Abstract
Advances in reagents, methodologies, analytic platforms, and tools have resulted in a dramatic transformation of the research pathology laboratory. These advances have increased our ability to efficiently generate substantial volumes of data on the expression and accumulation of mRNA, proteins, carbohydrates, signaling pathways, cells, and structures in healthy and diseased tissues that are objective, quantitative, reproducible, and suitable for statistical analysis. The goal of this review is to identify and present how to acquire the critical information required to measure changes in tissues. Included is a brief overview of two morphometric techniques, image analysis and stereology, and the use of artificial intelligence to classify cells and identify hidden patterns and relationships in digital images. In addition, we explore the importance of preanalytical factors in generating high-quality data. This review focuses on techniques we have used to measure proteoglycans, glycosaminoglycans, and immune cells in tissues using immunohistochemistry and in situ hybridization to demonstrate the various morphometric techniques. When performed correctly, quantitative digital pathology is a powerful tool that provides unbiased quantitative data that are difficult to obtain with other methods.
Keywords: artificial intelligence, asthma, digital pathology, extracellular matrix, glycosaminoglycans, image analysis, immunohistochemistry, influenza, in situ hybridization, machine learning, proteoglycans, stereology
Introduction
Advances in reagents, methodologies, analytic platforms, and tools have resulted in a dramatic transformation of the research pathology laboratory.1,2 The development of digital cameras, computer hardware and software, and microscopes that convert stained tissue sections on glass slides into high-resolution whole slide digital images (WSDIs) has been central to these advances. Access to WSDIs, which can be viewed over the Internet using virtual microscopy, has resulted in the growth of the subfield of digital pathology, a paradigm shift in the pathology laboratory.
The focus of digital pathology is the analysis and management of digitized glass slides of either clinical or research specimens.3 Two ways to perform analysis of WSDIs are human-supervised analysis and computer analysis. Human-supervised analysis began when scientists first introduced contrast to tissues using histochemical techniques such as hematoxylin and eosin (H&E) stain to identify cellular subsets, anatomic structures, and pathological alterations in tissue (Fig. 1A and B). Computer analysis of WSDIs has allowed researchers to efficiently obtain substantial volumes of data that are objective, quantitative, reproducible, and suitable for statistical analysis. Several companies, such as Visiopharm (Hoersholm, Denmark), Indica Labs (Albuquerque, NM; Halo), and Definiens (Munich, Germany), have developed advanced morphometric analysis software. Open-source software is also available for computer analysis, including ImageJ, Fiji, Icy, and CellProfiler. More recently, QuPath became the first open-source image analysis software capable of analyzing WSDIs.4 Throughout this review, methods we have used to measure the chondroitin sulfate proteoglycan (PG), versican, and the glycosaminoglycan (GAG), hyaluronan (HA), in tissues are presented to illustrate how to perform quantitative digital pathology using immunohistochemistry (IHC), affinity histochemistry, and in situ hybridization (ISH).
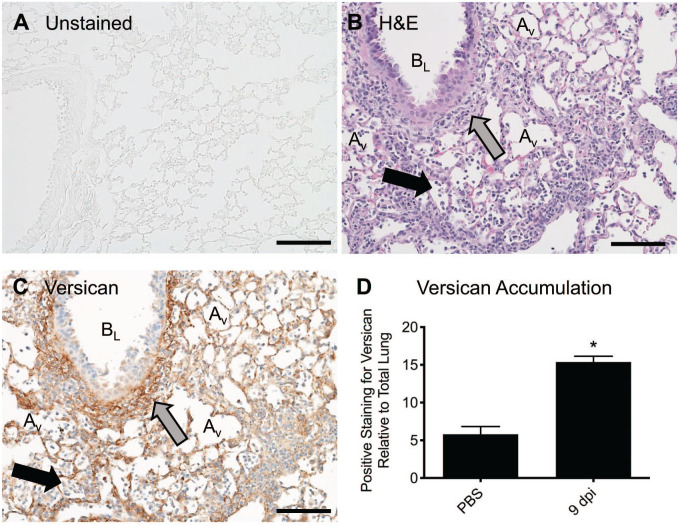
Figure 1.
Histochemical stains provide contrast to tissues, which is required to visualize cells, structures, and molecules. (A) The unstained lung tissue from a mouse is transparent, making it difficult to visualize structures and cells. (B) An H&E-stained lung tissue obtained from a mouse 9 dpi with influenza virus has contrast, allowing for the visualization of neutrophils and macrophages (black arrow) in alveoli. An accumulation of lymphocytes and macrophages (gray arrow) is observed in the peribronchiolar space around a bronchiole. (C) A tissue section adjacent to the section shown in (B) with positive immunostaining for the chondroitin sulfate proteoglycan, versican (brown). The gray arrow highlights an accumulation of versican in the peribronchiolar space of the same bronchiole shown in (A). Positive staining for versican on alveolar septa is also observed. Hematoxylin staining of nuclei (blue) provides the morphological details required to identify neutrophils and macrophages (black arrow) in alveoli. (D) Quantitative digital pathology was performed on WSDIs of lung tissue obtained from mice after oropharyngeal treatment with PBS (vehicle control) or 9 dpi with influenza virus (PR/8). The analysis showed a significant increase in versican accumulation in lungs at 9 dpi using the Mann–Whitney test, *p<0.03 with n=4 mice/group, values are mean ± SEM. Scale bar (A, B, and C), 100 μm. Abbreviations: AV, alveolus; BL, bronchiole lumen; dpi, days post-infection; H&E, hematoxylin and eosin; PBS, phosphate-buffered saline; WSDI, whole slide digital image.
Proteoglycans are dynamic molecules with complex macromolecular structures that are essential in biology.5–8 The basic structure of PGs includes a core protein with one or more covalently attached GAG side chain(s). GAGs are long linear negatively charged polymers of repeating disaccharides classified into four groups: HA, chondroitin sulfate/dermatan sulfate, heparan sulfate/heparin, and keratan sulfate. Most GAGs are synthesized and attached to a core protein in the Golgi; the exception is HA, which is synthesized at the plasma membrane.9–11 The PG family of proteins encompasses 43 distinct genes with numerous alternatively spliced variants divided into four major classes: intracellular, cell surface, pericellular, and extracellular PGs.5,9 Cell-associated PGs include intracellular and cell surface PGs. For example, serglycin is an intracellular PG found in hematopoietic and endothelial cells. Cell surface PGs include syndecans, which are single transmembrane domain proteins, and glypicans, which are proteins that are attached to the cell membrane through a glycosylphosphatidylinositol anchor.9,12–14 Pericellular PGs, such as perlecan, agrin, and collagen XVIII, are found in the basement membrane zone of cells.15 The extracellular PGs include the small leucine-rich PGs, such as biglycan, decorin, and lumican, and the HA- and lectin-binding PGs (hyalectans), such as versican and aggrecan.5,9,16,17
Gaining histological insight into how PGs and GAGs modulate cellular mechanisms requires efficient analysis tools. Manual and semiquantitative analysis techniques tend to be tedious and less precise than desired, resulting in the potential for introducing unintended errors into a study. The goal of this review is to address the critical steps and information required to objectively measure changes to PG and GAG expression and accumulation in tissues. To accomplish this goal, we provide a brief overview of image analysis, an analytical technique that measures objects in two-dimensional (2D) tissue sections, and stereology, an analytical technique that uses the spatial interpretation of 2D tissue sections to measure objects in three-dimensional (3D) tissues or organs. The application of artificial intelligence (AI) algorithms to automatically segment images, classify cells, and identify hidden patterns and relationships in digital images is also discussed. This is followed by a discussion of critical preanalytical considerations required to obtain robust and reproducible data when performing quantitative digital pathology. We also explore important elements of the preanalytical tests, which include protocols for validation and optimization of antibodies for IHC assays and appropriate positive and negative controls for IHC and ISH. This review focuses on the use of formalin-fixed paraffin-embedded (FFPE) tissue, which is often used for quantitative IHC and ISH studies.
Quantitative Digital Pathology
Digital Imaging and Whole Slide Scanning
Digital image acquisition is the conversion of an object by a sensor into a digital signal that can be processed by a computer and visualized on a computer screen. The development of whole slide images means that digital slides can be viewed, managed, and shared from the Cloud and analyzed on a computer monitor. The development of automated software to extract relevant data from whole slide images has resulted in the rapid expansion of computerized analysis, often referred to as quantitative digital pathology.18,19 Whole slide images offer an advantage as they are amenable to precise and reproducible data extraction that can be efficiently analyzed with automated open-source and commercially available software developed for image analysis and stereology. For example, the accumulation of versican was measured in tissue samples obtained from the lungs of mice exposed to either vehicle control or H1N1 influenza virus strain, A/PR/8/34 (PR/8), using WSDI and image analysis (Fig. 1C and D). This study showed a significant increase in the accumulation of versican in the lungs of mice 9 days post-infection (dpi) with PR/8 when compared with controls (15.37 ± 0.78%, vs 5.80 ± 1.01%, p<0.03, respectively). An important advantage of using tissues and histochemical stains to measure alterations in PGs is the ability to acquire morphological and contextual details from tissues such as information about physiological processes occurring in the lungs infected with influenza virus—for example, the migration of immune cells into the airways of the lungs of mice infected with influenza virus, which on day 9 after infection most often occurs in regions staining positive for versican (Fig. 1B and C). Generation of quality WSDIs is central to all quantitative digital analysis workflows and heavily impacted by preanalytical factors, as shown in Fig. 2.
