Abstract
Microfluidic platforms offer a drastic increase in throughput while minimizing sample usage and hands-on time, which make them important tools for large-scale biological studies. A range of such systems have been developed for enzyme activity studies, although their complexity largely hinders their application to the wider scientific community. Here, we present adaptation of an easy-to-use commercial microfluidic qPCR system for performing enzyme kinetic studies. We demonstrate the functionality of the Fluidigm Biomark HD system (the Fluidigm system) by determining the kinetic properties of three oxidases in a resorufin-based fluorescence assay. The results obtained in the microfluidic system proved reproducible and comparable to the ones obtained in a standard microplate-based assay. With a wide range of easy-to-use, off-the-shelf components, the microfluidic system presents itself as a simple and customizable platform for high-throughput enzyme activity studies.
Introduction
In the past decade, we have witnessed a drastic increase in genomic data being deposited in public databases, which provides the potential for the discovery of new enzymatic activities and a deeper understanding of the known ones.1−3 However, the current experimental methods cannot keep up with the characterization of novel genes, resulting in a chronic lack of validated data in public databases.4 Currently, the most common methods for in-depth enzyme characterization rely on manual preparation of enzyme–substrate mixtures in a microplate, which is time-consuming and requires large amounts of proteins and substrates. This usually result in a limited range of conditions and substrates being tested and hinder the exploration of enzymatic potential of a protein. A number of high-throughput methods have been developed for both end-point and kinetic measurements of enzyme activity;5−7 however, their adaptation by the wider scientific community proves difficult, as it requires specialized knowledge and access to microfluidic manufacturing facilities.8 A simple method for performing enzyme kinetic studies in readily available high-throughput off-the-shelf devices would help to address this problem.
Although no high-throughput microfluidic devices for measuring enzyme kinetics are commercially available, such instruments were developed for real-time qPCR applications.9,10 One such system was established by Fluidigm Corporation, where gene expression analysis is carried out in integrated fluidic circuit chips, with a setup that offers an increase in throughput and reduction in sample volumes, yet remaining user-friendly.11 In the Fluidigm system, samples and reagents are first pressure-loaded into nanoliter-sized reaction chambers of the chip, which is then transferred to a real-time PCR instrument designed to thermal cycle the microfluidic chips and image the data in real time. The Fluidigm ecosystem, including a number of different chips and a range of available fluorescent filters (Table S1), indicates its potential for novel applications, in addition to qPCR. In this work, we show that a qPCR microfluidic system can be easily adapted for enzyme kinetic studies with improved throughput and decreased sample usage over conventional methods.
Results
Characterization of Experimental Platform: a Microfluidic qPCR Device for Enzymatic Assays
