Abstract
Protein myristoylation, the addition of a 14-carbon saturated acyl group, is an abundant modification implicated in biological events as diverse as development, immunity, oncogenesis, and infections. N-myristoyltransferase (NMT) is the enzyme that catalyzes this modification. Many elegant studies have established the rules guiding the catalysis including substrate amino acid sequence requirement with the indispensable N-terminal glycine, and a co-translational mode of action. Recent advances in technology such as the development of fatty acid analogs, small molecule inhibitors, and new proteomic strategies, allowed a deeper insight into the NMT activity and function. Here we focus on discussing recent work demonstrating NMT as also a lysine myristoyltransferase, the enzyme’s regulation by a previously unnoticed solvent channel, and mechanism of NMT regulation by protein-protein interactions. We also summarize recent findings of NMT’s role in cancer, immunity and infections, and the advances in pharmacological targeting of myristoylation. Our analyses highlight opportunities for further understanding and discoveries.
Graphical Abstract
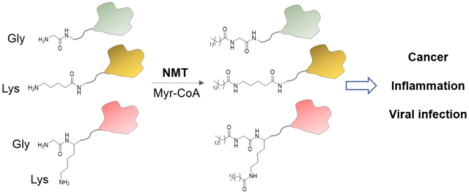
Introduction
N-myristoyltransferase (NMT) is a ubiquitous eukaryotic enzyme that for decades has been known to have a single role – myristylation of the amino group of the N-terminal glycine on proteins. This function is of high specificity guided by several factors such as substrate sequence requirements, protein interactions, and expression levels. N-myristoylation is the addition of a 14-carbon chain to the alpha amine (Nα) of the N-terminal glycine on proteins, exposed after the initiator methionine is cleaved by methionine aminopeptidase during translation, or after caspase cleavage during proteolytic events such as apoptosis. This modification often regulates protein-membrane or protein partner binding and is thought to be irreversible although there is some evidence of an ATP-dependent demyristoylase in the cytoplasmic fraction of the brain synaptosomes.1 Recent findings implicate N-myristoylation in protein stability, where free N-terminal glycine is recognized by E3 ubiquitin ligases leading to degradation of unmyristoylated proteins.2 NMT is essential for the survival of many eukaryotic organisms such as S. cerevisiae3 and pathogens C. albicans, T. brucei,4 C. neoformans5, and many viruses utilize host NMT for replication and infectivity,6 which made the enzyme an attractive therapeutic target. Furthermore, NMT knockout causes embryonic lethality in mice and Drosophila, pointing to its role in develepment.7, 8 Here we discuss the recent advances in understanding the NMT catalysis, especially its activity on lysine residues, and the enzyme’s potential for therapeutic use in cancer, immunity and infectious diseases, including viral infections. We note the areas that warrant further investigation such as reevaluation of eukaryotic and viral proteomes for lysine substrates of NMT and provide a brief update on the progress in the development of NMT-selective small molecule inhibitors.
Structure and regulation of NMT catalysis
Several studies have elucidated that the catalytic mechanism of NMT follows an ordered bi-bi reaction where myristoyl-CoA binds to the enzyme in a bent fashion resembling a “question mark”, which causes a conformational change allowing the peptide substrate to bind (Figure 1A). The interaction of the thioester carbonyl with the backbone amides of the enzyme (Phe 247 and Leu 248 in human NMT1) forms an oxy-anion hole activating the carbonyl for the nucleophilic attack by the Gly2 amine of the peptide substrate.9, 10 This reaction was thought to occur only with the N-terminal glycine, not any other amino acids because the absence of a side chain allows free rotation of the N-terminal amine necessary for the nucleophilic attack on the carbonyl carbon of the thioester bond of myristoyl-CoA.11 Later we will discuss recent studies that elegantly demonstrated the activity of NMT on lysine. Recently, a detailed reaction mechanism revealed that the C-terminus carboxylate of Gln496 acts as an indirect catalytic base initiating the water-mediated deprotonation of the N-terminus amino group. The carboxylate is located at the end of a 22 Å long previously unnoticed solvent channel that is thought to be important for the deprotonation.10 After catalysis, CoA is released followed by the myristoylated product.12 In addition to the N-terminal Gly, the peptide substrate specificity is dictated by other residues near the N-terminus. Although not an absolute requirement, NMT often prefers substrates with the sequence GXXXSK.13–16 Ser 6 interacts with a small hydrophilic pocket explaining the enzyme’s preference for this position.13, 14, 16 Lys 7 of the peptide substrate allows a tight binding and occupies a distinct negatively charged pocket with three aspartate residues that stabilize the substrate binding via a salt bridge between the Asp carboxylate and the epsilon amino group (Nɛ) of the lysine.14, 16 The sequence recognition is rather mosaic suggesting that NMT can accommodate various substrates and that other factors such as interacting partners might regulate substrate specificity in cells.15 For instance, the known NMT substrate ARF1 has the N-terminal sequence GNIFAN, which does not conform to the NMT preference. This example emphasizes the need to keep an open mind while examining potential NMT substrates. Furthermore, there are variations in sequence preference across species,14, 16, 17 suggesting the need for species-specific predictive tools of N-terminal myristoylation, which can be of use for developing NMT-targeted therapies.
Figure 1.
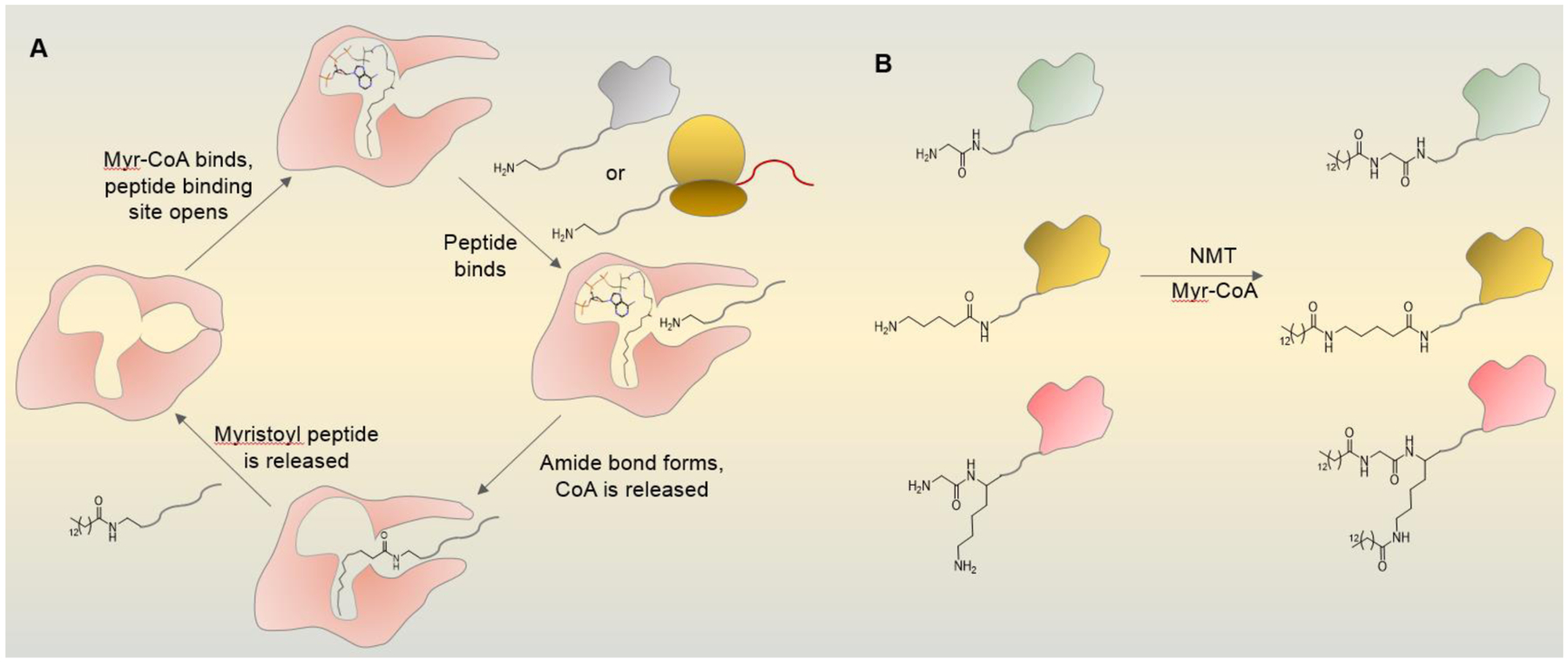
a) Mechanism of NMT-catalyzed myristoylation. Myristoyl-CoA binds to NMT in a question mark-like fashion. This causes a conformational change that opens the peptide-binding site. Peptide substrate of a folded protein or nascent peptide with a sterically unhindered N-terminal amino group binds to NMT allowing amide bond formation between the amino group and the myristoyl group and release of CoA. The modified peptide exits the active site and the enzyme is ready for the next catalytic cycle. b) NMT can myristoylate the amino groups of N-terminal glycine, lysine, or both.