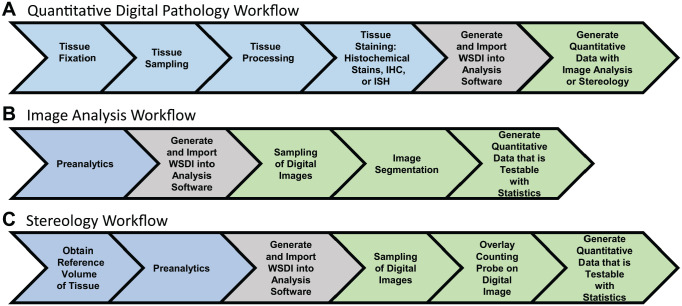
Figure 2.
(A) Workflow for performing quantitative digital pathology on 2D tissue sections using image analysis and stereology. Blue shading designates preanalytical factors, which are procedures a tissue undergoes before quantitative analysis. Gray shading designates generation of whole slide digital images, and green shading designates quantitative analysis of digital images. (B) Workflow for image analysis, a 2D analysis technique used to obtain quantitative data from tissue sections stained with histochemical stains, IHC, or ISH. (C) Workflow for stereology, a 3D analysis technique that is considered the gold standard for quantitative analysis of tissue sections. A critical preanalytical step often required when performing stereology is the need to obtain a reference volume, which is the volume of tissue, organ, or biopsy before processing of the tissue. Abbreviations: 2D, two-dimensional; 3D, three-dimensional; IHC, immunohistochemistry; ISH, in situ hybridization.
Two-dimensional Image Analysis
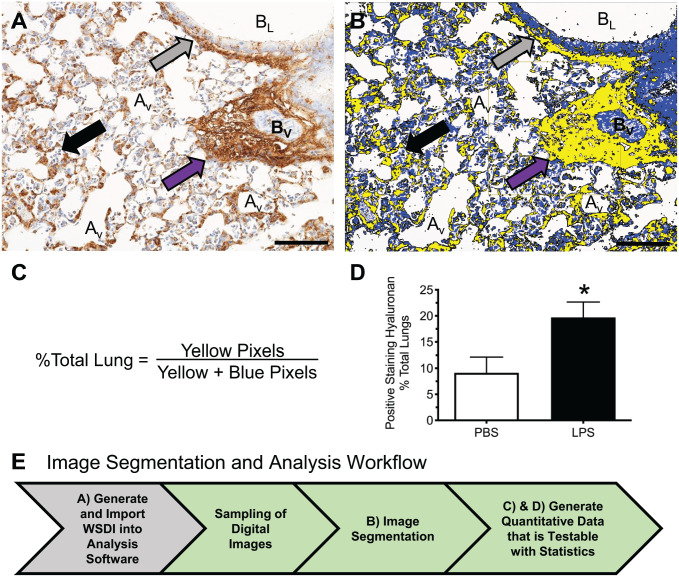
Image analysis is a morphometric technique for extracting data from digital images. Guidelines for performing image analysis have been described in detail.20–22 Image analysis is based on the evaluation of individual pixels or groups of pixels that meet specified criteria defining the target objects for analysis, enabling image segmentation.22 The segmented image is then used to quantitate an area (defined by the known area of the individual pixels involved) of specific features, such as the area of positive staining for the antigen of interest in tissue (Fig. 2B). For example, affinity histochemistry was performed using biotinylated HA-binding protein to detect and quantitate the accumulation of HA in the lungs of a mouse treated with lipopolysaccharide (LPS), a component of the cell wall of gram-negative bacteria.23 WSDIs of lung tissue stained for HA (Fig. 3A) were segmented using Visiopharm software (Visiopharm A/S) to segregate HA-stained regions as brown pixels from unstained lung tissue (Fig. 3B). The segmented image was then used to calculate the relative area of HA staining in mouse lungs using thresholding, so pixels above a specific threshold are equally counted (Fig. 3C and D). The relative area of staining or, in this case, the %Total lung is calculated using the HA-stained pixels (yellow pixels) as the numerator and the total pixels measured, which is the HA-positive pixels (yellow pixels) plus the unstained tissue (blue pixels), as the denominator (Fig. 3C). The significant increase in HA accumulation in lungs following exposure to LPS compared with control (19.71 ± 2.94% vs 9.14 ± 3.01%, p<0.04, respectively) provides evidence that the increased expression and accumulation of HA is an integral part of the immune response to lung infection caused by gram-negative bacteria.23,24 The advantage of performing image analysis is that once the protocols for segmenting an image are developed, one can rapidly obtain quantitative data using automated imaging software that performs analysis on WSDIs (Fig. 3E).
Figure 3.
Image analysis using whole slide digital imaging and automated image analysis software provides quantitative data on the relative amount of positive staining for HA in lungs of mice treated with PBS or LPS. (A) Positive immunostaining for HA (brown) in lung tissue obtained from a mouse 48 hr after oropharyngeal instillation of LPS. HA accumulation is observed in alveolar septa, peribronchiolar (gray arrow), and perivascular (purple arrow) spaces. Hematoxylin (blue) is the counterstain used to provide contrast, which allows for visualization of neutrophils within an alveolus (black arrow). (B) Segmentation of the digital image shown in (A) using Visiopharm Image Analysis Software where yellow pixels designate lung tissue that stains positive for HA and blue pixels define unstained lung tissue. (C) Formula used to determine the relative area of lung tissue stained positive for HA. (D) Accumulation of HA is significantly increased in the lungs of mice treated with LPS. Values are mean ± SEM with n=4 for each group. *Significantly different (p<0.04) than mice treated with PBS using the Mann–Whitney test and GraphPad Prism. (E) Workflow for quantification of HA in WSDIs using image analysis. Scale bar (A and B), 100 μm. Abbreviations: Av, alveolus; BL, bronchiole lumen; BV, blood vessel; HA, hyaluronan; LPS, lipopolysaccharide; PBS, phosphate-buffered saline; WSDI, whole slide digital image.
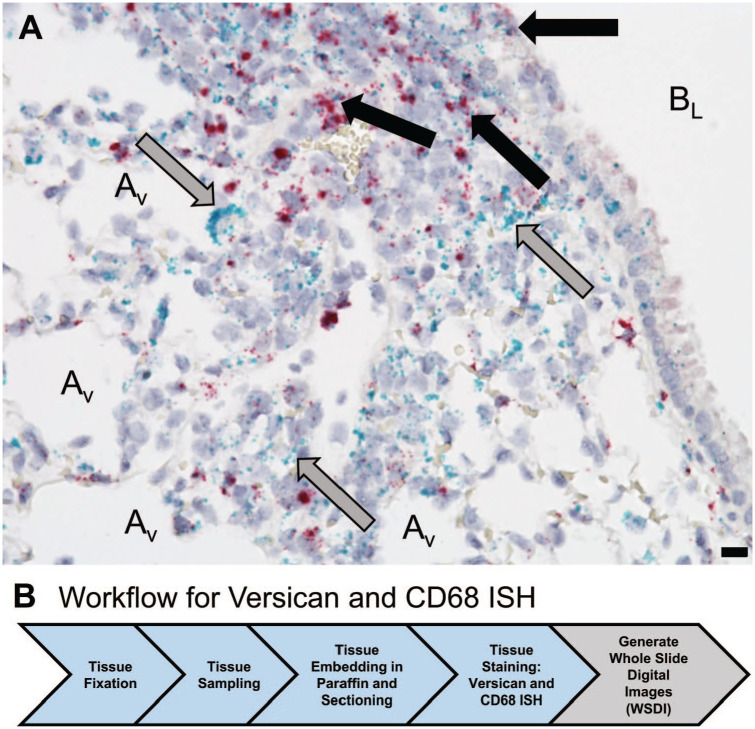
An important often overlooked element of image analysis is that it provides spatial information about cellular subsets, anatomic structures, and pathological alterations in tissue and has been used to describe the striking complexity of spatial interactions among immune cells in healthy and diseased tissues.25–29 To better understand how PGs and GAGs shape cellular phenotype in healthy and diseased tissues, the spatial relationships between cellular subsets and PGs using multiplex IHC or ISH can be defined. For example, ISH performed with probes for CD68, a marker for macrophages and dendritic cells, and versican mRNA, shows colocalization of versican in a subset of CD68-positive cells in lungs of mice infected with influenza type A virus (IAV) at 9 dpi (Fig. 4). The colocalization of versican in a subset of CD68-positive cells provides supporting evidence that increased expression of versican by macrophages or dendritic cells is a component of the host response to viral infection in the epithelial and peribronchiolar compartments of lungs. This image-based approach provides important spatial information required to understand complex biological processes occurring in tissues. It will, therefore, complement single-cell transcriptomics, a technique that performs RNA sequencing of dissociated cells. In Fig. 4B, the preanalytical workflow for performing ISH highlights the need for proper tissue sampling, which is discussed in a later section. The ability to measure alterations to mRNA in tissue using ISH and image analysis is discussed in the section on AI/machine learning.30
Figure 4.
(A) In situ hybridization provides evidence of colocalization of versican (red) and CD68 mRNA (green) in cells in the lungs obtained from a C57BL/6J male mouse 9 dpi with influenza virus. Black arrows identify cells positive for CD68 and versican mRNA among bronchiolar epithelial cells and in the peribronchiolar space. Gray arrows identify cells stained positive only for CD68 in the peribronchiolar space and adjacent to alveoli. (B) Workflow for in situ hybridization includes fixation in formalin and proper sampling using a cutting instrument that sections lung tissue into 2 mm sections that were then processed into 4 mm sections. In situ hybridization was performed on these tissue sections using RNAscope kits (Advanced Cell Diagnostics [ACD]; Newark, CA) on a Leica Bond Rx (Leica Biosystems; Nussloch, Germany). The Hamamatsu-HT 9600 Nanozoomer Digital Pathology System (Hamamatsu Photonics; Hamamatsu City, Japan) converted the stained tissue section into a WSDI. Scale bar, 10 µm. Abbreviations: AV, alveolus; BL, bronchiole lumen; dpi, days post-infection; WSDI, whole slide digital images.