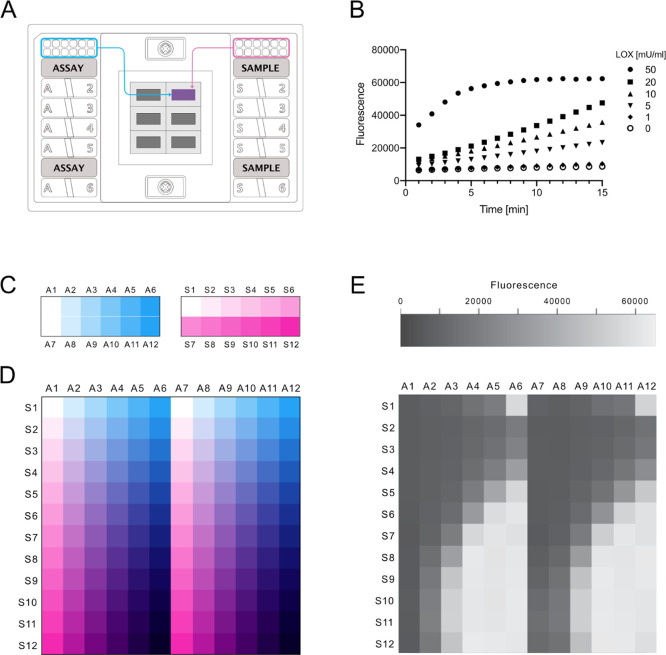
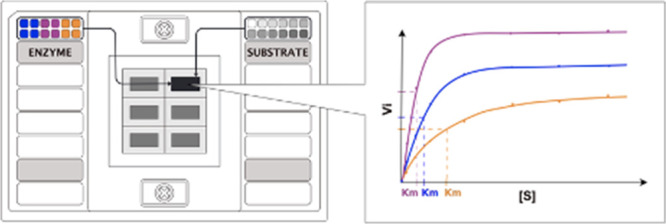
To assess the suitability of the Fluidigm system for enzyme activity screening, we chose a FlexSix Gene Expression IFC chip which contains six partitions, each with 12 wells on the assay side and 12 wells on the sample side (Figure 1A). The chip provides a medium range of throughput: from 144 up to 864 reactions per chip, depending on how samples are routed in the chip. As a model enzyme, we selected the hydrogen peroxide-producing enzyme lactate oxidase (LOX, EC 1.1.3.2). The activity of lactate oxidase can be detected using a simple coupled fluorescence assay, in which a nonfluorescent probe reacts with hydrogen peroxide to produce a fluorescent product, resorufin (Figure S1).
Figure 1.
Exploration of the Fluidigm gene expression system as a platform for enzyme activity measurements. (A) Schematic representation of a FlexSix microfluidic chip. (B) Example of the obtained initial reaction rates of lactate oxidase in reaction with lactate in a buffer with composition 0.01% Triton-X-100 and Tween 20 and addition of BSA. (C) Experimental setup for measurements of enzyme kinetic parameters in the Biomark HD system. Schematic overview of enzyme loading with 6 different concentrations (blue) in the “assay” input wells and loading of the substrate with 12 different concentrations (magenta) in the “sample” input wells. (D) Schematic overview of the sample/assay gradient that results from the all vs all mixing that occurs in the FlexSix chip. All enzyme concentrations are mixed with all substrate concentrations. In the illustrated scheme, two technical duplicates of each enzyme concentration are run. (E) Example of resorufin fluorescence intensity values recorded in the run. Here, for lactate oxidase, it is carried out at cycle 60.
We first tested over what range resorufin fluorescence was linear in the Fluidigm system by measuring a range of resorufin concentrations. Our results indicated that it is possible to capture the linear response for up to 5 μM of resorufin, which is comparable to data obtained in a microplate reader (Figure S2). Next, we asked whether enzyme activity could be captured in the system and measured the increase of fluorescence over time for different enzyme concentrations with 5 mM lactate. To lower the adhesion of protein to the chip’s channels and minimize protein precipitation, we tested two concentrations of nonionic detergents in a buffer, in the presence or absence of BSA. Additionally, we used the fluorescein dye as a loading control to inspect whether the mixing of the sample is consistent throughout the chip. The results showed that we were able to not only detect the activity of the enzyme but also capture its initial reaction rates (Figure 1B). Addition of BSA did not influence the results, while a higher concentration of detergents provided more consistent signal readouts throughout replicates but resulted in much higher overall background fluorescence (Figure S3). Readouts of the fluorescent signal from the fluorescein loading control indicate equal sample loading throughout the chip, with higher detergent concentrations leading to more reproducible loading (Figure S4). Overall, the results of the initial screen show the potential of the Fluidigm system for measuring the enzyme activity.
