Abstract
The determination of cause of death (COD) is one of the most important tasks in forensic practice and is mainly based on macroscopical and microscopical morphological signatures. However, some CODs are hard to determine because the significant morphological signatures can be nonspecific, variable, subjective, or even absent in the real world. In this study, gas chromatography coupled with high-resolution mass spectrometry (GC–HRMS)-based untargeted metabolomics was employed to obtain plasma metabolic profiles of rats that died from anaphylactic shock (AS), mechanical asphyxia (MA), or sudden cardiac death (SCD). The metabolic alterations of each COD group compared to the control group were investigated using a principal component analysis, partial least-squares discriminant analysis, the Wilcoxon test, and fold change analysis. A range of differential features was screened, and 11, 8, and 7 differential metabolites were finally verified for the AS, MA, and SCD groups, respectively. We proposed some explanations that may account for these metabolic differences, including glucose metabolism, the tricarboxylic acid cycle, glycolysis, lipid metabolism, creatinine catabolism, and purine metabolism. Next, for each COD, we used its differential metabolites, which were obtained through comparisons of each COD group to the control group and represented the metabolic changes of the individual COD, to perform a receiver operating characteristic (ROC) analysis to preliminarily evaluate their ability to discriminate each COD group from the other COD groups. We found that creatinine in the AS group and malic acid and uric acid in the MA group might represent some specific metabolic changes for the relevant COD with high areas under the curve in the ROC curve analysis. Moreover, the combination panel for AS or MA also showed a good ability to discriminate it from the others. However, SCD had fewer metabolic signatures and was relatively harder to discriminate from the other CODs in our work. The preliminary study demonstrates the feasibility of GC–HRMS-based untargeted metabolomics as a promising tool to reveal metabolic changes in different death processes and to determine the complex CODs.
Introduction
In forensic medicine, the current understanding of the mechanism of death in most death processes is still preliminary. The determination or diagnosis of most causes of death (CODs) is mainly at the histological level by examining significant morphological lesions or changes during autopsy and tissue section observation with microscopy.1 However, morphological signatures vary widely in the real world due to the variability of individuals, circumstances, postmortem intervals, and so on. It leads the morphological signatures to be nonspecific, variable, subjective, or even absent in forensic practice. For example, in sudden cardiac death (SCD), coronary atherosclerotic heart disease (CAHD) is one of the most common diseases. In CAHD, when atherosclerosis of the coronary artery is not serious (such as stenosis less than 50%), it is difficult to determine the COD as SCD because the morphological changes of acute myocardial infarction or ischemia resulting from a sudden blockage of the coronary artery are nonspecific. Actually, the determination of SCD is mainly based on an exclusion strategy. Therefore, many techniques from other fields have been introduced by forensic pathologists to offer a quantifiable or objective evidence, such as imageology,2−4 histochemistry,5−7 and molecular genetics.8−11 However, these techniques still have limitations due to their complexity and variability; therefore, they have not been applied in forensic pathology practice. In addition to SCD, the same is true for anaphylactic shock (AS), manual strangulation, and smothering. They are common CODs that are difficult to determine and are also based on an exclusion strategy because their morphological findings obtained from an autopsy or microexamination may not be clear and undisputed, and a clear clinical history may be lacking in some cases. Therefore, a new method and strategy are needed here to help in the determination of the three CODs. Manual strangulation and smothering have a similar death process and both belong to mechanical asphyxia (MA). Thus, we use MA to refer to manual strangulation and smothering in the text.
Untargeted metabolomics is an unbiased global method that investigates low-molecular-weight metabolites to profile endogenous metabolic alterations resulting from pathological and physiological changes.12,13 As many metabolites as possible, including chemical unknowns, are detected simultaneously after minimal and nonselective sample pretreatment.14,15 Untargeted metabolomics can serve as a promising tool to investigate metabolic responses to various diseases, identify potential biomarkers for diagnosis or prognosis purposes, and discover toxicity-related or drug-related metabolic pathways.16−19 Mass spectrometry (MS)-based methods are commonly used analytical techniques in metabolomics research because of their highly efficient separation and sensitive quantitative ability and MS databases for metabolite identification; these MS-based methods include gas chromatography coupled with MS (GC–MS), liquid chromatography coupled with MS (LC–MS), and capillary electrophoresis coupled with MS (CE-MS).20,21 Among these, GC–MS has been widely used due to its widespread availability, high resolution, and reproducibility.22,23 Moreover, high-resolution MS (HRMS) is increasingly utilized to produce metabolomics data with improvements in sensitivity, reliability, and quantity.24,25 Then, multivariate statistical analysis and chemometrics methods are used to handle the complex data for analyzing the metabolic variations and highlighting the discriminated metabolites.26
In the forensic science field, metabolomics has mainly been applied to drug-related cases, fingerprinting, and the estimation of postmortem interval in some studies,27 and a few studies have focused on exploring the metabolic changes in some diseases that can lead to death. Xia Hu et al. reported the serum metabolic profiling analysis of anaphylactic guinea pigs and identified nine potential biomarkers for clinical diagnosis purposes.28 Guillaume Rousseau et al. studied the postmortem human serum metabolomics of hypothermia fatalities to reveal the pathophysiological mechanisms and discover novel biomarkers for the diagnosis of hypothermia.29 Dian Wang et al. investigated metabolic alterations correlated with acute myocardial ischemia in a rat heart muscle and found several dangerous metabolites to offer novel clues for prevention or treatment.30 Therefore, one can see that different CODs, that is, different death mechanisms and processes may lead to different panels of metabolites. On this basis, GC–HRMS plasma metabolic profiles for AS, MA, and SCD rat models were generated, and the differences between the three CODs and control groups were explored in this study. Classification models were also established and validated to preliminarily evaluate the discriminative ability of the differential metabolites. We aimed to investigate the metabolic alterations of the three CODs and evaluate the potential of the untargeted metabolomics strategy in helping the determination of COD.
