Abstract
Natural products and their structural analogues have historically made a major contribution to pharmacotherapy, especially for cancer and infectious diseases. Nevertheless, natural products also present challenges for drug discovery, such as technical barriers to screening, isolation, characterization and optimization, which contributed to a decline in their pursuit by the pharmaceutical industry from the 1990s onwards. In recent years, several technological and scientific developments — including improved analytical tools, genome mining and engineering strategies, and microbial culturing advances — are addressing such challenges and opening up new opportunities. Consequently, interest in natural products as drug leads is being revitalized, particularly for tackling antimicrobial resistance. Here, we summarize recent technological developments that are enabling natural product-based drug discovery, highlight selected applications and discuss key opportunities.
Subject terms: Transferases, Chemical biology
Natural products have historically made a major contribution to pharmacotherapy, but also present challenges for drug discovery, such as technical barriers to screening, isolation, characterization and optimization. This Review discusses recent technological developments — including improved analytical tools, genome mining and engineering strategies, and microbial culturing advances — that are enabling a revitalization of natural product-based drug discovery.
Introduction
Historically, natural products (NPs) have played a key role in drug discovery, especially for cancer and infectious diseases1,2, but also in other therapeutic areas, including cardiovascular diseases (for example, statins) and multiple sclerosis (for example, fingolimod)3–5.
NPs offer special features in comparison with conventional synthetic molecules, which confer both advantages and challenges for the drug discovery process. NPs are characterized by enormous scaffold diversity and structural complexity. They typically have a higher molecular mass, a larger number of sp3 carbon atoms and oxygen atoms but fewer nitrogen and halogen atoms, higher numbers of H-bond acceptors and donors, lower calculated octanol–water partition coefficients (cLogP values, indicating higher hydrophilicity) and greater molecular rigidity compared with synthetic compound libraries1,6–9. These differences can be advantageous; for example, the higher rigidity of NPs can be valuable in drug discovery tackling protein–protein interactions10. Indeed, NPs are a major source of oral drugs ‘beyond Lipinski’s rule of five’11. The increasing significance of drugs not conforming to this rule is illustrated by the increase in molecular mass of approved oral drugs over the past 20 years12. NPs are structurally ‘optimized’ by evolution to serve particular biological functions1, including the regulation of endogenous defence mechanisms and the interaction (often competition) with other organisms, which explains their high relevance for infectious diseases and cancer. Furthermore, their use in traditional medicine may provide insights regarding efficacy and safety. Overall, the NP pool is enriched with ‘bioactive’ compounds covering a wider area of chemical space compared with typical synthetic small-molecule libraries13.
Despite these advantages and multiple successful drug discovery examples, several drawbacks of NPs have led pharmaceutical companies to reduce NP-based drug discovery programmes. NP screens typically involve a library of extracts from natural sources (Fig. 1), which may not be compatible with traditional target-based assays14. Identifying the bioactive compounds of interest can be challenging, and dereplication tools have to be applied to avoid rediscovery of known compounds. Accessing sufficient biological material to isolate and characterize a bioactive NP may also be challenging15. Furthermore, gaining intellectual property (IP) rights for (unmodified) NPs exhibiting relevant bioactivities can be a hurdle, since naturally occurring compounds in their original form may not always be patented (legal frameworks vary between countries and are evolving)16, although simple derivatives can be patent-protected (Box 1). An additional layer of complexity relates to the regulations defining the need for benefit sharing with countries of origin of the biological material, framed in the United Nations 1992 Convention on Biological Diversity and the Nagoya Protocol, which entered into force in 2014 (ref.17), as well as recent developments concerning benefit sharing linked to use of marine genetic resources18.
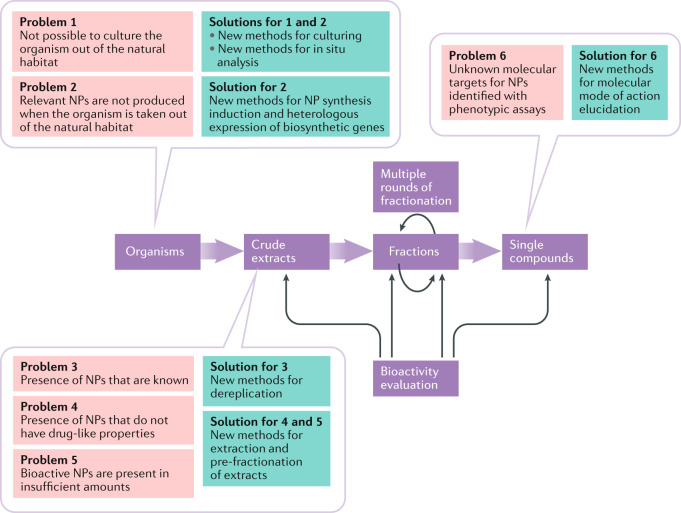
Fig. 1. Outline of traditional bioactivity-guided isolation steps in natural product drug discovery.
Steps in the process are shown in purple boxes, with associated key limitations shown in red boxes and advances that are helping to address these limitations in modern natural product (NP)-based drug discovery shown in green boxes. The process begins with extraction of NPs from organisms such as bacteria. The choice of extraction method determines which compound classes will be present in the extract (for example, the use of more polar solvents will result in a higher abundance of polar compounds in the crude extract). To maximize the diversity of the extracted NPs, the biological material can be subjected to extraction with several solvents of different polarity. Following the identification of a crude extract with promising pharmacological activity, the next step is its (often multiple) consecutive bioactivity-guided fractionation until the pure bioactive compounds are isolated. A key limitation for the potential of this approach to identify novel NPs is that many potential source organisms cannot be cultured or stop producing relevant NPs when taken out of their natural habitat. These limitations are being addressed through development of new methods for culturing, for in situ analysis, for NP synthesis induction and for heterologous expression of biosynthetic genes. At the crude extract step, challenges include the presence in the extracts of NPs that are already known, NPs that do not have drug-like properties or insufficient amounts of NPs for characterization. These challenges can be addressed through the development of methods for dereplication, extraction and pre-fractionation of extracts. Finally, at the last stage, when bioactive compounds are identified by phenotypic assays, significant time and effort are typically needed to identify the affected molecular targets. This challenge can be addressed by the development of methods for accelerated elucidation of molecular modes of action, such as the nematic protein organization technique (NPOT), drug affinity responsive target stability (DARTS), stable isotope labelling with amino acids in cell culture and pulse proteolysis (SILAC-PP), the cellular thermal shift assay (CETSA) and an extension known as thermal proteome profiling (TPP), stability of proteins from rates of oxidation (SPROX), the similarity ensemble approach (SEA) and bioinformatics-based analysis of connectivity (connectivity map, CMAP)23,189–192.
Although the complexity of NP structures can be advantageous, the generation of structural analogues to explore structure–activity relationships and to optimize NP leads can be challenging, particularly if synthetic routes are difficult. Also, NP-based drug leads are often identified by phenotypic assays, and deconvolution of their molecular mechanisms of action can be time-consuming19. Fortunately, there have been substantial advances20 both in the development of screening assays (for example, harnessing the potential of induced pluripotent stem cells and gene editing technologies) and in strategies to identify the modes of action of active compounds (reviewed previously21–23).
Here, we discuss recent technological and scientific advances that may help to overcome challenges in NP-based drug discovery, with an emphasis on three areas: analytical techniques, genome mining and engineering, and cultivation systems. In the concluding section, we highlight promising future directions for NP drug discovery.
Box 1 Natural products that activate the KEAP1/NRF2 pathway.
An example of a pathway affected by diverse natural products (NPs) is the KEAP1/NRF2 pathway. This pathway regulates the expression of networks of genes encoding proteins with versatile cytoprotective functions and has essential roles in the maintenance of redox and protein homeostasis, mitochondrial biogenesis and the resolution of inflammation196–199.
Activation of this pathway can protect against damage by most types of oxidants and pro-inflammatory agents, and it restores redox and protein homeostasis200. The pathway has therefore attracted attention for the development of drugs for the prevention and treatment of complex diseases, including neurological conditions such as relapsing–remitting multiple sclerosis201 and autism spectrum disorder202.
Dimethyl fumarate (DMF), the methyl ester of the NP fumarate (a tricarboxylic acid (TCA) cycle intermediate that is found in both animals and plants), is one of the earliest discovered inducers of the KEAP1/NRF2 pathway203,204. The origins of the development of DMF as a drug date back to the use in traditional medicine of the plant Fumaria officinalis. Initially, fumaric acid derivatives were used for the treatment of psoriasis as it was thought that psoriasis is caused by a metabolic deficiency in the TCA cycle that could be compensated for by repletion of fumarate205. Despite this erroneous assumption, DMF is effective in treating psoriasis, both topically and orally, and is the active principle of Fumaderm, which has been used clinically for several decades in the treatment of plaque psoriasis in Germany. More recently, a DMF formulation developed by Biogen has been tested in other immunological disorders, with successful phase III trials in multiple sclerosis206,207 leading to its approval by the FDA and EMA in 2013.
The isothiocyanate sulforaphane, isolated from broccoli (Brassica oleracea)208, is among the most potent naturally occurring inducers of the KEAP1/NRF2 pathway209 and has protective effects in animal models of Parkinson210, Huntington211 and Alzheimer212 diseases, traumatic brain injury213, spinal cord contusion injury214, stroke215, depression216 and multiple sclerosis217. Sulforaphane-rich broccoli extract preparations are being developed as preventive interventions in areas of the world with unavoidable exposure to environmental pollutants, such as China; the initial results of a randomized clinical trial showed rapid and sustained, statistically significant increases in the levels of excretion of the glutathione-derived conjugates of benzene and acrolein218, and a follow-up trial (NCT02656420) also demonstrated dose–response-dependent benzene detoxification219. In a placebo-controlled, double-blind, randomized clinical trial in young individuals (age 13–27 years) with autism spectrum disorder, sulforaphane reversed many of the clinical abnormalities202; these encouraging findings led to a recently completed clinical trial in children (age 3–12 years) (NCT02561481; results of the trial are not yet publicly available). An α-cyclodextrin complex of sulforaphane known as SFX-01 (developed by Evgen Pharma) is being clinically studied for its potential to reverse resistance to endocrine therapies in patients with ER+HER2- metastatic breast cancer (phase II trial completed220) and in patients with subarachnoid haemorrhage (phase II trial NCT02614742 recently completed; results not yet publicly available). Currently, a clinical trial of SFX-01 in patients hospitalized with COVID-19 is in its final stages of preparation.
Finally, the pentacyclic triterpenoids bardoxolone methyl (also known as RTA 402) and omaveloxolone (RTA 408), which are semi-synthetic derivatives of the NP oleanolic acid, are the most potent (active at nanomolar concentrations) activators of the KEAP1/NRF2 pathway known to date221. These compounds have shown protective effects in numerous animal models of chronic disease222, and are currently in clinical trials for a wide range of indications, such as chronic kidney disease in type 2 diabetes, pulmonary arterial hypertension, melanoma, radiation dermatitis, ocular inflammation and Friedreich’s ataxia200. Most recently, bardoxolone methyl has entered a clinical trial in patients hospitalized with confirmed COVID-19 (NCT04494646).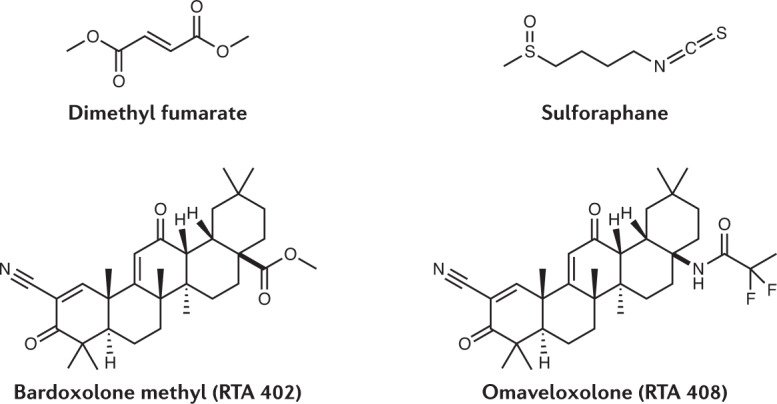
Application of analytical techniques
Classical NP-based drug research starts with biological screening of ‘crude’ extracts to identify a bioactive ‘hit’ extract, which is further fractionated to isolate the active NPs. Bioactivity-guided isolation is a laborious process with a number of limitations, but various strategies and technologies can be used to address some of them (Fig. 2). For example, to create libraries that are compatible with high-throughput screening, crude extracts can be pre-fractionated into sub-fractions that are more suitable for automated liquid handling systems. In addition, fractionation methods can be adjusted so that sub-fractions preferentially contain compounds with drug-like properties (typically moderate hydrophilicity). Such approaches can increase the number of hits compared with using crude extracts, as well as enabling more efficient follow-up of promising hits24.
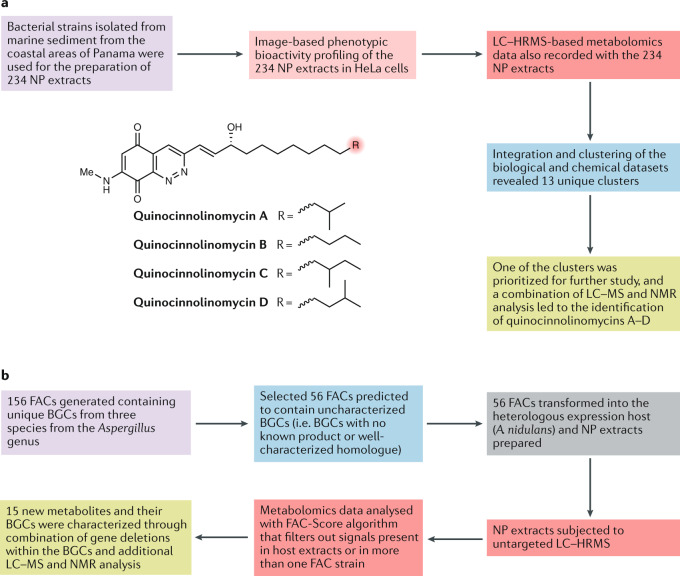
Fig. 2. Applications of advanced analytical technologies empowering modern natural product-based drug discovery.
a | An illustrative example of the application of liquid chromatography–high-resolution mass spectrometry (LC–HRMS) metabolomics in the screening of natural product (NP) extracts is the work of Kurita et al.58, in which 234 bacterial extracts were subjected to image-based phenotypic bioactivity screening and LC–HRMS metabolomics. Clustering of the resulting data allowed prioritization of promising extracts for further analysis, resulting in the discovery of the new NPs, quinocinnolinomycins A–D. b | Another illustrative example of LC–HRMS screening of NP extracts is the work of Clevenger et al.85, who obtained novel NP extracts through heterologous expression of fungal artificial chromosomes (FACs) containing uncharacterized biosynthetic gene clusters (BGCs) from diverse fungal species in Aspergillus nidulans. Analysis of the LC–HRMS metabolomics data with a FAC-Score algorithm directed the simultaneous discovery of 15 new NPs and the characterization of their BGCs.