Measurement of Enzyme Kinetics in the Fluidigm Microfluidic System
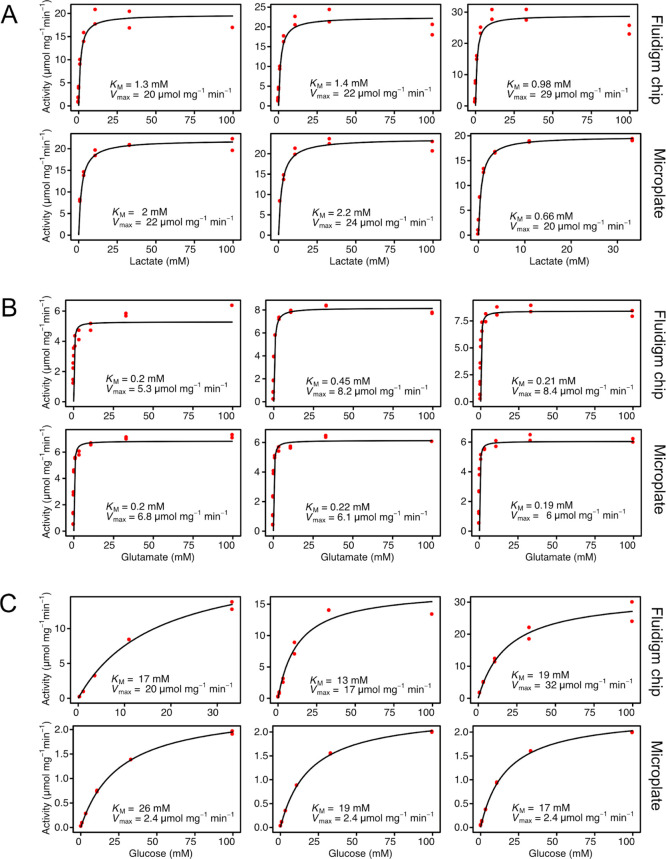
Next, we evaluated whether the Fluidigm system is reliable for studying enzymatic Michaelis–Menten kinetics. To test this, we measured the initial reaction rates of three hydrogen peroxide-producing enzymes: lactate oxidase, glucose oxidase (EC 1.1.3.4), and glutamate oxidase (EC 1.4.3.11). For each of the three oxidases, 5 different enzyme concentrations were placed in duplicate on the assay side of the FlexSix chip and 11 substrate concentrations were placed on the sample side (Figure 1C). During sample loading, in each partition, all enzyme samples were separately mixed with all substrate samples, creating an enzyme–substrate gradient (Figure 1D,E). The use of different enzyme concentrations enabled capturing kinetic information in one run only, without the need of previous knowledge of enzyme activity. The final enzyme concentrations were between 9.6 ng/mL and 6 μg/mL and the final substrate concentrations were between 1.7 μM and 100 mM. To enable rapid analysis of this data, we developed R-scripts which automatically identify the enzyme concentration for which initial rates are linear, fit a linear regression model to those data points, and extract the slope.
When establishing a new method, it is of key importance to validate it. We therefore set out to test the system’s reproducibility as well as accuracy. In order to test the reproducibility of the system, we repeated the assay for each enzyme three times using three different FlexSix chips. The obtained kinetic parameters of the three replicates were highly similar (Table 1, Figure 2), indicating outstanding reproducibility of the method. As a validation of system’s accuracy, we compared the results obtained in the Fluidigm system with the ones obtained from assays performed in a standard 384-well plate with the signal recorded in a fluorescent plate reader (Table 1, Figure 2). The values obtained in the microfluidic chip are comparable with the ones obtained in a plate reader and reported in the literature (Table 1). The largest difference in the obtained results is between Vmax values of glucose oxidase (microplate: 2.4 μmol mg–1 min–1; chip: 23 μmol mg–1 min–1), most likely due to enzyme stock dependency, which is supported by the fact that the range of Vmax reported in the literature is very broad (6–170 μmol mg–1 min–1). Overall, the results show that the performance of the Fluidigm system in measuring enzyme kinetics is on par, in terms of data quality, with that of a standard plate system used routinely today.