Results
Metabolic Profiles of Different CODs
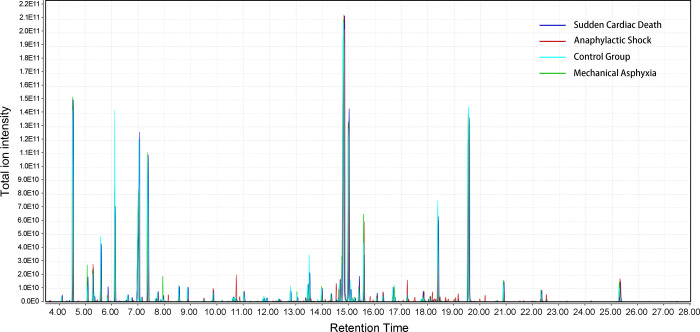
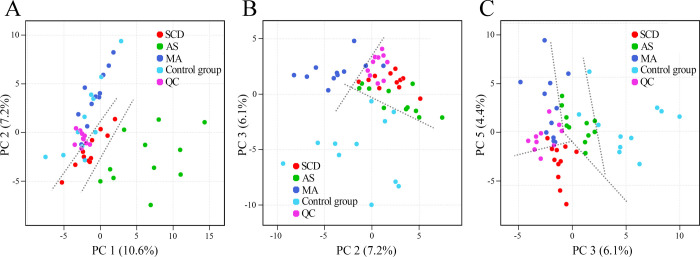
Rat plasma metabolic profiles of the three target COD groups and the control group were obtained by GC–HRMS-based metabolomics. The representative total ion chromatograms (TICs) were plotted first to find the potential metabolic differences in the rat plasma among these different CODs, as displayed in Figure 1. The data obtained through the above process were considered reliable, with a near-perfect overlay and minimal drift in the retention time of all the peaks. The metabolite compositions of the four groups were similar, while the intensity levels of some metabolites were different. After data filtering, data normalization according to the IS, and data scaling, the PCA was performed to separate the samples and visualize the overall differences in the data set. The PCA score plots are shown in Figure 2. Through the score plots based on different PC combinations, the slightly distinguishing distribution of the four groups was observed, though overlaps also existed. However, every group had a clear distinction against the others, as shown in Figure S1 (Supporting Information). The unsupervised pattern recognition results indicated that the metabolic profile of each COD group was different from the control group and from the other COD groups. In addition, all of the QC samples clustered together and did not show a separation trend in the PCA, which suggested that the metabolite extraction, data collection, and data analysis were performed stably and correctly.
Figure 1.
Representative total ion chromatograms of rat plasma samples from the three target COD groups and the control group.
Figure 2.
PCA score plots of the rat plasma GC–HRMS data derived from the three target COD groups and the control group. A: PC 1 vs PC 2; B: PC 2 vs PC 3; and C: PC 3 vs PC 5. QC: quality control samples. The dotted lines were set to help to show the slightly distinguishing distribution.
Differential Metabolite Analysis
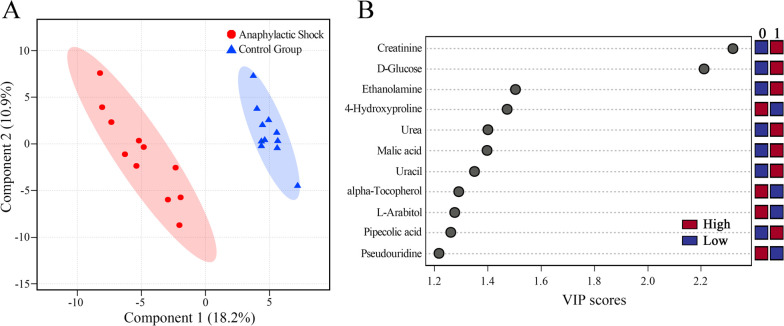
The discriminative features between the three target COD groups and the control group were then searched using VIP scores in the PLS-DA model, fold change, and the Wilcoxon test. PLS-DA is a supervised pattern recognition method that can discriminate samples according to predefined class labels.31 Taking the AS group as an example, the PLS-DA model is shown in Figure 3A in which a distinct separation trend between the AS group and the control group can be observed along with PC 1 (R2 = 0.999 and Q2 = 0.890). Then, the dataset was evaluated using 1000 permutation testing, which represented R2 as 0.996 and Q2 as 0.897 (p < 0.001), as shown in Figure S2 (Supporting Information). The results showed a low trend of overfitting. In the PLS-DA model, the importance or contribution degree of a feature can be measured by VIP scores, as shown in Figure 3B. The PLS-DA models with the permutation results and VIP plots of MA and SCD groups are presented in Figures S3–S6 (Supporting Information). The fold change was calculated to describe a level at which the relative content of a feature was increased or decreased, and the Wilcoxon test was performed to check whether there was a statistically significant difference. In total, 47, 40, and 37 differential features met the threshold requirements and were selected for the AS, MA, and SCD groups, respectively.