The exceptional preference of NMT for myristoyl-CoA remains puzzling given that in cells myristic acid comprises less than 1% of total fatty acids compared to about 20 % for palmitic acid.18 Structural work suggests that the length of the NMT hydrophobic pocket is best suited for a myristoyl chain,9 which partially explains this selectivity. Yet under some conditions, NMT can efficiently utilize other acyl-CoAs. For instance, the introduction of a triple bond at C6 of the acyl chain of palmitoyl-CoA results in a strong enhancement of the activity of S. cerevisiae Nmt, which is thought to result from inducing a bend in palmitoyl-CoA where it would naturally occur in myristoyl-CoA bound to NMT.19 It was also shown that on the peptide GARASVLS-NH2 derived from the HIV Gag protein the myristoyl-peptide formation was progressively decreased as the acyl-CoA chain was shortened with nearly no acylation upon shortening to 7 carbons.20 Whether these trends of acyl-CoA utilization efficiency depend on the peptide substrate in vitro and in cells is unclear. Yet some reports suggest a similar binding affinity for myristoyl-CoA and palmitoyl-CoA21, 22 and explain that NMT is “protected” from other acyl-CoAs by the acyl-CoA binding protein ACBD6.22–24 The ACBD6 binding to NMT via its ankyrin-repeat motif (ANK) allosterically activates the enzyme for myristoyl-CoA acceptance independently from the acyl-CoA binding domain of ACBD6. The ANK domain alone is able to stimulate NMT and it produces the same effect when attached via the linker domain to ACBD1 protein, which otherwise does not regulate NMT.24 At the same time, myristoyl-CoA bound to ACBD6 can be transferred to NMT. When NMT is introduced to myristoyl-CoA bound ACBD6, the formation of myristoyl-peptide is observed suggesting that ACBD6 hands myristoyl-CoA over to NMT; and this process is potentiated by ACBD6 phosphorylation at Ser106 and Ser108.23 Of note is that ACBD6 binds to NMT2 about 10-fold better than to NMT1 potentially acting as a regulator of the differences between the two NMTs.22 While it is clear that ACBD6 acts as an activator of NMT, the underlying mechanism warrants clarification on whether and how it guards the acyl-CoA specificity or whether it allosterically promotes peptide substrate binding. NMT does not seem to efficiently utilize acyl-CoAs with shorter chains such as the far more abundant acetyl-CoA, likely because the binding affinities of these short-chain acyl-CoA are too low.
There is also evidence for substrate-specific regulation of NMT activity. The cellular senescence-inhibited gene (CSIG) was shown to enhance the interaction between NMT1 and the serine/threonine protein phosphatase PPM1A promoting PPM1A myristoylation, which enhances its phosphatase activity on SMAD2 to inhibit TFG-β signaling.25 Other interactions might regulate NMT localization. While NMT is thought to be largely cytosolic, its interactions with calnexin26 and ribosomes27 are thought to keep some of the enzyme at the endoplasmic reticulum.
In vitro, a truncation of 28 amino acids from the N-terminus of NMT1 catalytic domain resulted in a ~3 fold increase in the enzyme activity suggesting that additional post-translational modifications can further modulate the enzyme function.28 During apoptosis, proteolytic cleavage regulates NMT localization. Both human NMT1 and NMT2 are cleaved in a time-dependent manner upon induction of apoptosis with Staurosporine and anti-Fas antibody in Hela and Jurkat T cells.29, 30 NMT1 is cleaved by Caspases-3 and −8 at Asp81, while NMT2 is cleaved at Asp 25 by caspase-3. This leads to the re-localization of NMT1 from the membrane to the cytosol, while the opposite is observed for NMT2.30 The truncated NMTs appear to remain the same activity level, but are thought to recognize substrates that can act either in a pro-survival or pro-apoptosis manner. At the same time proteolysis generates new substrates of NMT.29 For example, a pro-apoptotic protein BID is cleaved by caspase 8, which exposes an N-terminal glycine on a 15 kDa fragment that gets myristoylated by NMT.31 This targets the cleaved BID to the outer mitochondrial membrane to promote cytochrome c release and subsequent cell death. The discovery of this sophisticated molecular switch regulating apoptosis opened an avenue for understanding the mechanism of posttranslational myristoylation by NMT.
Human NMT1 and NMT2 are 77% identical at the amino acid sequence with the greatest divergence at their N-termini. In basal conditions they appear to have largely overlapping substrates, so their differential processing during proteolytic stress might result in more divergent substrate profiles, yet this warrants more investigation. Interestingly, NMT2 knockdown in SKOV-3 cells induced apoptosis to a greater extent than NMT1 knockdown hinting that the two human NMT enzymes might have different functions in regulating cell survival.32 The catalytic domains of NMT1 and NMT2 are nearly identical and the most divergence comes from the N-terminus. There is no structure reported for the N-terminus of NMT1 or NMT2, which hinders the understanding of this region. The two NMTs have nonoverlapping roles as NMT2 is typically unable to compensate for NMT1 depletion. Further investigation into the potential regulation of this region by metabolite binding, protein or nucleic acid interactions and post-translational modification might shed light on the need for the two NMT enzymes in higher eukaryotes.
NMT is the first mammalian lysine fatty acyl transferase
It was recently established that human NMT1 and NMT2 also act as mammalian lysine fatty acyl transferases. While lysine fatty acylation has been known to occur on mammalian proteins for nearly two decades33, the enzymes catalyzing such lysine modifications remained unknown. The amino group of the N-terminal glycine and that of the lysine side chain are chemically similar displaying similar lengths and steric properties where both are free to rotate, hinting that both could react in the active site of NMT. The modification of a Lys3 residue would be similar to the modification of a Gly2 (Figure 2). In vitro and in cells NMT can myristoylate lysine residues positioned near the N-terminus with decreased catalytic ability as the lysine moves away from the N-terminus.34 The overall catalytic mechanism appears similar to that of the N-terminal glycine, yet the structural data suggest that the solvent channel may not facilitate the reaction on lysine as the C-terminus catalytic base interacts directly with the Nɛ in contrast to the indirect interaction through a water molecule with the Nα of the N-terminal glycine.10 The first cellular lysine myristoylation substrate is the small GTPase ADP-ribosylation factor 6 (ARF6), which has a Lys3 following the N-terminal Gly2. After the reaction at the N-terminus, the Gly-myristoyl might move into the solvent channel positioning Lys3 into the catalytic center. These studies expand the repertoire of the NMT substrates: it can act on glycine, lysine, or both when located at the N-terminus (Figure 1B).
Figure 2.
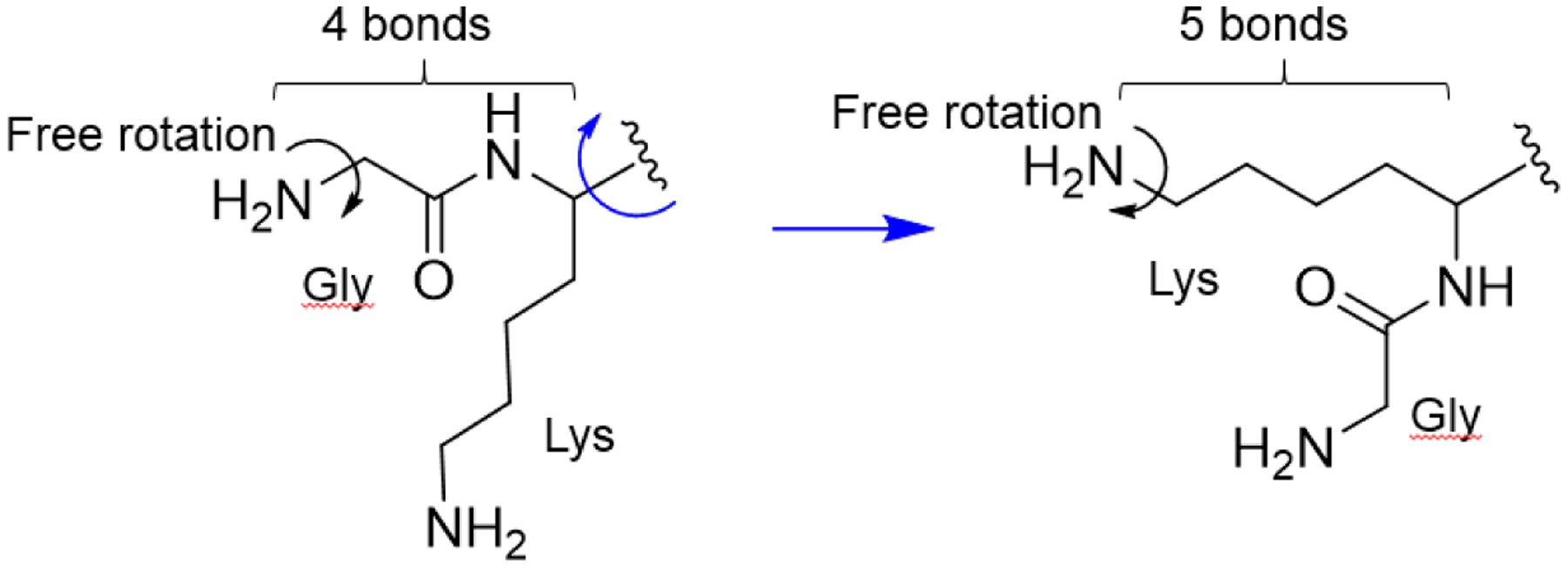
Similarities between the N-terminal glycine and lysine. In both cases, the amino groups are free to rotate and are a similar distance away from the Cα of lysine.
Interestingly, unlike other GNAT family enzymes, NMT contains two GNAT domains thought to result from gene duplication35 with the solvent channel in the second domain. This channel is lined with hydrophobic residues and is of a similar length to the canonical myristoyl pocket allowing the speculation that while the second domain lost its catalytic function, it might have retained its ability to hold a myristoyl group, which can explain the di-myristoylation ability of NMT. The second pocket, therefore, may serve as both the facilitator of the deprotonation event during Gly myristylation and the binding pocket for the Gly-myristoyl moiety during di-myristoylation. While the NMT sequence preference for lysine myristoylation is similar to that of glycine, in vitro and cellular studies suggest that the reaction on lysine might be regulated by the 3D structure of the substrate protein. Interestingly, in cells, NMT2 appears a better di-myristoyltransferase than NMT1, which again suggests an additional level of cellular regulation of this function.
Unlike glycine myristoylation, lysine myristoylation is reversible. SIRT2 is the eraser of ARF6 lysine myristoylation which is important for its localization and activation. Interestingly, it appears that SIRT2 prefers the GDP-bound form of ARF6 as the substrate, while NMT preferentially acts on ARF6-GTP. This substrate specificity allows the lysine myristoylation-demyristoylation cycle to connect to and drive the GTPase cycle (Figure 3). The newly identified reversible lysine myristoylation-demyristoylation cycle opens an avenue for future studies to further characterize the new function of NMT, identify additional substrates, and understand its other functional consequences.
Figure 3.