Stereology
Stereology is an analytical technique that is design-based and, in contrast to image analysis, is divorced from the limitations imposed by 2D assumption-based analysis techniques. Its potential and applications to biological structures have been described in detail.21,31–38 Design-based stereology relies on (1) uniform random sampling of the entire volume of concern and (2) unbiased questioning using geometric probes that question their target tissue for the presence or absence of an object. Geometric probes are always paired with their target objects such that the number of hits of the probe on the target object invariably sum to three dimensions. The probes are superimposed over digital images to enable extrapolation of quantitative 3D data on structures, cells, and molecules in an organ or tissue biopsy. These design-based stereological probes allow for the measurement of several parameters, including the total volume of an organ, local volumes, surface area or length of structures in an organ, cross-sectional area, and diameter.34,35,39 Design-based stereology accurately measures cell size and number within an organ.31,32,36 For example, counting objects in tissues, a zero-dimensional task, is performed using the design-based 3D volumetric probe, the disector, which is based on two tissue sections within a single block; the volume of the probe can then be defined by the area of the sections that were sampled and the distance between the sections. More specifically, the disector comprises three elements: (1) a counting rule that uses a pair of optical or physical tissue sections that are separated by a defined distance (h) determined by the height of the object being counted; (2) an integral test system with test points; and (3) an unbiased 2D counting frame with a known area.35,40,41
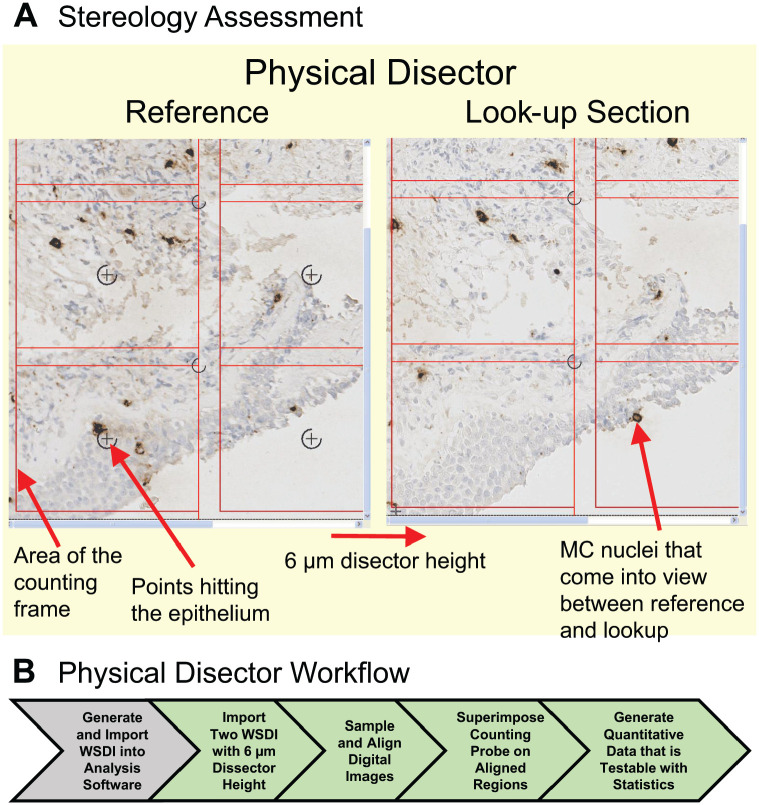
A recent design-based stereology study used the physical disector and tissue sections from airway biopsies to gain a better understanding of the mechanisms regulating airway hyperresponsiveness in patients with asthma.42 For these studies, the location of mast cells was defined and measured in two compartments, the submucosal and epithelial compartments. The analysis was performed on airway biopsies obtained from normal human subjects and patients with asthma. These tissue samples were fixed in methyl Carnoy’s solution before embedding in paraffin. To ensure that all portions of the airway biopsies were sampled, design-based stereology sampling techniques were used to obtain two serial tissue sections from the block that were separated by a distance of 6 µm. WSDIs of the two tissue sections were obtained using the Hamamatsu-HT 9600 Nanozoomer Digital Pathology System (Hamamatsu Photonics; Hamamatsu City, Japan). The WSDIs of the two tissue sections were imported into and aligned using the Visiopharm Autodisector module (Visiopharm A/S). Uniform random sampling of the two WSDIs was performed, and a counting probe was superimposed on the two aligned digital images. To estimate the number and location of mast cells in the tissue biopsies, the two digital images with a disector height of 6 µm (i.e. physical distance between the two sections) were defined as the “reference” and “look-up” sections (Fig. 5). Mast cell nuclei coming into view in the “reference” were counted if they were not present in the “look-up” section. A point associated with each counting frame was used to determine the reference volume of the submucosa and the epithelium. For this example, there were no mast cells in the initial look-up section. To increase efficiency, the “reference” and “look-up” sections can be reversed as demonstrated in Fig. 5, where a mast cell nucleus is present in the new reference section, which is now the right panel, and not in the corresponding “look-up” section, the left panel. Proper sampling techniques of both the airway biopsy and digital images ensured that accurate measurements of mast cells were made even though these cells may not be uniformly distributed in airways. Using this approach, the physical disector provided measurements of the volume density of mast cells in the submucosal and epithelial compartments of airway biopsies and showed a shift in mast cells from the submucosa to the airway epithelium. These data provided new information showing that mast cells are strongly associated with increased airway hyperresponsiveness and identified mast cells as regulators of type 2 inflammation in patients with asthma.42 This approach could be easily adapted to measure the association of immune cells with specific PGs in a tissue section to provide quantitative information about changes in PG volume and distribution within a section and how that change is correlated with alterations in the spatial location of select immune cells. Guidelines for performing design-based stereology using stereological probes have been described in detail.21,31–34,36,42–48
Figure 5.
The physical disector provides accurate measurement of cell numbers and/or volume in tissues. (A) Two WSDIs from two tissue sections approximately 6 µm apart (i.e., disector height) were processed using the Visiopharm Autodisector module, which sampled, aligned, and labeled the adjacent sections as the “reference” and “look-up” sections. A two-dimensional counting frame is shown superimposed on these images with inclusion lines (left and bottom) and exclusion lines (right and top) of the counting frame. A hematoxylin-stained nucleus (blue) in a mast cell (MC) identified using a murine monoclonal anti-tryptase antibody (brown stain) is counted if it is in focus, if it is inside the counting frame, or if it touches the inclusion lines but does not touch the exclusion lines. MC nuclei that were in focus in the “reference” were counted if they were not present in the “look-up” sections. For efficiency, this process was reversed so that the Look-up section became the Reference section. In this case, the MC nuclei shown by red arrow in the lower right-hand counting frame in the Look-up section are counted because they are not observed in the counting frame of the adjacent Reference section. A point associated with each counting frame was used to determine the reference volume by enumerating points hitting the submucosa and the epithelium. The point in the lower left-hand counting frame, highlighted by red arrow, is hitting the epithelium, so it is counted. (B) Workflow for performing analysis using the physical disector on two WSDIs to measure the volume of MCs in tissue biopsies. (A) is a supplemental figure from Altman et al.42 Abbreviation: WSDI, whole slide digital image.
When designing experiments using stereology, preplanning is essential for studies so that a reference volume of the tissue to be studied can be obtained before tissue processing (Fig. 2C). Preplanning is also required to ensure proper sampling protocols such as uniform random sampling are implemented to make sure that all components of a 3D organ have an equal chance of being sampled.31,32,34,45–47,49,50 When performed correctly, stereology is more precise and requires minimal or no assumptions to obtain quantitative measurements from tissues, thus enabling unbiased data. For these reasons, stereology has become the gold standard for quantitative digital pathology (Table 1).
Table 1.
Comparisons Between Stereology and Image Analysis.
| Stereology | Image Analysis | |
|---|---|---|
| Advantages | 1. Gold standard for quantitative analysis of tissues 2. Minimal to no assumptions 3. Provides precise and unbiased measurements 4. Able to accurately measure volumes, number of cells, surface area, etc. |
1. Easily automated 2. Quantitative results are obtained more rapidly than with stereology, so more amenable to high throughput 3. Excellent when used for IHC, ISH, and special stains when contrast allows easy segmentation of an image |
| Limitations | 1. Considerable amount of time is required to perform stereology 2. Typically requires the investigator to obtain an initial reference volume and monitor changes to the volume during tissue processing to account for shrinkage (Fig. 2C). |
1. Less rigorous and assumptions are required in many instances, which introduces bias 2. This technique provides measurements of relative change (%), which is typically a ratio of the ROI to the total tissue or other denominator 3. Counts 2D profiles, which overestimates the number of objects counted; is influenced by orientation and distribution of cells in a 3D tissue; and counts larger cells more than smaller cells. 4. Less precise than stereology |
Abbreviations: 2D, two-dimensional; 3D, three-dimensional; IHC, immunohistochemistry; ISH, in situ hybridization; ROI, region of interest.
Stereology vs Image Analysis
Whereas image analysis is easily automated and amenable to high-throughput analysis, a limitation of stereology is the amount of time required to correctly perform a study even with computational advances such as whole slide digital imaging and automated analysis software (Table 1). Nevertheless, image analysis is less rigorous and only provides representative 2D measurements of relative changes for antigens, mRNA, cells, or structures in tissues. Image analysis also requires assumptions which may introduce bias into a study (Table 1).51 A common assumption that is often made when performing image analysis is that a single 5-µm tissue section represents the entire organ. If the object of interest (e.g., PGs, GAGs, anatomic structures, or cells) is not uniformly distributed throughout a tissue or organ, obtaining only one tissue section from an organ has the potential to introduce sampling bias.51 Therefore, it is highly recommended that proper sampling techniques be incorporated into the experimental design of image analysis protocols to minimize the introduction of this type of bias.