Figure 3.
Screening differential metabolites for the anaphylactic shock group. A: Score plot of the PLS-DA model with R2 = 0.999 and Q2 = 0.890. The 1000 permutation testing resulted in R2 = 0.996 and Q2 = 0.897. B: VIP score plot of metabolites verified in the PLS-DA model between the anaphylactic shock group (0) and the control group (1). Red and blue represent the high and low relative contents of the corresponding metabolites, respectively.
Subsequently, we searched the NIST library and HMDB to identify the potential metabolites that corresponded to these differential features. According to preliminary searching candidates, commercial standard compounds were purchased to further confirm them by comparing the MS peaks and retention time. Finally, 11, 8, and 7 metabolites were verified for AS, MA, and SCD groups, respectively. The details of the differential metabolites associated with the three CODs are shown in Table 1.
Table 1. Differential Metabolites Associated with the Anaphylactic Shock, Mechanical Asphyxia, and Sudden Cardiac Death Groups Compared to the Control Group. RT: retention time. FC: fold change. VIP: variable importance in the projection.
| metabolites | chemical class | RT (min) | p value | log2 FC | VIP | trenda | HMDB ID | |
|---|---|---|---|---|---|---|---|---|
| anaphylactic shock | ethanolamine | amines | 4.18 | 0.004585 | –2.1378 | 1.480 | down | 0149 |
| urea | organic acid | 6.13 | 0.009060 | –0.6272 | 1.383 | down | 0294 | |
| uracil | pyrimidine | 8.26 | 0.012361 | –0.8234 | 1.334 | down | 0300 | |
| pipecolic acid | organic acid | 8.65 | 0.020402 | 1.0231 | 1.250 | up | 0070 | |
| malic acid | organic acid | 10.19 | 0.009107 | –0.8243 | 1.405 | down | 0156 | |
| 4-hydroxyproline | amino acid | 10.67 | 0.005830 | –0.6724 | 1.476 | down | 0725 | |
| creatinine | amino acid | 11.02 | 5.87E-08 | –1.5970 | 2.246 | down | 0562 | |
| l-Arabitol | carbohydrate | 12.89 | 0.019033 | 0.8294 | 1.276 | up | 1851 | |
| d-Glucose | carbohydrate | 14.52 | 8.86E-07 | 4.2540 | 2.145 | up | 0122 | |
| pseudouridine | nucleoside | 18.80 | 0.025718 | –0.6859 | 1.208 | down | 0767 | |
| α-tocopherol | quinone | 25.14 | 0.017142 | 1.0663 | 1.280 | up | 1893 | |
| mechanical asphyxia | 3-aminoisobutanoate | amino acid | 6.55 | 0.048807 | –0.9515 | 1.115 | down | 3911 |
| malic acid | organic acid | 10.19 | 0.001978 | 0.7442 | 1.635 | up | 0156 | |
| pyroglutamic acid | amino acid | 10.61 | 2.66E-06 | 2.0613 | 2.160 | up | 0267 | |
| l-Arabitol | carbohydrate | 12.89 | 0.003037 | 1.0107 | 1.581 | up | 1851 | |
| glycrol 3-phosphate | glycerophosphates | 13.35 | 0.004219 | 0.7721 | 1.537 | up | 0126 | |
| uric acid | purine | 16.74 | 7.38E-05 | 1.3556 | 1.952 | up | 0289 | |
| stearic acid | fatty acid | 18.08 | 0.005544 | 0.6950 | 1.499 | up | 0827 | |
| cholesterol | steroid | 25.28 | 0.008134 | 0.6220 | 1.442 | up | 0067 | |
| sudden cardiac death | ethanolamine | amines | 4.18 | 0.001777 | –1.2608 | 1.647 | down | 0149 |
| uracil | pyrimidine | 8.26 | 0.003214 | –0.9119 | 1.573 | down | 0300 | |
| pyroglutamic acid | amino acid | 10.61 | 0.000092 | 2.8968 | 1.935 | up | 0267 | |
| l-Aspartic acid | amino acid | 10.73 | 0.000018 | –1.6836 | 2.052 | down | 0191 | |
| 3-phosphoglyceric acid | carbohydrate | 13.83 | 0.000020 | –1.6492 | 2.044 | down | 0807 | |
| d-glucose | carbohydrate | 14.52 | 0.000042 | 2.8271 | 1.993 | up | 0122 | |
| α-tocopherol | quinone | 25.14 | 0.001376 | 1.3718 | 1.677 | up | 1893 |
The trend means that the relative content of the corresponding metabolite was increased or decreased compared to the control group.