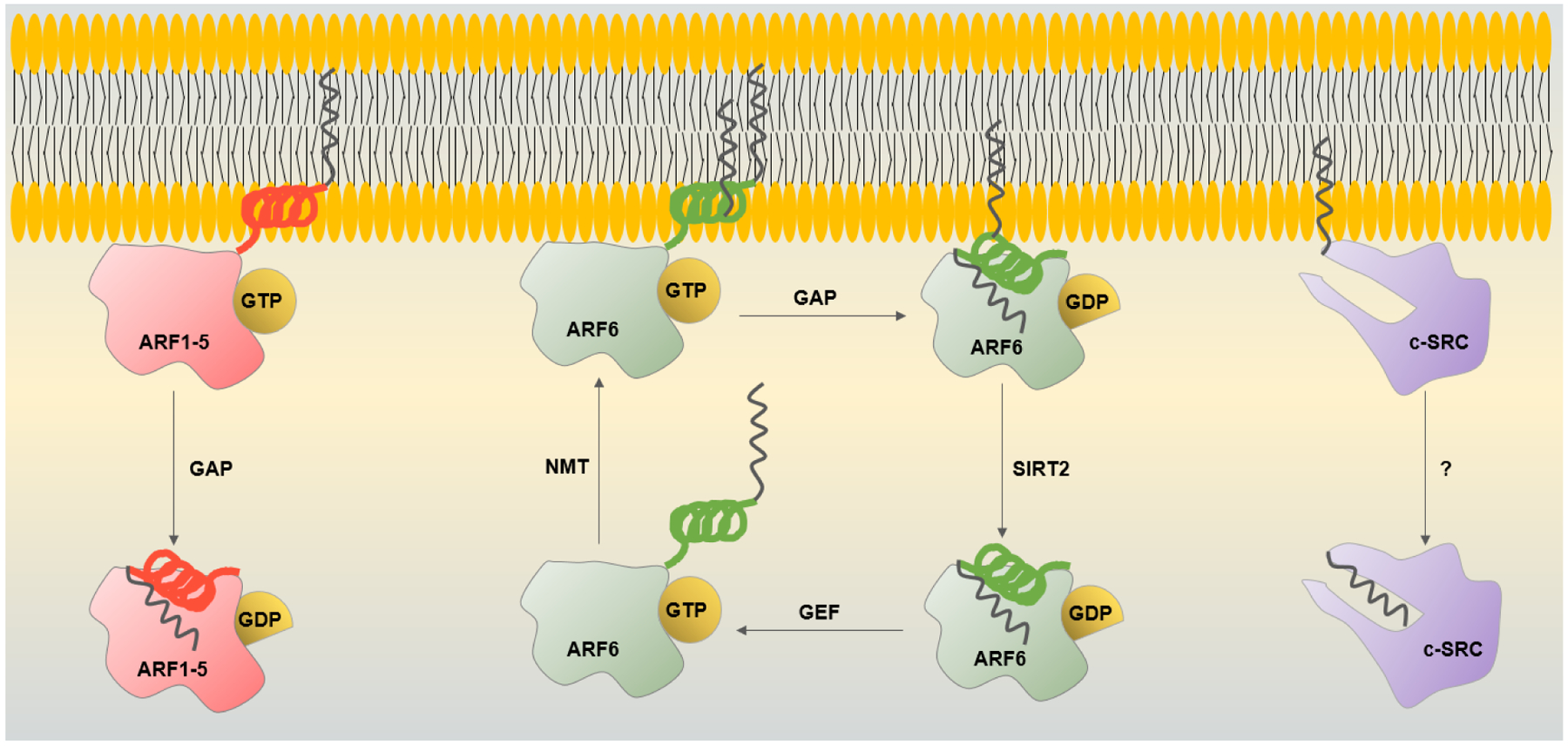
Myristoyl switches of ARF GTPases and c-Src. ARF1–5 are membrane-bound in the GTP-bound state via their amphipathic helix with one myristoyl group. The GTP hydrolysis facilitated by GAP causes a conformational change that sequesters the myristoylated helix releasing ARF1–5 from membranes. ARF6 has an additional myristoyl chain on lysine 3 that helps to retain ARF6-GDP on the plasma membrane and endomembranes. SIRT2 removes lysine myristoylation allowing ARF6-GDP to be activated by GEF. NMT myristoylates lysine 3 of ARF6-GTP, promoting its plasma membrane localization and completing the cycle. c-Src contains a hydrophobic pocket in its SH3 domain, that sequesters the myristoyl chain, but what causes the switch is unclear.
Functional effects of N-myristoylation
N-myristoylation most often serves as a protein membrane targeting signals such as that in the ARF, Src, and G protein families. The hydrophobic myristoyl chain can insert into the lipid bilayer anchoring proteins to membranes where they get activated and perform their roles. The G2A mutants of N-terminally myristoylated proteins are typically inactive. To date, no erasers of N-terminal glycine myristoylation have been identified, but the membrane-binding cycle is known to be regulated by other switch mechanisms. Ca2+-dependent myristoyl switches have been described for several proteins: among them are hippocalcin,36 recoverin37, and visin-like protein 3.38 The ARF proteins, like ARF1, use nucleotide binding to turn the myristoyl switch on and off. In the GDP bound state, the N-terminal helix along with its myristoyl chain is sequestered into a hydrophobic pocket, but the exchange to GTP facilitated by GEFs causes a conformation change that extrudes the myristoylated N-terminal helix allowing its insertion into membranes. Upon GTP hydrolysis accelerated by GTPase activating proteins (GAPs), the myristoyl moiety is again sequestered releasing the ARF protein from the membrane (Figure 3).39, 40 A myristoyl binding site was recently identified in the SH3 domain of c-Src. In the presence of lipid membranes, the myristoyl group is released and inserted into the lipid layer allowing c-Src activation and transforming ability.41 How c-Src switches between membrane-bound and cytosolic forms is an area of active research. In contrast to c-Src, the elegant study by Hantschel et al. demonstrated that the N-terminal myristoylation of another tyrosine kinase c-Abl does not regulate its membrane binding and is inhibitory to the enzyme’s kinase activity.42 The G2A mutation abrogates myristoylation leading to a dramatic increase in phosphotyrosine levels and kinase activity of c-Abl. Surprisingly, the mutation does not affect c-Abl localization as demonstrated by subcellular fractionation and macroscopy studies. This is a rare case of a myristoylation control of an enzyme activity that is independent of membrane binding.
A second membrane targeting signal such as a basic patch or palmitoylation of a nearby cysteine is often necessary for an efficient plasma membrane binding. Such mechanisms are utilized by the Src and G proteins where cysteine palmitoylation is catalyzed by one of the 23 human DHHC enzymes.43, 44 The lysine myristoylation on ARF6 discussed above also represents a second membrane targeting signal that is regulated by the enzyme SIRT2. ARFs 1–5 reside in the Golgi and regulate the ER-Golgi transport while ARF6 is found at the plasma membrane and endosomes and, unlike other ARFs tends to remain membrane-bound during its GTPase cycle (Figure 3).45 The second acylation on lysine allows efficient membrane anchoring of ARF6 and membrane retention even when it is inactive.34 Further studies are needed to understand how widespread this mode of regulation is in the mammalian proteome.
N-myristoylation promotes cancer progression
Targeting human NMT has recently emerged as a therapeutic strategy to treat cancer. Colon, gallbladder, and brain tumors have been shown to have elevated levels of NMT suggesting the reliance of cancer cells on protein myristoylation.46–48 The elevation of NMT2 in osteosarcoma might be contributing to the chemoresistance of this disease.49 Similarly, a down-regulation of miR-181c that among other genes targets NMT2 is found in imatinib-resistant chronic myeloid leukemia.50 This suggests that NMT2 levels can be used as a predictor of response to chemotherapeutics. Interestingly, an NMT inhibitor PCLX-001, which is also called DDD8648151 (Figure 4), is effective against NMT2 deficient blood cancers and is being advanced to clinical trials.52 The fact that it works well in NMT2-deficient blood cancers could be related to the chemoresistance effect of NMT2. Alternatively, it could be that lower levels of NMT2 make the cells more easily to be inhibited by PCLX-001.
Figure 4:
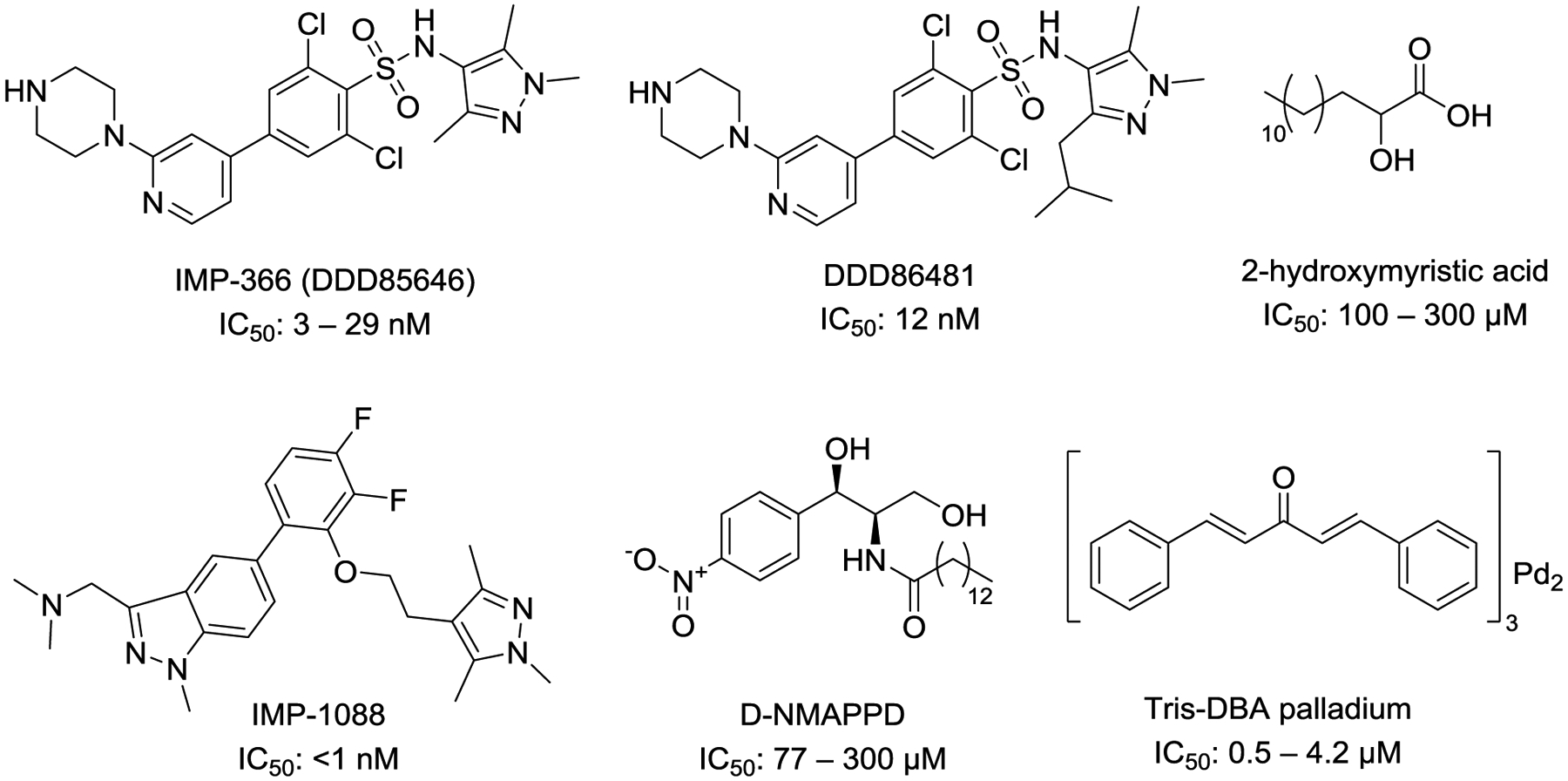
Structures and IC50 of small molecule NMT inhibitors.