Table 1. Comparison of Kinetic Values of Three Oxidases Obtained in the Fluidigm System and a Microplate Reader.
| enzyme | system | KM [mM]a | Vmax [μmol mg–1 min–1]a |
|---|---|---|---|
| lactate oxidase | Microplate | 1.62 ± 0.48 | 22.00 ± 1.15 |
| Chip | 1.23 ± 0.12 | 23.67 ± 2.73 | |
| Literatureb | 0.5–1 | 114–270 | |
| glutamate oxidase | Microplate | 0.20 ± 0.01 | 6.30 ± 0.25 |
| Chip | 0.29 ± 0.08 | 7.30 ± 1.00 | |
| Literatureb | 0.17–0.3 | 6–55 | |
| glucose oxidase | Microplate | 20.67 ± 2.72 | 2.40 ± 0.00 |
| Chip | 16.33 ± 1.76 | 23.00 ± 4.58 | |
| Literatureb | 22–32 | 6–170 |
Values for the microplate and the chip represent the mean average (±standard error of mean; n = 3).
Literature values ranges, as reported for wild-type enzymes in BRENDA DB.12
Figure 2.
Comparison of kinetic values obtained in the Fluidigm system and a plate reader. Michaelis–Menten curves of three repeats in each system for the three tested oxidases: (A) Lactate oxidase. (B) Glutamate oxidase. (C) Glucose oxidase.
Discussion
In this work, we show that a microfluidic qPCR system can be applied in biochemistry without modification and is suitable for determining enzyme kinetic parameters. Although we have only tested one microfluidic system in this work, the method can likely be adapted to systems from other manufacturers. The Fluidigm system offers the well-known advantages of microfluidic devices over conventional methods: increase of throughput and drastic reduction in both reagent volumes used, as well as manual handling time involved in performing experiments.5 Using only half the capacity of the smallest Fluidigm chip, it was possible to establish reliable kinetic values for three different enzymes in one run (432 reactions), whereas three separate runs had to be performed using microplates for validation comparison. Mixing of the enzyme and the substrate is performed automatically in the microfluidic chip, which reduces the manual handling required, while the reaction volume also decreased over 2000-fold (from 20 μL to 8.9 nL).
Like many microfluidic devices, the Fluidigm system used here comes with the drawback of only being suitable for fluorescence-based assays, which limits the type of enzyme activities that could potentially be monitored. This, however, is becoming less of an issue with an increase of new fluorescent enzyme screens being developed.13 The majority of microfluidic systems for measuring enzyme kinetics allow creating a wide concentration gradient of substrate in a programmable manner,7 which results in much higher precision in comparison to standard methods. In the Fluidigm system, a limited number of substrate dilutions are prepared by hand, similarly to a standard microplate assay, which lowers the precision and increases the chance of manual-handling error. However, the system presented in this work provides a clear advantage over the previously described methods: low system complexity, which allows users to operate it with commercially available equipment and software, without a fluid handling expertise. This is an important advantage, as the lack of easy to use, standardized systems is a major cause for low adaptation of microfluidics by nonspecialists such as biochemists.8 Although high-throughput qPCR systems such as Fluidigm are not yet common in a standard laboratory, they are becoming increasingly accessible as the need for gene expression analysis continues to grow and new genomic core facilities are being established.
A further advantage of the system over the existing microfluidic devices is that it allows parallelization of many enzyme kinetic measurements at the same time, with the possibility of testing different enzymes, substrates, and conditions in a single run. Availability of many easy to use, off-the-shelf chips makes the Fluidigm system a good platform for different experiment designs for enzyme screening. The system allows for both smaller scale experiments in FlexSix IFCs and larger scale ones, all-versus-all studies using the larger 96.96 IFC; where up to 9216 separate reactions can be performed simultaneously. In our study, we present the successful use of one type of chip and two fluorescent probes: resorufin and fluorescein, which present a potential of the system for testing a wide range of enzymes such as oxidoreductases, phosphatases, proteases, galactosidases, or deadenylases. However, in order to explore the full potential of the system, additional experiments with a wider range of chips, enzymatic activities, and substrates would be required. We believe that adapting a microfluidic gene expression system for measuring enzyme activity can facilitate the growing need for in-depth, yet high-throughput characterization of enzymes.