Moreover, the pathway analysis was employed to perform the pathway topology analysis with the MetaboAnalyst 4.0 tool to identify the relevant pathways involved in different CODs. The biological pathway analysis results for the AS, MA, and SCD groups showed the most influenced metabolic pathways, as presented in Figures S7–S9 (Supporting Information).
Classification Ability Evaluation
The ROC curve analysis was conducted to further examine the classification ability of these differential metabolites to determine complex CODs. It should be emphasized here that a classifier that could distinguish the target COD from the control COD lacks practical significance. Actually, AS, MA, and SCD are the three causes of death that would be confusing in a real case. Therefore, a strategy of building a classification model between a COD and a non-COD was executed to preliminarily evaluate the potential classification ability of these metabolites in the pilot study.
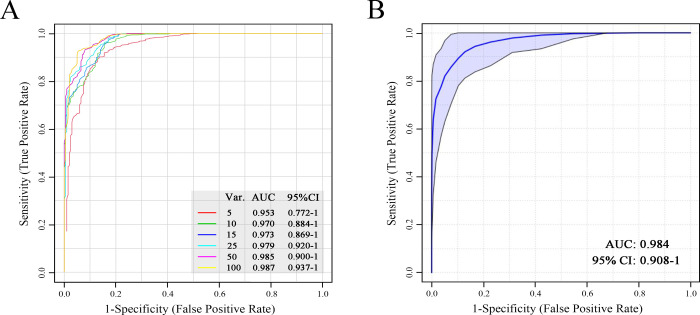
First, the entire data set (features without being selected) of the training set was put into the ROC curve analysis based on a random forest (RF) algorithm. Six ROC curves were generated using the top 5–100 features (“top” means higher VIP values), by 100 times of the Monte Carlo cross-validation procedure. The ROC results of the AS group are shown in Figure 4A. The AUC of the six curves ranged from 0.953 to 0.987, indicating an excellent classification performance between the AS and the non-AS groups.
Figure 4.
Evaluation of the classification ability using all the unselected data (A) and a combination of creatinine, pipecolic acid, and ethanolamine (B) via the ROC curve analysis for the anaphylactic shock group. Var.: the number of variables or features used in the model. The six ROC curves were generated using the top 5–100 features (“top” means higher VIP values). AUC: area under the curve; CI: confidence interval.
Next, the classification ability of the verified differential metabolites was evaluated individually, as shown in Figure S10 (Supporting Information). Most of them showed a poor classification ability with low AUCs; however, the creatinine showed good discriminative ability with a high AUC (0.955). To improve the discriminative ability and robustness, we combined creatinine, pipecolic acid, and ethanolamine to build a classification model using an RF model, and a good classifier was obtained in Figure 4B (AUC: 0.984 and 95% CI: 0.908–1). The classification model was also evaluated through permutation testing for predictive accuracy, as shown in Figure S11 (p < 0.001).
The same analysis was also performed for MA and SCD groups, as shown in Figures S12–S17 (Supporting Information). We found that malic acid, uric acid, and a combination (malic acid, uric acid, 3-aminoisobutanoic acid, and cholesterol) all showed good discriminative abilities between the MA group and the non-MA group. However, there was no single metabolite displaying a good classification ability between the SCD group and the non-SCD group. Although the combination of uracil, pyroglutamic acid, and α-tocopherol reached a better result with a higher AUC value (0.817), the classification model did not pass the permutation testing (p = 0.069, see Figure S17).
Additionally, an external validation was performed with the combination panel for AS and MA groups by the use of the test set. The confusion matrices are presented in Figure 5. Good prediction performances were observed for the AS and MA groups with prediction accuracies of 0.944 and 0.889, respectively. Moreover, the predictive capacities of the combination panels for AS and MA groups were also evaluated using the independent data from the test set, as shown in Figures S18 and S19 (Supporting Information). There was a decrease in the predictive abilities, but it is still good.
Figure 5.
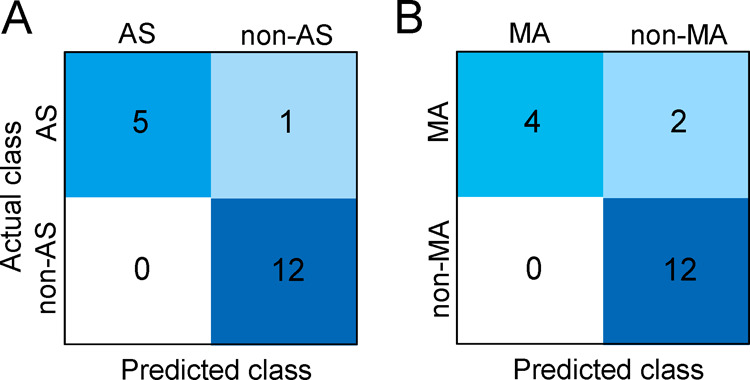
Confusion matrices obtained by the external validation for the anaphylactic shock (AS) group (A) and the mechanical asphyxia (MA) group (B). Non-AS represents MA and sudden cardiac death (SCD) groups. Non-MA represents AS and SCD groups.