There could be multiple mechanisms for the anticancer effects of NMT inhibition. In breast cancer, including triple-negative breast cancer, the knockdown of NMT1 suppresses tumor initiation, proliferation, and invasion by promoting oxidative and ER stress, which in turn activates autophagy via the JNK pathway.53 Quantitative proteomics revealed that the ER stress followed by cell cycle arrest and apoptosis was also induced in breast and colon cancer cell models upon small molecule-mediated NMT inhibition.54 The inhibition of the cell cycle was also observed in prostate cancer upon NMT knockdown or inhibition with a myristoyl-CoA analog B13. This cell cycle arrest led to the suppression of malignant growth and invasion, which occurred via the inhibition of Src myristoylation and subsequent downregulation of its oncogenic signaling.55 In MCF7 estrogen receptor-positive breast cancer cells, mTOR inhibition with rapamycin leads to NMT1 upregulation.56 NMT1 promotes lung cancer cell viability by supporting mitophagy. This occurs via the AMPKβ myristoylation that is required for the AMPK recruitment to the mitochondria where it physically associates with ATG16 and ATG5–12 and recruits VPS34 and ATG16 during mitochondrial damage. NMT1 mediated targeting to mitochondria is sufficient to initiate mitophagy and to promote cancer cell survival.57 Thus, NMT plays an important role in multiple cancers. However, given the NMT regulation of different substrate proteins in both normal cells and cancer cells, anti-cancer targeting of NMT would benefit from selective inhibition of NMT in cancer cells over normal cells.
NMT in immune responses
It is becoming more evident that NMT acts as a guardian of the immune response by regulating myelopoiesis, lymphopoiesis, innate immune response, and the immunological synapse.58 NMT1 is essential to monocytic differentiation during which the NMT activity is modulated by the expression levels of the enzyme and its inhibitor protein HSC70. NMT1 deficient mouse embryonic fibroblasts are strongly impaired in differentiating into macrophages.59 N-myristoylation also appears essential for thymocyte development and function. In a mouse model with a lineage-specific T cell deficiency in NMT1 and NMT2, T cell development and activation are severely impaired suggesting that N-myristoylation positively regulates immune response. Unlike the NMT1 thymocyte mutant mice, the NMT2 mutants demonstrate a largely wild type phenotype, but the double mutation strongly enhances the inhibition of T cell development and reduction in T cell numbers in the thymus, blood, lymph nodes, and spleen. The NMT deficiency leads to increased apoptosis during all stages of T cell development, decreased Erk phosphorylation, and mislocalization of myristoylated proteins.60 Interestingly, in rheumatoid arthritis (RA) NMT appears to play an anti-inflammatory role. T cells from RA patients have low NMT1 levels which suppress the myristoylation-dependent AMPK lysosomal translocation and activation. This leads to hyperactivation of mTORC1 signaling in RA T cells promoting their differentiation into pro-inflammatory Th1 and Th17 cells aggravating RA. In a humanized mouse model where synovitis was induced by transferring human peripheral blood mononuclear cells and ingrafting human synovial tissue, NMT1 overexpression had a strong anti-inflammatory effect.61 The inefficient mitophagy due to impaired AMPK function57 may reconcile this observation in RA with the necessity for NMT activity during T cell development.60 Mitochondrial dysfunction is the hallmark of RA and is thought to promote inflammation,62, 63 which might be aggravated by the impaired clearance of damaged mitochondria as a result of suppressed AMPK myristoylation in RA.64
T cell maturation depends on the Notch1 signaling pathway.65, 66 This might occur through the myristoylated Neutralized like-1 E3 ubiquitin ligase that facilitates the turnover and trafficking of Jagged, the Notch receptor ligand.67, 68 In developed T cells, myristoylation of LCK governs an intricate trafficking mechanism of the kinase and is necessary for the LCK localization to the T cell immune synapse, the interface between the target cell and the T cell. The myristoyl group anchors LCK to membranes and acts as a signal for UNC119A binding that allows LCK extraction from the membranes and delivery to the synapse where it is released with the help of ARL3 and ARL13B.69 LCK phosphorylates immunoreceptor tyrosine-based activation motifs (ITAMs) that regulate signaling cascades essential for T cell activation. Myristoylated Fyn and c-Src orchestrate centrosome and actin-dependent movements during immune synapse.58, 70 These reports point to a multifaceted and context-dependent role of NMT in immune response which warrants further work to aid informed therapeutic interventions.
Parasitic NMT as a therapeutic target in malaria and sleeping sickness
Eukaryotic parasites have their own single NMT, which is being explored as a potential therapeutic target in infectious diseases like malaria and sleeping sickness. Malaria is caused by the infections with the species of the genus Plasmodium such as Plasmodium falciparum and leads to nearly a million deaths worldwide annually. The low vaccination efficacy and development of resistance to available therapies call for new therapeutic strategies. In 2014, over 30 NMT substrates were identified in P. falciparum using chemical proteomics. These proteins are involved in a range of functions such as protein trafficking, migration, development, and signaling pathways, suggesting that NMT is necessary for the parasite viability. This study identified a promising small molecule displaying selectivity towards PfNMT over hNMT and the ability to inhibit parasite viability and invasion of red blood cells. Excitingly, this agent caused a strong reduction in the rodent malaria parasite P. berghei in mice without acute toxicity to the animals.71 A more recent high-throughput screening identified 23 chemical classes of inhibitors that were selected for Plasmodium NMT over the human NMT enzymes, but further lead optimization is needed for in vivo testing.72 To address a potential resistance of the parasite to NMT inhibition, another study identified the PfNMT G386E mutant that conferred resistance to the NMT inhibitor IMP-1002. DDD85646 could overcome this resistance suggesting that a combinatorial approach might be of use when targeting NMT in this disease.73
Sleeping sickness is another deadly infection caused by the Trypanosoma brucei parasites. Similar to P. falciparum, T. brucei has its own single NMT with more than 60 predicted substrates, some of which have been experimentally validated, and is being explored as a target against sleeping sickness. Administration of the NMT inhibitor DDD86546 cures trypanosomiasis in mice potentially through disruption of the endocytic pathway by inhibiting TbARF1 myristoylation.74 A recent chemical proteomics study in the clinically relevant bloodstream form of the parasite identified 53 high confidence and 10 medium confidence NMT substrate hits, many of which overlapped with the predicted group. Among these substrates were phosphatases, ARF GTPases, calpain-like proteins, and several uncharacterized proteins.75 T. brucei infection proceeds in two stages where it dwells in the bloodstream during the first stage and infects the nervous system in the second stage. This indicates the need for drugs able to cross the blood-brain barrier and a few promising leads were recently found.76, 77 While it is clear that NMT is a promising therapeutic target in malaria and sleeping sickness, the understanding of the underlying mechanisms is lacking. Further identification of specific NMT-regulated substrates and pathways essential to parasite viability might facilitate the pathogen-specific therapeutic intervention.