Metabolomics was developed as an approach to simultaneously analyse multiple metabolites in biological samples. Enabled by technological developments in chromatography and spectrometry, metabolomics was historically applied first in other research fields, such as biomedical and agricultural sciences2. Advances in the analytical instrumentation used in NP research25,26, coupled with computational approaches that can generate plausible NP analogue structures and their respective simulated spectra27, have also enabled application of ‘omics’ approaches such as metabolomics in NP-based drug discovery. Metabolomics can provide accurate information on the metabolite composition in NP extracts, thus helping to prioritize NPs for isolation, to accelerate dereplication28,29 and to annotate unknown analogues and new NP scaffolds. Moreover, metabolomics can detect differences between metabolite compositions in various physiological states of producing organisms and enable the generation of hypotheses to explain them, and can also provide extensive metabolite profiles to underpin phenotypic characterization at the molecular level30. Both options are very useful in understanding the molecular mechanisms of action of NPs.
For metabolite profiling, NP extracts are analysed by NMR spectroscopy or high-resolution mass spectrometry (HRMS), or respective combined methods involving upstream liquid chromatography (LC)31,32, such as LC–HRMS, which can separate numerous isomers present in NP extracts33. Moreover, such combined methods might integrate HRMS and NMR, allowing the simultaneous use of the advantages of both techiques34,35. NMR analysis of NP extracts is simple and reproducible, and provides direct quantitative information and detailed structural information, although it has relatively low sensitivity, meaning that it generally enables profiling only of major constituents33. The applications of NMR in NP research are versatile36 and the technique is used both directly for metabolomics of unfractionated NP extracts and for structural characterization of compounds and fractions obtained with appropriate separation methods, most often LC. HRMS is the gold standard for qualitative and quantitative metabolite profiling33 and is most commonly applied in combination with LC. HRMS can also be used in the direct infusion mode (called DIMS)37, whereby samples are directly profiled by MS without a chromatography step, or in MS imaging (MSI)38, which enables determination of the spatial distribution of NPs within living organisms. HRMS enables routine acquisition of accurate molecular mass information, which together with appropriate heuristic filtering can provide unambiguous assignment of molecular formulae for hundreds to thousands of metabolites within a single extract over a dynamic range that may exceed five orders of magnitude31,39. However, challenges remain in data mining and in the unambiguous identification of the metabolites using various workflows relying on open web-based tools40.
Dereplication of secondary metabolites in bioactive extracts includes the determination of molecular mass and formula and cross-searching in the literature or structural NP databases with taxonomic information, which greatly assists the identification process. Such metadata, which are difficult to query in the literature, are often compiled in proprietary databases, such as the Dictionary of Natural Products, which encompasses all NP structures reported with links to their biological sources (see Related links). However, a comprehensive experimental tandem mass spectrometry (MS/MS) database of all NPs reported to date does not exist, and a search for experimental spectra across various platforms is hindered by the lack of standardized collision energy conditions for fragmentation in LC–MS/MS25.
In this respect, the Global Natural Products Social (GNPS) molecular networking platform developed in the Dorrestein laboratory is an important addition to the toolbox41. Molecular networking organizes thousands of sets of MS/MS data recorded from a given set of extracts and visualizes the relationship of the analytes as clusters of structurally related molecules. This improves the efficiency of dereplication by enabling annotation of isomers and analogues of a given metabolite in a cluster42. The recorded experimental spectra can be searched against putative structures and their corresponding predicted MS/MS spectra generated by tools such as competitive fragmentation modelling (CFM-ID)43. Based on such approaches, vast databases of theoretical NP spectra have been created and applied in dereplication44. The GNPS molecular networking approach has limitations, however, such as better applicability to some classes of NPs than others and the uncertainty of structural assignment among possible predicted candidates. Efforts to address such issues are ongoing45–47, including overlaying molecular networks of large NP extract libraries with taxonomic information to improve the confidence of annotation48. Overall, molecular networking mainly allows better prioritization of the isolation of unknown compounds by strengthening the dereplication process and elucidating relationships between NP analogues, and rigorous structure elucidation for NPs of interest should not be neglected.
Another useful platform for metabolite identification is METLIN49, which includes a high-resolution MS/MS database with a fragment similarity search function that is useful for identification of unknown compounds. Other databases and in silico tools such as Compound Structure Identification (CSI): FingerID and Input Output Kernel Regression (IOKR) can be used to search available fragment ion spectra, as well as to generate predicted spectra of fragment ions not present in current databases50. A novel computational platform for predicting the structural identity of metabolites derived from any identified compound has also been recently reported51, which should increase the searchable chemical space of NPs.
To accelerate the identification of bioactive NPs in extracts, metabolomics data can be matched to the biological activities of these extracts52. Various chemometric methods such as multivariate data analysis can correlate the measured activity with signals in the NMR and MS spectra, enabling the active compounds to be traced in complex mixtures with no need for further bioassays53–55. Furthermore, several analytical modules involving different bioassays and detection technologies can be linked to allow simultaneous bioactivity evaluation and identification of compounds present in small amounts (analytical scale) in complex compound mixtures34,35.
Metabolomics data can be integrated with data obtained by other omics techniques such as transcriptomics and proteomics and/or with imaging-based screens. For example, Acharya et al. used this approach to characterize NP-mediated interactions between a Micromonospora species and a Rhodococcus species56. In another interesting example, Kurita et al. developed a compound activity mapping platform for the prediction of identities and mechanisms of action of constituents from complex NP extract libraries by integrating cytological profiling57 with untargeted metabolomics data from a library of extracts58, and identified quinocinnolinomycins as a new family of NPs causing endoplasmic reticulum stress58 (Fig. 2a).
Analytical advances that enable the profiling of responses to bioactive molecules at the single-cell level can also accelerate NP-based drug discovery. Irish, Bachmann, Earl and colleagues developed a high-throughput platform for metabolomic profiling of bioactivity by integrating phospho-specific flow cytometry, single-cell chemical biology and cellular barcoding with metabolomic arrays (characterized chromatographic microtitre arrays originating from biological extracts)59. Using this platform, the authors studied the single-cell responses of bone marrow biopsy samples from patients with acute myeloid leukaemia following exposure to microbial metabolomic arrays obtained from extracts of biosynthetically prolific bacteria, which enabled the identification of new bioactive polyketides59.
Finally, advances in analytical technologies continue to support the rigorous structure determination of NPs of interest. The progressive development of higher-field NMR instruments and probe technology60,61 has enabled NP structure determination from very small quantities (below 10 µg)62,63, which is important, as the available quantities of NPs are often limited. In addition, microcrystal electron diffraction (MicroED) has recently emerged as a cryo-electron microscopy-based technique for unambiguous structure determination of small molecules64 and is already finding important applications in NP research65. The increased resolution and sensitivity of analytical equipment can also help address problems associated with ‘residual complexity’ of isolated NPs; that is when biologically potent but unidentified impurities in an isolated NP sample (which could include structurally related metabolites or conformers) lead to an incorrect assignment of structure and/or activity66,67. To avoid futile downstream development efforts, Pauli and colleagues recommended that lead NPs should undergo advanced purity analysis at an early stage using quantitative NMR and LC–MS67.
Genome mining and engineering
Advances in knowledge on biosynthetic pathways for NPs and in developing tools for analysing and manipulating genomes are further key drivers for modern NP-based drug discovery. Two key characteristics enable the identification of biosynthetic genes in the genomes of the producing organisms. First, these genes are clustered in the genomes of bacteria and filamentous fungi. Second, many NPs are based on polyketide or peptide cores, and their biosynthetic pathways involve enzymes — polyketide synthases (PKSs) and nonribosomal peptide synthetases (NRPSs), respectively — that are encoded by large genes with highly conserved modules68.
‘Genome mining’ is based on searches for genes that are likely to govern biosynthesis of scaffold structures, and can be used to identify NP biosynthetic gene clusters69–71. Prioritization of gene clusters for further work is facilitated by advances in biosynthetic knowledge and predictive bioinformatics tools, which can provide hints about whether the metabolic products of the clusters have chemical scaffolds that are new or known, thereby supporting dereplication72,73. Such predictive tools for gene cluster analysis can be applied in combination with spectroscopic techniques to accelerate the identification of NPs65 and determine the stereochemistry of metabolic products66. Furthermore, to extend genome mining from a single genome to entire genera, microbiomes or strain collections, computational tools have been developed, such as BiG-SCAPE, which enables sequence similarity analysis of biosynthetic gene clusters, and CORASON, which uses a phylogenomic approach to elucidate evolutionary relationships between gene clusters74.
Phylogenetic studies of known groups of talented secondary metabolite producers can also empower discovery of novel NPs. Recently, a study comparing secondary metabolite profiles and phylogenetic data in myxobacteria demonstrated a correlation between the taxonomic distance and the production of distinct secondary metabolite families75. In filamentous fungi, it was likewise shown that secondary metabolite profiles are closely correlated with their phylogeny76. These organisms are rich in secondary metabolites, as demonstrated by LC–MS studies of their extracts under laboratory conditions77. Concurrent genomic and phylogenomic analyses implied that even the genomes of well-studied organism groups harbour many gene clusters for secondary metabolite biosynthesis with as yet unknown functions78. The phylogeny of biosynthetic gene clusters, together with analysis of the absence of known resistance determinants, was recently used to prioritize members of the glycopeptide antibiotic family that could have novel activities. This led to the identification of the known antibiotic complestatin and the newly discovered corbomycin as compounds that act through a previously uncharacterized mechanism involving inhibition of peptidoglycan remodelling79.
Many microorganisms cannot be cultured, or tools for their genetic manipulation are not sufficiently developed, which makes it more challenging to access their NP-producing potential. However, biosynthetic gene clusters for NPs can be cloned and heterologously expressed in organisms that are well-characterized and easier to culture and to genetically manipulate (such as Streptomyces coelicolor, Escherichia coli and Saccharomyces cerevisiae)80. The aim is to achieve higher production titres in the heterologous hosts than in wild-type strains, improving the availability of lead compounds80–82. Vectors that can carry large DNA inserts are needed for the cloning of complete NP biosynthetic gene clusters. Cosmids (which can have inserts of 30–40 kb), fosmids (which can harbour 40–50 kb) and bacterial artificial chromosomes (BACs; which can have inserts of 100 kb to >300 kb) have been developed83. For fungal gene clusters, self-replicating fungal artificial chromosomes (FACs) have been developed, which can have inserts of >100 kb (ref.84). FACs in combination with metabolomic scoring were used to develop a scalable platform, FAC-MS, allowing the characterization of fungal biosynthetic gene clusters and their respective NPs at unprecedented scale85. The application of FAC-MS for the screening of 56 biosynthetic gene clusters from different fungal species yielded the discovery of 15 new metabolites, including a new macrolactone, valactamide A85 (Fig. 2b).
Even in culturable microorganisms, many biosynthetic gene clusters may not be expressed under conventional culture conditions, and these silent clusters could represent a large untapped source of NPs with drug-like properties86. Several approaches can be pursued to identify such NPs. One approach is sequencing, bioinformatic analysis and heterologous expression of silent biosynthetic gene clusters, which has already led to the discovery of several new NP scaffolds from cultivable strains87. Direct cloning and heterologous expression was also used to discover the new antibiotic taromycin A, which was identified upon the transfer of a silent 67 kb NRPS biosynthetic gene cluster from Saccharomonospora sp. CNQ-490 into S. coelicolor88. To transfer a biosynthetic gene cluster of such size, a platform based on transformation-associated recombination (TAR) cloning was developed. This platform enables direct cloning and manipulation of large biosynthetic gene clusters in S. cerevisiae, maintenance and manipulation of the vector in E. coli, and heterologous expression of the cloned gene clusters in Actinobacteria (such as S. coelicolor) following chromosomal integration88, and is an alternative to BACs for heterologous expression of large biosynthetic gene clusters.
Heterologous expression has limitations, such as the need to clone and manipulate very large genome regions occupied by biosynthetic gene clusters and the difficulty of identifying a suitable host that provides all conditions necessary for the production of the corresponding NPs. These limitations can be circumvented by activating biosynthetic gene clusters directly in the native microorganism through targeted genetic manipulations, generally involving the insertion of activating regulatory elements or deletion of inhibitory elements such as repressors or their binding sites. For example, a derepression strategy of deleting gbnR, a gene for a transcriptional repressor in Streptomyces venezuelae ATCC 10712 was used by Sidda et al. in the discovery of gaburedins, a family of γ-aminobutyrate-derived ureas89. An example of the activator-based strategy is the constitutive expression of the samR0484 gene in Streptomyces ambofaciens ATCC 23877, which led to the discovery of stambomycins A–D, 51-membered cytotoxic glycosylated macrolides72. Alternatively, silent biosynthetic gene clusters can be activated using repressor decoys90, which have the same DNA nucleotide sequence as the binding sites for the repressors that prevent the expression of the clusters. When these decoys are introduced into the bacteria, they sequester the respective repressors, and the ‘endogenous’ binding sites in the genome remain unoccupied, leading to derepression of the previously silent biosynthetic genes and production of the corresponding NPs. This approach has been applied to activate eight silent biosynthetic gene clusters in multiple streptomycetes and led to the characterization of a novel NP, oxazolepoxidomycin A90. The repressor decoy strategy is simpler, easier and faster to perform than the deletion of genes encoding regulatory factors. However, it has the same limitation as other approaches that rely on the introduction of recombinant DNA molecules into cells: it is necessary to develop protocols for efficient introduction of DNA into the targeted host strain, and the decoy must be maintained on a high-copy plasmid to ensure efficient repressor sequestration.