Discussion
In this study, the brainstem injury was set as the control group, which has a fast death process and experiences the least biochemical stress, best reflecting the normal metabolic level of live rats. Additionally, the rats in the control group were also anesthetized to eliminate potential metabolic changes associated with the anesthesia. Thus, the differential metabolites that we screened and verified could represent the metabolic alterations in the death process for the relevant COD. They mainly belong to amino acids, organic acids, and carbohydrates; and they revealed some shared and specific metabolic changes in response to AS, MA, and SCD. Unfortunately, there are few previous studies on the metabolomics of COD. The supporting literature of metabolic changes in different death processes is lacking, but we hope this preliminary work might offer some new knowledge.
Metabolic Changes Related to AS
Anaphylaxis is a type I hypersensitivity reaction, and shock can happen and lead to death when the clinical symptoms are severe. The main pathophysiological features of AS are a profound reduction in the venous tone and fluid extravasation, causing a reduced venous return and depressed myocardial function.32 According to the classification results between the AS and non-AS groups, the decrease in creatinine may be a specific metabolic change when AS occurred. Creatinine is the end product of creatinine catabolism. It suggests that the process of the dehydration of creatinine was altered in AS.
Glucose is the major energy source for the biological activity. Malic acid is an intermediate in the tricarboxylic acid (TCA) cycle. The increase in the blood glucose level and the lower level of malic acid might be a result of the increased production of glucose and high energy expenditure through the TCA cycle in response to AS stress. Moreover, pipecolic acid is a metabolite of lysine, which also participates in energy metabolism.33 The results indicated that energy metabolism is a significant aspect of AS. Chan Seo et al. studied plasma GC–MS metabolomics in mice with asthma and found that the pipecolic acid content increased and the malic acid content decreased.34 This observation was consistent with our findings because there is a potential relationship between asthma and AS.
4-Hydroxyproline (Hyp) is a major component of collagen, which is used as a parameter of collagen catabolism. Urea is the end product of protein catabolism, and uracil may be an indicator of a pyrimidine metabolism disorder.35 The alteration of Hyp, urea, and uracil indicated the potential disorder of collagen catabolism, the deamination of amino acids, and the alteration of pyrimidine metabolism, respectively. Moreover, ethanolamine is the head group of phosphatidylethanolamine, which is a typical and abundant phospholipid in biological membranes. The decrease in ethanolamine in AS might be due to its increased consumption in the conformation or function of biological membranes. α-Tocopherol is traditionally recognized as the most active form of vitamin E in humans and is a powerful biological antioxidant.36 Its increase might suggest the active antioxidation in AS.
Metabolic Changes Related to MA
Ligature strangulation leads to the failure of body cells to either receive or utilize oxygen by neck compression. According to the classification results between the MA and non-MA groups, the accumulations of malic acid and uric acid may represent some specific metabolic change when MA occurred. The content of malic acid increased during MA, unlike that it decreased in AS. It suggests that the TCA cycle was altered in different ways. Unfortunately, we did not find possible explanations when reviewing the literature. In addition, uric acid is the final oxidation product of purine metabolism. Hsing-Jin Chen et al. found that uric acid can serve as a potential biomarker with a higher ratio of uric acid to creatinine in perinatal asphyxia.37 This finding supported our result that the uric acid content increased significantly in the MA group. Therefore, the alteration of purine metabolism may be a specific metabolic change in MA.
Glycerol 3-phosphate is a chemical intermediate in the glycolysis metabolic pathway. The increased level of glycerol 3-phosphate indicated that anaerobic glycolysis was activated due to the shortage of oxygen. Stearic acid is a long-chain fatty acid, and the increase in stearic acid suggested the activation of energy metabolism in MA. When severe hypoxia occurred, metabolic acidosis occurred. Pyroglutamic acid (PGA) is an organic acid, and the accumulation of PGA in blood results in metabolic acidosis.38−40 This finding suggests that the increase in PGA may be one way of causing metabolic acidosis during MA. Moreover, cholesterol has vital structural roles in membranes and lipid metabolism and is a biosynthetic precursor of steroid hormones. It also has significant functions in signal transduction. The level of cholesterol increased in the MA group, suggesting the possible alteration of lipid metabolism and signal transduction.