Materials
The Flex Six Gene Expression IFC (product number 100-6308) and Control Line Fluid Kit (product number 89000021) were purchased from Fluidigm Corporation, United States. All enzymes and chemicals were purchased from Sigma Aldrich, unless stated otherwise.
Methods
Fluidigm System Setup
The Flex Six Gene Expression IFC was used for all microfluidic experiments. The Flex Six chip has a total of six partitions which can be run either independently or simultaneously. Each partition has a 12 × 12 format, in which solutions from 12 assay inlets are mixed with solutions from 12 sample inlets in a 1:9 ratio. The final reaction volume is 8.9 nL. Samples placed in the assay inlets were prepared as 10× concentrations and solutions placed in the sample inlets were prepared as 1.1× concentrations. Before the first use of each IFC, the chips were primed with the Control Line Fluid Kit in Juno System (Fluidigm, United States) according to manufacturer’s guidelines. To load the IFC, barrier plugs were removed from the partitions in use, and 3 μL each of assay and sample was pipetted into their respective inlets. Caution was taken not to introduce air bubbles while pipetting. Next, the IFC was placed in the Juno machine and the “Load Mix Flex Six GE” was run. Immediately after the loading script was finished, the IFC was transferred to a Biomark HD machine (Fluidigm, United States) and data collection protocol was set up using Fluidigm’s Data Collection Software. After selecting partitions for the run, the “Gene Expression” application type was chosen, ROX was chosen as the “passive reference” (excitation filter: 575 nm; emission filter: 630 nm) and FAM-MGB as the “probe” (excitation filter: 475 nm; emission filter: 525 nm). The ROX channel was used to collect the signal from resorufin, and FAM-MGB was used to collect the signal from fluorescein-loading control, which is the inverse of what is done in a typical qPCR experiment. The chip run protocol was loaded and exposure was set to 0.03 s for ROX and 2 s for FAM. The chip run protocol was created using Fluidigm’s Real-Time PCR Analysis Software: measurements were taken every 30 s for 30 min, at 25 °C. After the run was finished, the IFC was placed in Juno and the “Post Run Flex Six GE’’ script was run in order to relax the valves.
Enzyme Assays in the Fluidigm Platform
All enzyme assays were performed in a buffer containing a final concentration of 20 mM HEPES pH 7.4, 50 μM Ampliflu Red, 0.1 U/mL horseradish peroxidase (HRP), 0.01% Tween 20, 0.01% Triton X 100, 0.01 mg/mL BSA, and 0.1 μM fluorescein, unless stated otherwise. However, based on the microfluidic chip function, solutions from assay and sample sides are mixed at a ratio of 1 to 9. One must therefore make use of suitably higher initial concentrations in the assay and sample wells to ensure correct final concentrations inside the chip. 10× concentrated Ampliflu Red, HRP, Tween 20, Triton X 100, BSA, fluorescein, and enzyme were placed in the assay inlet, whereas 1.1× concentrated fluorescein along with substrates were placed in the sample inlets.
For capturing enzyme initial rates (Figure 1), five concentrations of lactate oxidase (1, 5, 10, 20, and 50 mU/mL) were assayed with 5 mM lactate in the buffer listed above as well as buffers containing no BSA and 0.1% of the detergents Tween 20 and Triton-X-100.
For obtaining kinetic constants, lactate oxidase from Aerococcus viridans (product no. L9795), glucose oxidase from Aspergillus niger (product no. G2133), and glutamate oxidase from Streptomyces sp. (product no. G5921) were assayed with lactate, glucose, and glutamate, respectively. The final enzyme concentrations used, in duplicate, are as follows: 6 μg/mL, 1.2 μg/mL, 240 ng/mL, 48 ng/mL, 9.6 ng/mL, and 0 ng/mL. The following are the final substrate concentrations used (inside reaction chambers): 100 mM, 33.33 mM, 11.11 mM, 3.7 mM, 1.23 mM, 412 μM, 137 μM, 47 μM, 15 μM, 5 μM, 1.7 μM, and 0 μM.