Another approach focused on exchange of regulatory elements is based on the CRISPR–Cas9 technology. The promise of this technique is exemplified in a recent work by Zhang et al., which demonstrated that CRISPR–Cas9-mediated targeted promoter introduction can efficiently activate diverse biosynthetic gene clusters in multiple Streptomyces species, leading to the production of unique metabolites, including a novel polyketide in Streptomyces viridochromogenes91. The CRISPR–Cas9 technology was also used to knock out genes encoding two well-known and frequently rediscovered antibiotics in several actinomycete strains, which led to the production of different rare and previously unknown variants of antibiotics that were otherwise obscured, including amicetin, thiolactomycin, phenanthroviridin and 5-chloro-3-formylindole92.
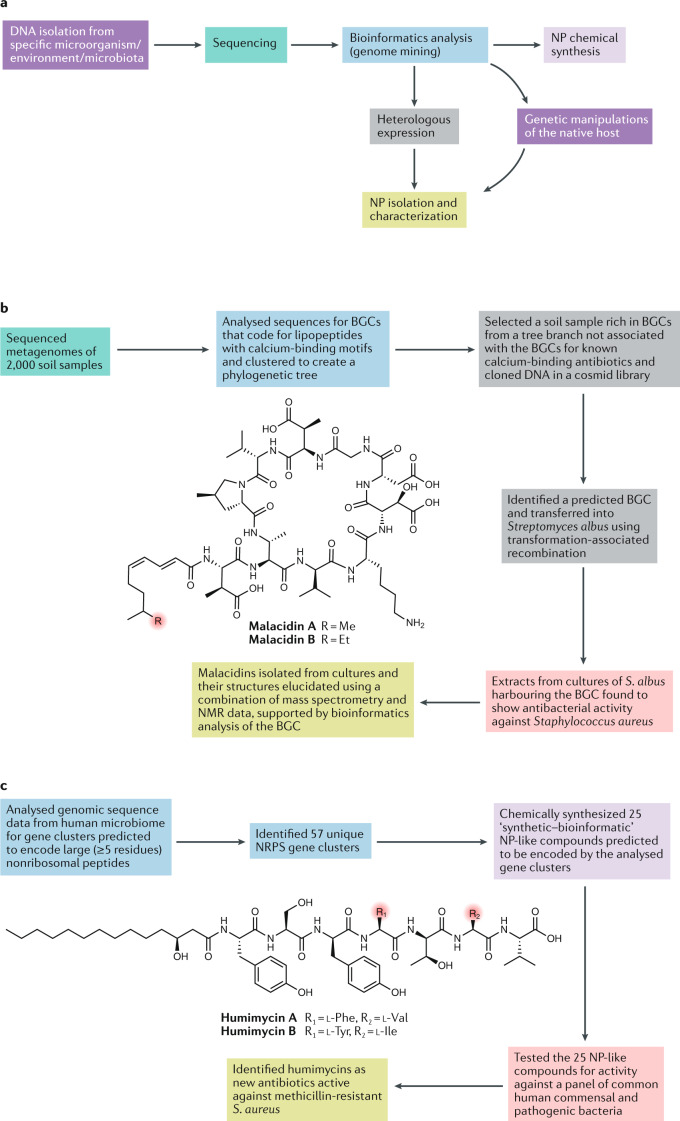
Approaches that rely on sequencing, bioinformatics and heterologous expression can also enable the identification of novel NPs from bacterial strains that have not yet been cultivated (Fig. 3a). For example, Hover et al. searched the metagenomes of 2,000 soil samples for biosynthetic gene clusters for lipopeptides with calcium-binding motifs. This led to the discovery of malacidins, members of the calcium-dependent antibiotic family, via heterologous expression of a 72 kb biosynthetic gene cluster from a desert soil sample in a Streptomyces albus host strain93 (Fig. 3b). However, in comparison with some of the other above-discussed strategies72,89,90, this metagenome-based discovery approach is more suited to finding new members of known NP classes rather than discovery of entirely new classes. In another study, Chu et al. developed a human microbiome-based approach that identified nonribosomal linear heptapeptides called humimycins as novel antibiotics active against methicillin-resistant Staphylococcus aureus (MRSA)94 (Fig. 3c). The structure of the NPs was predicted via bioinformatics analysis of gene clusters found in human commensal bacteria, followed by their chemical synthesis. A major strength of this innovative approach is that it is entirely independent of microbial cultivation and heterologous gene expression. Nevertheless, there are limitations related to the accuracy of computational chemical structure predictions and the feasibility of total chemical synthesis if structures are complex.
Fig. 3. Strategies for genome mining-driven discovery of natural products and natural product-like compounds.
a | Genome mining-based approaches to explore the biosynthetic capacity of microorganisms rely on DNA extraction, sequencing and bioinformatics analysis. The vast majority of microbes from different environments and microbiota communities have not been cultured, and their capacity to produce natural products (NPs) was largely inaccessible until recently. In the case of unculturable microorganisms, the bioinformatics analysis step can be followed by either targeted heterologous expression of biosynthetic gene clusters (BGCs) prioritized as being likely to yield relevant new NPs or direct chemical synthesis of ‘synthetic–bioinformatic’ NP-like compounds. b,c | These two approaches are exemplified by the recent discoveries of malacidins (panel b) and humimycins (panel c), respectively93,94. A major strength of the ‘synthetic–bioinformatic’ approach is that it is entirely independent of microbial culture and gene expression. Its limitations are the accuracy of computational chemical structure predictions and the feasibility of total chemical synthesis. NRPS, nonribosomal peptide synthetase.
The genomes of plants or animals can also be mined for novel NPs. For example, mining of 116 plant genomes enabled by identification of a precursor gene for the biosynthesis of lyciumins, a class of branched cyclic ribosomal peptides with hypotensive action produced by Lycium barbarum (popularly known as goji), identified diverse novel lyciumin chemotypes in seven other plants, including crops such as soybean, beet, quinoa and eggplant95. Genome mining in the animal kingdom is exemplified by the work of Dutertre et al., which used an integrated transcriptomics and proteomics approach to discover thousands of novel venom peptides from Conus marmoreus snails96. Proteomics analysis revealed that the vast majority of the conopeptide diversity was derived from a set of ~100 genes through variable peptide processing96.
Some bioactive compounds initially isolated from marine organisms might be products of symbionts, and genome mining can facilitate the characterization of such NPs. For example, it has been shown that bioactive compounds from the sponge Theonella swinhoei are produced by bacterial symbionts97, and characterization of the symbiont ‘Candidatus Entotheonella serta’ using single-cell genomics led to the discovery of gene clusters for misakinolide and theonellamide biosynthesis98. Another example of a marine NP produced by a bacterial symbiont is ET-743 (trabectedin), originally isolated from the tunicate Ecteinascidia turbinate. A meta-omics approach developed by Rath et al. revealed that the producer of this clinically used anticancer agent is the bacterial symbiont ‘Candidatus Endoecteinascidia frumentensis’99.
Similarly, plant microbiomes also represent a large reservoir for the identification of novel bioactive NPs (such as the antitumour agents maytansine, paclitaxel and camptothecin, which were initially isolated from plants and later shown to be produced by microbial endophytes)100 that can be tapped by genome mining approaches. An illustrative example is a recent work by Helfrich et al. that identified hundreds of novel biosynthetic gene clusters by genome mining of 224 bacterial strains isolated from Arabidopsis thaliana leaves101. A combination of bioactivity screening and imaging mass spectrometry was used to select a single species for further genomic analysis and led to the isolation of a NP with an unprecedented structure, the trans-acyltransferase PKS-derived antibiotic macrobrevin101.
Targeted genetic engineering of NP biosynthetic gene clusters can be of high value if the producing organism is difficult to cultivate or the yield of a NP is too low to allow comprehensive NP characterization. Rational genetic engineering and heterologous expression contributed to increase the production of vioprolides, a depsipeptide class of anticancer and antifungal NPs in the myxobacterium Cystobacter violaceus Cb vi35, by several orders of magnitude. In addition, non-natural vioprolide analogues were generated by this approach102. Similarly, promoter engineering and heterologous expression of biosynthetic gene clusters was reported to result in a 7-fold increase in the production of the cytotoxic NP disorazol103, and a 328-fold increase in the production of spinosad, an insecticidal macrolide produced by the bacterium Saccharopolyspora spinosa104.
Besides increasing NP yields, targeted gene manipulation can also be used to alter biosynthetic pathways in a predictable manner to produce new NP analogues with improved pharmacological properties, such as higher specific activity, lower toxicity and better pharmacokinetics. Such biosynthetic engineering approaches depend on a solid understanding of the biosynthetic pathway leading to a specific NP, access to the genes specifying this pathway and the ability to manipulate them in either the original or a heterologous host. Recent advances in biosynthetic engineering have enabled faster and more efficient production of NP analogues, including the development of methods for accelerated engineering and recombination of modules of PKS gene clusters105, NRPSs106,107 and NRPS–PKS assembly lines108, as well as elucidation of mechanisms for polyketide chain release that are contributing to NP structural diversification109,110. Examples of biosynthetic engineering applied to several important NPs include the generation of analogues of the immunosuppressant rapamycin111, the antitumour agents mithramycin112 and bleomycin113, and the antifungal agent nystatin114.
It should be noted that biosynthetic engineering has limitations regarding the parts of the NP molecule that can be targeted for modifications, and the chemical groups that can be introduced or removed. Considering the complexity of many NPs, however, total synthesis may be prohibitively costly, and a combined approach of biosynthetic engineering and chemical modification can provide a viable alternative for identifying improved drug candidates. For example, biosynthetic engineering may create a ‘handle’ for addition of a beneficial chemical group by synthetic chemistry, as demonstrated for the biosynthetically engineered analogues of nystatin mentioned above; further synthetic chemistry modifications resulted in compounds with improved in vivo pharmacotherapeutic characteristics compared with amphotericin B115,116.
Advances in microbial culturing systems
The complex regulation of NP biosynthesis in response to the environment means that the conditions under which producing organisms are cultivated can have a major impact on the chance of identifying novel NPs87. Several strategies have been developed to improve the likelihood of identifying novel NPs compared with monoculture under standard laboratory conditions and to make ‘uncultured’ microorganisms grow in a simulated natural environment117 (Fig. 4).
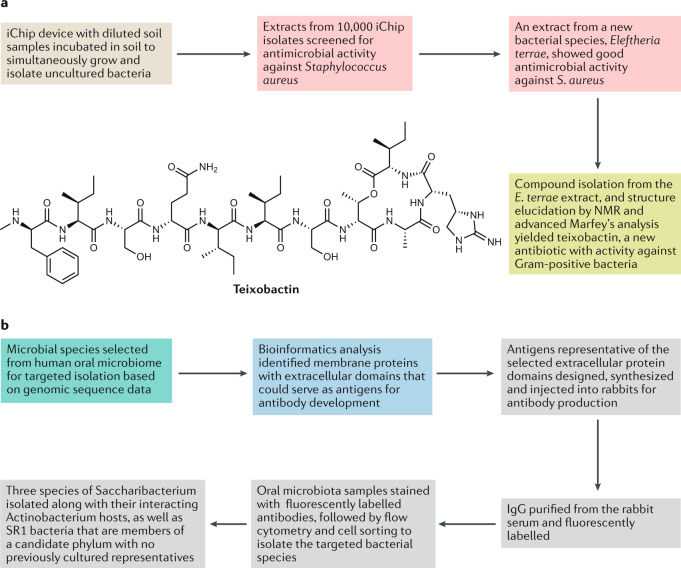
Fig. 4. Application of advanced microbial culturing approaches to identify new natural products.
New strategies for isolating previously uncultured microorganisms can enable access to new natural products (NPs) produced by them. a | To recapitulate the effect of complex signals coming from the native environment, microorganisms can be cultivated directly in the environment from which they were isolated. This concept is used with the iChip platform, in which diluted environmental samples are seeded in multiple small chambers separated from the native environment with a semipermeable membrane. The potential of this approach is illustrated by the recent discovery of teixobactin, a new antibiotic with activity against Gram-positive bacteria134,135. b | Another important recent development involves obtaining information from environmental samples using omics techniques such as metagenomics to identify and partially characterize microorganisms present in a specific environment before culturing. An approach relying on such preliminary information was recently used to engineer the capture of antibodies based on genetic information, which resulted in the successful cultivation of previously uncultured bacteria from the human mouth145. This reverse genomics workflow was validated by the isolation and cultivation of three species of Saccharibacteria (TM7) along with their interacting Actinobacteria hosts, as well as SR1 bacteria that are members of a candidate phylum with no previously cultured representatives.
One well-established approach to promote the identification of novel NPs is the modulation of culture conditions such as temperature, pH and nutrient sources. This strategy may lead to activation of silent gene clusters, thereby promoting production of different NPs. The term ‘One Strain Many Compounds’ (OSMAC) was coined for this approach about 20 years ago118, but the concept has a longer history119, with its use being routine in industrial microbiology since the 1960s120.
While OSMAC is still widely used for the identification of new bioactive compounds121,122, this approach has limited capacity to mimic the complexities of natural habitats. It is difficult to predict the combination of cues (which might also involve metabolites secreted by other members of the microbial community) to which the microorganism has evolved to respond by switching metabolic programmes. To account for such kinds of interactions, co-culturing using ‘helper’ strains can be applied123. This can enable the production and identification of new NPs, as illustrated by recent studies in which particular fungi were co-cultured with Streptomcyes species124,125.
Study of the molecular mechanisms underlying the ability of helper strains to increase the cultivability of previously uncultured microbes can lead to the identification of specific growth factors, allowing expansion of the number of species that can be successfully cultured. This strategy was used by D’Onofrio et al. for the identification of new acyl-desferrioxamine siderophores (iron-chelating compounds) as growth factors produced by helper strains promoting the growth of previously uncultured isolates from marine sediment biofilm117,126. The siderophore-assisted growth is based on the property of these compounds to provide iron for microbes unable to autonomously produce siderophores themselves, and the application of this approach led to the isolation of previously uncultivated microorganisms126. The development of strategies to cultivate microbial symbionts that produce NPs only upon interaction with their hosts can promote access to new NPs. Microbial symbionts interacting with insects or other organisms are a highly promising reservoir for the discovery of novel bioactive NPs produced in a unique ecological context127–130. To stimulate NP production, culturing strategies can be developed that better mimic the native environment of microbial symbionts of insects, including the use of media containing either lyophilized dead insects131 or l-proline, a major constituent of insect haemolymph132.