Metabolic Changes Related to SCD
CAHD is one of the most common causes of SCD, which results in acute myocardial infarction or ischemia, arrhythmias, and cardiac dysfunction. The potential mechanism also involves the stimulation of the stress response of the sympathetic nervous system and the subsequent elevation of circulating catecholamines.41 Unfortunately, we failed to find some specific metabolic changes for SCD compared to the other CODs according to the ROC curve analysis.
l-aspartic acid plays a role in translocating nicotinamide adenine dinucleotide (NADH) into mitochondria across the inner mitochondrial membrane for the oxidative phosphorylation by the aspartate–malate shuttle and is a major excitatory neurotransmitter.42 The decreased level of aspartic acid might be a result of its consumption, indicating more energy supply or the disorder of signal transduction. 3-phosphoglyceric acid (3-PG) is an important biochemical intermediate of lipid metabolism as the phosphate derivative of glyceric acid and can further participate in glycolysis.43 The decreased level of 3-PG suggested its consumption and the activation of glycolysis in SCD. The decrease in l-aspartic acid and 3-PG and the increase in glucose may indicate more energy supply in SCD. Moreover, the alterations in ethanolamine, uracil, PGA, and α-tocopherol might be related to the disorder of the conformation or function of biological membranes, pyrimidine metabolism disorders, the induction of metabolic acidosis, and active antioxidation, similarly.
The Potential of GC–HRMS-Based Metabolomics for the Determination of CODs
After verifying and explaining the differential metabolites, the potential ability of GC–HRMS data to determine the complex CODs was further evaluated. First, the entire data set with all features was used in the ROC curve analysis. Although we failed to annotate and verify some of them as specific metabolites, the excellent classification performance (with top 100 features, AUC > 0.95) indicated the advantages of untargeted metabolomics to discriminate complex CODs based on metabolic differences.
Next, each verified metabolite was subjected to the ROC curve analysis. The majority showed a poor classification efficiency with low AUCs. Nevertheless, it was known in advance because, take the AS group as an example, these differential metabolites were determined by comparing the AS group and the control group. They represented the metabolic alterations resulting from the pathophysiological changes in AS, but the other CODs can definitely share the metabolic alterations with similar pathophysiological processes. So it is reasonable that some of them cannot discriminate between the AS and the non-AS groups. Therefore, creatinine that showed a good discriminative ability may suggest some unique changes that occur in AS. Similarly, malic acid and uric acid may suggest some unique changes that occur in MA. Good classification abilities were obtained with high AUCs using the combination of some verified metabolites for AS and MA. In addition, the results of external validations demonstrated that the classification models were successfully established with good robustness. However, it is a pity that SCD had fewer metabolic signatures and we did not find any metabolites that may represent some unique metabolic changes.
We have examined the metabolic changes of the three CODs and preliminarily evaluated the determination potential of GC–HRMS-based untargeted metabolomics, but there are some limitations in our work. First, in the real world, the sample collection time could vary greatly and there might be postmortem-interval-dependent changes on the metabolome.44 Thus, the influence of decomposition needs further investigation for the postmortem determination. Second, we employed only the GC–HRMS technique due to equipment limitations. We will combine high-performance LC–MS with improvements in the metabolite detection range to enrich this study in the next. Third, when considering applying this strategy to human samples in a real case, the sample variability increases, which turns out to be more difficult to properly interpret the metabolic changes that resulted from the cause of death and to build classification models. Therefore, the potential effect of age, gender, underlying disease, and living habit should be systematically studied to develop the application value of this strategy.
Conclusions
The plasma GC–HRMS data showed that metabolic changes resulting from different death mechanisms and processes do exist. A range of differential features was screened, and 11, 8, and 7 differential metabolites were finally verified for the AS, MA, and SCD groups, respectively. We proposed some explanations that may account for these metabolic differences, including glucose metabolism, the TCA cycle, glycolysis, lipid metabolism, creatinine catabolism, and purine metabolism. Next, for each target COD, the differential metabolites’ ability to discriminate the COD from the other CODs was further evaluated by the ROC curve analysis. We found that creatinine may represent some specific metabolic changes in AS. Similarly, malic acid and uric acid may represent some specific metabolic changes in MA. Moreover, the combination panels displayed good classification abilities, and the external validation results demonstrated that the classification models were successfully established. In this study, SCD had fewer metabolic signatures and was relatively harder to discriminate than the other CODs. Our results showed the potential of untargeted metabolomics for the screening of metabolic alterations and the determination of different CODs. Furthermore, untargeted metabolomics may facilitate a better understanding of pathophysiological changes in these CODs.
Material and Methods
Chemicals
All chemicals and solvents were of analytical or HPLC grade. Methanol, pyridine, methoxylamine hydrochloride, and N,O-bis(trimethylsilyl) trifluoroacetamide (BSTFA) containing 1% trimethylchlorosilane (TMCS) were purchased from Sigma-Aldrich (MO, USA). l-2-Chlorophenylalanine was purchased from Aladdin Chemistry (Shanghai, China). Ultrapure water was produced by a Millipore Milli-Q Reagent Water System (MA, USA). The potential standard compounds used for verification were mainly purchased from Aladdin Chemistry (Shanghai, China), Yuanye Bio-Technology (Shanghai, China), Topscience (Shanghai, China), and the National Institutes for Food and Drug Control (Beijing, China).
