Abstract
Glycosaminoglycans (GAGs) are heterogeneous, negatively charged, macromolecules that are found in animal tissues. Based on the form of component sugar, GAGs have been categorized into four different families: heparin/heparan sulfate, chondroitin/dermatan sulfate, keratan sulfate, and hyaluronan. GAGs engage in biological pathway regulation through their interaction with protein ligands. Detailed structural information on GAG chains is required to further understanding of GAG–ligand interactions. However, polysaccharide sequencing has lagged behind protein and DNA sequencing due to the non-template-driven biosynthesis of glycans. In this review, we summarize recent progress in the analysis of GAG chains, specifically focusing on techniques related to mass spectroscopy (MS), including separation techniques coupled to MS, tandem MS, and bioinformatics software for MS spectrum interpretation. Progress in the use of other structural analysis tools, such as nuclear magnetic resonance (NMR) and hyphenated techniques, is included to provide a comprehensive perspective.
Keywords: glycosaminoglycans, mass spectroscopy, NMR, sequencing, structural analysis
Introduction
Glycosaminoglycans (GAGs) are biological macromolecules ubiquitously present in animal tissues. GAG chains bound to a core protein constitute a proteoglycan (PG), which is considered among the most structurally complex glycoconjugates. The core protein of a PG is synthesized by a template-driven process, after which GAG chains are added in a non-template-driven synthesis, resulting in the remarkable structural and functional diversity of PGs. The heterogeneity of the GAG chains greatly contributes to PG complexity. GAGs are unbranched, linear anionic polysaccharides composed of 10 to 200 repeating disaccharide units.1 Each disaccharide building block consists of an amino sugar (i.e., N-acetyl-glucosamine [GlcNAc], or N-acetyl-galactosamine [GalNAc]) and a uronic acid (i.e., glucuronic acid [GlcA], or iduronic acid [IdoA]) or D-galactose. These disaccharide units can be O- or N- substituted with sulfo groups at different positions. Based on the form of component sugar, GAGs have been classified into four different families: heparin/heparan sulfate (HP/HS), chondroitin/dermatan sulfate (CS/DS), keratan sulfate (KS), hyaluronan (HA). Among the four families of GAGs, HP/HS, CS/DS, and KS are all conjugated to core proteins as PGs and are synthesized principally in the Golgi apparatus of cells, whereas the HA is synthesized in the cellular plasma membrane unlinked to a protein core.2
The HP/HS are bioactive GAGs O-glycosidically linked to a serine residue of the core protein through a tetrasaccharide linkage region. The disaccharide building block of this family is →4) α-L-IdoA /β-D-GlcA (1→4) α-D-GlcNS/-D-GlcNAc (1→. In HP, IdoA accounts for >70% of the hexuronic acid. The IdoA residue in HP is generally 2-sulfated and the glucosamine can be N- and/or 6-sulfated. In contrast to HP, HS is majorly composed of GlcA as its hexuronic acid, and generally contains one sulfo group per disaccharide repeating unit. HS can be N-sulfated, 6-sulfated, and 3-sulfated on its glucosamine residue and 2-sulfated on its uronic acid residue.3 While HS is widely distributed on membrane and extracellular PGs, the HP PG is found primarily localized to intracellular granules of mast cells. HP has multifaceted biological functions, including its well-known anticoagulant activity and other bioactivities such as anti-inflammatory and anticancer activities.2,4,5 The complex biological activity of HS-GAGs includes their roles in developmental biology and cell signaling.4 HS PGs engage in various physiological and pathological processes, including embryonic development, adult tissue homeostasis, aging, and infection.6,7
The CS/DS GAGs also occur as PGs O-glycosidically linked to the serine residues of their core proteins. The GAGs of this family contain repeating disaccharide unit represented as →4) β-D-GlcA /α-L-IdoA (1→3)β-D-GalNAc(1→. The GlcA is the hexuronic acid found in CS, and IdoA is the hexuronic acid found in DS.8 Unlike heparin, the CS and DS have no N-sulfo-group substitution. The GalNAc can be sulfated at 4-O and/or 6-O positions. The hexuronic acid residues are sulfated at the 2-O, and the IdoA residues are rarely sulfated at 3-O positions. CS PGs serve as one of the major barrier-forming molecules in the central nervous system. CS is often found as a hybrid structure with DS and is involved in the formation of the neural network by capturing and presenting growth factors to stem cells or neuronal cells.9
KS has a galactose residue as its major backbone constitute instead of uronic acid. KS is 6-O sulfated GAGs with the disaccharide building block, →3) β-D-Gal(1→4) β-D-GlcNAc (1→. KS is subdivided into three classes according to the ways it is linked to the core protein.2 KS I (corneal type) is N-glycosidically conjugated through a biantennary branched linkage region to an asparagine residue of its core protein. KS II (skeletal type) is O-glycosidically linked to a serine or threonine residue of its core protein. K III is O-linked through mannose to serine, and is preferentially found in PG of brain and nervous tissue.10 Generally, the GAG chains of KS I are longer and less sulfated than that of KSII. Also, KS I can be distinguished from KS II by its capped non-reducing end.1 KS is believed to be the newest GAG from an evolutionary perspective and its biological functions are the least understood. A growing amount of evidence suggested potential roles for KS in cell-signaling processes.11
HA is the only GAG family that is neither sulfated nor linked to a protein core. However, HA is typically bound to matrix proteins in the extracellular space.12 It has a simple repeating disaccharide unit of →4) β-D-GlcA(1→3)β-D-GlcNAc(1→. HA is a significant biocompatible support material used in wound healing or as growth scaffolds in surgery. HA has also been widely used for providing lubrication and mechanical support for the joints, or serving as a drug delivery material.13
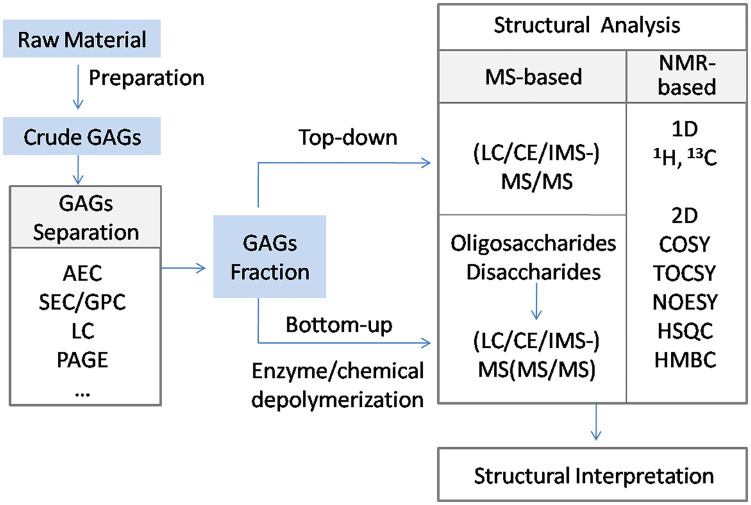
The numerous physiological functions of GAGs are carried out by their interaction with protein ligand.14 These interactions are often specific between GAGs and ligands, requiring a defined GAG sequence or domain. Therefore, structural elucidation of GAG chains is a prerequisite for exploring the mechanisms of GAG–ligand interactions. Due to the non-template-driven biosynthesis of GAGs, the development of GAG sequencing has lagged behind the sequencing of other biological macromolecular counterparts, protein and DNA. Thus far, there have been only two cases reporting GAG sequencing and both relatively simple GAGs, bikunin15 and decorin.16 Currently, the prevailing method for GAG chain determination is mass spectrometry (MS). Nuclear magnetic resonance (NMR) also represents an option for establishing GAG structure. As summarized in Fig. 1, GAG samples are prepared through extensive separation processes and then subjected to MS and/or NMR analysis using top-down or bottom-up sequencing strategies. This review focuses on recent developments of GAG analysis, including separation techniques coupled to MS, tandem MS, MS data interpretation, NMR, and other novel techniques applied for the characterization of GAGs with the purpose of providing useful information for glycomics research.
Figure 1.
A scheme for GAGs chain analysis. Abbreviations: GAGs, glycosaminoglycans; AEC, anion-exchange chromatography; SEC, size-exclusion chromatography; GPC, gel permeation chromatography; LC, liquid chromatography (other than AEC and SEC, including reverse phase liquid chromatography, reverse phase ion-pairing liquid chromatography, and hydrophilic interaction chromatography); PAGE, polyacrylamide gel electrophoresis; CE, capillary electrophoresis; MS, mass spectroscopy; IMS, ion mobility spectrometry; NMR, nuclear magnetic resonance; COSY, correlation spectroscopy; TOCSY, total correlation spectroscopy; NOESY, nuclear Overhauser effect spectroscopy; HSQC, heteronuclear single-quantum coherence; HMBC, heteronuclear multiple bond coherence.