Another assumption that is often made is that one can count objects (e.g., cells, glomeruli) in a 2D tissue section. Whereas it is tempting to perform counts of objects when using image analysis, it is important to remember that image analysis visualizes profiles of 3D objects, such as cells in 2D tissue sections. The counting of profiles is highly biased because counts are influenced by the height of the object perpendicular to the section plane as well as its overall 3D shape.31,32,51 Therefore, the use of image analysis to count objects in tissue results in geometric bias in which the number of objects in a tissue/organ can be misrepresented and, due to differences in height and complexity of volumetric shapes, larger and more complex objects are overrepresented when compared with smaller, simpler objects.31,32 Proper sampling of tissues is also required to obtain accurate counts, especially when cells are not uniformly distributed in an organ. The importance of proper sampling techniques is discussed in detail in section “Preanalytical Elements of Tissue Processing.” If the goal of a study is to obtain the number of cells or structures in an organ or biopsy, then stereology is required. Also, when confronted with small intergroup differences in a study, stereology might be a better tool compared with image analysis, which is less precise and may not be able to identify biologically relevant differences as significant (Table 1).
Finally, the following principle needs to be incorporated into all quantitative digital pathology studies, whether stereology or image analysis: “Be as accurate as possible and as precise as necessary.”52 This principle is based on the idea that accuracy, or the absence of bias, is fundamental, but precision can be adjusted or improved as needed. Such adjustments can be made at the many levels of sampling, from the number of animals per group, to the number of sections per tissue block, to the number of counts per field of view. (This will be discussed in more detail in section “Sampling of Tissues and Digital Images.”) Although these adjustments improve statistical precision, they are meaningless if the fundamental assumptions are biased or inaccurate.53
When performed correctly, stereology and image analysis are powerful tools that provide unbiased quantitative data from IHC and ISH experiments that are difficult to obtain with other methods. Quantitative data obtained using these analytical techniques can then be correlated with other measurements such as clinical data or data collected with flow cytometry or quantitative PCR.25,29,54,55 More importantly, these morphometric techniques provide spatial information that is lost when tissues are disrupted to obtain cells for flow cytometry and proteins or mRNA for proteomics and transcriptomics studies, respectively.
Artificial Intelligence/Machine Learning
The integration of AI with image analysis for detection, segmentation, feature extraction, and tissue classification of digital images has resulted in an even more rapid evolution and expansion of digital pathology.18,19,56–58 Although the promise of AI has garnered substantial attention and excitement, it is important to point out that the use of AI to obtain objective data from cells and tissues dates back over 40 years.59,60
AI, computer-based systems that automate decision-making processes, was initially defined as the overarching concept of the thinking machine.19,58 When first described, machine learning was defined as giving computers the ability to learn without having to be specifically programmed to provide an output.19,58 Frequently, this is accomplished through supervised learning, wherein the AI system uses algorithms to analyze training sets of data (e.g. digital images), learns from those data and adjusts its algorithms, and then based on that learning interprets or makes predictions about new data. Through an iterative process of training, the accuracy of the computer algorithms improves. Machine learning has tremendous utility due to its ability to automatically segment images, classify cells, and identify hidden patterns and relationships in digital images that humans might miss.61 For example, a study undertaken by Gross et al. classified cells in the heart based on their nuclei as cardiac myocytes vs non-cardiac myocytes.62 Using 5-ethynyl-2′-deoxyuridine (EdU), a thymidine analogue, and the TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) assay to label cells in the heart, the authors were then able to quantitate cardiac myocytes undergoing cell proliferation and apoptosis, respectively. Using multiplex IHC combined with similar machine learning models, one could also quantitate the degree of colocalization of specific PGs and GAGs to select cells and cellular processes in a tissue.
An emerging area within machine learning is deep learning, which is a form of machine learning that relies on both supervised and unsupervised learning. Deep learning applied to digital pathology uses artificial neural networks (ANN) to determine whether the output or interpretation of a digital image is correct.18,58 ANN use multiple layers of calculations imitating the complex network of neurons in the human brain to provide an analysis or output from the input data.56,58 A number of commercial software vendors have begun to incorporate deep learning modules into their image analysis software programs. These deep learning programs provide additional tools for sophisticated image segmentation.
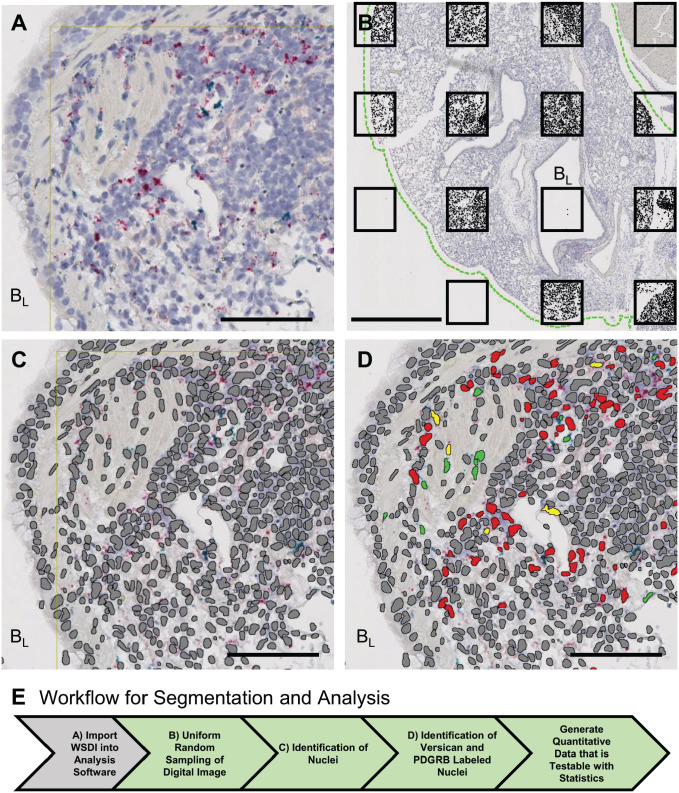
For example, using tissue obtained from a mouse 9 dpi with influenza virus (PR/8 strain), a deep learning module (Visiopharm) was used to identify nuclei in mouse lung tissue sections in which versican and platelet-derived growth factor receptor beta (PDGFRB) mRNA were detected using ISH (RNAscope technology; Advanced Cell Diagnostics [ACD], Newark, CA) (Fig. 6A). Recognition of PDGFRB-positive nuclei was used to identify fibroblasts and pericytes in the lung tissue.63 The challenges of segmenting nuclei using the deep learning module included prolonged processing times and difficulty in correctly determining nuclear borders.2,3 To decrease up to 1 hr per digital image processing time required for deep learning segmentation, uniform random sampling of the WSDI was performed (Fig. 6B). The difficulty in correctly identifying nuclear borders was due to poor nuclear morphology and clumping of nuclei. The poor nuclear morphology was the result of repeated heating of the tissue section when performing ISH. However, repeat annotation of nuclei in tissue sections followed by training generated a segmentation protocol that accurately identified the nuclei in ISH-stained tissues (Fig. 6C). An image analysis protocol was then developed and used to identify nuclei that were positively stained for versican mRNA alone (red), PDGFRB mRNA alone (green), or versican and PDGFRB mRNA colocalized in the same nuclei (yellow) (Fig. 6D). To determine the appropriate amount of tissue on a WSDI that needs to be sampled, we undertook a small pilot project that compared analysis of 20%, 30%, and 100% of lung tissue on the digital image. The colocalization of PDGFRB and versican mRNA was found in 18% of the nuclei for the 20% and 30% analyses compared with 19% in the 100% analysis. This proof-of-concept study shows how proper sampling of even 20% of the WSDI is able to provide similar information compared with sampling of the entire digital image.
Figure 6.
Deep learning algorithms and image analysis protocols were used to perform image segmentation and analysis to measure the colocalization of versican and PDGFRB mRNA in the nuclei of lung tissue obtained from an influenza virus–infected mouse. (A) Digital image of the tissue section stained for versican (red) and PDGFRB (green) mRNA in tissues adjacent to a bronchiole (BL) in lung tissue from a 9 dpi with influenza virus mouse. (B) To decrease nuclear identification processing time, uniform random sampling was performed before running the deep learning module. The green dashed line defines lung tissue and the black boxes show the uniform sampling pattern that was laid down with a random start on the WSDI. The analysis software identifies and only performs analysis on lung tissue. This is illustrated by the black fill in each of the boxes. (C) The Visiopharm deep learning module was used to train the computer to accurately identify nuclei (gray) for nuclear segmentation. (D) Image analysis protocols were used to identify nuclei that were positively stained for versican mRNA alone (red), PDGFRB mRNA alone (green) or versican and PDGFRB mRNA in the same nuclei (yellow). (E) Workflow for generating objective data on the colocalization of versican and PDGFRB. Scale bar (A, C, D), 85 μm; (B), 1 mm. Abbreviations: AV, alveolus; BL, bronchiole lumen; dpi, days post-infection; PDGFRB, platelet-derived growth factor receptor beta; WSDI, whole slide digital images
Preanalytical Elements of Tissue Processing
The quality and reproducibility of the data collected when performing stereology and image analysis rely directly on the quality of the tissue samples used for a study. Therefore, preanalytical elements need to be considered and integrated into each study’s experimental design. Careful consideration must be made of the following: (1) proper sampling of tissues and digital images, and (2) tissue fixation and processing.