Strategies to mimic the natural environment even more closely by harnessing in situ incubation in the environment from which the microorganism is sampled have been developed, dating back to more than 20 years ago with the biotech companies OneCell and Diversa. They developed platforms that allowed the growth of some previously uncultivated microbes from various environments based on diluting out and suspension in a single drop of medium120,133. More recently, such strategies have been highlighted by the development and application of a platform dubbed the iChip, in which diluted soil samples are seeded in multiple small chambers separated from the environment with a semipermeable membrane134. After seeding, the iChip is placed back into the soil from which the sample was taken for an in situ incubation period, allowing the cultured microorganisms to be exposed to influences from their native environment. The power of this culturing approach was demonstrated by the discovery of a new antibiotic, teixobactin, produced by a previously uncultured soil bacterium135,136 (Fig. 4a). This platform may be of great significance for NP drug discovery, given that it has been estimated that only 1% of soil organisms have so far been successfully cultured using traditional culturing techniques137.
The omics strategies discussed in previous sections can complement efforts to explore NPs produced upon microbial interactions. The application of such a strategy is illustrated in the work of Derewacz et al., who analysed the metabolome of a genome-sequenced Nocardiopsis bacterium upon co-culture with bacteria of the genera Escherichia, Bacillus, Tsukamurella and Rhodococcus138. Around 14% of the metabolomic features found in co-cultures were undetectable in monocultures, with many of those being unique to specific co-culture genera, and the previously unreported polyketides ciromicin A and B, which possess an unusual pyrrolidinol substructure and displayed moderate and selective cytotoxicity, were identified138. Other examples include a ‘culturomics’ approach that combines multiple culture conditions with MS profiling and 16S rRNA-based taxonomy to identify prokaryotic species from the human gut139, and an ultrahigh-throughput screening platform based on microfluidic droplet single-cell encapsulation and cultivation followed by next-generation sequencing and LC–MS, which allows investigation of pairwise interactions between target microorganisms140. The latter approach enabled identification of a slow-growing oral microbiota species that inhibits the growth of S. aureus140.
Historically early-adopted microbial culturing approaches led to a bias reflected in the predominant discovery of NPs from microorganisms that are easy to cultivate (such as streptomycetes and some common filamentous fungi). As a result, a vast number of NPs from such ‘easy to culture’ microbes have already been characterized, and conventional screening efforts tend to yield disappointing returns associated with frequent rediscovery of known NPs and their closely related congeners. Therefore, culturing strategies aimed at previously unexplored (or under-investigated) microbial groups, with the potential to produce NPs with entirely new scaffolds and bioactivities (such as Burkholderia, Clostridium and Xenorhabdus) are of high interest141,142. Closthioamide, the first secondary metabolite from a strictly anaerobic bacterium, was discovered from Clostridium cellulolyticum by this approach143. Targeted isolation of such species is important, and a genome-guided approach to achieve this goal has recently been demonstrated for Burkholderia strains in environmental samples144. Another highly innovative approach to the isolation and cultivation of previously uncultured bacteria was recently reported by Cross et al.145, who used genomic information to engineer antibodies predicted to target selected microorganisms and to specifically capture these microorganisms from complex communities and to isolate them in pure cultures. This approach was validated by isolation and cultivation of previously uncultured bacteria from the human oral cavity145 (Fig. 4b), and it could be applicable to a wide range of target organisms if suitable cultivation conditions can be identified for the isolated cells.
Despite these advances in culturing strategies, artificial conditions still do not fully represent the complex environment of natural habitats. To circumvent this problem, microbial and NP diversity can also be accessed via extraction of organisms and/or their NPs in situ. To directly gain compounds produced in the natural marine environment (which may be missed otherwise), resin capture technology can be used to capture compounds on inert sorbent supports ready to be desorbed, analysed and tested for biological activity146. Sustainable approaches for in situ extraction with green solvents, such as glycerol or natural deep eutectic and ionic solvents (NADES), could be used directly during field work147,148. To improve dereplication, analytical equipment miniaturization is also facilitating in situ analysis; examples include the introduction of devices for physicochemical data analysis, such as micro-MS and portable near infrared spectroscopy149,150.
Outlook for NPs in drug discovery
The technological advances discussed above have the potential to reinvigorate NP-based drug discovery in both established and emerging areas. NPs have long been the key source of new drugs against infectious diseases, especially antibiotics (reviewed elsewhere151,152). Selected NPs with antimicrobial properties discovered by leveraging advances discussed in the sections above, including strategies to exploit the human microbiome for novel NPs94,153 are highlighted in Figs 3,4. Along with the search for new NPs with antimicrobial activities, researchers are continuing to develop and optimize already known NP classes, making use of advances in biosynthetic engineering154, total synthesis155 or semi-synthetic strategies156,157. In addition, antivirulence strategies could represent an alternative approach to fighting infections158, for which NPs targeting bacterial quorum sensing could be of interest159.
NPs also have a successful history as cancer therapeutics, which has been well covered in other reviews160–163. An important new opportunity in this field is the capacity of some NPs to trigger a selective yet potent host immune reaction against cancer cells, particularly given the intense interest at present in strategies that could improve response rates to immune checkpoint inhibitors by turning ‘cold’ tumours ‘hot’164. For example, NPs such as cardiac glycosides165 can increase the immunogenicity of stressed and dying cancer cells by triggering immunogenic cell death, characterized by the release of damage-associated molecular patterns (DAMPs), which could open new avenues for drug discovery or repurposing166–168.
Botanical therapies containing complex mixtures of NPs have long attracted interest owing to the potential for synergistic therapeutic effects of components within the mixture169,170. However, the variability of the NP composition in the starting plant material owing to factors such as environmental variations in the location at which the plants were collected is a major challenge for the development of botanical drugs1. With the advances in technology for their characterization, such as metabolomics discussed above, as well as development of regulatory guidance for complex mixtures of NPs (see Related links), it is becoming more feasible to develop such mixtures as therapeutics, rather than to identify and purify a single active ingredient171.
Since gut microbiota are considered to play a major role in health and disease172–174, and NPs are known to affect the gut microbiome composition175–178, this area is an emerging opportunity for NP-based drug discovery. However, drug discovery efforts in this area are still in their infancy, with many open questions remaining179. A future direction may be the characterization of single microbiota-derived species for particular therapeutic applications, and the advances in culturing strategies, genome mining and analytics discussed above will be of great importance in this respect.
Many advances discussed above are supported by computational tools including databases (such as genomic, chemical or spectral analysis data; see ref.180 for a recent review on NP databases) and tools that enable the analysis of genetic information, the prediction of chemical structures and pharmacological activities181, the integration of data sets with diverse information (such as tools for multi-omics analysis)182 and machine learning applications183.
Although this Review focuses on technologies that enable the discovery of novel NPs, it is important to acknowledge that unmodified NPs may possess suboptimal efficacy or absorption, distribution, metabolism, excretion and toxicity (ADMET) properties. So, for development of NP hits into leads and ultimately into successful drugs, chemical modification may be required. In addition, bringing a compound into clinical development requires a sustainable and economically viable supply of sufficient quantities of the compound. Total chemical synthesis, semi-synthesis using a NP as a starting point for analogue generation and biosynthetic engineering modifying biosynthetic pathways of the producing organism will be of great importance in this context (Fig. 5). Recent advances in chemical synthesis and biosynthetic engineering technologies are strongly empowering NP-based drug discovery and development by enabling property optimization of complex NP scaffolds that were previously regarded as inaccessible. This allows the enrichment of screening libraries with NPs, NP hybrids, NP analogues and NP-inspired molecules, as well as superior structure functionalization approaches (including late-stage functionalization) for optimization of NP leads94,105–108,184–188.
Fig. 5. Strategies to obtain natural product analogues with superior properties.
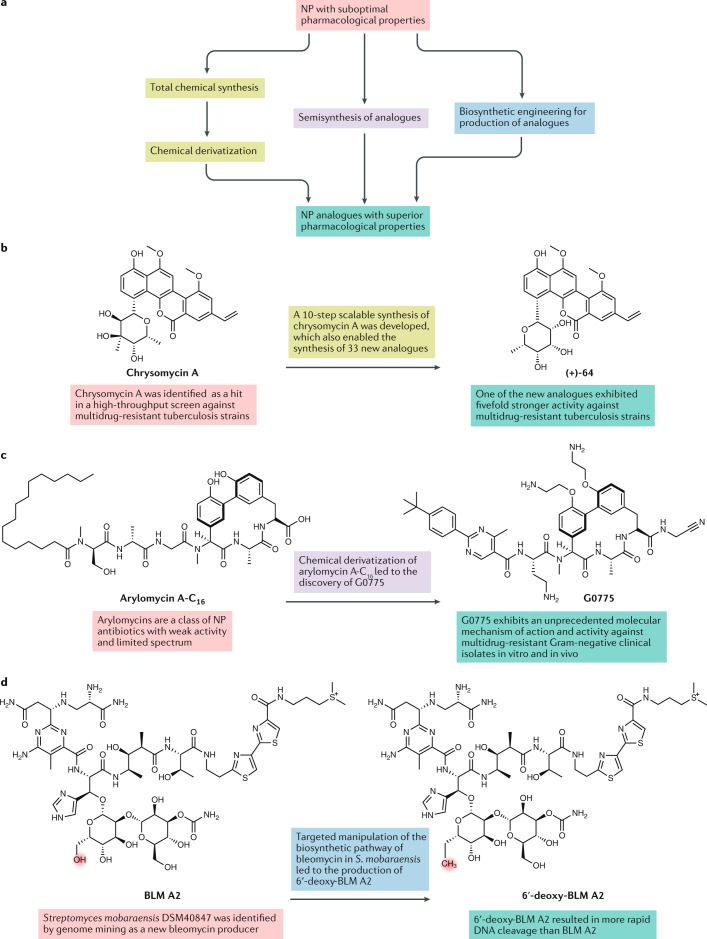
Unmodified natural products (NPs) often possess suboptimal properties, and superior analogues need to be obtained in order to yield valuable new drugs. a | NP analogues can be accessed through the development of total chemical synthesis followed by chemical derivatization, through semisynthesis using a NP as a starting point for the introduction of chemical modifications, and through biosynthetic engineering using manipulations of biosynthetic pathways of the producing organism to generate NP analogues. b,c | Tetracyclines are an example of NP-derived antibiotics that have already yielded several generations of successfully marketed semisynthetic and synthetic derivatives. The first generation of tetracyclines (such as chlortetracycline and tetracycline) were unmodified NPs, while the two subsequent generations of analogues with optimized properties were semisynthetic (second-generation, doxycycline, minocycline; third-generation, tigecycline) and the most recently developed fourth-generation analogues (eravacycline) are entirely synthetic, accessed via total synthesis193,194. More recent examples of property optimization of other classes of NPs through total chemical synthesis followed by chemical derivatization or through semisynthesis are illustrated by studies focused on analogues of chrysomycin A (panel b)195 and arylomycins (panel c)157, respectively. d | The biosynthetic engineering approach has also shown potential; for example, in the generation of analogues of rapamycin111, bleomycin113 (panel d) and nystatin114. 6′-deoxy-BLM A2, 6′-deoxy-bleomycin A2; BLM A2, bleomycin A2.
Finally, although NP-based drug discovery offers a unique niche for diverse forms of academia–industry collaboration, a key challenge is that scientific and technological expertise is often scattered over many academic institutions and companies. Focused efforts are needed to support translational NP research in academia, which has become more difficult in recent years given the decline in the number of large companies actively engaged in NP research. A conventional solution to improve academia–industry interaction is to focus the relevant expertise under one umbrella and in close spatial proximity. For example, the Phytovalley Tirol, centred in Innsbruck, Austria, brings together several research institutions and companies (among others, the Austrian Drug Screening Institute (ADSI), the Michael Popp Research Institute for New Phyto-Entities, Bionorica Research and Biocrates Life Sciences AG) with the aim of accelerating NP-based drug discovery. Another solution could be virtual consortia, such as the International Natural Product Sciences Taskforce (INPST) that we have recently established (see Related links), which provides a platform for integration of expertise, technology and materials from the participating academic and industrial entities.
In conclusion, NPs remain a promising pool for the discovery of scaffolds with high structural diversity and various bioactivities that can be directly developed or used as starting points for optimization into novel drugs. While drug development overall continues to be challenged by high attrition rates, there are additional hurdles for NPs due to issues such as accessibility, sustainable supply and IP constraints. However, we believe that the scientific and technological advances discussed in this Review provide a strong basis for NP-based drug discovery to continue making major contributions to human health and longevity.
