training Center, University of California, San Diego, La Jolla, California.
Literature Cited
- 1. Lindahl U, Couchman J, Kimata K, Esko JD. Proteoglycans and sulfated glycosaminoglycans. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, Seeberger PH, editors. Essentials of glycobiology. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2015. p. 207–21. [Google Scholar]
- 2. Braga T, Grujic M, Lukinius A, Hellman L, Abrink M, Pejler G. Serglycin proteoglycan is required for secretory granule integrity in mucosal mast cells. Biochem J. 2007;403:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ringvall M, Ronnberg E, Wernersson S, Duelli A, Henningsson F, Abrink M, Garcia-Faroldi G, Fajardo I, Pejler G. Serotonin and histamine storage in mast cell secretory granules is dependent on serglycin proteoglycan. J Allergy Clin Immunol. 2008;121:1020–6. [DOI] [PubMed] [Google Scholar]
- 4. Casu B, Naggi A, Torri G. Re-visiting the structure of heparin. Carbohydr Res. 2015;403:60–8. [DOI] [PubMed] [Google Scholar]
- 5. Shing Y, Folkman J, Haudenschild C, Lund D, Crum R, Klagsbrun M. Angiogenesis is stimulated by a tumor-derived endothelial cell growth factor. J Cell Biochem. 1985;29:275–87. [DOI] [PubMed] [Google Scholar]
- 6. Hauschka PV, Mavrakos AE, Iafrati MD, Doleman SE, Klagsbrun M. Growth factors in bone matrix. Isolation of multiple types by affinity chromatography on heparin-Sepharose. J Biol Chem. 1986;261:12665–74. [PubMed] [Google Scholar]
- 7. Klagsbrun M, Shing Y. Heparin affinity of anionic and cationic capillary endothelial cell growth factors: analysis of hypothalamus-derived growth factors and fibroblast growth factors. Proc Natl Acad Sci U S A. 1985;82:805–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shing Y, Folkman J, Sullivan R, Butterfield C, Murray J, Klagsbrun M. Heparin affinity: purification of a tumor-derived capillary endothelial cell growth factor. Science. 1984;223:1296–9. [DOI] [PubMed] [Google Scholar]
- 9. Maciag T, Mehlman T, Friesel R, Schreiber AB. Heparin binds endothelial cell growth factor, the principal endothelial cell mitogen in bovine brain. Science. 1984;225:932–5. [DOI] [PubMed] [Google Scholar]
- 10. Brigstock DR, Steffen CL, Kim GY, Vegunta RK, Diehl JR, Harding PA. Purification and characterization of novel heparin-binding growth factors in uterine secretory fluids. Identification as heparin-regulated Mr 10,000 forms of connective tissue growth factor. J Biol Chem. 1997;272:20275–82. [DOI] [PubMed] [Google Scholar]
- 11. Pellegrini L, Burke DF, von Delft F, Mulloy B, Blundell TL. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature. 2000;407:1029–34. [DOI] [PubMed] [Google Scholar]
- 12. Rapraeger AC, Krufka A, Olwin BB. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science. 1991;252:1705–8. [DOI] [PubMed] [Google Scholar]
- 13. Bjarnadottir SG, Flengsrud R. Affinity chromatography, two-dimensional electrophoresis, adapted immunodepletion and mass spectrometry used for detection of porcine and piscine heparin-binding human plasma proteins. J Chromatogr B. 2014;944:107–13. [DOI] [PubMed] [Google Scholar]
- 14. Saito A, Munakata H. Analysis of plasma proteins that bind to glycosaminoglycans. BBA-Gen Subjects. 2007;1770:241–6. [DOI] [PubMed] [Google Scholar]
- 15. Thacker BE, Seamen E, Lawrence R, Parker MW, Xu Y, Liu J, Vander Kooi CW, Esko JD. Expanding the 3-O-sulfate proteome—enhanced binding of neuropilin-1 to 3-O-sulfated heparan sulfate modulates its activity. ACS Chem Biol. 2016;11:971–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gesslbauer B, Derler R, Handwerker C, Seles E, Kungl AJ. Exploring the glycosaminoglycan-protein interaction network by glycan-mediated pull-down proteomics. Electrophoresis. 2016;37:1437–47. [DOI] [PubMed] [Google Scholar]