Animal Experiments
Sixty-two Sprague–Dawley rats (male, weight 230–270 g) were purchased from the Animal Center of Xi’an Jiaotong University and then housed under a 12 h light/dark cycle environment with food and water ad libitum. Forty-four rats of the 62 were randomly divided into four groups of different CODs (three targets and one control, 11 rats per group). The rest 18 rats were treated as the test set (three target groups and six rats per group). It should be emphasized that this study was approved by the Committee on the Ethics of Animal Experiments of Xi’an Jiaotong University and was conducted strictly according to the recommendations in the Guide for the Care and Use of Laboratory Animals Committee of Xi’an Jiaotong University. Urethane (20%, 1.0–1.2 g/kg) was intraperitoneally injected to anesthetize each rat well for minimal pain and discomfort before they were sacrificed.
For the AS group, rats were sensitized by intraperitoneal injection with ovalbumin as an allergen (10 mg/100 g, dissolved in 0.5 mL of 0.9% sodium chloride solution) on the first, third, and fifth days. The sensitized rats were then challenged intravenously with ovalbumin (100 mg, dissolved in 1.0 mL of 0.9% sodium chloride solution) on the 18th day. The rats would experience AS and die in approximately 15–30 min. For the MA group, rats were modeled using strangulation. A noose made of a cotton thread was placed around the neck, and a small stick was inserted into the noose from the back of the neck. Then, the noose was tightened by rotating the stick to asphyxiate the rat under a steady pressure until the rat died (approximately 4–5 min). For the SCD group, the most common CAHD was chosen to model SCD. Briefly, the thorax and pericardium of a rat were carefully cut to expose the heart. The left anterior descending coronary artery was ligatured tightly, and the rat died in approximately 10 min. Hemostatic measures were taken, and the wound was sewn up. In addition, the last group that experienced a brainstem injury by cervical dislocation was set as the control group. A whole blood sample was collected from the inferior vena cava when the death of each rat was confirmed. Then, the plasma was obtained by centrifugation at 3000 rpm for 15 min at 4 °C. These plasma samples were stored at −80 °C until use.
Sample Preparation
Plasma samples were thawed at room temperature, and 100 μL of each sample was added to a 1.5 mL Eppendorf tube. A total of 300 μL of ice cold methanol solution (for the removal of protein) and 20 μL of l-2-chlorophenylalanine (0.3 mg/mL), as the internal standard (IS), were added to each tube. The mixture was vortexed and then incubated on ice for 10 min. Subsequently, the samples were centrifuged at 13 000 rpm for 15 min at −4 °C, and 150 μL of supernatant of each sample was transferred to a glass sampling vial. The quality control (QC) sample was prepared by mixing aliquots of all supernatants to form a pooled sample. The collected supernatants were evaporated to dryness using a vacuum concentrator.
Next, the samples were chemically derivatized for the GC–HRMS analysis. For each sample, 80 μL of methoxylamine hydrochloride (15 mg/mL in pyridine) was added to the residue. The resultant mixture was vortex-mixed for 2 min and incubated for 90 min at 37 °C. Next, 100 μL of BSTFA (with 1% TMCS) was added into the mixture, which was vortexed for 2 min and then derivatized for 60 min at 70 °C. Following derivatization and cooling to room temperature, 1 μL of the sample was injected for the GC–HRMS analysis.
Acquisition of the GC–HRMS Data
The metabolic profiles were acquired using a Q Exactive GC Orbitrap GC–HRMS system (Thermo Fisher Scientific, USA) equipped with a TG-5SILMS capillary column (30 m × 0.25 mm × 0.25 μm, Thermo Fisher Scientific, USA). The equipment conditions were as follows: helium (>99.999%) was used as the carrier gas at a constant flow rate of 1.0 mL/min. The injector temperature was maintained at 250 °C. Samples (1 μL) were injected in a split mode (1:15). The solvent delay time was set to 3.5 min. The initial oven temperature was kept at 80 °C for 1.0 min, then increased from 80 to 300 °C at a rate of 10 °/min, and finally held at 300 °C for 5.0 min. The total run time was 28.0 min. The ion-source temperature and transfer line temperatures were 250 and 280 °C, respectively. The full scan mode was used in positive polarity with a scan range from 60 to 500 (m/z). The full MS resolution was 60 000 (FWHM) with a 1e6 ion AGC (automatic gain control) target. Moreover, the QC samples were injected at intervals of every five samples throughout the whole run.
Data Processing and Statistical Analysis
The open-source software MZmine245 was utilized to process the raw GC–HRMS data, including the baseline correction, peak detection, peak deconvolution, and peak alignment. Next, the resultant comma-separated value (CSV) files were further analyzed using the web-based tool MetaboAnalyst 4.0.46 Data filtering was performed first to remove the features that showed low repeatability by measuring the relative standard deviation (RSD = SD/average) using QC samples (the threshold for GC–MS-untargeted metabolomics analysis is 30%, and the RSD values of some features that were successfully verified in the next are listed in Table S1). Then, the data were normalized according to the feature intensity of IS. Next, the preprocessed data were log-transformed to make individual features more comparable.