Acknowledgements
This paper is affectionately dedicated in memory of Dr Mariola Macías (1984–2020) M.D., Ph.D. in Immunology, Emergency Physician at Hospital Punta Europa, Algeciras (Cadiz), Spain and active member of a research team working against SARS-CoV-2. An excellent professional and a better person. Her humanity, kindness, special and unmistakable smile, generosity, dedication and professionalism will never be forgotten. The authors are grateful to P. Kirkpatrick for his editorial contribution, which resulted in a greatly improved manuscript. A.G.A. acknowledges support from the Austrian Science Fund (FWF) project P25971-B23 (‘Improved cholesterol efflux by natural products’). R.B. acknowledges support by a grant from the Austrian Science Fund (FWF) P27505. V.B. acknowledges support by a grant from the Austrian Science Fund (FWF) P27682-B30. N.B. is recipient of an Australian Research Council DECRA Fellowship. A.C. and E.I. thank the Ministerio de Ciencia, Innovación y Universidades, Spain (Project AGL2017-89417-R) for support. M. Diederich is supported by the National Research Foundation (NRF) (grant number 019R1A2C1009231), by a grant from the MEST of Korea for Tumour Microenvironment Global Core Research Center (GCRC) (grant number NRF-2011-0030001), by the Creative-Pioneering Researchers Program through Seoul National University (Funding number: 370C-20160062), by the Brain Korea 21 (BK21) PLUS programme, by the ‘Recherche Cancer et Sang’ foundation, by the ‘Recherches Scientifiques Luxembourg’ association, by the ‘Een Häerz fir kriibskrank Kanner’ association, by the Action LIONS ‘Vaincre le Cancer’ association and by Télévie Luxembourg. The research work of A.T.D.-K. is funded by Cancer Research UK (C20953/A18644), the Biotechnology and Biological Sciences Research Council (BB/L01923X/1), Reata Pharmaceuticals, and Tenovus Scotland (T17/T14). B.L.F. acknowledges BMBF (TUNGER 036/FUCOFOOD) and AIF (AGEsense) for supporting his research. M.I.G. acknowledges financial support from the European Union’s Horizon 2020 research and innovation programme, project PlantaSYST (SGA No 739582 under FPA No. 664620) and the BG05M2OP001-1.003-001-C01 project, financed by the European Regional Development Fund through the ‘Science and Education for Smart Growth’ Operational Programme. K.M.G. is supported by the UK Medical Research Council (MC_UU_12011/4), the National Institute for Health Research (NIHR Senior Investigator (NF-SI-0515-10042) and the NIHR Southampton Biomedical Research Centre), the European Union (Erasmus+ Capacity-Building ENeA SEA Project and Seventh Framework Programme (FP7/2007-2013), projects EarlyNutrition and ODIN (grant agreements 289346 and 613977), the US National Institute On Ageing of the National Institutes of Health (award no. U24AG047867) and the UK ESRC and BBSRC (award no. ES/M00919X/1). Research in the laboratory of C.W.G. is supported by the Austrian Science Fund (FWF) through project P32109 and a NATVANTAGE grant 2019 by the Wilhelm Doerenkamp-Stiftung. A.K. acknowledges support by national funds through FCT-Foundation for Science and Technology of Portugal within the scope of UIDB/04423/2020 and UIDP/04423/2020. A.L. acknowledges HKBU SDF16-0603-P02 for supporting this research. F.A.M. acknowledges the support by Ministerio de Economia y Competitividad, Spain (project AGL2017-88083-R). A.M. acknowledges the support by a grant of the Romanian Ministry of Research and Innovation, CNCS – UEFISCDI, project number PN-III-P1-1.1-PD-2016-1900 – ‘PhytoSal’, within PNCDI III. G.P. acknowledges the support by NIH G12-MD007591, Kleberg Foundation and NIH R01-AG066749. M.R. acknowledges support by the Swiss National Science Foundation (Schweizerischer Nationalfonds, SNF), and by the Horizon 2020 programme of the European Union. J.M.R. acknowledges the support from the Austrian Science Fund (FWF: P24587), the Natvantage grant 2018 and the University of Vienna, Austria. G.L.R. acknowledges the group of Cellular and Molecular Nutrition (BJ-Lab) at the Institute of Food Sciences, National Research Council, Avellino, Italy. A.S.S. acknowledges the support by UIDB/00211/2020 with funding from FCT/MCTES through national funds. D.S. acknowledges the support by FWF S10711. D.S. is an Ingeborg Hochmair Professor at the University of Innsbruck. K.S.W. is supported by the National Centre for Research and Development (4/POLTUR-1/2016) and the National Science Centre (2017/27/B/NZ4/00917) and Medical University of Lublin, Poland. E.S.S. thanks Universidad Central de Chile, through Dirección de Investigación y Postgrado, for supporting this research. H. Stuppner acknowledges support by the Austrian Research Promotion Agency (FFG), the Austrian Science Fund (FWF) and the Horizon 2020 programme of the European Union (RISE, 691158). A.S. was granted by Instituto de Salud Carlos III, CIBEROBN (CB12/03/30038) and EU-COST Action (CA16112). M.W. acknowledges the support by DFG, BMBF, EU, CSC, DAAD, AvH and Land Baden Württemberg. J.L.W. is grateful to the Swiss National Science Foundation (SNF) for supporting its natural product metabolomics projects (grants nos. 310030E-164289, 31003A_163424 and 316030_164095). S.B.Z. acknowledges the support by University of Vienna, Vienna, Austria. M.H. acknowledges an EPSRC CASE Award (with Pukka Herbs Ltd, UK as industrial partner). I.B.-N. acknowledges the support of Competitivity Operational Program, 2014–2020, entitled ‘Clinical and economical impact of personalized targeted anti-microRNA therapies in reconverting lung cancer chemoresistance’ — CANTEMIR, No. 35/01.09.2016, MySMIS 103375; project PNCDI III 2015-2020 entitled ‘Increasing the performance of scientific research and technology transfer in translational medicine through the formation of a new generation of young researchers’ — ECHITAS, no. 29PFE/18.10.2018. This work was also funded by the Italian Ministry for University and Research (MIUR), grant PRIN: rot. 2017XYBP2R (to C.T.S).
Glossary
- sp3 carbon atoms
Tetravalent carbon atoms forming single covalent bonds with other atoms within the molecular structure. A higher fraction of sp3 carbons within molecules is a descriptor that indicates more complex 3D structures.
- Lipinski’s rule of five
This guideline for the likelihood of a compound having oral bioavailability is based on several characteristics containing the number 5. It predicts that a molecule is likely to have poor absorption or permeation if it has more than one of the following characteristics: there are >5 H-bond donors and >10 H-bond acceptors; the molecular weight is >500; or the partition coefficient LogP is >5. Notably, natural products were identified as common exceptions at the time of publication in 1997.
- Dereplication
Pharmacological screening of natural product extracts yields hits potentially containing multiple natural products that need to be considered for further study to identify the bioactive compounds. Dereplication is the process of recognizing and excluding from further study such hit mixtures that contain already known bioactive compounds.
- Phenotypic assays
Assays that rely on the ability of tested compounds to exert desired phenotypic changes in cells, isolated tissues, organs or animals. They offer a complementary strategy to target-based assays for identifying new potential drugs.
- Phylogenomic approach
The use of genomic data to reveal evolutionary relationships. In the context of natural product drug discovery, the use of phylogenomics is based on the assumption that organisms that have closer evolutionary relationships are more likely to produce similar natural products.
- Taxonomic distance
The distance of compared taxa on a constructed phylogenetic tree (also known as an evolutionary tree). Closer distance of compared taxa indicates a closer evolutionary relationship.
Competing interests
A.G.A. is executive administrator of the International Natural Product Sciences Taskforce (INPST) and Digital Health and Patient Safety Platform (DHPSP). M. Banach has served on the speakers’ bureau of Abbott/Mylan, Abbott Vascular, Actavis, Akcea, Amgen, Biofarm, KRKA, MSD, Novo-Nordisk, Novartis, Sanofi-Aventis, Servier and Valeant, has served as a consultant to Abbott Vascular, Akcea, Amgen, Daichii Sankyo, Esperion, Freia Pharmaceuticals, Lilly, MSD, Novartis, Polfarmex, Resverlogix, Sanofi-Aventis, and has received grants from Amgen, Mylan, Sanofi and Valeant. R.B. collaborates with Bayer Consumer Health and Dr Willmar Schwabe GmbH & Co. KG, and is scientific advisory committee member of PuraPharm International (HK) Limited and ISURA. G.K.B. is a board member of Bionorica SE. M. Daglia has received consultancy honoraria from Pfizer Italia and Mylan for training courses for chemists, and is a member of the INPST board of directors. A.T.D.-K. is a member of the Scientific and Medical Advisory Board of Evgen Pharma plc. I.E.O. is Dean of Faculty of Pharmacy, Gazi University, Ankara, Turkey, member of the Traditional Chinese Medicine Experts Group in European Pharmacopeia, and principal member of Turkish Academy of Sciences (TUBA). B.L.F. is a member of the INPST Board of Directors and has received research funding from Dr Willmar Schwabe GmbH & Co. KG. K.M.G. has received reimbursement for speaking at conferences sponsored by companies selling nutritional products and is part of an academic consortium that has received research funding from Abbott Nutrition, Nestec and Danone. C.W.G. is chairman of the scientific advisory board of Cyxone AB, SE. M.H.’s research group has received charitable donations from Dr Willmar Schwabe GmbH & Co. KG and recently completed a research project sponsored by Pukka Herbs, UK. A.L. is a member of the board of directors of Kaisa Health. M.J.S.M. is president of Kaiviti Consulting and consults for Gnosis by LeSaffre. F.N. is cofounder and shareholder of OncoNox and Aura Biopharm. G.P. is on the board of Neurotez and Neurotrope. M.R. serves as an adviser for the Nestlé Institute of Health Sciences. G.L.R. is a member of the board of directors of INPST. N.T.T. is Founder and CEO of NTZ Lab Ltd and advisory board member of INPST. M.W. collaborates with Finzelberg GmbH and Schwabe GmbH. J.L.W. collaborates with Nestlé and Firmenich. M.A.P. is CEO and owner of Bionorica SE. J.H. is an employee of and holds shares in UCB Pharma Ltd. M.M. is Founder and Chairman of Sami–Sabinsa Group of Companies. D.S.B. is an employee of Janssen R&D. M. Bodkin is an employee of Evotec (UK) Ltd.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Dictionary of Natural Products: http://dnp.chemnetbase.com/faces/chemical/ChemicalSearch.xhtml
FDA botanical drug development guidance for industry: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/botanical-drug-development-guidance-industry
INPST: https://inpst.net/
A full list of members and affiliations is presented at the end of the paper.
Contributor Information
Atanas G. Atanasov, Email: a.atanasov.mailbox@gmail.com
Claudiu T. Supuran, Email: claudiu.supuran@unifi.it
the International Natural Product Sciences Taskforce:
Ilkay Erdogan Orhan, Maciej Banach, Judith M. Rollinger, Davide Barreca, Wolfram Weckwerth, Rudolf Bauer, Edward A. Bayer, Muhammed Majeed, Anupam Bishayee, Valery Bochkov, Günther K. Bonn, Nady Braidy, Franz Bucar, Alejandro Cifuentes, Grazia D’Onofrio, Michael Bodkin, Marc Diederich, Albena T. Dinkova-Kostova, Thomas Efferth, Khalid El Bairi, Nicolas Arkells, Tai-Ping Fan, Bernd L. Fiebich, Michael Freissmuth, Milen I. Georgiev, Simon Gibbons, Keith M. Godfrey, Christian W. Gruber, Jag Heer, Lukas A. Huber, Elena Ibanez, Anake Kijjoa, Anna K. Kiss, Aiping Lu, Francisco A. Macias, Mark J. S. Miller, Andrei Mocan, Rolf Müller, Ferdinando Nicoletti, George Perry, Valeria Pittalà, Luca Rastrelli, Michael Ristow, Gian Luigi Russo, Ana Sanches Silva, Daniela Schuster, Helen Sheridan, Krystyna Skalicka-Woźniak, Leandros Skaltsounis, Eduardo Sobarzo-Sánchez, David S. Bredt, Hermann Stuppner, Antoni Sureda, Nikolay T. Tzvetkov, Rosa Anna Vacca, Bharat B. Aggarwal, Maurizio Battino, Francesca Giampieri, Michael Wink, Jean-Luc Wolfender, Jianbo Xiao, Andy Wai Kan Yeung, Gérard Lizard, Michael A. Popp, Michael Heinrich, Ioana Berindan-Neagoe, Marc Stadler, Maria Daglia, and Robert Verpoorte
References
- 1.Atanasov AG, et al. Discovery and resupply of pharmacologically active plant-derived natural products: a review. Biotechnol. Adv. 2015;33:1582–1614. doi: 10.1016/j.biotechadv.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015;14:111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 3.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 4.Waltenberger B, Mocan A, Šmejkal K, Heiss EHEH, Atanasov AAGAG. Natural products to counteract the epidemic of cardiovascular and metabolic disorders. Molecules. 2016;21:807. doi: 10.3390/molecules21060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tintore M, Vidal-Jordana A, Sastre-Garriga J. Treatment of multiple sclerosis — success from bench to bedside. Nat. Rev. Neurol. 2019;15:53–58. doi: 10.1038/s41582-018-0082-z. [DOI] [PubMed] [Google Scholar]
- 6.Feher M, Schmidt JM. Property distributions: differences between drugs, natural products, and molecules from combinatorial chemistry. J. Chem. Inf. Comput. Sci. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- 7.Barnes EC, Kumar R, Davis RA. The use of isolated natural products as scaffolds for the generation of chemically diverse screening libraries for drug discovery. Nat. Prod. Rep. 2016;33:372–381. doi: 10.1039/C5NP00121H. [DOI] [PubMed] [Google Scholar]
- 8.Li JW-H, Vederas JC. Drug discovery and natural products: end of an era or an endless frontier? Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 9.Clardy J, Walsh C. Lessons from natural molecules. Nature. 2004;432:829–837. doi: 10.1038/nature03194. [DOI] [PubMed] [Google Scholar]
- 10.Lawson ADG, MacCoss M, Heer JP. Importance of rigidity in designing small molecule drugs to tackle protein–protein interactions (PPIs) through stabilization of desired conformers. J. Med. Chem. 2018;61:4283–4289. doi: 10.1021/acs.jmedchem.7b01120. [DOI] [PubMed] [Google Scholar]
- 11.Doak BC, Over B, Giordanetto F, Kihlberg J. Oral druggable space beyond the rule of 5: insights from drugs and clinical candidates. Chem. Biol. 2014;21:1115–1142. doi: 10.1016/j.chembiol.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Shultz MD. Two decades under the influence of the rule of five and the changing properties of approved oral drugs. J. Med. Chem. 2019;62:1701–1714. doi: 10.1021/acs.jmedchem.8b00686. [DOI] [PubMed] [Google Scholar]
- 13.Lachance H, Wetzel S, Kumar K, Waldmann H. Charting, navigating, and populating natural product chemical space for drug discovery. J. Med. Chem. 2012;55:5989–6001. doi: 10.1021/jm300288g. [DOI] [PubMed] [Google Scholar]