- 17. Kumar V, Hassan MI, Kashav T, Singh TP, Yadav S. Heparin-binding proteins of human seminal plasma: purification and characterization. Mol Reprod Dev. 2008;75:1767–74. [DOI] [PubMed] [Google Scholar]
- 18. Zhang Y, Jiang N, Lu H, Hou N, Piao X, Cai P, Yin J, Wahlgren M, Chen Q. Proteomic analysis of Plasmodium falciparum schizonts reveals heparin-binding merozoite proteins. J Proteome Res. 2013;12:2185–93. [DOI] [PubMed] [Google Scholar]
- 19. Counihan NA, Kalanon M, Coppel RL, de Koning-Ward TF. Plasmodium rhoptry proteins: why order is important. Trends Parasitol. 2013;29:228–36. [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y, Jiang N, Jia B, Chang Z, Zhang Y, Wei X, Zhou J, Wang H, Zhao X, Yu S, Song M, Tu Z, Lu H, Yin J, Wahlgren M, Chen Q. A comparative study on the heparin-binding proteomes of Toxoplasma gondii and Plasmodium falciparum. Proteomics. 2014;14:1737–45. [DOI] [PubMed] [Google Scholar]
- 21. Hsiao FS, Sutandy FR, Syu GD, Chen YW, Lin JM, Chen CS. Systematic protein interactome analysis of glycosaminoglycans revealed YcbS as a novel bacterial virulence factor. Sci Rep. 2016;6:28425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guyot N, Labas V, Harichaux G, Chesse M, Poirier JC, Nys Y, Rehault-Godbert S. Proteomic analysis of egg white heparin-binding proteins: towards the identification of natural antibacterial molecules. Sci Rep. 2016;6:27974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paes Leme AF, Kitano ES, Furtado MF, Valente RH, Camargo AC, Ho PL, Fox JW, Serrano SM. Analysis of the subproteomes of proteinases and heparin-binding toxins of eight Bothrops venoms. Proteomics. 2009;9:733–45. [DOI] [PubMed] [Google Scholar]
- 24. Xu D, Young J, Song D, Esko JD. Heparan sulfate is essential for high mobility group protein 1 (HMGB1) signaling by the receptor for advanced glycation end products (RAGE). J Biol Chem. 2011;286:41736–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ori A, Wilkinson MC, Fernig DG. A systems biology approach for the investigation of the heparin/heparan sulfate interactome. J Biol Chem. 2011;286:19892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sandoval DR, Gomez Toledo A, Painter CD, Tota EM, Sheikh MO, West AMV, Frank MM, Wells L, Xu D, Bicknell R, Corbett KD, Esko JD. Proteomics-based screening of the endothelial heparan sulfate interactome reveals that C-type lectin 14a (CLEC14A) is a heparin binding protein. J Biol Chem. 2020;295:2804-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li W, Johnson DJD, Esmon CT, Huntington JA. Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin. Nat Struct Mol Biol. 2004;11:857–62. [DOI] [PubMed] [Google Scholar]
- 28. Dementiev A, Petitou M, Herbert JM, Gettins PGW. The ternary complex of antithrombin-anhydrothrombin heparin reveals the basis of inhibitor specificity. Nat Struct Mol Biol. 2004;11:863–7. [DOI] [PubMed] [Google Scholar]
- 29. Chleir F. Enoxaparin: a pharmacologic and clinical review. Phlebol-Ann Vasc. 2011;64:48–9. [Google Scholar]
- 30. Zong C, Venot A, Li X, Lu W, Xiao W, Wilkes JL, Salanga CL, Handel TM, Wang L, Wolfert MA, Boons GJ. Heparan sulfate microarray reveals that heparan sulfate-protein binding exhibits different ligand requirements. J Am Chem Soc. 2017;139:9534–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Paz JL, Spillmann D, Seeberger PH. Microarrays of heparin oligosaccharides obtained by nitrous acid depolymerization of isolated heparin. Chem Commun. 2006;29:3116-3118. [DOI] [PubMed] [Google Scholar]