Preparation of GAGs for Analysis
The native GAG is generally a component of a PG found in a biological sample. Thus, a procedure is needed for the extraction and purification to obtain GAG samples of sufficient purity for subsequent structure analysis. Detailed methods for preparing PGs and GAGs from biological origins are extensively described in several review articles.1,4,17 PGs that are present in physiological fluids or secreted into the fermentation media can directly be obtain by dialysis, ultrafiltration, and/or applying the sample to a fractionation column. However, preparing PGs and GAGs from large amounts of animal tissues can be very time-consuming and labor-intensive. A particularly complex protocol was described to obtain decorin PGs and GAGs from porcine skin for use in sequencing studies.18 In brief, tissues or cells were defatted, sliced, ground, or homogenized, and a buffered solution of chaotropic agent was used to extract the PGs. The extracts were dialyzed or applied to an anion-exchange column (AEC), the PGs were treated with nonspecific proteases, and the GAGs were released from core peptides using controlled β-elimination. The resulting crude GAGs can be collected by ultrafiltration or through solvent precipitation.1,17
The crude GAG extract, containing impurities and chains of many sizes, needs to be properly enriched and purified before it can be successfully interrogated by MS or NMR. There are a variety of separation techniques that can be used to purify and fractionate GAGs, including AEC, size-exclusion chromatography (SEC), reverse phase liquid chromatography (RP-LC), reverse phase ion-pairing LC (RPIP-LC), and hydrophilic interaction chromatography (HILIC). RP-LC, RPIP-LC, and HILIC methods for GAG separations are often applied as a hyphenated technique using an HPLC-MS platform. These methods will be introduced later in MS section. In addition to the above-mentioned chromatographic techniques, preparative polyacrylamide gel electrophoresis (PAGE) represents another good option for the purification of GAGs. The GAG fractions obtained from PAGE can often be directly applied to MS analysis for sequencing.16,19
GAG Chain Analysis
MS-based Analysis
General Introduction
Mass spectrometry plays a leading role in glycomics. Although other tools have been used in the early stages of glycan analysis, such as NMR or the gel electrophoresis–based sequencing, mass spectroscopy has become preeminent due to its high resolution, efficiency in providing detailed structural information, adaptability for coupling separation techniques, and a reduced demand for sample quantity.
The invention of soft-ionization techniques has tremendously facilitated the application of MS for the analysis of biological macromolecules. The two major soft-ionization techniques are matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI). The application of MALDI in GAG analysis has, however, been limited due to the propensity for labile sulfo groups to undergo decomposition. Thus, ESI is most frequently applied to GAG analysis. ESI in the negative-ion mode has been shown to be suitable for analyzing negatively charged GAGs as well as preserving their sulfo groups.20 ESI is quite adaptable and can be coupled to a variety of separation techniques, including LC, and capillary electrophoresis (CE).
Another major MS breakthrough facilitating glycomics research is the widespread availability of Fourier transform mass spectrometry (FTMS), including both Fourier transform ion cyclotron resonance (FT-ICR) and Orbitrap mass spectrometers.21 FTMS provides high resolution and mass accuracy, allowing unambiguous peak assignments that are essential in analyzing heterogeneous glycans.
A mass spectrum can provide general information about the GAG composition, such as degree of polymerization and number of sulfo-group substitutions. However, such information is insufficient to fully characterize a GAG chain. Tandem mass spectrometry is required to gain a deeper insight of the precise structure of GAG chains. Electron-based ion activation techniques, such as electron detachment dissociation (EDD) and negative electron transfer dissociation (NETD), are capable of generating extensive cross-ring cleavages, allowing identification of epimers, distinguishing between IdoA and GlcA, and determining the position of sulfo-group substitutions.22 Also, electron-based ion activation largely prevents the loss of sulfo group.23 Although the loss of sulfo groups can also be prevented by deprotonation of sulfo groups or addition of metal adducts.21 Moreover, the application of EDD or NETD can provide information of the same and different ions to aid the deconvolution of the oligosaccharide structure for GAG sequencing.
State-of-the-art MS and tandem MS techniques have been major technical achievements responsible for much of the success in GAG analysis. It is expected that the future will bring more progress in glycomic analysis as new analytical instruments and methods are developed.
Top-down and Bottom-up Strategies
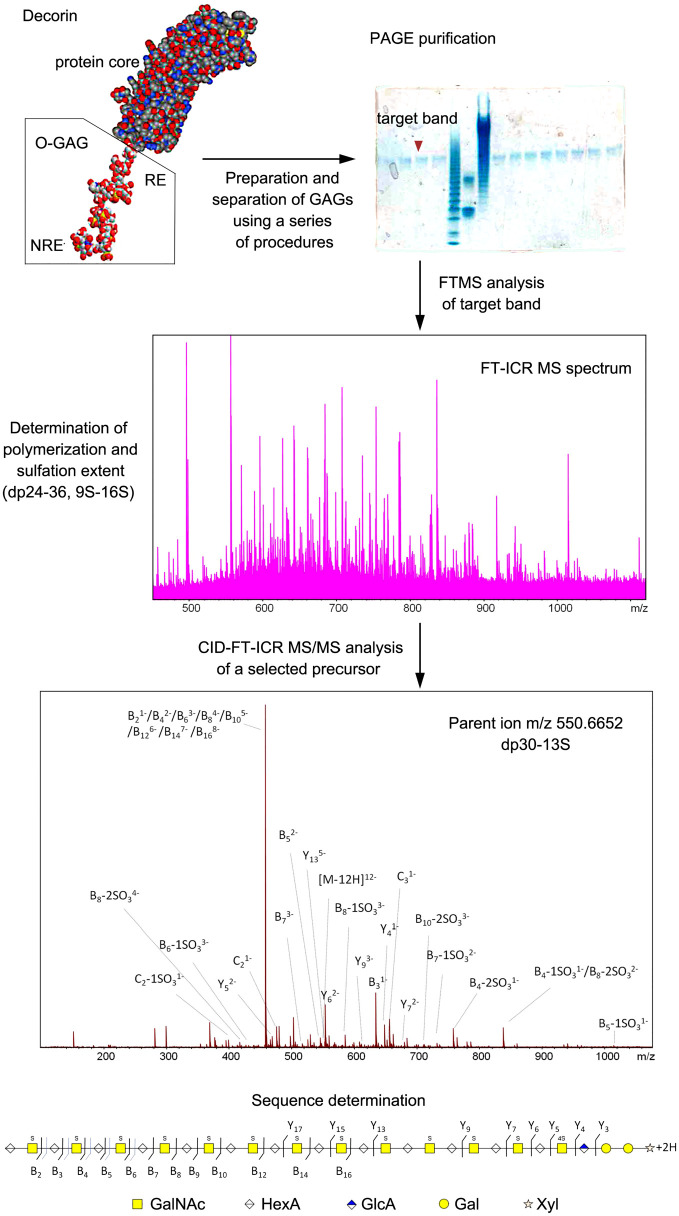
The structural characterization of GAGs can be achieved through different types of instrumental analysis, but such characterization basically follows two major strategies, referred to as top-down (Fig. 2) and bottom-up (Fig. 3). Applying a top-down strategy means a purified intact GAG chain is directly subjected to structural analysis without a pre-depolymerization step.15,16,18,24 The development of MS techniques, especially the emergence of FTMS that provides high spectral resolution and high mass accuracy, has facilitated the application of top-down MS strategies. A top-down strategy affords detailed information on the sequence of an intact GAG chain with reduced requirements for sample preparation steps.25
Figure 2.
A mainframe of a typical top-down GAG analysis. Decorin GAGs were purified and subjected to FTMS and MS/MS analysis for sequencing. Abbreviations: GAGs, glycosaminoglycans; FTMS, Fourier transform mass spectrometry; MS, mass spectroscopy; PAGE, polyacrylamide gel electrophoresis; FT-ICR, Fourier transform ion cyclotron resonance; CID, collision-induced dissociation. (Adapted from: Yu Y, Duan J, Leach IIIFE, Toida T, Higashi K, Zhang H, Zhang F, Amster IJ, Linhardt RJ. Sequencing the dermatan sulfate chain of decorin. J Am Chem Soc. 2017;139(46):16986–16995).