- 14.Henrich CJ, Beutler JA. Matching the power of high throughput screening to the chemical diversity of natural products. Nat. Prod. Rep. 2013;30:1284. doi: 10.1039/c3np70052f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cragg GM, Schepartz SA, Suffness M, Grever MR. The taxol supply crisis. New NCI policies for handling the large-scale production of novel natural product anticancer and anti-HIV agents. J. Nat. Prod. 1993;56:1657–1668. doi: 10.1021/np50100a001. [DOI] [PubMed] [Google Scholar]
- 16.Harrison C. Patenting natural products just got harder. Nat. Biotechnol. 2014;32:403–404. doi: 10.1038/nbt0514-403a. [DOI] [PubMed] [Google Scholar]
- 17.Burton G, Evans-Illidge EA. Emerging R and D law: the Nagoya Protocol and its implications for researchers. ACS Chem. Biol. 2014;9:588–591. doi: 10.1021/cb500045t. [DOI] [PubMed] [Google Scholar]
- 18.Heffernan O. Why a landmark treaty to stop ocean biopiracy could stymie research. Nature. 2020;580:20–22. doi: 10.1038/d41586-020-00912-w. [DOI] [PubMed] [Google Scholar]
- 19.Corson TW, Crews CM. Molecular understanding and modern application of traditional medicines: triumphs and trials. Cell. 2007;130:769–774. doi: 10.1016/j.cell.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moffat JG, Vincent F, Lee JA, Eder J, Prunotto M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 2017;16:531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- 21.Shi Y, Inoue H, Wu JC, Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 2017;16:115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fellmann C, Gowen BG, Lin P-C, Doudna JA, Corn JE. Cornerstones of CRISPR–Cas in drug discovery and therapy. Nat. Rev. Drug Discov. 2017;16:89–100. doi: 10.1038/nrd.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schirle M, Jenkins JL. Identifying compound efficacy targets in phenotypic drug discovery. Drug Discov. Today. 2016;21:82–89. doi: 10.1016/j.drudis.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Wagenaar MM. Pre-fractionated microbial samples-the second generation natural products library at Wyeth. Molecules. 2008;13:1406–1426. doi: 10.3390/molecules13061406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolfender J-L, Nuzillard J-M, van der Hooft JJJ, Renault J-H, Bertrand S. Accelerating metabolite identification in natural product research: toward an ideal combination of liquid chromatography–high-resolution tandem mass spectrometry and nmr profiling, in silico databases, and chemometrics. Anal. Chem. 2019;91:704–742. doi: 10.1021/acs.analchem.8b05112. [DOI] [PubMed] [Google Scholar]
- 26.Stuart KA, Welsh K, Walker MC, Edrada-Ebel RA. Metabolomic tools used in marine natural product drug discovery. Expert Opin. Drug Discov. 2020;15:499–522. doi: 10.1080/17460441.2020.1722636. [DOI] [PubMed] [Google Scholar]
- 27.Allard P-M, Genta-Jouve G, Wolfender J-L. Deep metabolome annotation in natural products research: towards a virtuous cycle in metabolite identification. Curr. Opin. Chem. Biol. 2017;36:40–49. doi: 10.1016/j.cbpa.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Allard P-M, et al. Pharmacognosy in the digital era: shifting to contextualized metabolomics. Curr. Opin. Biotechnol. 2018;54:57–64. doi: 10.1016/j.copbio.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubert J, Nuzillard J-M, Renault J-H. Dereplication strategies in natural product research: How many tools and methodologies behind the same concept? Phytochem. Rev. 2017;16:55–95. doi: 10.1007/s11101-015-9448-7. [DOI] [Google Scholar]
- 30.Liu X, Locasale JW. Metabolomics: a primer. Trends Biochem. Sci. 2017;42:274–284. doi: 10.1016/j.tibs.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eugster PJ, et al. Ultra high pressure liquid chromatography for crude plant extract profiling. J. AOAC Int. 2011;94:51–70. doi: 10.1093/jaoac/94.1.51. [DOI] [PubMed] [Google Scholar]
- 32.Stavrianidi A. A classification of liquid chromatography mass spectrometry techniques for evaluation of chemical composition and quality control of traditional medicines. J. Chromatogr. A. 2020;1609:460501. doi: 10.1016/j.chroma.2019.460501. [DOI] [PubMed] [Google Scholar]
- 33.Wolfender J-L, Marti G, Thomas A, Bertrand S. Current approaches and challenges for the metabolite profiling of complex natural extracts. J. Chromatogr. A. 2015;1382:136–164. doi: 10.1016/j.chroma.2014.10.091. [DOI] [PubMed] [Google Scholar]
- 34.Tahtah Y, et al. High-resolution PTP1B inhibition profiling combined with high-performance liquid chromatography–high-resolution mass spectrometry–solid-phase extraction–nuclear magnetic resonance spectroscopy: proof-of-concept and antidiabetic constituents in crude extract of Eremophila lucida. Fitoterapia. 2016;110:52–58. doi: 10.1016/j.fitote.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Chu C, et al. Antidiabetic constituents of Dendrobium officinale as determined by high-resolution profiling of radical scavenging and α-glucosidase and α-amylase inhibition combined with HPLC-PDA-HRMS-SPE-NMR analysis. Phytochem. Lett. 2019;31:47–52. doi: 10.1016/j.phytol.2019.03.002. [DOI] [Google Scholar]
- 36.Garcia-Perez I, et al. Identifying unknown metabolites using NMR-based metabolic profiling techniques. Nat. Protoc. 2020;15:2538–2567. doi: 10.1038/s41596-020-0343-3. [DOI] [PubMed] [Google Scholar]
- 37.Giavalisco P, et al. High-resolution direct infusion-based mass spectrometry in combination with whole 13C metabolome isotope labeling allows unambiguous assignment of chemical sum formulas. Anal. Chem. 2008;80:9417–9425. doi: 10.1021/ac8014627. [DOI] [PubMed] [Google Scholar]
- 38.Covington BC, McLean JA, Bachmann BO. Comparative mass spectrometry-based metabolomics strategies for the investigation of microbial secondary metabolites. Nat. Prod. Rep. 2017;34:6–24. doi: 10.1039/C6NP00048G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fontana A, Iturrino L, Corens D, Crego AL. Automated open-access liquid chromatography high resolution mass spectrometry to support drug discovery projects. J. Pharm. Biomed. Anal. 2020;178:112908. doi: 10.1016/j.jpba.2019.112908. [DOI] [PubMed] [Google Scholar]
- 40.Kind T, et al. Identification of small molecules using accurate mass MS/MS search. Mass. Spectrom. Rev. 2018;37:513–532. doi: 10.1002/mas.21535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang M, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang JY, et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013;76:1686–1699. doi: 10.1021/np400413s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen F, Greiner R, Wishart D. Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics. 2015;11:98–110. doi: 10.1007/s11306-014-0676-4. [DOI] [Google Scholar]
- 44.Allard P-M, et al. Integration of molecular networking and in-silico MS/MS fragmentation for natural products dereplication. Anal. Chem. 2016;88:3317–3323. doi: 10.1021/acs.analchem.5b04804. [DOI] [PubMed] [Google Scholar]
- 45.da Silva RR, et al. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 2018;14:e1006089. doi: 10.1371/journal.pcbi.1006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Randazzo GM, et al. Prediction of retention time in reversed-phase liquid chromatography as a tool for steroid identification. Anal. Chim. Acta. 2016;916:8–16. doi: 10.1016/j.aca.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Z, Xiong X, Zhu Z-J. MetCCS predictor: a web server for predicting collision cross-section values of metabolites in ion mobility-mass spectrometry based metabolomics. Bioinformatics. 2017;33:2235–2237. doi: 10.1093/bioinformatics/btx140. [DOI] [PubMed] [Google Scholar]
- 48.Rutz A, et al. Taxonomically informed scoring enhances confidence in natural products annotation. Front. Plant. Sci. 2019;10:1329. doi: 10.3389/fpls.2019.01329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guijas C, et al. METLIN: a technology platform for identifying knowns and unknowns. Anal. Chem. 2018;90:3156–3164. doi: 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aksenov AA, da Silva R, Knight R, Lopes NP, Dorrestein PC. Global chemical analysis of biology by mass spectrometry. Nat. Rev. Chem. 2017;1:0054. doi: 10.1038/s41570-017-0054. [DOI] [Google Scholar]
- 51.Fox Ramos AE, et al. CANPA: computer-assisted natural products anticipation. Anal. Chem. 2019;91:11247–11252. doi: 10.1021/acs.analchem.9b02216. [DOI] [PubMed] [Google Scholar]
- 52.Wolfender J-L, Litaudon M, Touboul D, Queiroz EF. Innovative omics-based approaches for prioritisation and targeted isolation of natural products – new strategies for drug discovery. Nat. Prod. Rep. 2019;36:855–868. doi: 10.1039/C9NP00004F. [DOI] [PubMed] [Google Scholar]
- 53.Graziani V, et al. Metabolomic approach for a rapid identification of natural products with cytotoxic activity against human colorectal cancer cells. Sci. Rep. 2018;8:5309. doi: 10.1038/s41598-018-23704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grienke U, et al. 1H NMR-MS-based heterocovariance as a drug discovery tool for fishing bioactive compounds out of a complex mixture of structural analogues. Sci. Rep. 2019;9:11113. doi: 10.1038/s41598-019-47434-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aligiannis N, et al. Heterocovariance based metabolomics as a powerful tool accelerating bioactive natural product identification. ChemistrySelect. 2016;1:2531–2535. doi: 10.1002/slct.201600744. [DOI] [Google Scholar]
- 56.Acharya D, et al. Omics technologies to understand activation of a biosynthetic gene cluster in Micromonospora sp. WMMB235: deciphering keyicin biosynthesis. ACS Chem. Biol. 2019;14:1260–1270. doi: 10.1021/acschembio.9b00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schulze CJ, et al. ‘Function-first’ lead discovery: mode of action profiling of natural product libraries using image-based screening. Chem. Biol. 2013;20:285–295. doi: 10.1016/j.chembiol.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kurita KL, Glassey E, Linington RG. Integration of high-content screening and untargeted metabolomics for comprehensive functional annotation of natural product libraries. Proc. Natl Acad. Sci. USA. 2015;112:11999–12004. doi: 10.1073/pnas.1507743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Earl DC, et al. Discovery of human cell selective effector molecules using single cell multiplexed activity metabolomics. Nat. Commun. 2018;9:39. doi: 10.1038/s41467-017-02470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wishart DS. NMR metabolomics: a look ahead. J. Magn. Reson. 2019;306:155–161. doi: 10.1016/j.jmr.2019.07.013. [DOI] [PubMed] [Google Scholar]
- 61.Berlinck RGS, et al. Approaches for the isolation and identification of hydrophilic, light-sensitive, volatile and minor natural products. Nat. Prod. Rep. 2019;36:981–1004. doi: 10.1039/C9NP00009G. [DOI] [PubMed] [Google Scholar]
- 62.Hilton BD, Martin GE. Investigation of the experimental limits of small-sample heteronuclear 2D NMR. J. Nat. Prod. 2010;73:1465–1469. doi: 10.1021/np100481m. [DOI] [PubMed] [Google Scholar]
- 63.Sultan S, et al. Evolving trends in the dereplication of natural product extracts. 3: Further lasiodiplodins from Lasiodiplodia theobromae, an endophyte from Mapania kurzii. Tetrahedron Lett. 2014;55:453–455. doi: 10.1016/j.tetlet.2013.11.060. [DOI] [Google Scholar]
- 64.Jones CG, et al. The CryoEM method MicroED as a powerful tool for small molecule structure determination. ACS Cent. Sci. 2018;4:1587–1592. doi: 10.1021/acscentsci.8b00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ting CP, et al. Use of a scaffold peptide in the biosynthesis of amino acid-derived natural products. Science. 2019;365:280–284. doi: 10.1126/science.aau6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ganesh T, et al. Evaluation of the tubulin-bound paclitaxel conformation: synthesis, biology, and SAR studies of C-4 to C-3′ bridged paclitaxel analogues. J. Med. Chem. 2007;50:713–725. doi: 10.1021/jm061071x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choules MP, et al. Residual complexity does impact organic chemistry and drug discovery: the case of rufomyazine and rufomycin. J. Org. Chem. 2018;83:6664–6672. doi: 10.1021/acs.joc.8b00988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ziemert N, Alanjary M, Weber T. The evolution of genome mining in microbes - a review. Nat. Prod. Rep. 2016;33:988–1005. doi: 10.1039/C6NP00025H. [DOI] [PubMed] [Google Scholar]
- 69.Viehrig K, et al. Structure and biosynthesis of crocagins: polycyclic posttranslationally modified ribosomal peptides from Chondromyces crocatus. Angew. Chem. Int. Ed. Engl. 2017;56:7407–7410. doi: 10.1002/anie.201612640. [DOI] [PubMed] [Google Scholar]
- 70.Surup F, et al. Crocadepsins-depsipeptides from the myxobacterium Chondromyces crocatus found by a genome mining approach. ACS Chem. Biol. 2018;13:267–272. doi: 10.1021/acschembio.7b00900. [DOI] [PubMed] [Google Scholar]
- 71.Kayrouz CM, Zhang Y, Pham TM, Ju KS. Genome mining reveals the phosphonoalamide natural products and a new route in phosphonic acid biosynthesis. ACS Chem. Biol. 2020;15:1921–1929. doi: 10.1021/acschembio.0c00256. [DOI] [PubMed] [Google Scholar]
- 72.Laureti L, et al. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc. Natl Acad. Sci. USA. 2011;108:6258–6263. doi: 10.1073/pnas.1019077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weber T, Kim HU. The secondary metabolite bioinformatics portal: Computational tools to facilitate synthetic biology of secondary metabolite production. Synth. Syst. Biotechnol. 2016;1:69–79. doi: 10.1016/j.synbio.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Navarro-Muñoz JC, et al. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 2020;16:60–68. doi: 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoffmann T, et al. Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria. Nat. Commun. 2018;9:803. doi: 10.1038/s41467-018-03184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Helaly SE, Thongbai B, Stadler M. Diversity of biologically active secondary metabolites from endophytic and saprotrophic fungi of the ascomycete order Xylariales. Nat. Prod. Rep. 2018;35:992–1014. doi: 10.1039/C8NP00010G. [DOI] [PubMed] [Google Scholar]
- 77.Dalinova A, et al. Isolation and bioactivity of secondary metabolites from solid culture of the fungus, Alternaria sonchi. Biomolecules. 2020;10:81. doi: 10.3390/biom10010081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zerikly M, Challis GL. Strategies for the discovery of new natural products by genome mining. ChemBioChem. 2009;10:625–633. doi: 10.1002/cbic.200800389. [DOI] [PubMed] [Google Scholar]
- 79.Culp EJ, et al. Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature. 2020;578:582–587. doi: 10.1038/s41586-020-1990-9. [DOI] [PubMed] [Google Scholar]