Viral utilization of the host NMT
NMT has been widely explored as a therapeutic target for viral infections because many viruses use host NMT for increased pathogenesis (Table 1). Some of the known myristoylated viral proteins are the VP4 of poliovirus, hepatitis B virus pre-S1 protein and Gag and Nef of simian and human immunodeficiency viruses (SIV and HIV).78 Myristoylation of Nef and Gag are essential to HIV type-1 replication and virulence. Nef myristoylation induces endocytosis of CD4 on the surface of T cells, which prevents superinfections detrimental to cell survival,79 and interestingly it is preferentially myristoylated by NMT2.80 Myristoylation of Gag is necessary for its membrane association followed by the assembly and budding of new viral particles.81 NMT1 enhances the replication of HIV type-1 by increasing the expression of viral RNA.82, 83
Table 1:
Myristoylated viral proteins. The table was adapted from Maurer-Stroh and Frank Eisenhaber, Trends in Microbiology, 2004.6
| Protein | Virus | Function | N-terminal sequence | Ref. |
|---|---|---|---|---|
| Nef | Lentiviruses (HIV, SIV) | Membrane targeting to downregulate immune cell surface molecules | GGKWSKSSI | 88 |
| Gag | Retroviruses, Poxviruses (HIV) | Binds plasma membrane to initiate viral particle assembly | ||
| VP4 coat protein | Picornaviridae [rhino-, cardio-, aphtho-, entero- (echo-, coxsackie-, polio-) viruses] | Virion assembly | GAQVSTQKS | 93 |
| Large surface antigen | Hepadnaviridae (hepatitis B virus) | Essential for viral infectivity but not assembly | GQNLSTSNP | 94 |
| L1R envelope protein | Poxviridae [Chordopoxvirinae (e.g. variola, vaccinia)and Enteropoxvirinae], Asfiviridae, Iridoviridae (lymphocysti-, rana-, iridovirus), Ascoviridae (Ascovirus) | Membrane targeting and virion assembly | GAAASIQTT | 95, 96 |
| v-src | Rous sarcoma virus | Membrane binding necessary for oncogenic transformation | GSSKSKPKD | 97, 98 |
| A16L late protein | Poxviridae [Chordopoxvirinae (e.g. variola, vaccinia) and Enteropoxvirinae] | Protein associates with viral, not cellular, structures | GAAVTLNRI | 99, 100 |
| VP2 coat protein | Polyomaviruses | Viral assembly and infectivity | GAALALLGD | 101, 102 |
| G9R | Poxviridae [Chordopoxvirinae (e.g. variola, vaccinia)] | Protein associates with viral, not cellular, structures | GGGVSVELP | 99 |
| UL11 | Alphaherpesvirinae (simplex-, varicello-, Marek’s disease-like and infectious laryngotracheitis-like viruses) | Association with lipid rafts | GLSFSGARP | 103, 104 |
| UL99, U71, BBLF1 | Betaherpesvirinae (roseolo-, cytomegalo-, muromegalovirus), Gammaherpesvirinae (e.g. lymphocryptovirus), Alphaherpesvirinae | Association with ERGIC necessary for virion assembly | GAELCKRIC GAKCCKPVS GALWSLCRR |
105 |
| Major outer capsid protein μ1 | Ortho- and aquareoviruses | Cleavage of μ1 to μ1C | GNASSIVQT | 106 |
| E7R | Orthopoxviruses | Soluble protein | GTAATIQTP | 99 |
| Polyprotein pp220 | Asfivirus | Potential membrane-anchor regulating core assembly | GNRGSSTSS | 107, 108 |
| Membrane fusion protein p15 | Orthoreovirus | Required for membrane fusion activity | GQRHSIVQP | 109, 110 |
Most picornaviruses such as poliovirus and coxsackievirus utilize host NMT for capsid protein myristoylation. The capsid protein VP0 is cleaved to VP2 and VP4 and shares the same myristoylated N-terminus with VP4. Genetic knockout of NMT1 or NMT2 in HAP1 cells demonstrated that only NMT1 is necessary for viral replication. Pharmacological inhibition of NMT with DDD85646 led to a strong reduction of myristoylation of VP0 and suppression of particle infectivity. Surprisingly, electron microscopy studies revealed that these particles were identical to the control particles and did not display a defect in cell attachment suggesting that lack of myristoylation might cause a defect in the transfer of viral RNA into the host cell.84 However, the myristoylation of VP0 of rhinoviruses, the cause for respiratory diseases such as common cold, is necessary for the assembly of the viral capsid to support viral replication.85 Interestingly, parechoviruses and kobuviruses that do not undergo VP0 cleavage are not affected by NMT inhibition suggesting that VP0 processing confers reliance on NMT.84
Dengue virus, the cause of dangerous dengue fever, appears to rely on NMT as well. NMT is upregulated during the infection of dendritic cells and it interacts with the viral envelope protein, while NMT gene silencing significantly suppresses dengue virus replication.86 Thus far myristoylation of the Dengue viral proteins has only been predicted but lacks experimental proof, which is also true for many other viruses.6 Coronaviruses such as those that caused SARS and COVID-19 pandemics are thought to lack myristoylation because they do not contain the preferred NMT recognition sequence, however, this has no experimental evidence.78 Given the recent finding of the NMT activity towards lysine and the fluidity of NMT substrate recognition preference, it might be of use to reevaluate viral proteins for potential lysine substrate sites of NMT. For instance, the N-terminal sequence of SARS-CoV nsp4 protein is KIVSTCFK, and nsp4 of SARS-CoV2 and some bat viruses contain similar motifs with the N-terminal lysine, which raises the possibility that they are modified by NMT.
Viral protein myristoylation might be important in generating an immune response. X-ray crystallography analyses revealed that MHC class I proteins Mamu-B*05104 and Mamu-B*098 contain a large hydrophobic pocket that can bind a myristoylated glycine of the peptides C14-Gly-Gly-Ala-Ile and C14-Gly-Gly-Ala-Ile-Ser, respectively, which are derived from the viral Nef protein. Gly2 and Ala3 or Ile4 are exposed for the recognition by the cytotoxic T lymphocytes to initiate their activation.87 While myristoylation of viral proteins has been known for decades, its inhibition as a therapeutic strategy is still at the stage of exploration. Given the importance of NMT to the function of normal cells, its inhibition might be toxic, therefore small molecule inhibitors with increased specificity for viral protein myristylation are needed. This could be aided by the identification of regulatory proteins of NMT. For instance, it is tempting to speculate that targeting ACBD6-NMT2 interaction could preferentially inhibit myristoylation of Nef, a better substrate of NMT2 than NMT1, given the preference of ACBD6 for NMT2 over NMT1.
Pharmacological targeting of NMT
Because of the roles of NMT in cancer and parasitic and viral infections, inhibition of N-myristoylation has been explored as an attractive therapeutic strategy against these diseases. 2-hydroxymyristic acid, D-NMAPPD (B13), or Tris-DBA palladium, IMP-366 (DDD85646), PCLX-001(DDD86481) and IMP-1088 are most commonly used pan-N-myristoylation inhibitors (Figure 4). The high structural similarity between NMT1 and NMT2 poses a challenge for the design of NMT1/NMT2-selective inhibitors and there have been no reports of such selective small molecules so far. A recent comparison of the potency and selectivity of the widely used NMT inhibitors revealed that 2-hydroxymyristic acid, D-NMAPPD, and Tris-DBA palladium are poor inhibitors of NMT. D-NMAPPD and Tris-DBA palladium cause off-target mediated cell cytotoxicity. In the same study IMP-366 (DDD85646) and IMP-1088 appeared as highly specific and potent inhibitors of NMT1 and NMT2 with nanomolar IC50 values.111
Small molecule NMT inhibition has shown therapeutic potential in viral and parasitic infections and cancer. 2-Hydroxymyristic acid was shown to suppress replication of enterovirus 71,112 while the recently discovered IMP-1088 had a therapeutic potential against rhinovirus by inhibiting capsid protein myristoylation.85 DDD85646 is the most widely used NMT inhibitor that can suppress picornavirus replication via the host NMT inhibition,84 as well as malaria and sleeping sickness parasites by inhibiting their NMTs.73–75 In addition, DDD85646 suppressed breast and colon cancer cell growth by inducing ER stress and consequent cell cycle arrest and apoptosis.54 PCLX-001 (DDD86481) is being advanced to clinical trials for treating hematologic cancers by Pacylex, which represents the most promising NMT inhibitor in cancer treatment. An anticancer effect was also achieved with D-NMAPPD, a myristoyl-CoA analog that has been reported to suppress prostate cancer progression by inhibiting Src myristoylation,55 and Tris-DBA palladium that was effective against melanoma.113 Further understanding of the differences in structure and regulation between human and pathogenic NMTs and between human NMT1 and NMT2 could aid the development of small molecules with increased selectivity and decreased toxicity.
Conclusions and outstanding questions
In recent years much has been learned about the catalytic mechanism and physiological functions of NMT. This knowledge opened several exciting and important research avenues. Given the nearly identical catalytic domains of the two human NMT enzymes, the necessity for both remains elusive and is likely hidden in the divergence of their N-termini that could potentially differ in protein, metabolite, or nucleic acid binding and posttranslational modifications. This area remains underexplored perhaps in part due to the unstructured nature of these regions. Furthermore, additional work is needed to identify NMT regulators and to understand the NMT selectivity for myristoyl-CoA and its mechanism of activation by ACBD6.
NMT has recently emerged as the first mammalian lysine myristoyltransferase with so far a single protein substrate, ARF6. Interestingly, in cells, NMT2 appears as a more potent lysine transferase than NMT1 due to an unknown level of regulation. It is of interest to further characterize the differences in the two activities of NMT, identify new lysine substrates while including viral proteins in evaluation, and determine the physiological implications of the lysine modification. Exploring conditions of proteolysis could aid the identification of new substrates followed by finding lysine myristoylation erasers such as sirtuins or HDACs. In cancer, there is a need for more mechanistic understandings of how NMT controls the oxidative and ER stress and whether NMT regulates cancer stemness. The role of NMT in immunity appears context-dependent, which is important to consider in evaluating NMT as a modulatory node for inflammation. Having this knowledge could aid in the development of highly selective NMT inhibitors with therapeutic potential.
Acknowledgments
The work was supported by the NSF Graduate Research Fellowship Program and SUNY Diversity fellowship as well as NIH/NIDDK grant R01DK107868.