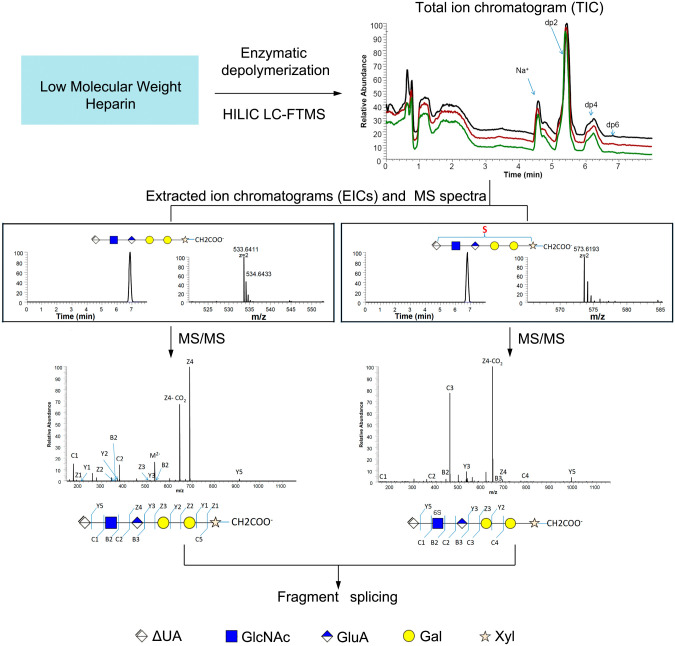
Figure 3.
A mainframe of a typical bottom-up GAG analysis. Low-molecular-weight heparins were depolymerized and subjected to LC-MS and MS/MS analysis. Abbreviations: GAGs, glycosaminoglycans; LC, liquid chromatography; MS, mass spectroscopy; HILIC, hydrophilic interaction chromatography; LC, liquid chromatography; FTMS, Fourier transform mass spectrometry (Adapted from: Li G, Steppich J, Wang Z, Sun Y, Xue C, Linhardt RJ, Li L. Bottom-up low molecular weight heparin analysis using liquid chromatography-Fourier transform mass spectrometry for extensive characterization. Anal Chem. 2014;86(13):6626–6632).
In contrast, in a bottom-up approach (Fig. 3), intact GAG chains are depolymerized using controlled chemical or enzymatic methods. Such an approach is essential for analyzing polysaccharides of very high molecular weight. The resulting oligosaccharides obtained on controlled depolymerization are usually then subjected to a separation procedure such as LC, CE, or ion mobility (IM) for structural characterization.24,26–28 Controlled chemical and enzymatic methods for GAG depolymerization have been described in detail.1 Compared with a top-down strategy, the bottom-up requires more steps to prepare samples, and the depolymerization procedure may result in the loss of substantial amounts of structural information.
Even when performing top-down sequencing, it is quite common to use a bottom-up strategy to provide additional supporting information on disaccharide units or domain structures. In this case, the GAGs are digested with a combination of different enzymes called GAG lyases.16 The determination of building block disaccharides can help narrow down peak assignments in the MS spectra, and enables more confident interpretation of MS data.
Separation Techniques
A sample needs to be properly purified before being examined by MS to undertake the sequencing of a GAG chain. A hyphenated purification method with MS detection can be achieved by using different separation techniques coupled with MS. The major separation techniques adapted to MS coupling are LC methods, including RP-LC, RPIP-LC, HILIC, SEC, porous graphitized carbon (PGC), and other separation techniques such as CE and IM. There are a considerable number of review articles on combining separations with MS in GAG analysis.28–37 Thus, the following section focuses on only work published in the past few years.
LC-based Separation
RP-LC is the most wildly used chromatography method. However, due to the polyanionic features of GAGs, this method is not directly suitable for GAG separations. This limitation can be addressed by modifying GAG oligosaccharides to make them amenable to RP-LC. A rapid separation and detection method for HP/HS disaccharide analysis, relying on 2-aminoacridone (AMAC) derivatization, was built to examine samples by an optimized selected ion recording (SIR) RP-ultraperformance liquid chromatography (UPLC)-MS38 without requiring the removal of excess unreacted reagent. Liu and coworkers39 also used AMAC to label disaccharides derived from CS and HS of human intervertebral disc samples. An RP-LC coupled with triple quadruple mass was used for online MS analysis done by multiple reaction monitoring (MRM). Principal components analysis identified clear separation of GAG profiles between nucleus pulposus and annulus fibrosus in specimens from young and old patients. In the research carried out by the Sharp group,40 a method using trideuteroacetyl derivatization and RP-LC-MS/MS was developed to sequence a complex mixture of HS tetrasaccharides. Liang and coworkers41 introduced a propionylation method to replace sulfo groups from oligosaccharide and used LC-MS/MS with coupled C18 columns to separate and fully sequence two mixed tetrasaccharide isomers, which differed solely by the order of their N-sulfation and N-acetylation. A stable isotope-labeled hydrazide tag (INLIGHT) was used for labeling oligosaccharides derived from digested heparin and enabled identification of a total amount of 116 unique oligosaccharides.42 Derivatization and labeling are frequently used in glycomics to enhance RP-LC separation for oligosaccharides and for a variety of other purposes, including stabilizing residues, serving as a linker for oligosaccharide attachment, facilitating ultraviolet/fluorescent/MS detection, and supporting detailed structural characterization by tandem mass.37 Recently published works using double labeling43 and sequential labeling44 both have demonstrated the virtue of labeling for the subsequent detection.
GAG oligosaccharides can be separated and analyzed in an RP-LC-MS platform without derivatization by adding ion-pairing reagent (IPR) to the mobile phase. An alkylamine ion-pairing reagent can essentially neutralize the ionic features of GAG oligomers. This offers the advantage of enhanced volatility, especially through a pH change above the pKa after the separation, facilitating coupled MS detection.45 Reagents such as tributylamine,24,45 n-pentylamine,46 n-hexylamine,47 and n-butylamine48 are often used as anion-pairing reagent to increase separation efficiency in RPIR-LC system for GAG analysis. Persson et al.48 developed a novel approach based on dibutylamine RPIP-LC-MS/MS platform and used in disaccharide fingerprinting, revealing differences in xyloside-primed CD/DC as well as sialylation of the linkage region from two different cell lines. The N-unsubstituted glucosamine residues in HP/HS were examined using RPIP-LC (reversed phase ion-pairing liquid chromatography)-MS method. By adding hexylamine47 and pentylamine,46 N-unsubstituted disaccharides/oligosaccharides were well separated and characterized.
HILIC is a preferential method for polar compound separations and it is also easily coupled with MS. Therefore, it has become a popular approach in GAG analysis. HILIC is efficiently coupled with LC-Orbitrap MS for oligosaccharide characterization.24,27 HILIC has also been coupled with triple quadrupole M/M with an MRM approach to analyze and quantify heparin building blocks.49,50 HILIC can be combined with weak anion exchange (WAX) method. Turiák and coworkers51 coupled self-packed HILIC-WAX capillary columns to a UPLC Q-Tof MS system, revealing that HS disaccharide composition varies in different grades of prostate cancer tissues. Zhang and coworkers used UPLC-HILIC/WAX MS/MS equipped with a commercial HILIC/WAX column to evaluate both the epimerization and composition of heparin and dalteparin.52 HILIC can also be used to separate derivatized oligosaccharides. Antia and coworkers used a HILIC-ESI-MS in positive-ion mode to investigate procainamide-derivatized HS disaccharides, completing separation of eight HS disaccharides together with an internal standard.53 A HILIC method was used in conjunction with a microfluidic chip LC-MS and provided an efficient separation of GAGs.54,55
Porous graphitized carbon (PGC) can be used as a stationary phase in LC. PGC is less durable compared with common stationary phase materials. In a PGC-LC experiment, longer equilibration time is required to produce repeatable results.25 However, PGC-LC-ESI-MS analysis is characterized by a high isomer separation power enabling a specific glycan compound analysis on the level of individual structures, and is well described in Stavenhagen et al.’s56 review. Turnbull and coworkers recently applied a commercial PGC column to an LC-MS system to investigate digested HS oligosaccharide mixtures. The method was effective and allowed the separation of oligosaccharide from tetrasaccharide to octasaccharide, enabling more confident interpretation of the MS/MS data.57
CE
In addition to LC-based methods, CE is another attractive option for GAG analysis amenable to MS coupling. CE has been shown to be superior in multiple aspects such as a high separation efficiency, short analysis time, low consumption of sample, and reproducibility.34,58 Linhardt and coworkers used a reversed-polarity CE coupled with LTQ Orbitrap MS method to analyze HP/HS disaccharides, Arixtra, and low-molecular-weight heparin (LMWH) building blocks. The results obtained were comparable or better than those obtained when these analyses were performed using LC-MS.59 This method could also separate oligosaccharides from tetrasaccharides to dodecasaccharides, elucidating sulfation position and epimeric structural differences, and could also determine more than 80 molecular compositions from complex GAG mixture.60 Recent work published by Stickney et al.61 combined, for the first time, negative electron transfer dissociation (NETD) and CE for assigning the structures of GAG oligomers. This approach allowed enoxaparin (a complex mixture of LMWHs) to be separated within 30 min, 37 unique molecular compositions be identified, and nine structures were assigned with tandem MS.