- 32. de Paz JL, Horlacher T, Seeberger PH. Oligosaccharide microarrays to map interactions of carbohydrates in biological systems. Method Enzymol. 2006;415:269–92. [DOI] [PubMed] [Google Scholar]
- 33. Noti C, de Paz JL, Polito L, Seeberger PH. Preparation and use of microarrays containing synthetic heparin oligosaccharides for the rapid analysis of heparin-protein interactions. Chemistry. 2006;12:8664–86. [DOI] [PubMed] [Google Scholar]
- 34. Pomin VH, Wang X. Synthetic oligosaccharide libraries and microarray technology: a powerful combination for the success of current glycosaminoglycan interactomics. ChemMedChem. 2018;13:648–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cardin AD, Weintraub HJ. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. [DOI] [PubMed] [Google Scholar]
- 36. Margalit H, Fischer N, Ben-Sasson SA. Comparative analysis of structurally defined heparin binding sequences reveals a distinct spatial distribution of basic residues. J Biol Chem. 1993;268:19228–31. [PubMed] [Google Scholar]
- 37. Fromm JR, Hileman RE, Caldwell EE, Weiler JM, Linhardt RJ. Pattern and spacing of basic amino acids in heparin binding sites. Arch Biochem Biophys. 1997;343:92–100. [DOI] [PubMed] [Google Scholar]
- 38. Torrent M, Nogues MV, Andreu D, Boix E. The “CPC clip motif”: a conserved structural signature for heparin-binding proteins. PLoS One. 2012;7:e42692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pulido D, Rebollido-Rios R, Valle J, Andreu D, Boix E, Torrent M. Structural similarities in the CPC clip motif explain peptide-binding promiscuity between glycosaminoglycans and lipopolysaccharides. J R Soc Interface. 2017;14:20170423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu D, Esko JD. Demystifying heparan sulfate-protein interactions. Ann Rev Biochem. 2014;83:129–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vives RR, Crublet E, Andrieu JP, Gagnon J, Rousselle P, Lortat-Jacob H. A novel strategy for defining critical amino acid residues involved in protein/glycosaminoglycan interactions. J Biol Chem. 2004;279:54327–33. [DOI] [PubMed] [Google Scholar]
- 42. Ori A, Free P, Courty J, Wilkinson MC, Fernig DG. Identification of heparin-binding sites in proteins by selective labeling. Mol Cell Proteomics. 2009;8:2256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bitomsky W, Wade RC. Docking of glycosaminoglycans to heparin-binding proteins: validation for aFGF, bFGF, and antithrombin and application to IL-8. J Am Chem Soc. 1999;121:3004–13. [Google Scholar]
- 44. Lortat-Jacob H, Grosdidier A, Imberty A. Structural diversity of heparan sulfate binding domains in chemokines. Proc Natl Acad Sci U S A. 2002;99:1229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mottarella SE, Beglov D, Beglova N, Nugent MA, Kozakov D, Vajda S. Docking server for the identification of heparin binding sites on proteins. J Chem Inform Model. 2014;54:2068–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Samsonov SA, Pisabarro MT. Computational analysis of interactions in structurally available protein-glycosaminoglycan complexes. Glycobiology. 2016;26:850–61. [DOI] [PubMed] [Google Scholar]