- 80.Zhang H, Boghigian BA, Armando J, Pfeifer BA. Methods and options for the heterologous production of complex natural products. Nat. Prod. Rep. 2011;28:125–151. doi: 10.1039/C0NP00037J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anyaogu DC, Mortensen UH. Heterologous production of fungal secondary metabolites in aspergilli. Front. Microbiol. 2015;6:77. doi: 10.3389/fmicb.2015.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sucipto H, Pogorevc D, Luxenburger E, Wenzel SC, Müller R. Heterologous production of myxobacterial α-pyrone antibiotics in Myxococcus xanthus. Metab. Eng. 2017;44:160–170. doi: 10.1016/j.ymben.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 83.Nora LC, et al. The art of vector engineering: towards the construction of next-generation genetic tools. Microb. Biotechnol. 2019;12:125–147. doi: 10.1111/1751-7915.13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bok JW, et al. Fungal artificial chromosomes for mining of the fungal secondary metabolome. BMC Genomics. 2015;16:343. doi: 10.1186/s12864-015-1561-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Clevenger KD, et al. A scalable platform to identify fungal secondary metabolites and their gene clusters. Nat. Chem. Biol. 2017;13:895–901. doi: 10.1038/nchembio.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mao D, Okada BK, Wu Y, Xu F, Seyedsayamdost MR. Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr. Opin. Microbiol. 2018;45:156–163. doi: 10.1016/j.mib.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015;13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 88.Yamanaka K, et al. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl Acad. Sci. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sidda JD, et al. Discovery of a family of γ-aminobutyrate ureas via rational derepression of a silent bacterial gene cluster. Chem. Sci. 2014;5:86–89. doi: 10.1039/C3SC52536H. [DOI] [Google Scholar]
- 90.Wang B, Guo F, Dong S-H, Zhao H. Activation of silent biosynthetic gene clusters using transcription factor decoys. Nat. Chem. Biol. 2019;15:111–114. doi: 10.1038/s41589-018-0187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang MM, et al. CRISPR–Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat. Chem. Biol. 2017;13:607–609. doi: 10.1038/nchembio.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Culp EJ, et al. Hidden antibiotics in actinomycetes can be identified by inactivation of gene clusters for common antibiotics. Nat. Biotechnol. 2019;37:1149–1154. doi: 10.1038/s41587-019-0241-9. [DOI] [PubMed] [Google Scholar]
- 93.Hover BM, et al. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 2018;3:415–422. doi: 10.1038/s41564-018-0110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chu J, et al. Discovery of MRSA active antibiotics using primary sequence from the human microbiome. Nat. Chem. Biol. 2016;12:1004–1006. doi: 10.1038/nchembio.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kersten RD, Weng J-K. Gene-guided discovery and engineering of branched cyclic peptides in plants. Proc. Natl Acad. Sci. USA. 2018;115:E10961–E10969. doi: 10.1073/pnas.1813993115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dutertre S, et al. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol. Cell. Proteom. 2013;12:312–329. doi: 10.1074/mcp.M112.021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilson MC, et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506:58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 98.Mori T, et al. Single-bacterial genomics validates rich and varied specialized metabolism of uncultivated Entotheonella sponge symbionts. Proc. Natl Acad. Sci. USA. 2018;115:1718–1723. doi: 10.1073/pnas.1715496115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rath CM, et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem. Biol. 2011;6:1244–1256. doi: 10.1021/cb200244t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Newman DJ. Are microbial endophytes the ‘actual’ producers of bioactive antitumor agents? Trends Cancer. 2018;4:662–670. doi: 10.1016/j.trecan.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 101.Helfrich EJN, et al. Bipartite interactions, antibiotic production and biosynthetic potential of the Arabidopsis leaf microbiome. Nat. Microbiol. 2018;3:909–919. doi: 10.1038/s41564-018-0200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yan F, et al. Biosynthesis and heterologous production of vioprolides: rational biosynthetic engineering and unprecedented 4-methylazetidinecarboxylic acid formation. Angew. Chem. Int. Ed. 2018;57:8754–8759. doi: 10.1002/anie.201802479. [DOI] [PubMed] [Google Scholar]
- 103.Tu Q, et al. Genetic engineering and heterologous expression of the disorazol biosynthetic gene cluster via Red/ET recombineering. Sci. Rep. 2016;6:21066. doi: 10.1038/srep21066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Song C, et al. Enhanced heterologous spinosad production from a 79-kb synthetic multioperon assembly. ACS Synth. Biol. 2019;8:137–147. doi: 10.1021/acssynbio.8b00402. [DOI] [PubMed] [Google Scholar]
- 105.Wlodek A, et al. Diversity oriented biosynthesis via accelerated evolution of modular gene clusters. Nat. Commun. 2017;8:1206. doi: 10.1038/s41467-017-01344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bozhüyük KAJ, et al. De novo design and engineering of non-ribosomal peptide synthetases. Nat. Chem. 2018;10:275–281. doi: 10.1038/nchem.2890. [DOI] [PubMed] [Google Scholar]
- 107.Bozhüyük KAJ, et al. Modification and de novo design of non-ribosomal peptide synthetases using specific assembly points within condensation domains. Nat. Chem. 2019;11:653–661. doi: 10.1038/s41557-019-0276-z. [DOI] [PubMed] [Google Scholar]
- 108.Awakawa T, et al. Reprogramming of the antimycin NRPS-PKS assembly lines inspired by gene evolution. Nat. Commun. 2018;9:3534. doi: 10.1038/s41467-018-05877-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Masschelein J, et al. A dual transacylation mechanism for polyketide synthase chain release in enacyloxin antibiotic biosynthesis. Nat. Chem. 2019;11:906–912. doi: 10.1038/s41557-019-0309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kosol S, et al. Structural basis for chain release from the enacyloxin polyketide synthase. Nat. Chem. 2019;11:913–923. doi: 10.1038/s41557-019-0335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gregory MA, et al. Structure guided design of improved anti-proliferative rapalogs through biosynthetic medicinal chemistry. Chem. Sci. 2013;4:1046–1052. doi: 10.1039/C2SC21833J. [DOI] [Google Scholar]
- 112.Méndez C, González-Sabín J, Morís F, Salas JA. Expanding the chemical diversity of the antitumoral compound mithramycin by combinatorial biosynthesis and biocatalysis: the quest for mithralogs with improved therapeutic window. Planta Med. 2015;81:1326–1338. doi: 10.1055/s-0035-1557876. [DOI] [PubMed] [Google Scholar]
- 113.Hindra, et al. Genome mining of Streptomyces mobaraensis DSM40847 as a bleomycin producer providing a biotechnology platform to engineer designer bleomycin analogues. Org. Lett. 2017;19:1386–1389. doi: 10.1021/acs.orglett.7b00283. [DOI] [PubMed] [Google Scholar]
- 114.Brautaset T, et al. Improved antifungal polyene macrolides via engineering of the nystatin biosynthetic genes in Streptomyces noursei. Chem. Biol. 2008;15:1198–1206. doi: 10.1016/j.chembiol.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 115.Preobrazhenskaya MN, et al. Synthesis and study of the antifungal activity of new mono- and disubstituted derivatives of a genetically engineered polyene antibiotic 28,29-didehydronystatin A1 (S44HP) J. Antibiot. 2010;63:55–64. doi: 10.1038/ja.2009.118. [DOI] [PubMed] [Google Scholar]
- 116.Tevyashova AN, et al. Structure-antifungal activity relationships of polyene antibiotics of the amphotericin B group. Antimicrob. Agents Chemother. 2013;57:3815–3822. doi: 10.1128/AAC.00270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lewis K, Epstein S, D’Onofrio A, Ling LL. Uncultured microorganisms as a source of secondary metabolites. J. Antibiot. 2010;63:468–476. doi: 10.1038/ja.2010.87. [DOI] [PubMed] [Google Scholar]
- 118.Schiewe H-J, Zeeck A. Cineromycins, γ-butyrolactones and ansamycins by analysis of the secondary metabolite pattern created by a single strain of Strepomyces. J. Antibiot. 1999;52:635–642. doi: 10.7164/antibiotics.52.635. [DOI] [PubMed] [Google Scholar]
- 119.Zähner H. Some aspects of antibiotics research. Angew. Chem. Int. Ed. Engl. 1977;16:687–694. doi: 10.1002/anie.197706871. [DOI] [PubMed] [Google Scholar]
- 120.Newman D. Screening and identification of novel biologically active natural compounds. F1000Research. 2017;6:783. doi: 10.12688/f1000research.11221.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hussain A, et al. Novel bioactive molecules from Lentzea violacea strain AS 08 using one strain-many compounds (OSMAC) approach. Bioorg. Med. Chem. Lett. 2017;27:2579–2582. doi: 10.1016/j.bmcl.2017.03.075. [DOI] [PubMed] [Google Scholar]
- 122.Hemphill CFP, et al. OSMAC approach leads to new fusarielin metabolites from Fusarium tricinctum. J. Antibiot. 2017;70:726–732. doi: 10.1038/ja.2017.21. [DOI] [PubMed] [Google Scholar]
- 123.Vartoukian SR, Palmer RM, Wade WG. Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol. Lett. 2010;309:1–7. doi: 10.1111/j.1574-6968.2010.02000.x. [DOI] [PubMed] [Google Scholar]
- 124.Moussa M, et al. Co-culture of the fungus Fusarium tricinctum with Streptomyces lividans induces production of cryptic naphthoquinone dimers. RSC Adv. 2019;9:1491–1500. doi: 10.1039/C8RA09067J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Abdel-Razek AS, Hamed A, Frese M, Sewald N, Shaaban M. Penicisteroid C: new polyoxygenated steroid produced by co-culturing of Streptomyces piomogenus with Aspergillus niger. Steroids. 2018;138:21–25. doi: 10.1016/j.steroids.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 126.D’Onofrio A, et al. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem. Biol. 2010;17:254–264. doi: 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Van Arnam EB, Currie CR, Clardy J. Defense contracts: molecular protection in insect-microbe symbioses. Chem. Soc. Rev. 2018;47:1638–1651. doi: 10.1039/C7CS00340D. [DOI] [PubMed] [Google Scholar]
- 128.Molloy EM, Hertweck C. Antimicrobial discovery inspired by ecological interactions. Curr. Opin. Microbiol. 2017;39:121–127. doi: 10.1016/j.mib.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 129.Tobias NJ, Shi YM, Bode HB. Refining the natural product repertoire in entomopathogenic bacteria. Trends Microbiology. 2018;26:833–840. doi: 10.1016/j.tim.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 130.Imai Y, et al. A new antibiotic selectively kills Gram-negative pathogens. Nature. 2019;576:459–464. doi: 10.1038/s41586-019-1791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bode E, et al. Biosynthesis and function of simple amides in Xenorhabdus doucetiae. Environ. Microbiol. 2017;19:4564–4575. doi: 10.1111/1462-2920.13919. [DOI] [PubMed] [Google Scholar]
- 132.Crawford JM, Kontnik R, Clardy J. Regulating alternative lifestyles in entomopathogenic bacteria. Curr. Biol. 2010;20:69–74. doi: 10.1016/j.cub.2009.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zengler K, et al. Cultivating the uncultured. Proc. Natl Acad. Sci. USA. 2002;99:15681–15686. doi: 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nichols D, et al. Use of ichip for high-throughput in situ cultivation of ‘uncultivable’ microbial species. Appl. Environ. Microbiol. 2010;76:2445–2450. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ling LL, et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Homma T, et al. Dual targeting of cell wall precursors by teixobactin leads to cell lysis. Antimicrob. Agents Chemother. 2016;60:6510–6517. doi: 10.1128/AAC.01050-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pham VHT, Kim J. Cultivation of unculturable soil bacteria. Trends Biotechnol. 2012;30:475–484. doi: 10.1016/j.tibtech.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 138.Derewacz DK, Covington BC, McLean JA, Bachmann BO. Mapping microbial response metabolomes for induced natural product discovery. ACS Chem. Biol. 2015;10:1998–2006. doi: 10.1021/acschembio.5b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lagier JC, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 140.Terekhov SS, et al. Microfluidic droplet platform for ultrahigh-throughput single-cell screening of biodiversity. Proc. Natl Acad. Sci. USA. 2017;114:2550–2555. doi: 10.1073/pnas.1621226114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Challinor VL, Bode HB. Bioactive natural products from novel microbial sources. Ann. NY Acad. Sci. 2015;1354:82–97. doi: 10.1111/nyas.12954. [DOI] [PubMed] [Google Scholar]
- 142.Pidot SJ, Coyne S, Kloss F, Hertweck C. Antibiotics from neglected bacterial sources. Int. J. Med. Microbiol. 2014;304:14–22. doi: 10.1016/j.ijmm.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 143.Lincke T, Behnken S, Ishida K, Roth M, Hertweck C. Closthioamide: an unprecedented polythioamide antibiotic from the strictly anaerobic bacterium Clostridium cellulolyticum. Angew. Chem. Int. Ed. 2010;49:2011–2013. doi: 10.1002/anie.200906114. [DOI] [PubMed] [Google Scholar]
- 144.Haeckl FPJ, et al. A selective genome-guided method for environmental Burkholderia isolation. J. Ind. Microbiol. Biotechnol. 2019;46:345–362. doi: 10.1007/s10295-018-02121-x. [DOI] [PubMed] [Google Scholar]
- 145.Cross KL, et al. Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat. Biotechnol. 2019;37:1314–1321. doi: 10.1038/s41587-019-0260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vlachou P, et al. Innovative approach to sustainable marine invertebrate chemistry and a scale-up technology for open marine ecosystems. Mar. Drugs. 2018;16:152. doi: 10.3390/md16050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zainal-Abidin MH, Hayyan M, Hayyan A, Jayakumar NS. New horizons in the extraction of bioactive compounds using deep eutectic solvents: a review. Anal. Chim. Acta. 2017;979:1–23. doi: 10.1016/j.aca.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 148.Dai Y, van Spronsen J, Witkamp G-J, Verpoorte R, Choi YH. Ionic liquids and deep eutectic solvents in natural products research: mixtures of solids as extraction solvents. J. Nat. Prod. 2013;76:2162–2173. doi: 10.1021/np400051w. [DOI] [PubMed] [Google Scholar]
- 149.Nemes P, Vertes A. Ambient mass spectrometry for in vivo local analysis and in situ molecular tissue imaging. Trends Analyt. Chem. 2012;34:22–34. doi: 10.1016/j.trac.2011.11.006. [DOI] [Google Scholar]