Keywords
- Myristoylation
addition of a 14-carbon saturated acyl chain to proteins
- N-myristoyltransferase or NMT
the enzyme that catalyzes N-terminal myristoylation
- Lysine fatty acylation
addition of long-chain fatty acyl groups to lysine residues
- Virus
an infective particle that replicates inside a host cell
- Solvent channel
structural feature of some enzymes thought to regulate deprotonation
- Membrane anchor
typically a lipid modification that facilitates protein membrane binding
- Inhibitor
small molecule that suppresses a function of a protein
- Cancer
a disease caused by uncontrolled proliferation of malignant cells in the body
- Parasite
disease-causing organism of protozoa, helminths, or arthropods
References
- 1.Manenti S, Sorokine O, Van Dorsselaer A, and Taniguchi H (1994) Demyristoylation of the major substrate of protein kinase C (MARCKS) by the cytoplasmic fraction of brain synaptosomes, J Biol Chem 269, 8309–8313. [PubMed] [Google Scholar]
- 2.Timms RT, Zhang Z, Rhee DY, Harper JW, Koren I, and Elledge SJ (2019) A glycine-specific N-degron pathway mediates the quality control of protein N-myristoylation, Science 365, eaaw4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duronio RJ, Towler DA, Heuckeroth RO, and Gordon JI (1989) Disruption of the yeast N-myristoyl transferase gene causes recessive lethality, Science 243, 796–800. [DOI] [PubMed] [Google Scholar]
- 4.Weinberg RA, McWherter CA, Freeman SK, Wood DC, Gordon JI, and Lee SC (1995) Genetic studies reveal that myristoylCoA:protein N-myristoyltransferase is an essential enzyme in Candida albicans, Mol Microbiol 16, 241–250. [DOI] [PubMed] [Google Scholar]
- 5.Lodge JK, Jackson-Machelski E, Toffaletti DL, Perfect JR, and Gordon JI (1994) Targeted gene replacement demonstrates that myristoyl-CoA: protein N-myristoyltransferase is essential for viability of Cryptococcus neoformans, Proc Natl Acad Sci U S A 91, 12008–12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maurer-Stroh S, and Eisenhaber F (2004) Myristoylation of viral and bacterial proteins, Trends Microbiol 12, 178–185. [DOI] [PubMed] [Google Scholar]
- 7.Yang SH, Shrivastav A, Kosinski C, Sharma RK, Chen MH, Berthiaume LG, Peters LL, Chuang PT, Young SG, and Bergo MO (2005) N-myristoyltransferase 1 is essential in early mouse development, J Biol Chem 280, 18990–18995. [DOI] [PubMed] [Google Scholar]
- 8.Ntwasa M, Aapies S, Schiffmann DA, and Gay NJ (2001) Drosophila embryos lacking N-myristoyltransferase have multiple developmental defects, Exp Cell Res 262, 134–144. [DOI] [PubMed] [Google Scholar]
- 9.Bhatnagar RS, Futterer K, Farazi TA, Korolev S, Murray CL, Jackson-Machelski E, Gokel GW, Gordon JI, and Waksman G (1998) Structure of N-myristoyltransferase with bound myristoylCoA and peptide substrate analogs, Nat Struct Biol 5, 1091–1097. [DOI] [PubMed] [Google Scholar]
- 10.Dian C, Perez-Dorado I, Riviere F, Asensio T, Legrand P, Ritzefeld M, Shen M, Cota E, Meinnel T, Tate EW, and Giglione C (2020) High-resolution snapshots of human N-myristoyltransferase in action illuminate a mechanism promoting N-terminal Lys and Gly myristoylation, Nat Commun 11, 1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Towler D, and Glaser L (1986) Protein fatty acid acylation: enzymatic synthesis of an N-myristoylglycyl peptide, Proc Natl Acad Sci U S A 83, 2812–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farazi TA, Waksman G, and Gordon JI (2001) Structures of Saccharomyces cerevisiae N-myristoyltransferase with bound myristoylCoA and peptide provide insights about substrate recognition and catalysis, Biochemistry 40, 6335–6343. [DOI] [PubMed] [Google Scholar]
- 13.Jiang H, Zhang X, Chen X, Aramsangtienchai P, Tong Z, and Lin H (2018) Protein Lipidation: Occurrence, Mechanisms, Biological Functions, and Enabling Technologies, Chemical reviews 118, 919–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castrec B, Dian C, Ciccone S, Ebert CL, Bienvenut WV, Le Caer JP, Steyaert JM, Giglione C, and Meinnel T (2018) Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern, Nat Chem Biol 14, 671–679. [DOI] [PubMed] [Google Scholar]
- 15.Towler DA, Adams SP, Eubanks SR, Towery DS, Jackson-Machelski E, Glaser L, and Gordon JI (1987) Purification and characterization of yeast myristoyl CoA:protein N-myristoyltransferase, Proc Natl Acad Sci U S A 84, 2708–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Towler DA, Adams SP, Eubanks SR, Towery DS, Jackson-Machelski E, Glaser L, and Gordon JI (1988) Myristoyl CoA:protein N-myristoyltransferase activities from rat liver and yeast possess overlapping yet distinct peptide substrate specificities, J Biol Chem 263, 1784–1790. [PubMed] [Google Scholar]
- 17.Traverso JA, Giglione C, and Meinnel T (2013) High-throughput profiling of N-myristoylation substrate specificity across species including pathogens, Proteomics 13, 25–36. [DOI] [PubMed] [Google Scholar]
- 18.Rioux V, and Legrand P (2007) Saturated fatty acids: simple molecular structures with complex cellular functions, Curr Opin Clin Nutr Metab Care 10, 752–758. [DOI] [PubMed] [Google Scholar]
- 19.Rudnick DA, Lu T, Jackson-Machelski E, Hernandez JC, Li Q, Gokel GW, and Gordon JI (1992) Analogs of palmitoyl-CoA that are substrates for myristoyl-CoA:protein N-myristoyltransferase, Proc Natl Acad Sci U S A 89, 10507–10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kishore NS, Wood DC, Mehta PP, Wade AC, Lu T, Gokel GW, and Gordon JI (1993) Comparison of the acyl chain specificities of human myristoyl-CoA synthetase and human myristoyl-CoA:protein N-myristoyltransferase, J Biol Chem 268, 4889–4902. [PubMed] [Google Scholar]
- 21.Bhatnagar RS, Jackson-Machelski E, McWherter CA, and Gordon JI (1994) Isothermal titration calorimetric studies of Saccharomyces cerevisiae myristoyl-CoA:protein N-myristoyltransferase. Determinants of binding energy and catalytic discrimination among acyl-CoA and peptide ligands, J Biol Chem 269, 11045–11053. [PubMed] [Google Scholar]
- 22.Soupene E, Kao J, Cheng DH, Wang D, Greninger AL, Knudsen GM, DeRisi JL, and Kuypers FA (2016) Association of NMT2 with the acyl-CoA carrier ACBD6 protects the N-myristoyltransferase reaction from palmitoyl-CoA, J Lipid Res 57, 288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soupene E, and Kuypers FA (2019) ACBD6 protein controls acyl chain availability and specificity of the N-myristoylation modification of proteins, J Lipid Res 60, 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soupene E, Schatz UA, Rudnik-Schoneborn S, and Kuypers FA (2020) Requirement of the acyl-CoA carrier ACBD6 in myristoylation of proteins: Activation by ligand binding and protein interaction, PLoS One 15, e0229718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu F, Xie N, Jiang Z, Li G, Ma L, and Tong T (2018) The Cellular Senescence-Inhibited Gene Is Essential for PPM1A Myristoylation To Modulate Transforming Growth Factor beta Signaling, Mol Cell Biol 38, e00414–00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dudek E, Millott R, Liu WX, Beauchamp E, Berthiaume LG, and Michalak M (2015) N-Myristoyltransferase 1 interacts with calnexin at the endoplasmic reticulum, Biochem Biophys Res Commun 468, 889–893. [DOI] [PubMed] [Google Scholar]
- 27.Glover CJ, Hartman KD, and Felsted RL (1997) Human N-myristoyltransferase amino-terminal domain involved in targeting the enzyme to the ribosomal subcellular fraction, J Biol Chem 272, 28680–28689. [DOI] [PubMed] [Google Scholar]
- 28.Kumar S, and Sharma RK (2015) N-terminal region of the catalytic domain of human N-myristoyltransferase 1 acts as an inhibitory module, PLoS One 10, e0127661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thinon E, Serwa RA, Broncel M, Brannigan JA, Brassat U, Wright MH, Heal WP, Wilkinson AJ, Mann DJ, and Tate EW (2014) Global profiling of co- and post-translationally N-myristoylated proteomes in human cells, Nat Commun 5, 4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perinpanayagam MA, Beauchamp E, Martin DD, Sim JY, Yap MC, and Berthiaume LG (2013) Regulation of co- and post-translational myristoylation of proteins during apoptosis: interplay of N-myristoyltransferases and caspases, FASEB J 27, 811–821. [DOI] [PubMed] [Google Scholar]
- 31.Zha J, Weiler S, Oh KJ, Wei MC, and Korsmeyer SJ (2000) Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis, Science 290, 1761–1765. [DOI] [PubMed] [Google Scholar]
- 32.Ducker CE, Upson JJ, French KJ, and Smith CD (2005) Two N-myristoyltransferase isozymes play unique roles in protein myristoylation, proliferation, and apoptosis, Mol Cancer Res 3, 463–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevenson FT, Bursten SL, Locksley RM, and Lovett DH (1992) Myristyl acylation of the tumor necrosis factor alpha precursor on specific lysine residues, J Exp Med 176, 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosciuk T, Price IR, Zhang X, Zhu C, Johnson KN, Zhang S, Halaby SL, Komaniecki GP, Yang M, DeHart CJ, Thomas PM, Kelleher NL, Fromme JC, and Lin H (2020) NMT1 and NMT2 are lysine myristoyltransferases regulating the ARF6 GTPase cycle, Nat Commun 11, 1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weston SA, Camble R, Colls J, Rosenbrock G, Taylor I, Egerton M, Tucker AD, Tunnicliffe A, Mistry A, Mancia F, de la Fortelle E, Irwin J, Bricogne G, and Pauptit RA (1998) Crystal structure of the anti-fungal target N-myristoyl transferase, Nat Struct Biol 5, 213–221. [DOI] [PubMed] [Google Scholar]