Enzyme Assays in Microplates
For kinetic constant calculations, the final concentrations of enzymes and substrates used were the same as for enzymatic assays in the Biomark HD system (see above). The assays were performed in low-volume 384-well black flat bottom plates (Greiner) and the reaction volume was 20 μL. The assay buffer contained 20 mM HEPES pH 7.4, 50 μM Ampliflu Red, and 0.1 U/mL HRP. The assays were started by the addition of substrate to the buffer-enzyme mix and the readouts were carried out every 30 s for 30 min with an excitation filter of 544 nm and an emission filter of 590 nm in a FLUOstar Omega microplate reader (BMG Labtech, Germany).
Standard Curves
For obtaining a resorufin standard curve for the Fluidigm system, nine resorufin dilutions were prepared in 20 mM HEPES buffer, pH 7.4 with 0.11 μM fluorescein and pipetted into the respective sample inlets, together with a no resorufin control. The buffer containing 20 mM HEPES pH 7.4, 1.1 μM fluorescein, 0.11% Tween 20, 0.11% Triton X 100, and 0.11 mg/mL BSA was placed in the assay inlets. Because of the all versus all mixing of samples and assays inside the chip, this results in multiple technical replicates for each concentration. The final resorufin concentrations in the microfluidic chambers were 10, 5, 2.5, 1.25, 0.625, 0.313, 0.156, 0.079, 0.04, and 0 μM.
For obtaining a resorufin standard curve using the FLUOstar Omega microplate reader, nine resorufin dilutions were prepared in triplicates in 20 mM HEPES buffer, pH 7.4, together with a no resorufin control. The final resorufin concentrations were 12.5, 6.25, 3.13, 1.56, 0.78, 0.39, 0.195, 0.097, 0.049, and 0 μM. Measurements were obtained using an excitation filter of 544 nm and an emission filter of 590 nm.
Data Analysis
Data obtained from the Fluidigm instrument
were analyzed as follows. After a finished run, corners of the chip
were adjusted manually in Fluidigm’s Real-Time PCR Analysis
Software and the quantification of fluorescence intensities in the
ROX and FAM channels was carried out automatically using software.
The run data containing fluorescence intensity values were exported
in a *.csv format. Two input files describing the sample layout on
both “assay” and “sample” sides were manually
created. Custom scripts in R version 3.4.4 (www.r-project.org) were used
to analyze data. Briefly, the resorufin fluorescence (ROX channel)
was background-subtracted and converted to product concentration using
a standard curve. Samples for which no activity was observed (typically
those with enzyme loading and low-substrate concentrations) were removed.
Regression models were fitted to the linear range of data and the
slopes were calculated to obtain resorufin production per minute (μM
min–1). The enzyme specific activity (μmol
mg–1 min–1) was finally calculated
using a reaction volume of 8.87 nL and the protein concentration used
in each reaction. Finally, the enzyme concentration yielding the best
linear model fit to the activity data (greatest R2), as
well as the highest Spearman correlation between the substrate concentration
and the reaction rate, was automatically selected and fitted to the
Michaelis–Menten equation: 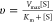 . Scripts
and raw data are available at
GitHub (https://github.com/EngqvistLab/biomark_assays).
. Scripts
and raw data are available at
GitHub (https://github.com/EngqvistLab/biomark_assays).
Data obtained from the FLUOstar Omega plate reader were analyzed in the same manner as data from the Fluidigm instrument. The only difference is that fluorescence intensities did not have to be extracted and the reaction volumes were 20 μL.
Acknowledgments
M.K.M.E. thanks Daniel Cook, Anette Støttrup, and Daniel Foldager Larsen for initial discussions on the feasibility of using the Biomark system for enzyme assays. The authors thank Daniel Dancer for crucial technical assistance and Petter Woll for graciously granting access to his Biomark HD system, thus enabling these experiments to be carried out.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c04918.