Proper Sampling of Tissues and Digital Images
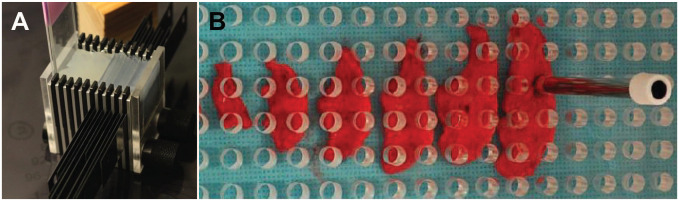
When designing studies to make quantitative assessments of PGs in a 3D organ, attention must be given to sampling because measurements are made using 2D tissue sections, which results in a loss of information. A goal of sampling is to ensure that all parts of an organ or a WSDI have the same probability of being represented in the analysis. To account for the uneven distribution of an object of interest in a 3D space, investigators designing stereology studies have developed rigorous sampling protocols using a number of statistical techniques to guarantee the unbiased selection of tissue sections and digital images to decrease variability and minimize bias.31,34,36,47,49,52 For example, the use of systematic uniform random sampling using a fixed, periodic sampling interval and a random starting point within the thickness of this interval ensures random sampling of tissues collected from mice (Fig. 7A). This becomes more difficult when working with larger animals but can be overcome with the use of plexiglass templates with evenly spaced 5 mm holes and a punch biopsy to collect tissues (Fig. 7B).47 The use of Plexiglas templates to perform uniform random sampling in non-human primates infected with Mycobacterium tuberculosis combined with quantitative analysis using stereology showed a significant correlation between bacterial numbers and the inflammatory response in samples collected from lungs.47
Figure 7.
Uniform random sampling of tissues minimizes bias and variability due to the multifocal distribution of cells and other analytes of interest in tissues. (A) Cutting instrument with trimming blades used to section mouse lungs into 2 mm sagittal sections to ensure adequate sampling. This cutting instrument was used to sample the lung tissues analyzed in Figs. 1, 3, 4, and 6. (B) Stratified uniform sampling of a lung lobe from a non-human primate that uses a plexiglass template with evenly spaced 5 mm holes that was randomly placed over section of a lung lobe. A punch biopsy was used to collect tissue for microbial cultures and histology, with 10% of the lung lobe collected for each test. (B) is adapted from Luciw et al.47
In their seminal paper on sampling, Gundersen et al. provided insight into considerations regarding sampling by measuring the variance at each level of sampling used in stereology and showed that the greatest contribution to variance was among animals (70%), followed by tissue blocks/tissue sections (19%), fields of view (8%), and intercepts + measuring (3%).46 Thus, the “Do More Less Well” subtitle of this work conveys that one should “do more” by increasing the number of subjects or blocks/sections studied to decrease variability and increase the rigor of a study. This concept is applicable to image analysis as well as stereology studies.51 Applying lessons learned from stereology regarding minimizing sampling bias to 2D image analysis studies will increase the likelihood that results are representative of the tissue as a whole.51,64
Tissue Processing
Tissue processing includes fixation, dehydration, and then the infiltration of tissues with paraffin. While this stabilizes tissues and preserves tissue morphology, it produces artifacts, including, but not limited to, non-uniform shrinkage of tissue, deformation of tissue structures, loss of tissue epitopes required for antibody recognition, and fragmentation of RNA.23,65 The work of Xie et al. showed that inadequate tissue processing resulted in the retention of water in tissue sections, which resulted in antigen degradation as measured by IHC, Western blots, and protein array technologies.65 Therefore, investigators need to carefully consider the requirements for tissue processing before starting a study. This is particularly important when planning studies for quantitative analysis because such artifacts will result in flawed results.
Although FFPE-processed tissues are most commonly used for IHC and ISH when quantitative analysis is performed, other methods are being established. A problem inherent to the use of FFPE is shrinkage, which for lung tissue can be up to 40% based on stereological measurements.23 When using stereology to measure the length, surface, or volume of objects in a tissue, one must correct for shrinkage to preserve unbiasedness.50 Tissue shrinkage is not a problem when using stereology to count objects.
Studies comparing snap-freezing to ethanol-based fixatives or aldehyde-based fixation show that although the amount of antigen (DNA, RNA or protein) recovery differs between the methods, the quality of antigen recovered is better in EtOH-based fixatives.66–68 Therefore, when planning a study, it is important to determine the best fixative for the biomolecules that will be studied. In addition, it is also important to follow guidelines set out for preserving physiological volumes in different tissues. Examples are the principles for standardization of study design outlined by the American Thoracic Society to promote comparability of morphometric studies of lungs.69
Histochemical Stains
Histochemical stains not only provide the ability to visualize biological structures, they also provide the contrast required to perform quantitative analysis. For example, the identification of hematoxylin-stained nuclei in tissue biopsies was used to measure the shift in mast cell infiltration to the airway epithelium using stereology (Fig. 5A). Hematoxylin staining of nuclei was also used for the deep learning segmentation protocol developed to identify the colocalization of versican and PDGFRB mRNA in nuclei (Fig. 6). Histochemical stains have been used for many years to define the location of PGs and GAGs in tissue. A number of these special stains, which are cationic and therefore bind to GAGs, include alcian blue, toluidine blue, and basic fuchsin.70–72 Movat’s pentachrome stain is another histochemical stain that is used for studies of the extracellular matrix as it stains elastic fibers black; collagen, yellow; and GAGs, blue.73,74 To define the nature of staining patterns observed with histochemical dyes such as alcian blue, enzymatic digestion of tissue sections with hyaluronidase, chondroitinase ABC, or heparinase can be performed to remove specific classes of GAGs and create negative controls. Several excellent references provide methods and technical descriptions of the many histochemical stains used on tissues.75,76
Preanalytical Elements for IHC
Validation of an Antibody for IHC
IHC takes advantage of the specificity inherent to antibodies to detect a vast number of antigens, including, but not limited to, proteins, peptides, carbohydrates including GAGs, lipids, amines, sugars, amino acids, drugs, phosphorylated proteins, and the inactive and active forms of proteases. All too often, researchers obtain an untested antibody and use it for IHC without validating and optimizing the antibody for its target antigen in tissue. This can cause erroneous interpretations if antibody staining reflects off-target binding, which is defined as binding of an antibody in tissues to sites other than the target antigen. Antibody validation and optimization for IHC will only be introduced here as there are additional guidelines for both described in more detail elsewhere.1,77–79 It is important to point out that one can never prove that an antibody specifically binds to its epitope in a tissue section using immunohistochemical techniques.80 Therefore, the purpose of antibody validation and optimization is to increase one’s confidence that the IHC protocol being developed is providing accurate and reproducible information.
The first step of antibody validation includes biochemical analysis, with the Western blot being the most commonly used method.79 When performing a Western blot, tissue homogenates are the most appropriate sample to use to validate an antibody. If tissue homogenates are not available, at minimum a cell lysate using a cell known to express the target antigen should be used. If an antibody specifically recognizes its target antigen, there should be a single band of the appropriate molecular weight on the Western blot. If multiple bands are observed on the Western blot, potential considerations include the presence of protein isoforms, posttranslational modifications, and proteolytic degradation or the nonspecific binding of the antibody to other proteins. If no bands are visualized on the Western blot, that could indicate that the antibody recognizes a conformational epitope that was lost during the protein denaturation protocol for Western blotting. If this is suspected, native gels or other assays, such as immunoprecipitation, dot blots, and ELISA, should be considered to confirm biochemical specificity. Finally, evidence of biochemical specificity of an antibody does not guarantee that the antibody will specifically identify the target antigen in preserved tissue.
Optimization of an Antibody for IHC
Evaluating whether antigen retrieval is required is an important step used to enhance the ability of an antibody to bind its target epitope in formalin FFPE. The two commonly used antigen retrieval protocols are heat-induced epitope retrieval (HIER) and proteinase digestion. The single most significant advance in IHC in the last 70 years was the development of HIER antigen retrieval techniques. These techniques have increased the likelihood that an antibody will detect its epitope in archived FFPE tissue and have greatly expanded the use of IHC in clinical and research pathology laboratories.81,82 Two commonly used HIER protocols include the use of buffers of citrate at pH 6.0 or EDTA at pH 9.0 with or without detergent additives heated to approximately 100C, with immersion of slides in the buffer typically for 10 to 30 min. The mechanisms by which HIER antigen retrieval increases the ability of the primary antibody to detect its epitope in tissue are still not known. However, one potential mechanism is that these techniques break aldehyde-induced protein–protein crosslinks, which dissociates irrelevant proteins from the target epitope and restores immunoreactivity.83 The second type, proteolytic-induced epitope retrieval (PIER), uses proteinases, such as proteinase K, trypsin, or pronase, to digest tissue sections either at room temperature or at 37C, exposing previously masked epitopes. For both HIER and PIER, it will be necessary to optimize conditions of time, temperature, and pH for each antibody. Interestingly, PGs are often less sensitive to differences in antigen retrieval.84
An antigen retrieval step often required when working with proteoglycans is the treatment of tissue sections with enzymes to allow the primary antibody to bind to their epitope. An example is the use of chondroitinase ABC on tissues to remove the chondroitin sulfate side chains of versican, which is required when performing IHC with the polyclonal rabbit anti-mouse versican antibody directed against a.a. 1360–1439 of mouse versican (EMD Millipore, cat. no. AB1033; Fig. 1C).85 An extensive review of immunohistochemical labeling methodologies for FFPE and cryopreserved tissues for studying PGs and GAGs is provided by Hayes et al.86
Performing a proper dilution series, typically over one to two logs of antibody concentrations, is a critical step in IHC protocol optimization. The goal of the antibody dilution series is to find the concentration of antibody at which high-affinity binding of an antibody to its epitope in tissue is maintained but low-affinity nonspecific interactions do not occur. Taking an antibody dilution out until the intensity of staining diminishes or is extinguished is essential. Many antibodies against PGs will provide a clean signal devoid of background across a broad working range; however, higher dilutions (less antibody) are preferred as they result in a cleaner cytomorphological pattern as well as decreased assay costs. Due to the lot-to-lot variability observed with antibodies, it is also recommended that a dilution series be performed when a new lot of a previously validated antibody is obtained.