- 47. Takaoka T, Mori K, Okimoto N, Neya S, Hoshino T. Prediction of the structure of complexes comprised of proteins and glycosaminoglycans using docking simulation and cluster analysis. J Chem Theory Comput. 2007;3:2347–56. [DOI] [PubMed] [Google Scholar]
- 48. Sankaranarayanan NV, Desai UR. Toward a robust computational screening strategy for identifying glycosaminoglycan sequences that display high specificity for target proteins. Glycobiology. 2014;24:1323–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Forster M, Mulloy B. Computational approaches to the identification of heparin-binding sites on the surfaces of proteins. Biochem Soc Trans. 2006;34:431–4. [DOI] [PubMed] [Google Scholar]
- 50. Sarkar A, Yu W, Desai UR, MacKerell AD, Mosier PD. Estimating glycosaminoglycan-protein interaction affinity: water dominates the specific antithrombin-heparin interaction. Glycobiology. 2016;26:1041–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Singh A, Tessier MB, Pederson K, Wang X, Venot AP, Boons GJ, Prestegard JH, Woods RJ. Extension and validation of the GLYCAM force field parameters for modeling glycosaminoglycans. Can J Chem. 2016;94:927–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gandhi NS, Mancera RL. Free energy calculations of glycosaminoglycan-protein interactions. Glycobiology. 2009;19:1103–15. [DOI] [PubMed] [Google Scholar]
- 53. Samsonov SA, Teyra J, Pisabarro MT. Docking glycosaminoglycans to proteins: analysis of solvent inclusion. J Comput Aid Mol Des. 2011;25:477–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4:P3. [PubMed] [Google Scholar]
- 56. Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech. 2008;10:P1008. [Google Scholar]
- 57. Monsinjon T, Gasque P, Chan P, Ischenko A, Brady JJ, Fontaine M. Regulation by complement C3a and C5a anaphylatoxins of cytokine production in human umbilical vein endothelial cells. FASEB J. 2003;17:1003–14. [DOI] [PubMed] [Google Scholar]
- 58. Jauneau AC, Ischenko A, Chan P, Fontaine M. Complement component anaphylatoxins upregulate chemokine expression by human astrocytes. FEBS Lett. 2003;537:17–22. [DOI] [PubMed] [Google Scholar]
- 59. Proudfoot AEI, Johnson Z, Bonvin P, Handel TM. Glycosaminoglycan interactions with chemokines add complexity to a complex system. Pharmaceuticals (Basel). 2017;10:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pangburn MK, Atkinson MA, Meri S. Localization of the heparin-binding site on complement factor H. J Biol Chem. 1991;266:16847–53. [PubMed] [Google Scholar]
- 61. Ormsby RJ, Jokiranta TS, Duthy TG, Griggs KM, Sadlon TA, Giannakis E, Gordon DL. Localization of the third heparin-binding site in the human complement regulator factor H1. Mol Immunol. 2006;43:1624–32. [DOI] [PubMed] [Google Scholar]
- 62. Loeven MA, Rops AL, Berden JH, Daha MR, Rabelink TJ, van der Vlag J. The role of heparan sulfate as determining pathogenic factor in complement factor H-associated diseases. Mol Immunol. 2015;63:203–8. [DOI] [PubMed] [Google Scholar]
- 63. Loeven MA, Rops AL, Lehtinen MJ, van Kuppevelt TH, Daha MR, Smith RJ, Bakker M, Berden JH, Rabelink TJ, Jokiranta TS, van der Vlag J. Mutations in complement factor H impair alternative pathway regulation on mouse glomerular endothelial cells in vitro. J Biol Chem. 2016;291:4974–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lehtinen MJ, Rops AL, Isenman DE, van der Vlag J, Jokiranta TS. Mutations of factor H impair regulation of surface-bound C3b by three mechanisms in atypical hemolytic uremic syndrome. J Biol Chem. 2009;284:15650–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Clark SJ, Higman VA, Mulloy B, Perkins SJ, Lea SM, Sim RB, Day AJ. His-384 allotypic variant of factor H associated with age-related macular degeneration has different heparin binding properties from the non-disease-associated form. J Biol Chem. 2006;281:24713–20. [DOI] [PubMed] [Google Scholar]