Ion Mobility Spectrometry (IMS)
IMS is a new technique, which emerged as a promising choice for GAG separation. Different from LC and CE, IMS is a postionization, gas-phase method, providing fast separation, and easy to be coupled to MS.23 IMS defines how an ion drifts through a gas buffer under the influence of a varying or fixed electric field. Different ions are separated according to their motion, which are affected by both the acceleration caused by the electric field and the deceleration caused by collisions with gas molecules. The ion’s motion is associated with molecular mass, charge, size, and shape. Thus, IMS is considered to be a powerful tool for separating isomers. A variety of IMS methods have been developed, such as field asymmetric IMS (FAIMS), trapped IMS (TIMS), traveling wave ion mobility (TWIMS), and drift Tube IMS (DTIMS). A combined FAIMS-FTICR-MS/MS was used in GAG analysis and was shown to be capable of resolving isomeric and isobaric GAG negative ions having the same mass-to-charge ratio, and it also facilitated MS/MS analysis.62 Wei and coworkers23 used gated-trapped ion mobility spectrometry (gated-TIMS), which allowed the ions of a given mobility selected by an electrical gate and accumulated in a low-pressure collision cell. Highly sulfated HP/HS isomers were separated by gated-TIMS and examined by coupling NETD MS/MS. The method was useful for both qualitative and quantitative GAG analysis. A TWIMS method was used to investigate changes in the gas-phase conformation of antithrombin III upon binding of Arixtra, validating the utility of IMS to measure protein conformational changes induced by the binding of GAG ligands.63 Miller and coworkers64 used ion mobility mass spectrometry and tandem MS to investigate heparin/HS-like isomers. Six synthetically produced octasaccharides, which are isomeric with regard to either glucuronic acid (GlcA) or iduronic acid (IdoA) residues at various positions, were analyzed and differentiated. Changing from IdoA to GlcA in specific locations resulted in strong conformational distortions, showing the importance of sequence to overall conformation.
In addition, hyphenated separations have also been used in research, such as combination of RPIP to HILIC,65 and RPIP44 or SEC66 coupled to AEC. These separation methods play significant roles in the analysis of glycan and GAG structures.
Tandem MS
Tandem MS plays a crucial role in characterization of GAG structure. It generates glycosidic and cross-ring cleavage, leading to fragmentation of precursor ions. Fragments, particularly cross-ring ones, can provide detailed information about the configuration and modification of each monosaccharide residue, allowing the GAG structures to be fully characterized.
There have been different kinds of ion activation methods applied in tandem MS, including collision-induced dissociation (CID),67 infrared multi-photon dissociation (IRMPD),68 electron-induced association (EID),69 EDD,22,62,68,70 and NETD.23,61,71,72 CID, an ionization method based on collision-activation, has been widely applied for characterizing GAG chains. Intact GAG chains have been sequenced using CID by Linhardt and coworkers.15,16
In recent years, the electron-based ionization methods such as EDD and NETD have become new trends in glycomics research. EDD can produce more informative spectra of both cross-ring and glycosidic cleavage product ions, while CID and IRMPD are not as efficient as EDD in causing cross-ring cleavages.68 EDD also minimizes sulfo-group loss, which is a major complication when using the CID method. Research carried out by Amster and coworkers has confirmed the benefit of using EDD for identification of C5 uronic acid epimers.22,62,70 According to these reports, EDD can generate different ion fragment patterns for 2-O-sulfo uronic acid epimeric tetrasaccharides.70 Further research confirmed the efficacy of using these ions for assigning the C-5 stereochemistry of the reducing end uronic acid in 33 HS tetrasaccharides.22 The combination with high-field asymmetric waveform ion mobility spectrometry (FAIMS) was described previously. EDD was able to distinguish the two sets of epimeric GAG tetramers separated in FAIMS method.62
Although EDD can provide rich informative fragments, it is a relatively slow technique intended for FT-ICR because it requires long acquisition times and is not conducive to a high-throughput platform. Another electron activation method NETD shows its promise in both generating sequence-informative fragmentation spectra and improving throughput rate (i.e., 1 s for EDD vs. 0.1 s for NETD).21 NETD is as excellent as EDD in generating extensive, structurally informative fragments and minimizing the occurrence of sulfate loss peaks.71 NETD can enable structural characterization of synthetic HS isomers containing 3-O-sulfation.72 In recent years, NETD has been applied in GAG analysis when coupled to new separation techniques such as CE61 and gated-TIMS.23 NETD was found well adapted for these separation approaches, providing efficient information for stereoisomer identification, and negligible sulfo losses.
Besides the electron-based ion activation, a photodissociation method, referring to ultraviolet photodissociation (UVPD), has been suggested for the analysis of GAG chains.21 UVPD has also been applied in proteomics and some preliminary studies targeting oligosaccharides.73,74 Further exploration is needed for its application in glycomics.
Structural Interpretation
Tandem MS is a powerful tool to reveal the complex structure of GAG chains. Fragments generated by tandem MS are crucial for establishing a precursor’s structure. The fragmentation of oligosaccharide follows a certain pattern, and a nomenclature has been established by Domon and Costello.21,75,76 The processing of an MS spectrum into meaningful information is a time-consuming task. Manual interpretation of tandem data sets required for GAG oligosaccharide analysis may take many hours or even days.17 Hence, suitable bioinformatics software is essential to assist data interpretation. Many labs have developed tools for automated or semi-automated interpretation of MS data for GAG analysis. The following section is an introduction to these programs.
HOST is a computational tool early developed to help HP/HS oligosaccharide sequencing using enzymatic digestion and ESI-MSn. Based on disaccharide composition analysis, the program scores and returns the most likely sequence.21,77 The widely used software, GlycoWorkbench is a package designed to semi-automatically annotate glycomics data. It can be used to annotate MS and MS/MS spectra of free oligosaccharides, N- and O-linked glycans, GAGs and glycolipids, as well as MS spectra of glycoproteins.78 GlycoWorkbench allows the user to draw a glycan structure and calculate a list of theoretical fragment ions.21 Another popular bioinformatics software called GlycReSoft was developed for automated recognition of glycans from LC/MS data.79 It is able to automatically assign structures within 5-ppm mass accuracy.80 The software GAD-ID is the first to automate the interpretation of mixtures when coupled to LC-MS/MS, but it requires derivatization to replace sulfo group with acetyl groups. This tool uses a scoring system based on peak intensities and could properly assign 21 synthetic tetrasaccharides in a defined mixture from a single LC-MS/MS run.81 Recently, a statistical model, which using a multivariate expectation maximization algorithm, was build to estimate the accuracy of derivatized HP/HS assignments to tandem mass (MS/MS) spectra made by GAG-ID. This analysis makes it possible to filter large MS/MS database search results with predictable false identification error rates.82 For HP/HS analysis, GlycCompSoft is designed to enable the comparison of top-down analytical glycomics data on two or more LWMHs. In a test based on three lots of Lovenox, Clexane, and three generic enoxaparin samples, the program proved its low error rate and good time efficiency when processing large data sets. Hu and coworkers developed an algorithm to convert the raw data from electron-based dissociation (ExD) tandem mass spectra into HS saccharide sequences.83 A genetic algorithm has been used by Duan and Amster to design a de novo approach for rapid, high-throughput GAG analysis. The program is coded in a software package using the MATLAB environment. By using a genetic algorithm, the search space of isomeric structures that are considered is greatly reduced, which enables the analysis time to be reduced to a maximum of a few minutes. This approach was tested and properly identified structures from MS2 data of GAG chain from bikunin.84 Machine learning strategy was introduced by Hong and coworkers to develop GlycoDeNovo. This is a novel algorithm that can automatically learn fragmentation patterns from real-world tandem MS data. It contains a data-driven IonClassifier, to which machine learning is applied to distinguish B and C ions from other ion types using experimental spectra of glycan standards.85 A tandem mass spectrum peak finding program was recently developed by Hogan and coworkers specifically for GAGs. The program being called GAGfinder uses precursor composition information to generate all theoretical fragments, which is a targeted, brute force approach to carry out spectrum interpretation.86
There have been major achievements in MS spectrum interpretation in the last few years. However, some of the tools are more suitable for specific types of GAGs and certain workflows. A more rapid, high-throughput, universal and user-friendly software will need to be developed for future GAG analysis.
NMR
In addition to MS-based techniques, NMR spectroscopy has become another powerful technique for determining GAG structures. NMR permits structural analysis directly on unmodified GAGs using isotopes 1H, 13C, and 15N.1,87 The most basic NMR approaches used for GAG analysis are 1H and 13C mono-dimensional NMR (1D NMR). A 1H-NMR spectrum is quite often used in distinguishing IdoA and GlcA residues, revealing monosaccharide compositions, and assigning anomeric configurations. 13C-NMR is less sensitive than 1H-NMR but offers more dispersion of chemical shift resulting in less signal overlap.113C-NMR spectroscopy is particularly useful in determining sulfation pattern and positional linkages of polysaccharides. However, due to the structural complexity or impurity of some GAG samples, the area of signals in 1D NMR spectra is often very complicated and affected by severe signal overlap. In this case, multidimensional NMR spectroscopy is required to help assign signals, complete structural characterization, or even quantify the composition of sugar residues.88 The commonly used multidimensional NMR in examining glycans is two-dimensional (2D) NMR both homonuclear and heteronuclear, including 1H–1H correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), nuclear Overhauser effect spectroscopy (NOESY), rotating frame Overhauser effect spectroscopy (ROESY), 1H–13C heteronuclear single-quantum coherence (HSQC), and heteronuclear multiple bond coherence (HMBC).