Other optimization steps for minimizing nonspecific binding can be included in staining protocols. Thorough washing of tissue sections at all stages of the IHC protocol helps to minimize hydrophobic or ionic interactions. Protein blocks using serum, antibodies, or casein reduce binding to nonspecific sites in tissues. Several of the detection systems rely on peroxidase, alkaline phosphatase, and biotin, all of which can be endogenous factors in tissue, which may add to nonspecific staining. To minimize the effect of these endogenous factors, hydrogen peroxide is used to quench endogenous peroxidases; levamisole is used to block endogenous alkaline phosphatases; and the avidin–biotin block method is often used to minimize the potential of endogenous biotin causing nonspecific staining.
Following the optimization of an IHC protocol, additional steps are often used to increase one’s confidence that the positive staining for an antibody in tissue is specific for recognizing the desired target antigen. (1) This includes demonstrating that two antibodies recognizing distinct epitopes in the target antigen have the same IHC staining pattern in a tissue section.87 The in situ proximity ligation assay extends this step by only detecting signals when two antibodies bind the target jointly, ruling out crossreactive off-target binding in tissue.88 (2) The use of genetic strategies such as tissues from mice that lack the target antigen as biological negative control tissue.89 (3) The use of recombinant strategies to overexpress or delete the target antigen in cells.1 (4) The use of statistically independent assays such as quantitative real-time PCR or flow cytometry to show that the data collected with these techniques correlate with changes measured with quantitative assessment of a target antigen in IHC-stained tissues.55,79 (5) The use of ISH to show that the mRNA and the IHC-positive staining for the analyte of interest are in the same location. Validation an antibody using ISH has limitations because the tissue half-life of a protein and mRNA may differ, especially for proteins such as PGs, which can have long tissue half-lives.
Controls
IHC is no different than any other biological assay in that appropriate negative and positive controls need to be included as part of each experiment.80 The goal of antibody and tissue controls is to provide evidence that the primary antibody is specifically binding its epitope in tissue. Controls also account for the day-to-day variability inherent to biological assays and can help to troubleshoot problems as they arise.
The simplest and probably most critical negative control for IHC is replacement of the primary antibody with a negative antibody control. Negative antibody controls are used because they do not detect the epitope of interest but maintain many of the nonspecific binding characteristics of primary antibodies. When using polyclonal antibodies for IHC protocols, one should use preimmune serum or commercial sera from the same species used to raise the primary antibody. When using monoclonal antibodies for IHC protocols, a negative control antibody of the same species, antibody isotype, and concentration matched need to be used. Replacement of the primary antibody with phosphate-buffered saline is not an acceptable negative antibody control and should only be used to identify the nonspecific binding of the secondary antibody in tissue. When using chromogenic stains that require peroxidases or alkaline phosphatases linked to the primary antibody, negative antibody controls help to identify staining caused by endogenous enzymes that maintain enzymatic activity even in FFPE tissues. Similarly, when performing IHC using fluorescent probes, the use of negative control antibodies assists in the identification of autofluorescence in tissue.90
The use of negative and/or positive tissue controls to identify nonspecific binding of the primary antibody in tissue is especially necessary when validating a new antibody.1,78 Positive and negative tissue controls are performed at the same time as the test tissues to help confirm that positive staining for the target antigen is valid. Negative tissue controls use cells or tissues known to lack the target antigen. Positive tissue controls use cells or tissues where the target antigen is known to be present or is overexpressed. When using cell pellets as a tissue control, the cell pellet should be fixed and processed into paraffin to allow antibodies to be evaluated under conditions similar to those of the tissue being studied. A commonly used negative tissue control is from mice lacking the target antigen. For example, negative tissue controls obtained from mice with a global deletion of versican (i.e., Vcan−/− mice) treated with polyinosinic-polycytidylic acid showed minimal positive immunostaining for versican in lungs compared with the wild-type controls.89 For many proteins, the cellular or subcellular location of the target antigen in tissue is known. Therefore, when using positive tissue controls, if positive staining for a target antigen is observed in a structure, cell, or subcellular compartment (e.g., plasma membrane, cytoplasm, or nucleus) where the target protein is known to be located, then confidence that the antibody binding is specific to its epitope in tissue is increased.
For studies where ISH is performed, appropriate positive and negative controls are required. When performing ISH using RNAscope reagents (ACD), the probes for the target RNA and the positive and negative controls are included with the assay. These are typically species and target tissue–matched probes for housekeeping genes for the positive control and probes for bacterial dapB for the negative control. As with IHC, positive and negative tissue controls for ISH are necessary to help assess the quality of the RNA in a tissue section and to assist in troubleshooting problems that arise during a study.
To conclude, advances in quantitative analysis of images have been made by leveraging the field of digital pathology, WSDI, and novel computational tools that have automated many of the processes required to obtain contextual data from histochemical, IHC, and ISH studies. When performed correctly, quantitative digital pathology generates unbiased data that are testable with statistics. It also allows for single-cell analysis within architectural tissue context. The ability to obtain spatial information is important because it provides increased knowledge about cellular phenotypes and cell-to-cell interactions in the context of their microenvironment, which includes specialized cells, structures—blood vessels and airways—and pathological changes. For individuals interested in PGs, GAGs, and the extracellular matrix, it enables investigation into how alterations in the extracellular microenvironment impact cellular phenotypes in situ. This high content analysis extends our ability to define mechanisms of action for specific molecules and cells to better understand biological processes occurring in health and disease.
Footnotes
Author Contributions: All authors contributed to the literature review and manuscript preparation.
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: 1R01AI136468-01 (C.W.F.), 1R01AI130280 (C.W.F.), SINGH19R0 (C.W.F.), 1P30DK089507 (C.W.F.), P30-DK17047 (C.W.F.), 1K24AI130263-04 (T.S.H.), Department of Diagnostic Medicine/Pathobiology, Kansas State University (A.S.D.), and HCS Capstone Grant (J.E.B.)
Contributor Information
A. Sally Davis, Department of Diagnostic Medicine/Pathobiology, College of Veterinary Medicine, Kansas State University, Manhattan, Kansas.
Mary Y. Chang, Department of Comparative Medicine, University of Washington, Seattle, Washington; Center for Lung Biology, University of Washington at South Lake Union, Seattle, Washington.
Jourdan E. Brune, Department of Comparative Medicine, University of Washington, Seattle, Washington; Center for Lung Biology, University of Washington at South Lake Union, Seattle, Washington.
Teal S. Hallstrand, Division of Pulmonary, Critical Care and Sleep Medicine, Department of Medicine, University of Washington, Seattle, Washington Center for Lung Biology, University of Washington at South Lake Union, Seattle, Washington.
Brian Johnson, Department of Comparative Medicine, University of Washington, Seattle, Washington.
Sarah Lindhartsen, Department of Comparative Medicine, University of Washington, Seattle, Washington.
Stephen M. Hewitt, Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesda, Maryland.
Charles W. Frevert, Department of Comparative Medicine, University of Washington, Seattle, Washington; Division of Pulmonary, Critical Care and Sleep Medicine, Department of Medicine, University of Washington, Seattle, Washington; Center for Lung Biology, University of Washington at South Lake Union, Seattle, Washington.