- 36.O’Callaghan DW, Tepikin AV, and Burgoyne RD (2003) Dynamics and calcium sensitivity of the Ca2+/myristoyl switch protein hippocalcin in living cells, J Cell Biol 163, 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ames JB, Ishima R, Tanaka T, Gordon JI, Stryer L, and Ikura M (1997) Molecular mechanics of calcium-myristoyl switches, Nature 389, 198–202. [DOI] [PubMed] [Google Scholar]
- 38.Li C, Lim S, Braunewell KH, and Ames JB (2016) Structure and Calcium Binding Properties of a Neuronal Calcium-Myristoyl Switch Protein, Visinin-Like Protein 3, PLoS One 11, e0165921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Randazzo PA, Terui T, Sturch S, Fales HM, Ferrige AG, and Kahn RA (1995) The myristoylated amino terminus of ADP-ribosylation factor 1 is a phospholipid- and GTP-sensitive switch, The Journal of biological chemistry 270, 14809–14815. [DOI] [PubMed] [Google Scholar]
- 40.Kahn RA, Randazzo P, Serafini T, Weiss O, Rulka C, Clark J, Amherdt M, Roller P, Orci L, and Rothman JE (1992) The amino terminus of ADP-ribosylation factor (ARF) is a critical determinant of ARF activities and is a potent and specific inhibitor of protein transport, J Biol Chem 267, 13039–13046. [PubMed] [Google Scholar]
- 41.Le Roux AL, Mohammad IL, Mateos B, Arbesu M, Gairi M, Khan FA, Teixeira JMC, and Pons M (2019) A Myristoyl-Binding Site in the SH3 Domain Modulates c-Src Membrane Anchoring, iScience 12, 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, and Superti-Furga G (2003) A myristoyl/phosphotyrosine switch regulates c-Abl, Cell 112, 845–857. [DOI] [PubMed] [Google Scholar]
- 43.Wedegaertner PB, Wilson PT, and Bourne HR (1995) Lipid modifications of trimeric G proteins, J Biol Chem 270, 503–506. [DOI] [PubMed] [Google Scholar]
- 44.Koegl M, Zlatkine P, Ley SC, Courtneidge SA, and Magee AI (1994) Palmitoylation of multiple Src-family kinases at a homologous N-terminal motif, Biochem J 303 (Pt 3), 749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters PJ, Hsu VW, Ooi CE, Finazzi D, Teal SB, Oorschot V, Donaldson JG, and Klausner RD (1995) Overexpression of wild-type and mutant ARF1 and ARF6: distinct perturbations of nonoverlapping membrane compartments, J Cell Biol 128, 1003–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raju RV, Moyana TN, and Sharma RK (1997) N-Myristoyltransferase overexpression in human colorectal adenocarcinomas, Exp Cell Res 235, 145–154. [DOI] [PubMed] [Google Scholar]
- 47.Rajala RV, Radhi JM, Kakkar R, Datla RS, and Sharma RK (2000) Increased expression of N-myristoyltransferase in gallbladder carcinomas, Cancer 88, 1992–1999. [PubMed] [Google Scholar]
- 48.Lu Y, Selvakumar P, Ali K, Shrivastav A, Bajaj G, Resch L, Griebel R, Fourney D, Meguro K, and Sharma RK (2005) Expression of N-myristoyltransferase in human brain tumors, Neurochem Res 30, 9–13. [DOI] [PubMed] [Google Scholar]
- 49.Walters DK, Steinmann P, Langsam B, Schmutz S, Born W, and Fuchs B (2008) Identification of potential chemoresistance genes in osteosarcoma, Anticancer Res 28, 673–679. [PubMed] [Google Scholar]
- 50.Mosakhani N, Mustjoki S, and Knuutila S (2013) Down-regulation of miR-181c in imatinib-resistant chronic myeloid leukemia, Mol Cytogenet 6, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fang W, Robinson DA, Raimi OG, Blair DE, Harrison JR, Lockhart DE, Torrie LS, Ruda GF, Wyatt PG, Gilbert IH, and van Aalten DM (2015) N-myristoyltransferase is a cell wall target in Aspergillus fumigatus, ACS Chem Biol 10, 1425–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berthiaume LG, and Beauchamp E (2018) Patent US 20180208990A1: Epigenetic Silencing of NMT2, (Inc, P. P., Ed.), pp 1–23, US. [Google Scholar]
- 53.Deng L, Gao X, Liu B, He X, Xu J, Qiang J, Wu Q, and Liu S (2018) NMT1 inhibition modulates breast cancer progression through stress-triggered JNK pathway, Cell Death Dis 9, 1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thinon E, Morales-Sanfrutos J, Mann DJ, and Tate EW (2016) N-Myristoyltransferase Inhibition Induces ER-Stress, Cell Cycle Arrest, and Apoptosis in Cancer Cells, ACS Chem Biol 11, 2165–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim S, Alsaidan OA, Goodwin O, Li Q, Sulejmani E, Han Z, Bai A, Albers T, Beharry Z, Zheng YG, Norris JS, Szulc ZM, Bielawska A, Lebedyeva I, Pegan SD, and Cai H (2017) Blocking Myristoylation of Src Inhibits Its Kinase Activity and Suppresses Prostate Cancer Progression, Cancer Res 77, 6950–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jacquier M, Kuriakose S, Bhardwaj A, Zhang Y, Shrivastav A, Portet S, and Varma Shrivastav S (2018) Investigation of Novel Regulation of N-myristoyltransferase by Mammalian Target of Rapamycin in Breast Cancer Cells, Sci Rep 8, 12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang J, Xu ZX, Ding Z, Lu Y, Yu Q, Werle KD, Zhou G, Park YY, Peng G, Gambello MJ, and Mills GB (2015) Myristoylation confers noncanonical AMPK functions in autophagy selectivity and mitochondrial surveillance, Nat Commun 6, 7926. [DOI] [PubMed] [Google Scholar]
- 58.Udenwobele DI, Su RC, Good SV, Ball TB, Varma Shrivastav S, and Shrivastav A (2017) Myristoylation: An Important Protein Modification in the Immune Response, Front Immunol 8, 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shrivastav A, Varma S, Lawman Z, Yang SH, Ritchie SA, Bonham K, Singh SM, Saxena A, and Sharma RK (2008) Requirement of N-myristoyltransferase 1 in the development of monocytic lineage, J Immunol 180, 1019–1028. [DOI] [PubMed] [Google Scholar]
- 60.Rampoldi F, Bonrouhi M, Boehm ME, Lehmann WD, Popovic ZV, Kaden S, Federico G, Brunk F, Grone HJ, and Porubsky S (2015) Immunosuppression and Aberrant T Cell Development in the Absence of N-Myristoylation, J Immunol 195, 4228–4243. [DOI] [PubMed] [Google Scholar]
- 61.Wen Z, Jin K, Shen Y, Yang Z, Li Y, Wu B, Tian L, Shoor S, Roche NE, Goronzy JJ, and Weyand CM (2019) N-myristoyltransferase deficiency impairs activation of kinase AMPK and promotes synovial tissue inflammation, Nat Immunol 20, 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valcarcel-Ares MN, Riveiro-Naveira RR, Vaamonde-Garcia C, Loureiro J, Hermida-Carballo L, Blanco FJ, and Lopez-Armada MJ (2014) Mitochondrial dysfunction promotes and aggravates the inflammatory response in normal human synoviocytes, Rheumatology (Oxford) 53, 1332–1343. [DOI] [PubMed] [Google Scholar]
- 63.Falconer J, Murphy AN, Young SP, Clark AR, Tiziani S, Guma M, and Buckley CD (2018) Review: Synovial Cell Metabolism and Chronic Inflammation in Rheumatoid Arthritis, Arthritis Rheumatol 70, 984–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krysko DV, Agostinis P, Krysko O, Garg AD, Bachert C, Lambrecht BN, and Vandenabeele P (2011) Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation, Trends Immunol 32, 157–164. [DOI] [PubMed] [Google Scholar]
- 65.Deftos ML, Huang E, Ojala EW, Forbush KA, and Bevan MJ (2000) Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes, Immunity 13, 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.MacDonald HR, Wilson A, and Radtke F (2001) Notch1 and T-cell development: insights from conditional knockout mice, Trends Immunol 22, 155–160. [DOI] [PubMed] [Google Scholar]
- 67.Koutelou E, Sato S, Tomomori-Sato C, Florens L, Swanson SK, Washburn MP, Kokkinaki M, Conaway RC, Conaway JW, and Moschonas NK (2008) Neuralized-like 1 (Neurl1) targeted to the plasma membrane by N-myristoylation regulates the Notch ligand Jagged1, J Biol Chem 283, 3846–3853. [DOI] [PubMed] [Google Scholar]
- 68.Lee HJ, Yoon JH, Ahn JS, Jo EH, Kim MY, Lee YC, Kim JW, Ann EJ, and Park HS (2015) Fe65 negatively regulates Jagged1 signaling by decreasing Jagged1 protein stability through the E3 ligase Neuralized-like 1, Biochim Biophys Acta 1853, 2918–2928. [DOI] [PubMed] [Google Scholar]
- 69.Stephen LA, ElMaghloob Y, McIlwraith MJ, Yelland T, Castro Sanchez P, Roda-Navarro P, and Ismail S (2018) The Ciliary Machinery Is Repurposed for T Cell Immune Synapse Trafficking of LCK, Dev Cell 47, 122–132 e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin-Cofreces NB, Sancho D, Fernandez E, Vicente-Manzanares M, Gordon-Alonso M, Montoya MC, Michel F, Acuto O, Alarcon B, and Sanchez-Madrid F (2006) Role of Fyn in the rearrangement of tubulin cytoskeleton induced through TCR, J Immunol 176, 4201–4207. [DOI] [PubMed] [Google Scholar]
- 71.Chan XW, Wrenger C, Stahl K, Bergmann B, Winterberg M, Muller IB, and Saliba KJ (2013) Chemical and genetic validation of thiamine utilization as an antimalarial drug target, Nat Commun 4, 2060. [DOI] [PubMed] [Google Scholar]
- 72.Harupa A, De Las Heras L, Colmenarejo G, Lyons-Abbott S, Reers A, Caballero Hernandez I, Chung CW, Charter D, Myler PJ, Fernandez-Menendez RM, Calderon F, Palomo S, Rodriguez B, Berlanga M, Herreros-Aviles E, Staker BL, Fernandez Alvaro E, and Kaushansky A (2020) Identification of Selective Inhibitors of Plasmodium N-Myristoyltransferase by High-Throughput Screening, J Med Chem 63, 591–600. [DOI] [PubMed] [Google Scholar]