- 66. Kelly U, Yu L, Kumar P, Ding JD, Jiang H, Hageman GS, Arshavsky VY, Frank MM, Hauser MA, Rickman CB. Heparan sulfate, including that in Bruch’s membrane, inhibits the complement alternative pathway: implications for age-related macular degeneration. J Immunol. 2010;185:5486–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yu H, Munoz EM, Edens RE, Linhardt RJ. Kinetic studies on the interactions of heparin and complement proteins using surface plasmon resonance. Biochim Biophys Acta. 2005;1726:168–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yamaoka T, Kusumoto S, Ando K, Ohba M, Ohmori T. Receptor tyrosine kinase-targeted cancer therapy. Int J Mol Sci. 2018;19:3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Goetz R, Mohammadi M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat Rev Mol Cell Bio. 2013;14:166–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lanzi C, Cassinelli G. Receptor tyrosine kinases and heparan sulfate proteoglycans: interplay providing anticancer targeting strategies and new therapeutic opportunities. Biochem Pharmacol. 2020;178:114084. [DOI] [PubMed] [Google Scholar]
- 71. Hu Z, Wang C, Xiao Y, Sheng N, Chen Y, Xu Y, Zhang L, Mo W, Jing N, Hu G. NDST1-dependent heparan sulfate regulates BMP signaling and internalization in lung development. J Cell Sci. 2009;122:1145–54. [DOI] [PubMed] [Google Scholar]
- 72. Gopal S, Sogaard P, Multhaupt HA, Pataki C, Okina E, Xian X, Pedersen ME, Stevens T, Griesbeck O, Park PW, Pocock R, Couchman JR. Transmembrane proteoglycans control stretch-activated channels to set cytosolic calcium levels. J Cell Biol. 2015;210:1199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mitsou I, Multhaupt HAB, Couchman JR. Proteoglycans, ion channels and cell-matrix adhesion. Biochem J. 2017;474:1965–79. [DOI] [PubMed] [Google Scholar]
- 74. Wei H, Cai H, Wu J, Wei Z, Zhang F, Huang X, Ma L, Feng L, Zhang R, Wang Y, Ragg H, Zheng Y, Zhou A. Heparin binds lamprey angiotensinogen and promotes thrombin inhibition through a template mechanism. J Biol Chem. 2016;291:24900–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang Y, Ragg H. An unexpected link between angiotensinogen and thrombin. FEBS Lett. 2011;585:2395–9. [DOI] [PubMed] [Google Scholar]
- 76. Stanford KI, Bishop JR, Foley EM, Gonzales JC, Niesman IR, Witztum JL, Esko JD. Syndecan-1 is the primary heparan sulfate proteoglycan mediating hepatic clearance of triglyceride-rich lipoproteins in mice. J Clin Invest. 2009;119:3236–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. MacArthur JM, Bishop JR, Stanford KI, Wang LC, Bensadoun A, Witztum JL, Esko JD. Liver heparan sulfate proteoglycans mediate clearance of triglyceride-rich lipoproteins independently of LDL receptor family members. J Clin Invest. 2007;117:153–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Foley EM, Esko JD. Hepatic heparan sulfate proteoglycans and endocytic clearance of triglyceride-rich lipoproteins. Prog Mol Biol Transl. 2010;93:213–33. [DOI] [PubMed] [Google Scholar]