Literature Cited
- 1. Goodwin P, Johnson B, Frevert C. Microscopy, immunohistochemistry, digital imaging, and quantitative microscopy. In: Treuting PM, Dintzis SM, Montine KS, editors. Comparative anatomy and histology: a mouse, rat, and human atlas. 2nd ed. Amsterdam; Boston: Elsevier/Academic Press; 2018. p. 55–66. [Google Scholar]
- 2. Brown C, Sekhavati F, Cardenes R, Windmueller C, Dacosta K, Rodriguez-Canales J, Steele KE. CTLA-4 immunohistochemistry and quantitative image analysis for profiling of human cancers. J Histochem Cytochem. 2019;67(12):901–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zarella MD, Bowman D, Aeffner F, Farahani N, Xthona A, Absar SF, Parwani A, Bui M, Hartman DJ. A practical guide to whole slide imaging: a white paper from the digital pathology association. Arch Pathol Lab Med. 2019;143(2):222–34. [DOI] [PubMed] [Google Scholar]
- 4. Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, James JA, Salto-Tellez M, Hamilton PW. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Iozzo RV, Schaefer L. Proteoglycan form and function: a comprehensive nomenclature of proteoglycans. Matrix Biol. 2015;42:11–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kang I, Chang MY, Wight TN, Frevert CW. Proteoglycans as immunomodulators of the innate immune response to lung infection. J Histochem Cytochem. 2018;66(4):241–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wight TN, Kinsella MG, Evanko SP, Potter-Perigo S, Merrilees MJ. Versican and the regulation of cell phenotype in disease. Biochim Biophys Acta. 2014;1840(8):2441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pataki CA, Couchman JR, Brabek J. Wnt signaling cascades and the roles of syndecan proteoglycans. J Histochem Cytochem. 2015;63(7):465–80. [DOI] [PubMed] [Google Scholar]
- 9. Lindahl U, Couchman J, Kimata K, Esko JD. Proteoglycans and sulfated glycosaminoglycans. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, Seeberger PH, editors. Essentials of glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2015. p. 207–21. [Google Scholar]
- 10. Hascall V, Esko JD. Hyaluronan. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, Seeberger PH, editors. Essentials of glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2015. p. 197–206. [PubMed] [Google Scholar]
- 11. Multhaupt HA, Couchman JR. Heparan sulfate biosynthesis: methods for investigation of the heparanosome. J Histochem Cytochem. 2012;60(12):908–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kolset SO, Tveit H. Serglycin—structure and biology. Cell Mol Life Sci. 2008;65(7–8):1073–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Couchman JR, Multhaupt H, Sanderson RD. Recent insights into cell surface heparan sulphate proteoglycans and cancer. F1000Res. 2016;5:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li N, Fu H, Hewitt SM, Dimitrov DS, Ho M. Therapeutically targeting glypican-2 via single-domain antibody-based chimeric antigen receptors and immunotoxins in neuroblastoma. Proc Natl Acad Sci U S A. 2017;114(32):E6623–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Erickson AC, Couchman JR. Still more complexity in mammalian basement membranes. J Histochem Cytochem. 2000;48(10):1291–306. [DOI] [PubMed] [Google Scholar]
- 16. Nastase MV, Janicova A, Roedig H, Hsieh LT, Wygrecka M, Schaefer L. Small leucine-rich proteoglycans in renal inflammation: two sides of the coin. J Histochem Cytochem. 2018;66(4):261–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wight TN. Provisional matrix: a role for versican and hyaluronan. Matrix Biol. 2017;60–1:38–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aeffner F, Zarella MD, Buchbinder N, Bui MM, Goodman MR, Hartman DJ, Lujan GM, Molani MA, Parwani AV, Lillard K, Turner OC, Vemuri VNP, Yuil-Valdes AG, Bowman D. Introduction to digital image analysis in whole-slide imaging: a white paper from the digital pathology association. J Pathol Inform. 2019;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Turner OC, Aeffner F, Bangari DS, High W, Knight B, Forest T, Cossic B, Himmel LE, Rudmann DG, Bawa B, Muthuswamy A, Aina OH, Edmondson EF, Saravanan C, Brown DL, Sing T, Sebastian MM. Society of toxicologic pathology digital pathology and image analysis special interest group article*: opinion on the application of artificial intelligence and machine learning to digital toxicologic pathology. Toxicol Pathol. 2020;48(2):277–94. [DOI] [PubMed] [Google Scholar]
- 20. Gonzalez RC, Woods RE. Digital image processing. New York: Pearson; 2018. xvi, 1168 p. [Google Scholar]
- 21. Russ JC. The image processing handbook. 5th ed. Boca Raton: CRC/Taylor and Francis; 2007. 817 p. [Google Scholar]
- 22. Grunkin M, Raundahl J, Foged NT. Practical considerations of image analysis and quantification of signal transduction IHC staining. Methods Mol Biol. 2011;717:143–54. [DOI] [PubMed] [Google Scholar]
- 23. Chang MY, Tanino Y, Vidova V, Kinsella MG, Chan CK, Johnson PY, Wight TN, Frevert CW. A rapid increase in macrophage-derived versican and hyaluronan in infectious lung disease. Matrix Biol. 2014;34:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bell TJ, Brand OJ, Morgan DJ, Salek-Ardakani S, Jagger C, Fujimori T, Cholewa L, Tilakaratna V, Östling J, Thomas M, Day AJ, Snelgrove RJ, Hussell T. Defective lung function following influenza virus is due to prolonged, reversible hyaluronan synthesis. Matrix Biol. 2019;80:14–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gerner MY, Casey KA, Kastenmuller W, Germain RN. Dendritic cell and antigen dispersal landscapes regulate T cell immunity. J Exp Med. 2017;214(10):3105–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity. 2012;37(2):364–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gerner MY, Torabi-Parizi P, Germain RN. Strategically localized dendritic cells promote rapid T cell responses to lymph-borne particulate antigens. Immunity. 2015;42(1):172–85. [DOI] [PubMed] [Google Scholar]
- 28. Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH, Balter A, Kawashima R, Choe G, Sauer D, El Rassi E, Clayburgh DR, Kulesz-Martin MF, Lutz ER, Zheng L, Jaffee EM, Leyshock P, Margolin AA, Mori M, Gray JW, Flint PW, Coussens LM. Quantitative multiplex immunohistochemistry reveals myeloid-inflamed tumor-immune complexity associated with poor prognosis. Cell Rep. 2017;19(1):203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Amodio D, Cotugno N, Macchiarulo G, Rocca S, Dimopoulos Y, Castrucci MR, De Vito R, Tucci FM, McDermott AB, Narpala S, Rossi P, Koup RA, Palma P, Petrovas C. Quantitative multiplexed imaging analysis reveals a strong association between immunogen-specific b cell responses and tonsillar germinal center immune dynamics in children after influenza vaccination. J Immunol. 2018;200(2):538–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang H, Su N, Wang LC, Wu X, Bui S, Nielsen A, Vo H-T, Luo Y, Ma X-J. Quantitative ultrasensitive bright-field RNA in situ hybridization with RNAscope. Methods Mol Biol. 2014;1211:201–12. [DOI] [PubMed] [Google Scholar]
- 31. Boyce RW, Dorph-Petersen KA, Lyck L, Gundersen HJ. Design-based stereology: introduction to basic concepts and practical approaches for estimation of cell number. Toxicol Pathol. 2010;38(7):1011–25. [DOI] [PubMed] [Google Scholar]
- 32. Brown DL. Practical stereology applications for the pathologist. Vet Pathol. 2017;54(3):358–68. [DOI] [PubMed] [Google Scholar]
- 33. Howard V, Reed MG. Unbiased stereology: three-dimensional measurement in microscopy. New York: Springer; 1998. xvii, 246 p. [Google Scholar]
- 34. Hyde DM, Tyler NK, Plopper CG. Morphometry of the respiratory tract: avoiding the sampling, size, orientation, and reference traps. Toxicol Pathol. 2007;35(1):41–8. [DOI] [PubMed] [Google Scholar]
- 35. Kristiansen SL, Nyengaard JR. Digital stereology in neuropathology. APMIS. 2012;120(4):327–40. [DOI] [PubMed] [Google Scholar]
- 36. Nyengaard JR. Stereologic methods and their application in kidney research. J Am Soc Nephrol. 1999;10(5):1100–23. [DOI] [PubMed] [Google Scholar]
- 37. Schneider JP, Ochs M. Stereology of the lung. Methods Cell Biol. 2013;113:257–94. [DOI] [PubMed] [Google Scholar]
- 38. West MJ. Introduction to stereology. In: West MJ, editor. Basic stereology for biologists and neuroscientists. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2012. p. 843–51. [Google Scholar]
- 39. Muhlfeld C, Ochs M. Quantitative microscopy of the lung: a problem-based approach. Part 2: stereological parameters and study designs in various diseases of the respiratory tract. Am J Physiol Lung Cell Mol Physiol. 2013;305(3):L205–21. [DOI] [PubMed] [Google Scholar]
- 40. Gundersen HJG. Notes on the estimation of the numerical density of arbitrary profiles—the edge effect. J Microsc. 1977;111:219–23. [Google Scholar]
- 41. Sterio DC. The unbiased estimation of number and sizes of arbitrary particles using the disector. J Microsc. 1984;134(Pt 2):127–36. [DOI] [PubMed] [Google Scholar]
- 42. Altman MC, Lai Y, Nolin JD, Long S, Chen CC, Piliponsky AM, Altemeier WA, Larmore M, Frevert CW, Mulligan MS, Ziegler SF, Debley JS, Peters MC, Hallstrand TS. Airway epithelium-shifted mast cell infiltration regulates asthmatic inflammation via IL-33 signaling. J Clin Invest. 2019;129(11):4979–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Baddeley A, Jensen EBV. Stereology for statisticians. Boca Raton, FL: Chapman & Hall/CRC Press; 2005. 395 p. [Google Scholar]
- 44. Evans SM, Janson AM, Nyengaard JR. Quantitative methods in neuroscience: a neuroanatomical approach. New York: Oxford University Press; 2004. xii, 327 p. [Google Scholar]
- 45. Cruz-Orive LM, Weibel ER. Sampling designs for stereology. J Microsc. 1981;122(Pt 3):235–57. [DOI] [PubMed] [Google Scholar]
- 46. Gundersen HJG, Osterby R. Optimizing sampling efficiency of stereological studies in biology: or “do more less well!” J Microsc. 1981;121:65–73. [DOI] [PubMed] [Google Scholar]
- 47. Luciw PA, Oslund KL, Yang XW, Adamson L, Ravindran R, Canfield DR, Tarara R, Hirst L, Christensen M, Lerche NW, Offenstein H, Lewinsohn D, Ventimiglia F, Brignolo L, Wisner ER, Hyde DM. Stereological analysis of bacterial load and lung lesions in nonhuman primates (rhesus macaques) experimentally infected with Mycobacterium tuberculosis. Am J Physiol Lung Cell Mol Physiol. 2011;301(5):L731–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hallstrand TS, Lai Y, Altemeier WA, Appel CL, Johnson B, Frevert CW, Hudkins KL, Bollinger JG, Woodruff PG, Hyde DM, Henderson WR, Gelb MH. Regulation and function of epithelial secreted phospholipase A2 group X in asthma. Am J Respir Crit Care Med. 2013;188(1):42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nyengaard JR, Gundersen HJG. Sampling for stereology in lungs. Eur Respir Rev. 2006;15(101):107–14. [Google Scholar]