- 73.Schlott AC, Mayclin S, Reers AR, Coburn-Flynn O, Bell AS, Green J, Knuepfer E, Charter D, Bonnert R, Campo B, Burrows J, Lyons-Abbott S, Staker BL, Chung CW, Myler PJ, Fidock DA, Tate EW, and Holder AA (2019) Structure-Guided Identification of Resistance Breaking Antimalarial NMyristoyltransferase Inhibitors, Cell Chem Biol 26, 991–1000 e1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frearson JA, Brand S, McElroy SP, Cleghorn LA, Smid O, Stojanovski L, Price HP, Guther ML, Torrie LS, Robinson DA, Hallyburton I, Mpamhanga CP, Brannigan JA, Wilkinson AJ, Hodgkinson M, Hui R, Qiu W, Raimi OG, van Aalten DM, Brenk R, Gilbert IH, Read KD, Fairlamb AH, Ferguson MA, Smith DF, and Wyatt PG (2010) N-myristoyltransferase inhibitors as new leads to treat sleeping sickness, Nature 464, 728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wright MH, Paape D, Price HP, Smith DF, and Tate EW (2016) Global Profiling and Inhibition of Protein Lipidation in Vector and Host Stages of the Sleeping Sickness Parasite Trypanosoma brucei, ACS Infect Dis 2, 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harrison JR, Brand S, Smith V, Robinson DA, Thompson S, Smith A, Davies K, Mok N, Torrie LS, Collie I, Hallyburton I, Norval S, Simeons FRC, Stojanovski L, Frearson JA, Brenk R, Wyatt PG, Gilbert IH, and Read KD (2018) A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors, J Med Chem 61, 8374–8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bayliss T, Robinson DA, Smith VC, Brand S, McElroy SP, Torrie LS, Mpamhanga C, Norval S, Stojanovski L, Brenk R, Frearson JA, Read KD, Gilbert IH, and Wyatt PG (2017) Design and Synthesis of Brain Penetrant Trypanocidal N-Myristoyltransferase Inhibitors, J Med Chem 60, 9790–9806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schoeman D, and Fielding BC (2019) Coronavirus envelope protein: current knowledge, Virol J 16, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aiken C, Konner J, Landau NR, Lenburg ME, and Trono D (1994) Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain, Cell 76, 853–864. [DOI] [PubMed] [Google Scholar]
- 80.Seaton KE, and Smith CD (2008) N-Myristoyltransferase isozymes exhibit differential specificity for human immunodeficiency virus type 1 Gag and Nef, J Gen Virol 89, 288–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bryant M, and Ratner L (1990) Myristoylation-dependent replication and assembly of human immunodeficiency virus 1, Proc Natl Acad Sci U S A 87, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ohta H, Takamune N, Kishimoto N, Shoji S, and Misumi S (2015) N-Myristoyltransferase 1 enhances human immunodeficiency virus replication through regulation of viral RNA expression level, Biochem Biophys Res Commun 463, 988–993. [DOI] [PubMed] [Google Scholar]
- 83.Bhargavan B, and Kanmogne GD (2019) Epigenetics, N-myrystoyltransferase-1 and casein kinase-2-alpha modulates the increased replication of HIV-1 CRF02_AG, compared to subtype-B viruses, Sci Rep 9, 10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Corbic Ramljak I, Stanger J, Real-Hohn A, Dreier D, Wimmer L, Redlberger-Fritz M, Fischl W, Klingel K, Mihovilovic MD, Blaas D, and Kowalski H (2018) Cellular N-myristoyltransferases play a crucial picornavirus genus-specific role in viral assembly, virion maturation, and infectivity, PLoS Pathog 14, e1007203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mousnier A, Bell AS, Swieboda DP, Morales-Sanfrutos J, Perez-Dorado I, Brannigan JA, Newman J, Ritzefeld M, Hutton JA, Guedan A, Asfor AS, Robinson SW, Hopkins-Navratilova I, Wilkinson AJ, Johnston SL, Leatherbarrow RJ, Tuthill TJ, Solari R, and Tate EW (2018) Fragment-derived inhibitors of human N-myristoyltransferase block capsid assembly and replication of the common cold virus, Nat Chem 10, 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suwanmanee S, Mahakhunkijcharoen Y, Ampawong S, Leaungwutiwong P, Misse D, and Luplertlop N (2019) Inhibition of N-myristoyltransferase1 affects dengue virus replication, Microbiologyopen 8, e00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamamoto Y, Morita D, Shima Y, Midorikawa A, Mizutani T, Suzuki J, Mori N, Shiina T, Inoko H, Tanaka Y, Mikami B, and Sugita M (2019) Identification and Structure of an MHC Class I-Encoded Protein with the Potential to Present N-Myristoylated 4-mer Peptides to T Cells, J Immunol 202, 3349–3358. [DOI] [PubMed] [Google Scholar]
- 88.Yu G, and Felsted RL (1992) Effect of myristoylation on p27 nef subcellular distribution and suppression of HIV-LTR transcription, Virology 187, 46–55. [DOI] [PubMed] [Google Scholar]
- 89.Ono A, and Freed EO (1999) Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus, J Virol 73, 4136–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rein A, McClure MR, Rice NR, Luftig RB, and Schultz AM (1986) Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus, Proc Natl Acad Sci U S A 83, 7246–7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hizi A, Henderson LE, Copeland TD, Sowder RC, Krutzsch HC, and Oroszlan S (1989) Analysis of gag proteins from mouse mammary tumor virus, J Virol 63, 2543–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shoji S, Tashiro A, Furuishi K, Takenaka O, Kida Y, Horiuchi S, Funakoshi T, and Kubota Y (1989) Antibodies to an NH2-terminal myristoyl glycine moiety can detect NH2-terminal myristoylated proteins in the retrovirus-infected cells, Biochem Biophys Res Commun 162, 724–732. [DOI] [PubMed] [Google Scholar]
- 93.Moscufo N, Simons J, and Chow M (1991) Myristoylation is important at multiple stages in poliovirus assembly, J Virol 65, 2372–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gripon P, Le Seyec J, Rumin S, and Guguen-Guillouzo C (1995) Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity, Virology 213, 292–299. [DOI] [PubMed] [Google Scholar]
- 95.Ravanello MP, Franke CA, and Hruby DE (1993) An NH2-terminal peptide from the vaccinia virus L1R protein directs the myristylation and virion envelope localization of a heterologous fusion protein, J Biol Chem 268, 7585–7593. [PubMed] [Google Scholar]
- 96.Ravanello MP, and Hruby DE (1994) Conditional lethal expression of the vaccinia virus L1R myristylated protein reveals a role in virion assembly, J Virol 68, 6401–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Garber EA, Cross FR, and Hanafusa H (1985) Processing of p60v-src to its myristylated membrane-bound form, Mol Cell Biol 5, 2781–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kamps MP, Buss JE, and Sefton BM (1986) Rous sarcoma virus transforming protein lacking myristic acid phosphorylates known polypeptide substrates without inducing transformation, Cell 45, 105–112. [DOI] [PubMed] [Google Scholar]
- 99.Martin KH, Grosenbach DW, Franke CA, and Hruby DE (1997) Identification and analysis of three myristylated vaccinia virus late proteins, J Virol 71, 5218–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ojeda S, Senkevich TG, and Moss B (2006) Entry of vaccinia virus and cell-cell fusion require a highly conserved cysteine-rich membrane protein encoded by the A16L gene, J Virol 80, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Streuli CH, and Griffin BE (1987) Myristic acid is coupled to a structural protein of polyoma virus and SV40, Nature 326, 619–622. [DOI] [PubMed] [Google Scholar]
- 102.Krauzewicz N, Streuli CH, Stuart-Smith N, Jones MD, Wallace S, and Griffin BE (1990) Myristylated polyomavirus VP2: role in the life cycle of the virus, J Virol 64, 4414–4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.MacLean CA, Dolan A, Jamieson FE, and McGeoch DJ (1992) The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle, J Gen Virol 73 (Pt 3), 539–547. [DOI] [PubMed] [Google Scholar]
- 104.Koshizuka T, Kawaguchi Y, Nozawa N, Mori I, and Nishiyama Y (2007) Herpes simplex virus protein UL11 but not UL51 is associated with lipid rafts, Virus Genes 35, 571–575. [DOI] [PubMed] [Google Scholar]
- 105.Sanchez V, Sztul E, and Britt WJ (2000) Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-golgi-intermediate compartment, J Virol 74, 3842–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tillotson L, and Shatkin AJ (1992) Reovirus polypeptide sigma 3 and N-terminal myristoylation of polypeptide mu 1 are required for site-specific cleavage to mu 1C in transfected cells, J Virol 66, 2180–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Simon-Mateo C, Andres G, and Vinuela E (1993) Polyprotein processing in African swine fever virus: a novel gene expression strategy for a DNA virus, EMBO J 12, 2977–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Andres G, Garcia-Escudero R, Salas ML, and Rodriguez JM (2002) Repression of African swine fever virus polyprotein pp220-encoding gene leads to the assembly of icosahedral core-less particles, J Virol 76, 2654–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dawe S, and Duncan R (2002) The S4 genome segment of baboon reovirus is bicistronic and encodes a novel fusion-associated small transmembrane protein, J Virol 76, 2131–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Top D, Read JA, Dawe SJ, Syvitski RT, and Duncan R (2012) Cell-cell membrane fusion induced by p15 fusion-associated small transmembrane (FAST) protein requires a novel fusion peptide motif containing a myristoylated polyproline type II helix, J Biol Chem 287, 3403–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kallemeijn WW, Lueg GA, Faronato M, Hadavizadeh K, Goya Grocin A, Song OR, Howell M, Calado DP, and Tate EW (2019) Validation and Invalidation of Chemical Probes for the Human N-myristoyltransferases, Cell Chem Biol 26, 892–900 e894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tan YW, Hong WJ, and Chu JJ (2016) Inhibition of enterovirus VP4 myristoylation is a potential antiviral strategy for hand, foot and mouth disease, Antiviral Res 133, 191–195. [DOI] [PubMed] [Google Scholar]
- 113.Bhandarkar SS, Bromberg J, Carrillo C, Selvakumar P, Sharma RK, Perry BN, Govindarajan B, Fried L, Sohn A, Reddy K, and Arbiser JL (2008) Tris (dibenzylideneacetone) dipalladium, a N-myristoyltransferase-1 inhibitor, is effective against melanoma growth in vitro and in vivo, Clin Cancer Res 14, 5743–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]