Fluorescent filters in the Fluidigm system, reaction scheme of fluorescence assay for hydrogen peroxide detection, resorufin fluorescence in the Fluidigm system and microplate reader, influence of buffer composition on fluorescence readout in the Fluidigm system, and distribution of fluorescent signal of the loading control fluorescein (PDF)
Author Contributions
M.K.M.E. conceptualized the project. E.R. and M.K.M.E. designed the experiments. E.R. carried out th experiments. M.K.M.E. conceived and implemented the data analysis pipeline. E.R. and M.K.M.E. performed the data analysis. E.R. wrote the draft manuscript. E.R. and M.K.M.E. carried out revisions on the initial draft and wrote the final version.
The authors declare no competing financial interest.
Notes
All raw data and computer code to process these are made freely available for re-use (https://github.com/EngqvistLab/biomark_assays).
Supplementary Material
References
- UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land M.; Hauser L.; Jun S.-R.; Nookaew I.; Leuze M. R.; Ahn T.-H.; Karpinets T.; Lund O.; Kora G.; Wassenaar T.; Poudel S.; Ussery D. W. Insights from 20 Years of Bacterial Genome Sequencing. Funct. Integr. Genomics 2015, 15, 141–161. 10.1007/s10142-015-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt J. A. The Need for Manuscripts To Include Database Identifiers for Proteins. Biochemistry 2018, 57, 4239–4240. 10.1021/acs.biochem.8b00585. [DOI] [PubMed] [Google Scholar]
- Furnham N.; Garavelli J. S.; Apweiler R.; Thornton J. M. Missing in Action: Enzyme Functional Annotations in Biological Databases. Nat. Chem. Biol. 2009, 5, 521–525. 10.1038/nchembio0809-521. [DOI] [PubMed] [Google Scholar]
- Bunzel H. A.; Garrabou X.; Pott M.; Hilvert D. Speeding up Enzyme Discovery and Engineering with Ultrahigh-Throughput Methods. Curr. Opin. Struct. Biol. 2018, 48, 149–156. 10.1016/j.sbi.2017.12.010. [DOI] [PubMed] [Google Scholar]
- Longwell C. K.; Labanieh L.; Cochran J. R. High-Throughput Screening Technologies for Enzyme Engineering. Curr. Opin. Biotechnol. 2017, 48, 196–202. 10.1016/j.copbio.2017.05.012. [DOI] [PubMed] [Google Scholar]
- Hess D.; Yang T.; Stavrakis S. Droplet-Based Optofluidic Systems for Measuring Enzyme Kinetics. Anal. Bioanal. Chem. 2020, 412, 3265–3283. 10.1007/s00216-019-02294-z. [DOI] [PubMed] [Google Scholar]
- Fernandes A. C.; Gernaey K. V.; Krühne U. Connecting Worlds - a View on Microfluidics for a Wider Application. Biotechnol. Adv. 2018, 36, 1341–1366. 10.1016/j.biotechadv.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Shembekar N.; Chaipan C.; Utharala R.; Merten C. A. Droplet-Based Microfluidics in Drug Discovery, Transcriptomics and High-Throughput Molecular Genetics. Lab Chip 2016, 16, 1314–1331. 10.1039/c6lc00249h. [DOI] [PubMed] [Google Scholar]
- Volpatti L. R.; Yetisen A. K. Commercialization of Microfluidic Devices. Trends Biotechnol. 2014, 32, 347–350. 10.1016/j.tibtech.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Spurgeon S. L.; Jones R. C.; Ramakrishnan R. High Throughput Gene Expression Measurement with Real Time PCR in a Microfluidic Dynamic Array. PLoS One 2008, 3, e1662 10.1371/journal.pone.0001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske L.; Placzek S.; Schomburg I.; Chang A.; Schomburg D. BRENDA in 2019: A European ELIXIR Core Data Resource. Nucleic Acids Res. 2019, 47, D542–D549. 10.1093/nar/gky1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acker M. G.; Auld D. S. Considerations for the Design and Reporting of Enzyme Assays in High-Throughput Screening Applications. Perspectives in Science 2014, 1, 56–73. 10.1016/j.pisc.2013.12.001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.