- 50. Knust J, Ochs M, Gundersen HJ, Nyengaard JR. Stereological estimates of alveolar number and size and capillary length and surface area in mice lungs. Anat Rec. 2009;292(1):113–22. [DOI] [PubMed] [Google Scholar]
- 51. Brown DL. Bias in image analysis and its solution: unbiased stereology. J Toxicol Pathol. 2017;30(3):183–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ochs M, Muhlfeld C. Quantitative microscopy of the lung: a problem-based approach. Part 1: basic principles of lung stereology. Am J Physiol Lung Cell Mol Physiol. 2013;305(1):L15–22. [DOI] [PubMed] [Google Scholar]
- 53. Mayhew TM. Taking tissue samples from the placenta: an illustration of principles and strategies. Placenta. 2008;29(1):1–14. [DOI] [PubMed] [Google Scholar]
- 54. Nolin JD, Lai Y, Ogden HL, Manicone AM, Murphy RC, An D, Frevert CW, Ghomashchi F, Naika GS, Gelb MH, Gauvreau GM, Piliponsky AM, Altemeier WA, Hallstrand TS. Secreted PLA2 group X orchestrates innate and adaptive immune responses to inhaled allergen. JCI Insight. 2017;2(21):e94929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Snyder JM, Washington IM, Birkland T, Chang MY, Frevert CW. Correlation of versican expression, accumulation, and degradation during embryonic development by quantitative immunohistochemistry. J Histochem Cytochem. 2015;63(12):952–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hinton G. Deep learning—a technology with the potential to transform health care. JAMA. 2018;320(11):1101–2. [DOI] [PubMed] [Google Scholar]
- 57. Wang S, Yang DM, Rong R, Zhan X, Xiao G. Pathology image analysis using segmentation deep learning algorithms. Am J Pathol. 2019;189(9):1686–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Handelman GS, Kok HK, Chandra RV, Razavi AH, Lee MJ, Asadi H. eDoctor: machine learning and the future of medicine. J Intern Med. 2018;284(6):603–19. [DOI] [PubMed] [Google Scholar]
- 59. Bartels PH, Bahr GF, Bibbo M, Wied GL. Objective cell image analysis. J Histochem Cytochem. 1972;20(4):239–54. [DOI] [PubMed] [Google Scholar]
- 60. Shapiro B, Lemkin P, Lipkin L. The application of artificial intelligence techniques to biologic cell identification. J Histochem Cytochem. 1974;22(7):741–50. [DOI] [PubMed] [Google Scholar]
- 61. Hewitt SM. Data, information, and knowledge. J Histochem Cytochem. 2019;67(4):227–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gross P, Honnorat N, Varol E, Wallner M, Trappanese DM, Sharp TE, Starosta T, Duran JM, Koller S, Davatzikos C, Houser SR. Nuquantus: machine learning software for the characterization and quantification of cell nuclei in complex immunofluorescent tissue images. Sci Rep. 2016;6:23431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wilson CL, Stephenson SE, Higuero JP, Feghali-Bostwick C, Hung CF, Schnapp LM. Characterization of PDGFR-β-positive pericytes from IPF and non-IPF lungs. Am J Physiol Lung Cell Mol Physiol. 2018;315(6):L991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. James NT. Common statistical errors in morphometry. Pathol Res Pract. 1989;185(5):764–8. [DOI] [PubMed] [Google Scholar]
- 65. Xie R, Chung JY, Ylaya K, Williams RL, Guerrero N, Nakatsuka N, Badie C, Hewitt SM. Factors influencing the degradation of archival formalin-fixed paraffin-embedded tissue sections. J Histochem Cytochem. 2011;59(4):356–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chung JY, Song JS, Ylaya K, Sears JD, Choi L, Cho H, Rosenberg AV, Hewitt SM. Histomorphological and molecular assessments of the fixation times comparing formalin and ethanol-based fixatives. J Histochem Cytochem. 2018;66(2):121–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Perry C, Chung JY, Ylaya K, Choi CH, Simpson A, Matsumoto KT, Smith WA, Hewitt SM. A buffered alcohol-based fixative for histomorphologic and molecular applications. J Histochem Cytochem. 2016;64(7):425–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gillespie JW, Best CJ, Bichsel VE, Cole KA, Greenhut SF, Hewitt SM, Ahram M, Gathright YB, Merino MJ, Strausberg RL, Epstein JL, Hamilton SR, Gannot G, Baibakova GV, Calvert VS, Flaig MJ, Chuaqui RF, Herring JC, Pfeifer J, Petricoin EF, Linehan WM, Duray PH, Bova GS, Emmert-Buck MR. Evaluation of non-formalin tissue fixation for molecular profiling studies. Am J Pathol. 2002;160(2):449–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hsia CC, Hyde DM, Ochs M, Weibel ER, ATS/ERS Joint Task Force on Quantitative Assessment of Lung Structure. An official research policy statement of the American Thoracic Society/European Respiratory Society: standards for quantitative assessment of lung structure. Am J Respir Crit Care Med. 2010;181(4):394–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen K, Wight TN. Proteoglycans in arterial smooth muscle cell cultures: an ultrastructural histochemical analysis. J Histochem Cytochem. 1984;32(4):347–57. [DOI] [PubMed] [Google Scholar]
- 71. van Kuppevelt TH, Veerkamp JH. Application of cationic probes for the ultrastructural localization of proteoglycans in basement membranes. Microsc Res Tech. 1994;28(2):125–40. [DOI] [PubMed] [Google Scholar]
- 72. Schumacher U, Adam E. Standardization of staining in glycosaminoglycan histochemistry: alcian blue, its analogues, and diamine methods. Biotech Histochem. 1994;69(1):18–24. [DOI] [PubMed] [Google Scholar]
- 73. McDonald TO, Gerrity RG, Jen C, Chen HJ, Wark K, Wight TN, Chait A, O’Brien KD. Diabetes and arterial extracellular matrix changes in a porcine model of atherosclerosis. J Histochem Cytochem. 2007;55(11):1149–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Theocharis AD, Karamanos NK. Decreased biglycan expression and differential decorin localization in human abdominal aortic aneurysms. Atherosclerosis. 2002;165(2):221–30. [DOI] [PubMed] [Google Scholar]
- 75. Horobin RW, Kiernan JA, Conn HJ, Biological Stain Commission. Conn’s biological stains: a handbook of dyes, stains and fluorochromes for use in biology and medicine. 10th ed. Oxford: Published for the Biological Stain Commission by BIOS; 2002. xv, 555 p. [Google Scholar]
- 76. Luna LG, Armed Forces Institute of Pathology (U.S.). Manual of histologic staining methods of the Armed Forces Institute of Pathology. 3rd ed. New York: Blakiston Division; 1968. xii, 258 p. [Google Scholar]
- 77. Frevert CW, Johnson B, Stahl WL, editors. Immunohistochemistry: antibody specificity. Kidlington; Oxford: Elsevier; 2014. [Google Scholar]
- 78. Manuel SL, Johnson BW, Frevert CW, Duncan FE. Revisiting the scientific method to improve rigor and reproducibility of immunohistochemistry in reproductive science. Biol Reprod. 2018;99(4):673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Edfors F, Hober A, Linderback K, Maddalo G, Azimi A, Sivertsson A, Tegel H, Hober S, Szigyarto CAK, Fagerberg L, von Feilitzen K, Oksvold P, Lindskog C, Forsström B, Uhlen M. Enhanced validation of antibodies for research applications. Nat Commun. 2018;9(1):4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hewitt SM, Baskin DG, Frevert CW, Stahl WL, Rosa-Molinar E. Controls for immunohistochemistry: the Histochemical Society’s standards of practice for validation of immunohistochemical assays. J Histochem Cytochem. 2014;62(10):693–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shi SR, Key ME, Kalra KL. Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J Histochem Cytochem. 1991;39(6):741–8. [DOI] [PubMed] [Google Scholar]
- 82. Shi SR, Liu C, Taylor CR. Standardization of immunohistochemistry for formalin-fixed, paraffin-embedded tissue sections based on the antigen-retrieval technique: from experiments to hypothesis. J Histochem Cytochem. 2007;55(2):105–9. [DOI] [PubMed] [Google Scholar]
- 83. Bogen SA, Vani K, Sompuram SR. Molecular mechanisms of antigen retrieval: antigen retrieval reverses steric interference caused by formalin-induced crosslinks. Biotech Histochem. 2009;84(5):207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Li D, Li N, Zhang YF, Fu H, Feng M, Schneider D, Su L, Wu X, Zhou J, Mackay S, Kramer J, Duan Z, Yang H, Kolluri A, Hummer AM, Torres MB, Zhu H, Hall MD, Luo X, Chen J, Wang Q, Abate-Daga D, Dropublic B, Hewitt SM, Orentas RJ, Greten TF, Ho M. Persistent polyfunctional chimeric antigen receptor T cells that target glypican 3 eliminate orthotopic hepatocellular carcinomas in mice. Gastroenterology. 2020;158(8):2250–65.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Evanko SP, Chan CK, Johnson PY, Frevert CW, Wight TN. The biochemistry and immunohistochemistry of versican. Methods Cell Biol. 2018;143:261–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hayes AJ, Hughes CE, Caterson B. Antibodies and immunohistochemistry in extracellular matrix research. Methods. 2008;45(1):10–21. [DOI] [PubMed] [Google Scholar]
- 87. Braunschweig T, Chung JY, Choi CH, Cho H, Chen QR, Xie R, Perry C, Khan J, Hewitt SM. Assessment of a panel of tumor markers for the differential diagnosis of benign and malignant effusions by well-based reverse phase protein array. Diagn Pathol. 2015;10:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lindskog C, Backman M, Zieba A, Asplund A, Uhlen M, Landegren U, Pontén F. Proximity ligation assay as a tool for antibody validation in human tissues. J Histochem Cytochem. 2020;68(7):515–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kang I, Harten IA, Chang MY, Braun KR, Sheih A, Nivison MP, Johnson PY, Workman G, Kaber G, Evanko SP, Chan CK, Merrilees MJ, Ziegler SF, Kinsella MG, Frevert CW, Wight TN. Versican deficiency significantly reduces lung inflammatory response induced by polyinosine-polycytidylic acid stimulation. J Biol Chem. 2017;292(1):51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Davis AS, Richter A, Becker S, Moyer JE, Sandouk A, Skinner J, Taubenberger JK. Characterizing and diminishing autofluorescence in formalin-fixed paraffin-embedded human respiratory tissue. J His-tochem Cytochem. 2014;62(6):405–23. [Google Scholar]