NMR is an attractive technique for GAG analysis because it directly interrogates a sample requiring no derivatization procedure and results in no sulfo loss. As NMR is nondestructive, it allows a sample to be further used for other analytical methods. Moreover, NMR is sensitive to conformational changes and can provide information on the secondary structure of GAGs and their complexes with proteins.1 However, the NMR spectroscopy is limited by its relatively low sensitivity, and usually requires milligram amounts of pure samples for glycan analysis. In a protocol published by Carnachan and Hinkley,89 a concentration of 17 mg/mL of GAG sample was the minimum for highly reproducible HS analysis using 2D NMR. Increasing the strength of the static magnetic field can help improve NMR sensitivity. However, it is hard to detect a minor monosaccharide residue in a long glycan chain. For a precise de novo structural characterization of unknown oligosaccharide using NMR, the size of GAG chains is limited to approximately an octadecasaccharide.1 Therefore, the NMR is often used alone in a bottom-up approach or as assistance for top-down MS analysis.
One-dimensional 1H-NMR has been used to help to determine the ratio of IdoA/IdoA2S: GlcA in DS chain, in support with MS analysis.16 One- and two-dimensional NMR were used to compare relative quantities of seven types of monosaccharide between parent and daughter heparins.24 NMR was also used to characterize structures of pure oligosaccharides derived in a bottom-up approach. A series of GAG oligosaccharides up to octasaccharides were separated from different kinds of sea cucumbers and characterized by 1D and 2D (COSY, TOCSY, ROESY, HSQC and HMBC) NMR.90 A bottom-up approach was used to analyze the central core of fucosylated CS from two species of sea cucumbers using COSY, TOCSY, and HSQC, and only slight differences were found between the two species.91 Guerrini and coworkers92 used HSQC for the structural characterization of three commercially available LMWHs of enoxaparin, dalteparin, and tinzaparin. In this research, the minor residues generated by a depolymerization procedure, as well as the relevant residues belonging to the parent heparin, have been characterized and quantified. HSQC has recently been considered a quantitative tool for GAG analysis. This can be achieved by proper selection of analytical signals among those with similar magnetic relaxation and one bond proton-carbon J-coupling. The method was elaborated by Torri and Guerrini88 that can be confidently used to compare different GAGs, by quantification of their various substituted monosaccharide components.
Hyphenated and Other Techniques
Although both MS and NMR show their specific advantages in characterizing GAG chain, in most cases, the combined application of both methods provides more informative details and allows a more thorough structural characterization. NMR can assist MS in identifying monosaccharides and quantifying IdoA/GlcA ratio. Beccati and coworkers93 reported an integrated approach using IPRP-HPLC, LC-MS, NMR, SEC-MS, and ESI-Q-TOF-MS to characterize bovine kidney HS, providing overlapping and confirmatory information from different perspectives for the determination of the GAG structure. Recently, some research carried out using hyphenated or individual spectroscopic techniques was also quite inspiring. Renois-Predelus and coworkers94 reported a novel approach coupling mass spectrometry with ion spectroscopy for the structural analysis of GAGs. Distinctive spectroscopic fingerprints of the monosaccharide standards GalNAc4S and GalNAc6S were found in the 3 μm range. Khanal and coworkers95 used IMS combined with cryogenic, messenger-tagging, infrared (IR) spectroscopy, and MS to identify different isomeric disaccharides of CS and HS origin. Disaccharide isomers can be uniquely distinguished by their vibrational spectrum between –3200 and 3700 cm–1 due to their different hydroxyl group hydrogen-bonding patterns. Another spectroscopic associated research method used a combination of gas-phase IRMPD (infrared multiple photon dissociation) spectroscopy with MS to address the differentiation of positional isomers of sulfated carbohydrates on monosaccharide standards and disaccharides of HP and CS. Mass spectrometric fingerprints and gas-phase vibrational spectra in the near and mid-IR regions were obtained, which validate the potential of the spectroscopic approach to resolve isomeric disaccharides.96 In a review published by Mohamed and coworkers,97 the application of vibrational spectroscopy for GAG analysis was introduced. By comparing the GAG IR and Raman spectral signatures of the media and live cells, Chinese hamster ovary (CHO) wild type cell line and its mutant counterpart, CHO-745, which lacks xylosyltransferase associated with low GAG synthesis, were discriminated. In recently published research, surface plasmon resonance imaging (SPRi) was coupled to an on-chip mass spectrometry. The thermodynamic parameters of the interactions between cytokines and GAGs were determined using SPRi monitoring, and the captured carbohydrates were directly examined on the biochip surface using MALDI-TOF MS.98
Perspectives
The future development of GAG analysis and sequencing will be facilitated by the emerging innovative techniques, materials, and methods. A “shotgun ion mobility mass spectrometry sequencing” (SIMMS2) method was recently be developed by Miller and coworkers.99 Intact HS saccharides are subjected to ion mobility mass spectrometry and MS2 analysis. Matching of data for intact and fragment ions against known values for defined HS oligosaccharide structures permits unambiguous sequence determination of validated standards as well as unknown natural saccharides, including variants with 3-O-sulfo groups. It is envisioned that by limited digestion with multiple site-specific enzymes, sets of partially overlapping fragments can be separated and sequenced by ion mobility mass spectrometry and MS2. Overlaps of such fragments may enable assembly in a similar manner to the shotgun sequencing approaches used in genomics and proteomics. A newly emerging technique, based on nanopore sensing, seems to represent a promising breakthrough. The nanopore can be made of either biological (such as aerolysin and α-hemolysin)100 or inorganic materials.101–103 The nanopore filled with electrolyte solution allows ions to pass through unless a charged polymer in the passage and disrupts ion flow, generating a blocked-pore current. Karawdeniya and coworkers102 reported this technique allowed easy differentiation between a clinical heparin sample and one contaminated with oversulfated chondroitin sulfate (OSCS). In other research carried out by Im and coworkers, solid-state nanopore device with a support vector machine (SVM) learning algorithm was used for GAG analysis. The nanopore/SVM technique was able to distinguish between monodisperse fragments of HP and CS with high accuracy (>90%), allowing the detection of as little as 0.8% (w/w) CS impurity in a heparin sample.103
The development of new materials will also promote progress in GAG-related research. Mothéré and coworkers used molecular imprinting technologies to prepare a library of polyethylene glycol acrylate functionalized hydrogels, which showed certain recognition specificity and selectivity to sulfated oligosaccharide.104 Also, GAGs are natural negatively charged biopolymers, which suggest they have great potential to be engaged in new material development.2,105
New methods that are fast, high throughput, and user-friendly are still needed for GAG analysis, such as a fast and sensitive detection of HP based on chemiluminescence sensing,106 high-throughput microarrays that enable in situ structural characterization of GAGs,98,107 and more easy-to-use structural interpretation software for MS will be important in advancing the field. In addition, due to the highly complex and heterogeneous nature of GAGs, it is quite common that multiple analytical techniques are used to complete GAG analysis. Therefore, a bioinformatics platform on which integrated information can be stored and crosschecked will no doubt be meaningful for glycomics scientists.25
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: YS contributed to the data searching and organizing, and draft the article. FZ assisted in writing and revising the manuscript. RJL designed the topic and assisted in writing and revising the manuscript.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Contributor Information
Yuefan Song, National R & D Branch Center for Seaweed Processing, College of Food Science and Engineering, Dalian Ocean University, Dalian, P.R. China; Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, New York.
Fuming Zhang, Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, New York.
Robert J. Linhardt, Center for Biotechnology and Interdisciplinary Studies, Rensselaer Polytechnic Institute, Troy, New York.