- 150.Pasquini C. Near infrared spectroscopy: a mature analytical technique with new perspectives–a review. Anal. Chim. Acta. 2018;1026:8–36. doi: 10.1016/j.aca.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 151.Hutchings M, Truman A, Wilkinson B. Antibiotics: past, present and future. Curr. Opin. Microbiol. 2019;51:72–80. doi: 10.1016/j.mib.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 152.Rossiter SE, Fletcher MH, Wuest WM. Natural products as platforms to overcome antibiotic resistance. Chem. Rev. 2017;117:12415–12474. doi: 10.1021/acs.chemrev.7b00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zipperer A, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016;535:511–516. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 154.Lešnik U, et al. Construction of a new class of tetracycline lead structures with potent antibacterial activity through biosynthetic engineering. Angew. Chem. Int. Ed. Engl. 2015;54:3937–3940. doi: 10.1002/anie.201411028. [DOI] [PubMed] [Google Scholar]
- 155.Kling A, et al. Antibiotics. Targeting DnaN for tuberculosis therapy using novel griselimycins. Science. 2015;348:1106–1112. doi: 10.1126/science.aaa4690. [DOI] [PubMed] [Google Scholar]
- 156.Shaeer KM, Zmarlicka MT, Chahine EB, Piccicacco N, Cho JC. Plazomicin: a next-generation aminoglycoside. Pharmacotherapy. 2019;39:77–93. doi: 10.1002/phar.2203. [DOI] [PubMed] [Google Scholar]
- 157.Smith PA, et al. Optimized arylomycins are a new class of Gram-negative antibiotics. Nature. 2018;561:189–194. doi: 10.1038/s41586-018-0483-6. [DOI] [PubMed] [Google Scholar]
- 158.Dickey SW, Cheung GYC, Otto M. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 2017;16:457–471. doi: 10.1038/nrd.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Park SR, et al. Discovery of cahuitamycins as biofilm inhibitors derived from a convergent biosynthetic pathway. Nat. Commun. 2016;7:10710. doi: 10.1038/ncomms10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mann J. Natural products in cancer chemotherapy: past, present and future. Nat. Rev. Cancer. 2002;2:143–148. doi: 10.1038/nrc723. [DOI] [PubMed] [Google Scholar]
- 161.Beck A, Goetsch L, Dumontet C, Corvaïa N. Strategies and challenges for the next generation of antibody–drug conjugates. Nat. Rev. Drug Discov. 2017;16:315–337. doi: 10.1038/nrd.2016.268. [DOI] [PubMed] [Google Scholar]
- 162.Pereira, R. B. et al. Marine-derived anticancer agents: clinical benefits, innovative mechanisms, and new targets. Mar. Drugs17 (2019). [DOI] [PMC free article] [PubMed]
- 163.Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020;83:770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 164.Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019;18:197–218. doi: 10.1038/s41573-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 165.Menger L, et al. Cardiac glycosides exert anticancer effects by inducing immunogenic cell death. Sci. Transl. Med. 2012;4:143ra99. doi: 10.1126/scitranslmed.3003807. [DOI] [PubMed] [Google Scholar]
- 166.Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat. Rev. Immunol. 2017;17:97–111. doi: 10.1038/nri.2016.107. [DOI] [PubMed] [Google Scholar]
- 167.Diederich M. Natural compound inducers of immunogenic cell death. Arch. Pharm. Res. 2019;42:629–645. doi: 10.1007/s12272-019-01150-z. [DOI] [PubMed] [Google Scholar]
- 168.Radogna F, Dicato M, Diederich M. Natural modulators of the hallmarks of immunogenic cell death. Biochem. Pharmacol. 2019;162:55–70. doi: 10.1016/j.bcp.2018.12.016. [DOI] [PubMed] [Google Scholar]
- 169.Schmidt BM, Ribnicky DM, Lipsky PE, Raskin I. Revisiting the ancient concept of botanical therapeutics. Nat. Chem. Biol. 2007;3:360–366. doi: 10.1038/nchembio0707-360. [DOI] [PubMed] [Google Scholar]
- 170.Schmidt B, et al. A natural history of botanical therapeutics. Metabolism. 2008;57:S3–S9. doi: 10.1016/j.metabol.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kellogg JJ, et al. Comparison of metabolomics approaches for evaluating the variability of complex botanical preparations: green tea (Camellia sinensis) as a case study. J. Nat. Prod. 2017;80:1457–1466. doi: 10.1021/acs.jnatprod.6b01156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Marchesi JR, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Abdollahi-Roodsaz S, Abramson SB, Scher JU. The metabolic role of the gut microbiota in health and rheumatic disease: mechanisms and interventions. Nat. Rev. Rheumatol. 2016;12:446–455. doi: 10.1038/nrrheum.2016.68. [DOI] [PubMed] [Google Scholar]
- 174.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N. Engl. J. Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 175.Scherlach K, Hertweck C. Mediators of mutualistic microbe-microbe interactions. Nat. Prod. Rep. 2018;35:303–308. doi: 10.1039/C7NP00035A. [DOI] [PubMed] [Google Scholar]
- 176.Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J. Clin. Invest. 2014;124:4212–4218. doi: 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Peterson CT, et al. Effects of turmeric and curcumin dietary supplementation on human gut microbiota: a double-blind, randomized, placebo-controlled pilot study. J Evid. Based Integr. Med. 2018;23:2515690X18790725. doi: 10.1177/2515690X18790725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Eid, H. M. et al. Significance of microbiota in obesity and metabolic diseases and the modulatory potential by medicinal plant and food ingredients. Front. Pharmacol. 8, (2017). [DOI] [PMC free article] [PubMed]
- 179.Valencia PM, Richard M, Brock J, Boglioli E. The human microbiome: opportunity or hype? Nat. Rev. Drug Discov. 2017;16:823–824. doi: 10.1038/nrd.2017.154. [DOI] [PubMed] [Google Scholar]
- 180.Sorokina M, Steinbeck C. Review on natural products databases: Where to find data in 2020. J. Cheminform. 2020;12:20. doi: 10.1186/s13321-020-00424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Schneider G, et al. Deorphaning the macromolecular targets of the natural anticancer compound doliculide. Angew. Chem. Int. Ed. 2016;55:12408–12411. doi: 10.1002/anie.201605707. [DOI] [PubMed] [Google Scholar]
- 182.Palazzotto E, Weber T. Omics and multi-omics approaches to study the biosynthesis of secondary metabolites in microorganisms. Curr. Opin. Microbiol. 2018;45:109–116. doi: 10.1016/j.mib.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 183.Dias T, Gaudêncio SP, Pereira F. A computer-driven approach to discover natural product leads for methicillin-resistant staphylococcus aureus infection therapy. Mar. Drugs. 2019;17:16. doi: 10.3390/md17010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Boström J, Brown DG, Young RJ, Keserü GM. Expanding the medicinal chemistry synthetic toolbox. Nat. Rev. Drug Discov. 2018;17:709–727. doi: 10.1038/nrd.2018.116. [DOI] [PubMed] [Google Scholar]
- 185.Zhao X, et al. A novel drug discovery strategy inspired by traditional medicine philosophies. Science. 2015;347:S38–S40. [Google Scholar]
- 186.Liao S, et al. Tanshinol borneol ester, a novel synthetic small molecule angiogenesis stimulator inspired by botanical formulations for angina pectoris. Br. J. Pharmacol. 2019;176:3143–3160. doi: 10.1111/bph.14714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bai Y, et al. Polygala tenuifolia-Acori tatarinowii herbal pair as an inspiration for substituted cinnamic α-asaronol esters: design, synthesis, anticonvulsant activity, and inhibition of lactate dehydrogenase study. Eur. J. Med. Chem. 2019;183:111650. doi: 10.1016/j.ejmech.2019.111650. [DOI] [PubMed] [Google Scholar]
- 188.Seiple IB, et al. A platform for the discovery of new macrolide antibiotics. Nature. 2016;533:338–345. doi: 10.1038/nature17967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wang L, et al. Novel interactomics approach identifies ABCA1 as direct target of evodiamine, which increases macrophage cholesterol efflux. Sci. Rep. 2018;8:11061. doi: 10.1038/s41598-018-29281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chang J, Kim Y, Kwon HJ. Advances in identification and validation of protein targets of natural products without chemical modification. Nat. Prod. Rep. 2016;33:719–730. doi: 10.1039/C5NP00107B. [DOI] [PubMed] [Google Scholar]
- 191.Adhikari J, Fitzgerald MC. SILAC-pulse proteolysis: a mass spectrometry-based method for discovery and cross-validation in proteome-wide studies of ligand binding. J. Am. Soc. Mass. Spectrom. 2014;25:2073–2083. doi: 10.1007/s13361-014-0992-y. [DOI] [PubMed] [Google Scholar]
- 192.Gregori-Puigjane E, et al. Identifying mechanism-of-action targets for drugs and probes. Proc. Natl Acad. Sci. USA. 2012;109:11178–11183. doi: 10.1073/pnas.1204524109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yñigez-Gutierrez AE, Bachmann BO. Fixing the unfixable: the art of optimizing natural products for human medicine. J. Med. Chem. 2019;62:8412–8428. doi: 10.1021/acs.jmedchem.9b00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Markley JL, Wencewicz TA. Tetracycline-inactivating enzymes. Front. Microbiol. 2018;9:1058. doi: 10.3389/fmicb.2018.01058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wu F, et al. Chrysomycin A derivatives for the treatment of multi-drug-resistant tuberculosis. ACS Cent. Sci. 2020;6:928–938. doi: 10.1021/acscentsci.0c00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Dayalan Naidu S, Kostov RV, Dinkova-Kostova AT. Transcription factors Hsf1 and Nrf2 engage in crosstalk for cytoprotection. Trends Pharmacol. Sci. 2015;36:6–14. doi: 10.1016/j.tips.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 197.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 198.Mills EL, et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Murphy KE, Park JJ. Can co-activation of Nrf2 and neurotrophic signaling pathway slow Alzheimer’s disease? Int. J. Mol. Sci. 2017;18:1168. doi: 10.3390/ijms18061168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Cuadrado A, et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 201.Linker RA, et al. Fumaric acid esters exert neuroprotective effects in neuroinflammation via activation of the Nrf2 antioxidant pathway. Brain. 2011;134:678–692. doi: 10.1093/brain/awq386. [DOI] [PubMed] [Google Scholar]
- 202.Singh K, et al. Sulforaphane treatment of autism spectrum disorder (ASD) Proc. Natl Acad. Sci. USA. 2014;111:15550–15555. doi: 10.1073/pnas.1416940111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Spencer SR, Wilczak CA, Talalay P. Induction of glutathione transferases and NAD(P)H:quinone reductase by fumaric acid derivatives in rodent cells and tissues. Cancer Res. 1990;50:7871–7875. [PubMed] [Google Scholar]
- 204.Soušek J, et al. Alkaloids and organic acids content of eight Fumaria species. Phytochem. Anal. 1999;10:6–11. doi: 10.1002/(SICI)1099-1565(199901/02)10:1<6::AID-PCA431>3.0.CO;2-0. [DOI] [Google Scholar]
- 205.Linker RA, Haghikia A. Dimethyl fumarate in multiple sclerosis: latest developments, evidence and place in therapy. Ther. Adv. Chronic Dis. 2016;7:198–207. doi: 10.1177/2040622316653307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Fox RJ, et al. Efficacy and tolerability of delayed-release dimethyl fumarate in Black, Hispanic, and Asian patients with relapsing-remitting multiple sclerosis: post hoc integrated analysis of DEFINE and CONFIRM. Neurol. Ther. 2017;6:175–187. doi: 10.1007/s40120-017-0077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Fernández Ó, et al. Efficacy and safety of delayed-release dimethyl fumarate for relapsing-remitting multiple sclerosis in prior interferon users: an integrated analysis of DEFINE and CONFIRM. Clin. Ther. 2017;39:1671–1679. doi: 10.1016/j.clinthera.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 208.Zhang Y, Talalay P, Cho CG, Posner GH. A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proc. Natl Acad. Sci. USA. 1992;89:2399–2403. doi: 10.1073/pnas.89.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Dinkova-Kostova AT, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl Acad. Sci. USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Morroni F, et al. Neuroprotective effect of sulforaphane in 6-hydroxydopamine-lesioned mouse model of Parkinson’s disease. Neurotoxicology. 2013;36:63–71. doi: 10.1016/j.neuro.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 211.Liu Y, et al. Sulforaphane enhances proteasomal and autophagic activities in mice and is a potential therapeutic reagent for Huntington’s disease. J. Neurochem. 2014;129:539–547. doi: 10.1111/jnc.12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kim HV, et al. Amelioration of Alzheimer’s disease by neuroprotective effect of sulforaphane in animal model. Amyloid. 2013;20:7–12. doi: 10.3109/13506129.2012.751367. [DOI] [PubMed] [Google Scholar]
- 213.Zhao J, Moore AN, Clifton GL, Dash PK. Sulforaphane enhances aquaporin-4 expression and decreases cerebral edema following traumatic brain injury. J. Neurosci. Res. 2005;82:499–506. doi: 10.1002/jnr.20649. [DOI] [PubMed] [Google Scholar]
- 214.Benedict AL, et al. Neuroprotective effects of sulforaphane after contusive spinal cord injury. J. Neurotrauma. 2012;29:2576–2586. doi: 10.1089/neu.2012.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Alfieri A, et al. Sulforaphane preconditioning of the Nrf2/HO-1 defense pathway protects the cerebral vasculature against blood-brain barrier disruption and neurological deficits in stroke. Free Radic. Biol. Med. 2013;65:1012–1022. doi: 10.1016/j.freeradbiomed.2013.08.190. [DOI] [PubMed] [Google Scholar]
- 216.Wu S, et al. Sulforaphane produces antidepressant- and anxiolytic-like effects in adult mice. Behav. Brain Res. 2016;301:55–62. doi: 10.1016/j.bbr.2015.12.030. [DOI] [PubMed] [Google Scholar]
- 217.Li B, et al. Sulforaphane ameliorates the development of experimental autoimmune encephalomyelitis by antagonizing oxidative stress and Th17-related inflammation in mice. Exp. Neurol. 2013;250:239–249. doi: 10.1016/j.expneurol.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 218.Egner PA, et al. Rapid and sustainable detoxication of airborne pollutants by broccoli sprout beverage: results of a randomized clinical trial in China. Cancer Prev. Res. 2014;7:813–823. doi: 10.1158/1940-6207.CAPR-14-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chen JG, et al. Dose-dependent detoxication of the airborne pollutant benzene in a randomized trial of broccoli sprout beverage in Qidong, China. Am. J. Clin. Nutr. 2019;110:675–684. doi: 10.1093/ajcn/nqz122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Howell SJ, et al. Final results of the STEM trial: SFX-01 in the treatment and evaluation of ER+ Her2– metastatic breast cancer (mBC) Ann. Oncol. 2019;30:v122. doi: 10.1093/annonc/mdz242.036. [DOI] [Google Scholar]
- 221.Dinkova-Kostova AT, et al. Extremely potent triterpenoid inducers of the phase 2 response: correlations of protection against oxidant and inflammatory stress. Proc. Natl Acad. Sci. USA. 2005;102:4584–4589. doi: 10.1073/pnas.0500815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Liby KT, Sporn MB. Synthetic oleanane triterpenoids: multifunctional drugs with a broad range of applications for prevention and treatment of chronic disease. Pharmacol. Rev. 2012;64:972–1003. doi: 10.1124/pr.111.004846. [DOI] [PMC free article] [PubMed] [Google Scholar]