- 79. Gonzales JC, Gordts PLSM, Foley EM, Esko JD. Apolipoproteins E and AV mediate lipoprotein clearance by hepatic proteoglycans. J Clin Invest. 2013;123:2742–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tran-Lundmark K, Tran PK, Paulsson-Berne G, Friden V, Soininen R, Tryggvason K, Wight TN, Kinsella MG, Boren J, Hedin U. Heparan sulfate in perlecan promotes mouse atherosclerosis: roles in lipid permeability, lipid retention, and smooth muscle cell proliferation. Circ Res. 2008;103:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kunjathoor VV, Chiu DSO, ’ Brien KD, LeBoeuf RC. Accumulation of biglycan and perlecan, but not versican, in lesions of murine models of atherosclerosis. Arterioscler Thromb Vasc Biol. 2002;22:462–8. [DOI] [PubMed] [Google Scholar]
- 82. Garl PJ, Wenzlau JM, Walker HA, Whitelock JM, Costell M, Weiser-Evans MCM. Perlecan-induced suppression of smooth muscle cell proliferation is mediated through increased activity of the tumor suppressor PTEN. Circ Res. 2004;94:175–83. [DOI] [PubMed] [Google Scholar]
- 83. Saxena U, Klein MG, Goldberg IJ. Identification and characterization of the endothelial cell surface lipoprotein lipase receptor. J Biol Chem. 1991;266:17516–21. [PubMed] [Google Scholar]
- 84. Davies BSJ, Beigneux AP, Barnes RH, Tu YP, Gin P, Weinstein MM, Nobumori C, Nyren R, Goldberg I, Olivecrona G, Bensadoun A, Young SG, Fong LG. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 2010;12:42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bishop JR, Passos-Bueno MR, Fong L, Stanford KI, Gonzales JC, Yeh E, Young SG, Bensadoun A, Witztum JL, Esko JD, Moulton KS. Deletion of the basement membrane heparan sulfate proteoglycan type XVIII collagen causes hypertriglyceridemia in mice and humans. PLoS One. 2010;5:e13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Allan CM, Larsson M, Jung RS, Ploug M, Bensadoun A, Beigneux AP, Fong LG, Young SG. Mobility of “HSPG-bound” LPL explains how LPL is able to reach GPIHBP1 on capillaries. J Lipid Res. 2017;58:216–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Elgundi Z, Papanicolaou M, Major G, Cox TR, Melrose J, Whitelock JM, Farrugia BL. Cancer metastasis: the role of the extracellular matrix and the heparan sulfate proteoglycan perlecan. Front Oncol. 2020;9:1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bottaro DP. The role of extracellular matrix heparan sulfate glycosaminoglycan in the activation of growth factor signaling pathways. Ann Ny Acad Sci. 2002;961:158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jhc-10.1177_0022155420988661 for A Systems View of the Heparan Sulfate Interactome by Liliana Schaefer, Charles W. Frevert, Alejandro Gómez Toledo, James T. Sorrentino, Daniel R. Sandoval, Johan Malmström, Nathan E. Lewis and Jeffrey D. Esko in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-xlsx-2-jhc-10.1177_0022155420988661 for A Systems View of the Heparan Sulfate Interactome by Liliana Schaefer, Charles W. Frevert, Alejandro Gómez Toledo, James T. Sorrentino, Daniel R. Sandoval, Johan Malmström, Nathan E. Lewis and Jeffrey D. Esko in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-xlsx-3-jhc-10.1177_0022155420988661 for A Systems View of the Heparan Sulfate Interactome by Liliana Schaefer, Charles W. Frevert, Alejandro Gómez Toledo, James T. Sorrentino, Daniel R. Sandoval, Johan Malmström, Nathan E. Lewis and Jeffrey D. Esko in Journal of Histochemistry & Cytochemistry
Supplemental material, sj-xlsx-4-jhc-10.1177_0022155420988661 for A Systems View of the Heparan Sulfate Interactome by Liliana Schaefer, Charles W. Frevert, Alejandro Gómez Toledo, James T. Sorrentino, Daniel R. Sandoval, Johan Malmström, Nathan E. Lewis and Jeffrey D. Esko in Journal of Histochemistry & Cytochemistry