Literature Cited
- 1. Li L, Ly M, Linhardt RJ. Proteoglycan sequence. Mol Biosyst. 2012;8(6):1613–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Köwitsch A, Zhou G, Groth T. Medical application of glycosaminoglycans: a review. J Tissue Eng Regen Med. 2018;12(1):e23–41. [DOI] [PubMed] [Google Scholar]
- 3. Zhao J, Zhu Y, Song X, Xiao Y, Su G, Liu X, Wang Z, Xu Y, Liu J, Eliezer D. 3-O-sulfation of heparan sulfate enhances tau interaction and cellular uptake. Angew Chem Int Ed Engl. 2020;59(5):1818–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fu L, Suflita M, Linhardt RJ. Bioengineered heparins and heparan sulfates. Adv Drug Deliv Rev. 2016;97:237–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yip GW, Smollich M, Götte M. Therapeutic value of glycosaminoglycans in cancer. Mol Cancer Ther. 2006;5(9):2139–48. [DOI] [PubMed] [Google Scholar]
- 6. De Pasquale V, Pavone LM. Heparan sulfate proteoglycans: the sweet side of development turns sour in mucopolysaccharidoses. Biochim Biophys Acta Mol Basis Dis. 2019;1865:165539. [DOI] [PubMed] [Google Scholar]
- 7. Kamhi E, Joo EJ, Dordick JS, Linhardt RJ. Glyco-saminoglycans in infectious disease. Biol Rev Camb Philos Soc. 2013;88(4):928–43. [DOI] [PubMed] [Google Scholar]
- 8. Volpi N. Chondroitin sulfate safety and quality. Molecules. 2019;24(8):1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sugahara K, Mikami T. Chondroitin/dermatan sulfate in the central nervous system. Curr Opin Struct Biol. 2007;17(5):536–45. [DOI] [PubMed] [Google Scholar]
- 10. Uchimura K. Keratan sulfate: biosynthesis, structures, and biological functions. Methods Mol Biol. 2015;1229:389–400. [DOI] [PubMed] [Google Scholar]
- 11. Caterson B, Melrose J. Keratan sulfate, a complex glycosaminoglycan with unique functional capability. Glycobiology. 2018;28(4):182–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Toole B. Hyaluronan and its binding proteins, the hyaladherins. Curr Opin Cell Biol. 1990;2(5):839–44. [DOI] [PubMed] [Google Scholar]
- 13. Tamer TM. Hyaluronan and synovial joint: function, distribution and healing. Interdiscip Toxicol. 2013;6(3):111–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang J, Chi L. Characterization of structural motifs for interactions between glycosaminoglycans and proteins. Carbohydr Res. 2017;452:54–63. [DOI] [PubMed] [Google Scholar]
- 15. Ly M, Leach FE, III, Laremore TN, Toida T, Amster IJ, Linhardt RJ. The proteoglycan bikunin has a defined sequence. Nat Chem Biol. 2011;7(11):827–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu Y, Duan J, Leach FE, III, Toida T, Higashi K, Zhang H, Zhang F, Amster IJ, Linhardt RJ. Sequencing the dermatan sulfate chain of decorin. J Am Chem Soc. 2017;139(46):16986–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ly M, Laremore TN, Linhardt RJ. Proteoglycomics: recent progress and future challenges. OMICS. 2010;14(4):389–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao X, Yang B, Solakylidirim K, Joo EJ, Toida T, Higashi K, Linhardt RJ, Li L. Sequence analysis and domain motifs in the porcine skin decorin glycosaminoglycan chain. J Biol Chem. 2013;288(13):9226–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bodet P-E, Salard I, Przybylski C, Gonnet F, Gomila C, Ausseil J, Daniel R. Efficient recovery of glycosaminoglycan oligosaccharides from polyacrylamide gel electrophoresis combined with mass spectrometry analysis. Anal Bioanal Chem. 2017;409(5):1257–69. [DOI] [PubMed] [Google Scholar]
- 20. Zaia J. Glycosaminoglycan glycomics using mass spectrometry. Mol Cell Proteomics. 2013;12(4):885–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Duan J, Amster IJ. Application of FTMS to the analysis of glycosaminoglycans. In: Schmitt-Kopplin P, Kanawati B, editors. Fundamentals and applications of Fourier transform mass spectrometry. Amsterdam: Elsevier; 2019. p. 623–49. [Google Scholar]
- 22. Agyekum I, Zong C, Boons G-J, Amster IJ. Single stage tandem mass spectrometry assignment of the C-5 uronic acid stereochemistry in heparan sulfate tetrasaccharides using electron detachment dissociation. J Am Soc Mass Spectrom. 2017;28(9):1741–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wei J, Wu J, Tang Y, Ridgeway ME, Park MA, Costello CE, Zaia J, Lin C. Characterization and quantification of highly sulfated glycosaminoglycan isomers by gated-trapped ion mobility spectrometry negative electron transfer dissociation MS/MS. Anal Chem. 2019;91(4):2994–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu X, St Ange K, Wang X, Lin L, Zhang F, Chi L, Linhardt RJ. Parent heparin and daughter LMW heparin correlation analysis using LC-MS and NMR. Anal Chim Acta. 2017;961:91–99. [DOI] [PubMed] [Google Scholar]
- 25. Solakyildirim K. Recent advances in glycosaminoglycan analysis by various mass spectrometry techniques. Anal Bioanal Chem. 2019;411:3731–41. [DOI] [PubMed] [Google Scholar]
- 26. Li G, Steppich J, Wang Z, Sun Y, Xue C, Linhardt RJ, Li L. Bottom-up low molecular weight heparin analysis using liquid chromatography-Fourier transform mass spectrometry for extensive characterization. Anal Chem. 2014;86(13):6626–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin L, Yu Y, Zhang F, Xia K, Zhang X, Linhardt RJ. Bottom-up and top-down profiling of pentosan polysulfate. Analyst. 2019;144(16):4781–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Volpi N, Linhardt RJ. High-performance liquid chromatography-mass spectrometry for mapping and sequencing glycosaminoglycan-derived oligosaccharides. Nat Protoc. 2010;5(6):993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zaia J. On-line separations combined with MS for analysis of glycosaminoglycans. Mass Spectrom Rev. 2009;28(2):254–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang B, Solakyildirim K, Chang Y, Linhardt RJ. Hyphenated techniques for the analysis of heparin and heparan sulfate. Anal Bioanal Chem. 2011;399(2):541–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zaia J. Mass spectrometry and the emerging field of glycomics. Chem Biol. 2008;15(9):881–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Korir AK, Larive CK. Advances in the separation, sensitive detection, and characterization of heparin and heparan sulfate. Anal Bioanal Chem. 2009;393(1):155–169. [DOI] [PubMed] [Google Scholar]
- 33. Fasciano JM, Danielson ND. Ion chromatography for the separation of heparin and structurally related glycoaminoglycans: a review. J Sep Sci. 2016;39(6):1118–29. [DOI] [PubMed] [Google Scholar]
- 34. Volpi N, Maccari F, Linhardt RJ. Capillary electrophoresis of complex natural polysaccharides. Electro-phoresis. 2008;29(15):3095–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sisu E, Flangea C, Serb A, Zamfir AD. Modern developments in mass spectrometry of chondroitin and dermatan sulfate glycosaminoglycans. Amino Acids. 2011;41(2):235–56. [DOI] [PubMed] [Google Scholar]
- 36. Laremore TN, Leach FE, III, Solakyildirim K, Amster IJ, Linhardt RJ. Glycosaminoglycan characterization by electrospray ionization mass spectrometry including Fourier transform mass spectrometry. Methods Enzymol. 2010;478:79–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruhaak L, Zauner G, Huhn C, Bruggink C, Deelder A, Wuhrer M. Glycan labeling strategies and their use in identification and quantification. Anal Bioanal Chem. 2010;397(8):3457–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Antia IU, Yagnik DR, Munoz LP, Shah AJ, Hills FA. Heparan sulfate disaccharide measurement from biological samples using pre-column derivatization, UPLC-MS and single ion monitoring. Anal Biochem. 2017;530:17–30. [DOI] [PubMed] [Google Scholar]
- 39. Liu X, Krishnamoorthy D, Lin L, Xue P, Zhang F, Chi L, Linhardt R, Iatridis J. A method for characterising human intervertebral disc glycosaminoglycan disaccharides using liquid chromatography-mass spectrometry with multiple reaction monitoring. Eur Cell Mater. 2018;35:117–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang R, Zong C, Venot A, Chiu Y, Zhou D, Boons G-J, Sharp JS. De novo sequencing of complex mixtures of heparan sulfate oligosaccharides. Anal Chem. 2016;88(10):5299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liang Q, Chopra P, Boons G-J, Sharp JS. Improved de novo sequencing of heparin/heparan sulfate oligosaccharides by propionylation of sites of sulfation. Carbohydr Res. 2018;465:16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mangrum JB, Mehta AY, Alabbas AB, Desai UR, Hawkridge AM. Comparative analysis of INLIGHT™-labeled enzymatically depolymerized heparin by reverse-phase chromatography and high-performance mass spectrometry. Anal Bioanal Chem. 2017;409(2):499–509. [DOI] [PubMed] [Google Scholar]
- 43. Przybylski C, Bonnet V, Vivès RR. A microscale double labelling of GAG oligosaccharides compatible with enzymatic treatment and mass spectrometry. Chem Commun (Camb). 2019;55(29):4182–85. [DOI] [PubMed] [Google Scholar]
- 44. Shioiri T, Tsuchimoto J, Watanabe H, Sugiura N. Sequence determination of synthesized chondroitin sulfate dodecasaccharides. Glycobiology. 2016;26(6):592–606. [DOI] [PubMed] [Google Scholar]
- 45. Yang B, Weyers A, Baik JY, Sterner E, Sharfstein S, Mousa SA, Zhang F, Dordick JS, Linhardt RJ. Ultra-performance ion-pairing liquid chromatography with on-line electrospray ion trap mass spectrometry for heparin disaccharide analysis. Anal Biochem. 2011;415(1):59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liang QT, Xiao XM, Lin JH, Wei Z. A new sequencing approach for N-unsubstituted heparin/heparan sulfate oligosaccharides. Glycobiology. 2015;25(7):714–25. [DOI] [PubMed] [Google Scholar]
- 47. Du JY, Chen LR, Liu S, Lin JH, Liang QT, Lyon M, Wei Z. Ion-pairing liquid chromatography with on-line electrospray ion trap mass spectrometry for the structural analysis of N-unsubstituted heparin/heparan sulfate. J Chromatogr B Analyt Technol Biomed Life Sci. 2016;1028:71–76. [DOI] [PubMed] [Google Scholar]
- 48. Persson A, Toledo AG, Vorontsov E, Nasir W, Willén D, Noborn F, Ellervik U, Mani K, Nilsson J, Larson G. LC–MS/MS characterization of xyloside-primed glycosaminoglycans with cytotoxic properties reveals structural diversity and novel glycan modifications. J Biol Chem. 2018;293(26):10202–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun X, Guo Z, Yu M, Lin C, Sheng A, Wang Z, Linhardt RJ, Chi L. Hydrophilic interaction chromatography-multiple reaction monitoring mass spectrometry method for basic building block analysis of low molecular weight heparins prepared through nitrous acid depolymerization. J Chromatogr A. 2017;1479:121–8. [DOI] [PubMed] [Google Scholar]
- 50. Sun X, Sheng A, Liu X, Shi F, Jin L, Xie S, Zhang F, Linhardt RJ, Chi L. Comprehensive identification and quantitation of basic building blocks for low-molecular weight heparin. Anal Chem. 2016;88(15):7738–44. [DOI] [PubMed] [Google Scholar]
- 51. Turiák L, Tóth G, Ozohanics O, Révész Ács ÁA, Vékey K, Zaia J, Drahos L. Sensitive method for glycosaminoglycan analysis of tissue sections. J Chromatogr A. 2018;1544:41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang T, Liu X, Li H, Wang Z, Chi L, Li J-P, Tan T. Characterization of epimerization and composition of heparin and dalteparin using a UHPLC-ESI-MS/MS method. Carbohydr Polym. 2019;203:87–94. [DOI] [PubMed] [Google Scholar]
- 53. Antia IU, Mathew K, Yagnik DR, Hills FA, Shah AJ. Analysis of procainamide-derivatised heparan sulphate disaccharides in biological samples using hydrophilic interaction liquid chromatography mass spectrometry. Anal Bioanal Chem. 2018;410(1):131–43. [DOI] [PubMed] [Google Scholar]
- 54. Staples GO, Bowman MJ, Costello CE, Hitchcock AM, Lau JM, Leymarie N, Miller C, Naimy H, Shi X, Zaia J. A chip-based amide-HILIC LC/MS platform for glycosaminoglycan glycomics profiling. Proteomics. 2009;9(3):686–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang Y, Shi X, Yu X, Leymarie N, Staples GO, Yin H, Killeen K, Zaia J. Improved liquid chromatography-MS/MS of heparan sulfate oligosaccharides via chip-based pulsed makeup flow. Anal Chem. 2011;83(21):8222–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stavenhagen K, Kolarich D, Wuhrer M. Clinical glycomics employing graphitized carbon liquid chromatography–mass spectrometry. Chromatographia. 2015;78(5–6):307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Miller RL, Guimond SE, Prescott M, Turnbull JE, Karlsson N. Versatile separation and analysis of heparan sulfate oligosaccharides using graphitized carbon liquid chromatography and electrospray mass spectrometry. Anal Chem. 2017;89(17):8942–50. [DOI] [PubMed] [Google Scholar]
- 58. Zamfir AD. Applications of capillary electrophoresis electrospray ionization mass spectrometry in glycosaminoglycan analysis. Electrophoresis. 2016;37(7–8):973–86. [DOI] [PubMed] [Google Scholar]
- 59. Lin L, Liu X, Zhang F, Chi L, Amster IJ, Leach FE, Xia Q, Linhardt RJ. Analysis of heparin oligosaccharides by capillary electrophoresis–negative-ion electrospray ionization mass spectrometry. Anal Bioanal Chem. 2017;409(2):411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sanderson P, Stickney M, Leach FE, III, Xia Q, Yu Y, Zhang F, Linhardt RJ, Amster IJ. Heparin/heparan sulfate analysis by covalently modified reverse polarity capillary zone electrophoresis-mass spectrometry. J Chromatogr A. 2018;1545:75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stickney M, Sanderson P, Leach FE, III, Zhang F, Linhardt RJ, Amster IJ. Online capillary zone electrophoresis negative electron transfer dissociation tandem mass spectrometry of glycosaminoglycan mixtures. Int J Mass Spectrom. 2019;445:116209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kailemia MJ, Park M, Kaplan DA, Venot A, Boons G-J, Li L, Linhardt RJ, Amster IJ. High-field asymmetric-waveform ion mobility spectrometry and electron detachment dissociation of isobaric mixtures of glycosaminoglycans. J Am Soc Mass Spectrom. 2013;25(2):258–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhao Y, Singh A, Li L, Linhardt RJ, Xu Y, Liu J, Woods RJ, Amster IJ. Investigating changes in the gas-phase conformation of Antithrombin III upon binding of Arixtra using traveling wave ion mobility spectrometry (TWIMS). Analyst. 2015;140(20):6980–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Miller RL, Wei W, Schwörer R, Zubkova OV, Tyler PC, Turnbull JE, Leary JA. Composition, sequencing and ion mobility mass spectrometry of heparan sulfate-like octasaccharide isomers differing in glucuronic and iduronic acid content. Eur J Mass Spectrom (Chichester). 2015;21(3):245–54. [DOI] [PubMed] [Google Scholar]
- 65. Liu H, Joshi A, Chopra P, Liu L, Boons G-J, Sharp JS. Salt-free fractionation of complex isomeric mixtures of glycosaminoglycan oligosaccharides compatible with ESI-MS and microarray analysis. Sci Rep. 2019;9(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Miller RL, Guimond SE, Shivkumar M, Blocksidge J, Austin JA, Leary JA, Turnbull JE. Heparin isomeric oligosaccharide separation using volatile salt strong anion exchange chromatography. Anal Chem. 2016;88(23):11542–50. [DOI] [PubMed] [Google Scholar]
- 67. Zaia J. Mass spectrometry of oligosaccharides. Mass Spectrom Rev. 2004;23(3):161–227. [DOI] [PubMed] [Google Scholar]
- 68. Wolff JJ, Amster IJ, Chi L, Linhardt RJ. Electron detachment dissociation of glycosaminoglycan tetrasaccharides. J Am Soc Mass Spectrom. 2007;18(2):234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wolff JJ, Laremore TN, Aslam H, Linhardt RJ, Amster IJ. Electron-induced dissociation of glycosaminoglycan tetrasaccharides. J Am Soc Mass Spectrom. 2008;19(10):1449–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Agyekum I, Patel AB, Zong C, Boons G-J, Amster IJ. Assignment of hexuronic acid stereochemistry in synthetic heparan sulfate tetrasaccharides with 2-O-sulfo uronic acids using electron detachment dissociation. Int J Mass Spectrom. 2015;390:163–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Leach FE, III, Riley NM, Westphall MS, Coon JJ, Amster IJ. Negative electron transfer dissociation sequencing of increasingly sulfated glycosaminoglycan oligosaccharides on an Orbitrap mass spectrometer. J Am Soc Mass Spectrom. 2017;28(9):1844–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wu J, Wei J, Hogan JD, Chopra P, Joshi A, Lu W, Klein J, Boons G-J, Lin C, Zaia J. Negative electron transfer dissociation sequencing of 3-O-sulfation-containing heparan sulfate oligosaccharides. J Am Soc Mass Spectrom. 2018;29(6):1262–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Devakumar A, Thompson MS, Reilly JP. Fragmentation of oligosaccharide ions with 157 nm vacuum ultraviolet light. Rapid Commun Mass Spectrom. 2005;19(16):2313–20. [DOI] [PubMed] [Google Scholar]
- 74. Ko BJ, Brodbelt JS. 193 nm ultraviolet photodissociation of deprotonated sialylated oligosaccharides. Anal Chem. 2011;83(21):8192–200. [DOI] [PubMed] [Google Scholar]
- 75. Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J. 1988;5(4):397–409. [Google Scholar]
- 76. Zhang Z, Linhardt RJ. Sequence analysis of native oligosaccharides using negative ESI tandem MS. Curr Anal Chem. 2009;5(3):225–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Saad OM, Leary JA. Heparin sequencing using enzymatic digestion and ESI-MS n with HOST: a heparin/HS oligosaccharide sequencing tool. Anal Chem. 2005;77(18):5902–11. [DOI] [PubMed] [Google Scholar]
- 78. Damerell D, Ceroni A, Maass K, Ranzinger R, Dell A, Haslam SM. The GlycanBuilder and GlycoWorkbench Glycoinformatics tools: updates and new developments. Biol Chem. 2012;393(11):1357–62. [DOI] [PubMed] [Google Scholar]
- 79. Maxwell E, Tan Y, Tan Y, Hu H, Benson G, Aizikov K, Conley S, Staples GO, Slysz GW, Smith RD. GlycReSoft: a software package for automated recognition of glycans from LC/MS data. PLOS ONE. 2012;7(9):e45474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li L, Zhang F, Zaia J, Linhardt RJ. Top-down approach for the direct characterization of low molecular weight heparins using LC-FT-MS. Anal Chem. 2012;84(20):8822–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chiu Y, Huang R, Orlando R, Sharp JS. GAG-ID: heparan sulfate (HS) and heparin glycosaminoglycan high-throughput identification software. Mol Cell Proteomics. 2015;14(6):1720–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chiu Y, Schliekelman P, Orlando R, Sharp JS. A multivariate mixture model to estimate the accuracy of glycosaminoglycan identifications made by tandem mass spectrometry (MS/MS) and database search. Mol Cell Proteomics. 2017;16(2):255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hu H, Mao Y, Huang Y, Lin C, Zaia J. Bioinformatics of glycosaminoglycans. Perspect Sci. 2017;11:40–44. [Google Scholar]
- 84. Duan J, Jonathan Amster I. An automated, high-throughput method for interpreting the tandem mass spectra of glycosaminoglycans. J Am Soc Mass Spectrom. 2018;29(9):1802–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hong P, Sun H, Sha L, Pu Y, Khatri K, Yu X, Tang Y, Lin C. GlycoDeNovo–an efficient algorithm for accurate de novo glycan topology reconstruction from tandem mass spectra. J Am Soc Mass Spectrom. 2017;28(11):2288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Hogan JD, Klein JA, Wu J, Chopra P, Boons G-J, Carvalho L, Lin C, Zaia J. Software for peak finding and elemental composition assignment for glycosaminoglycan tandem mass spectra. Mol Cell Proteomics. 2018;17(7):1448–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Soares PA, Queiroz IN, Pomin VH. NMR structural biology of sulfated glycans. J Biomol Struct Dyn. 2017;35(5):1069–84. [DOI] [PubMed] [Google Scholar]
- 88. Torri G, Guerrini M. Quantitative 2D NMR analysis of glycosaminoglycans. In: Wawer I, Diehl B, editors. NMR spectroscopy in pharmaceutical analysis. Amsterdam: Elsevier; 2008. p. 407–28. [Google Scholar]
- 89. Carnachan SM, Hinkley SF. Heparan sulfate identification and characterisation: method I. Heparan sulfate identification by NMR analysis. Bio Protoc. 2017;7(7) 1–7. doi:10.21769/BioProtoc.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Shang F, Gao N, Yin R, Lin L, Xiao C, Zhou L, Li Z, Purcell SW, Wu M, Zhao J. Precise structures of fucosylated glycosaminoglycan and its oligosaccharides as novel intrinsic factor Xase inhibitors. Eur J Med Chem. 2018;148:423–35. [DOI] [PubMed] [Google Scholar]
- 91. Santos GR, Porto AC, Soares PA, Vilanova E, Mourão PA. Exploring the structure of fucosylated chondroitin sulfate through bottom-up nuclear magnetic resonance and electrospray ionization-high-resolution mass spectrometry approaches. Glycobiology. 2017;27(7):625–34. [DOI] [PubMed] [Google Scholar]
- 92. Guerrini M, Guglieri S, Naggi A, Sasisekharan R, Torri G. Low molecular weight heparins: structural differentiation by bidimensional nuclear magnetic resonance spectroscopy. Semin Thromb Hemost; 2007;33(5):478–87. [DOI] [PubMed] [Google Scholar]
- 93. Beccati D, Lech M, Ozug J, Gunay NS, Wang J, Sun EY, Pradines JR, Farutin V, Shriver Z, Kaundinya GV. An integrated approach using orthogonal analytical techniques to characterize heparan sulfate structure. Glycoconj J. 2017;34(1):107–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Renois-Predelus G, Schindler B, Compagnon I. Analysis of sulfate patterns in glycosaminoglycan oligosaccharides by MSn coupled to infrared ion spectroscopy: the case of GalNAc4S and GalNAc6S. J Am Soc Mass Spectrom. 2018;29(6):1242–49. [DOI] [PubMed] [Google Scholar]
- 95. Khanal N, Masellis C, Kamrath MZ, Clemmer DE, Rizzo TR. Glycosaminoglycan analysis by cryogenic messenger-tagging IR spectroscopy combined with IMS-MS. Anal Chem. 2017;89(14):7601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Schindler B, Barnes L, Gray C, Chambert S, Flitsch S, Oomens J, Daniel R, Allouche A, Compagnon I. IRMPD spectroscopy sheds new (infrared) light on the sulfate pattern of carbohydrates. J Phys Chem A. 2017;121(10):2114–20. [DOI] [PubMed] [Google Scholar]
- 97. Mohamed HT, Untereiner V, Sockalingum GD, Brézillon S. Implementation of infrared and Raman modalities for glycosaminoglycan characterization in complex systems. Glycoconj J. 2017;34(3):309–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Przybylski C, Gonnet F, Saesen E, Lortat-Jacob H, Daniel R. Surface plasmon resonance imaging coupled to on-chip mass spectrometry: a new tool to probe protein-GAG interactions. Anal Bioanal Chem. 2020;412(2):507–19. [DOI] [PubMed] [Google Scholar]
- 99. Miller RL, Guimond SE, Schwörer R, Zubkova OV, Tyler PC, Xu Y, Liu J, Chopra P, Boons G-J, Grabarics M. Shotgun ion mobility mass spectrometry sequencing of heparan sulfate saccharides. Nat Commun. 2020;11(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Fennouri A, Ramiandrisoa J, Bacri L, Mathé J, Daniel R. Comparative biosensing of glycosaminoglycan hyaluronic acid oligo-and polysaccharides using aerolysin and alpha hemolysin nanopores. Eur Phys J E Soft Matter. 2018;41(10):127. [DOI] [PubMed] [Google Scholar]
- 101. Rivas F, Zahid OK, Reesink HL, Peal BT, Nixon AJ, DeAngelis PL, Skardal A, Rahbar E, Hall AR. Label-free analysis of physiological hyaluronan size distribution with a solid-state nanopore sensor. Nat Commun. 2018;9(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Karawdeniya BI, Bandara YND, Nichols JW, Chevalier RB, Dwyer JR. Surveying silicon nitride nanopores for glycomics and heparin quality assurance. Nat Commun. 2018;9(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Im J, Lindsay S, Wang X, Zhang P. Single Molecule Identification and Quantification of Glycosaminoglycans Using Solid-State Nanopores. ACS Nano. 2019;13(6):6308–18. [DOI] [PubMed] [Google Scholar]
- 104. Mothéré M, Singabraya D, Driguez PA, Siñeriz F, Papy Garcia D. Poly(ethylene glycol acrylate)-functionalized hydrogels for heparan sulfate oligosaccharide recognition. J Mol Recognit. 2017;30(3):e2584. [DOI] [PubMed] [Google Scholar]
- 105. Kemp MM, Kumar A, Clement D, Ajayan P, Mousa S, Linhardt RJ. Hyaluronan-and heparin-reduced silver nanoparticles with antimicrobial properties. Nano-medicine. 2009;4(4):421–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Qi Y, He J, Xiu F-R, Yu X, Li Y, Lu Y, Gao X, Song Z, Li B. A facile chemiluminescence sensing for ultrasensitive detection of heparin using charge effect of positively-charged AuNPs. Spectrochim Acta A Mol Biomol Spectrosc. 2019;216:310–18. [DOI] [PubMed] [Google Scholar]
- 107. Kunzke T, Balluff B, Feuchtinger A, Buck A, Langer R, Luber B, Lordick F, Zitzelsberger H, Aichler M, Walch A. Native glycan fragments detected by MALDI-FT-ICR mass spectrometry imaging impact gastric cancer biology and patient outcome. Oncotarget. 2017;8(40):68012. [DOI] [PMC free article] [PubMed] [Google Scholar]