Subsequently, the principal component analysis (PCA), which is a classic multivariate statistical method for reducing data dimensionality, was conducted to visualize the sample projection in three dimensions of the new uncorrelated variables (principal components, PCs). The partial least-squares discriminant analysis (PLS-DA), fold change (FC) analysis, and Wilcoxon test were then performed between experimental groups and control groups for the metabolic signature discovery. The 10-fold cross-validation and 1000 permutation testing were applied to check the risk of overfitting in PLS-DA models. The metabolic features whose variable importance in the projection (VIP) values greater than 1.0 in the PLS-DA model, FCs greater than 1.5 or less than 0.6667 (the FC cutoff is 1.5), and p values less than 0.05 were considered significantly different.47,48 The differential metabolites were first identified by searching the National Institute of Standards and Technology (NIST) library and Human Metabolome Database (HMDB) and were verified by comparing the MS spectra and retention time (within 2% error margin) of commercially available standard compounds. In addition, the areas under the curve (AUCs) of the receiver operating characteristic (ROC) curve analysis were calculated to evaluate the predictive effectiveness of the different panels of metabolites. The classification models were then validated using the data derived from the rats in the test set.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81730056, 81722027, 82072115, 81601645, and 81671869), the National Key R&D Program of China (2016YFC0800702), and the Science and Technology Committee of Shanghai Municipality (17DZ2273200 and 19DZ2292700).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c05178.
PCA score plots of every two groups; permutation testing results for PLS-DA models; PLS-DA models and VIP scores for MA and SCD groups; metabolic pathway analysis; ROC curves for each verified metabolite; ROC curves for all the features and metabolite combination in MA and SCD groups; external validation results; and RSD values of the verified metabolites (PDF)
Author Contributions
§ Kai Zhang and Hui Yan contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Prahlow J. A.Forensic Pathology for Police, Death Investigators, Attorneys, and Forensic Scientists; Springer Science & Business Media: 2010. [Google Scholar]
- Michaud K.; Grabherr S.; Doenz F.; Mangin P. Evaluation of Postmortem MDCT and MDCT-Angiography for the Investigation of Sudden Cardiac Death Related to Atherosclerotic Coronary Artery Disease. Int. J. Cardiovasc. Imaging 2012, 28, 1807–1822. 10.1007/s10554-012-0012-x. [DOI] [PubMed] [Google Scholar]
- Taylor A. M.; Sebire N. J.; Ashworth M. T.; Schievano S.; Scott R. J.; Wade A.; Chitty L. S.; Robertson N.; Thayyil S. Postmortem Cardiovascular Magnetic Resonance Imaging in Fetuses and Children. Circulation 2014, 129, 1937–1944. 10.1161/CIRCULATIONAHA.113.005641. [DOI] [PubMed] [Google Scholar]
- Michaud K.; Genet P.; Sabatasso S.; Grabherr S. Postmortem Imaging as a Complementary Tool for the Investigation of Cardiac Death. Forensic Sci. Res. 2019, 4, 211–222. 10.1080/20961790.2019.1630944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljakna A.; Fracasso T.; Sabatasso S. Molecular Tissue Changes in Early Myocardial Ischemia: From Pathophysiology to the Identification of New Diagnostic Markers. Int. J. Legal Med. 2018, 132, 425–438. 10.1007/s00414-017-1750-z. [DOI] [PubMed] [Google Scholar]
- Meng X.; Ming M.; Wang E. Heart Fatty Acid Binding Protein as a Marker for Postmortem Detection of Early Myocardial Damage. Forensic Sci. Int. 2006, 160, 11–16. 10.1016/j.forsciint.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Li Y.; Si R.; Feng Y.; Chen H. H.; Zou L.; Wang E.; Zhang M.; Warren H. S.; Sosnovik D. E.; Chao W. Myocardial Ischemia Activates an Injurious Innate Immune Signaling via Cardiac Heat Shock Protein 60 and Toll-like Receptor 4. J. Biol. Chem. 2011, 286, 31308–31319. 10.1074/jbc.M111.246124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K. S. The Promise of Molecular Autopsy in Forensic Pathology Practice. Acad. Forensic Pathol. 2017, 7, 551–566. 10.23907/2017.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio L. A.; Olivier M.; Hubacek J. A.; Cohen J. C.; Cox D. R.; Fruchart J.-C.; Krauss R. M.; Rubin E. M. An Apolipoprotein Influencing Triglycerides in Humans and Mice Revealed by Comparative Sequencing. Science 2001, 294, 169 LP–173. 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- Szalai C.; Keszei M.; Duba J.; Prohászka Z.; Kozma G. T.; Császár A.; Balogh S.; Almássy Z.; Fust G.; Czinner A. Polymorphism in the Promoter Region of the Apolipoprotein A5 Gene Is Associated with an Increased Susceptibility for Coronary Artery Disease. Atherosclerosis 2004, 173, 109–114. 10.1016/j.atherosclerosis.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Wang F.; Xu C.-Q.; He Q.; Cai J.-P.; Li X.-C.; Wang D.; Xiong X.; Liao Y.-H.; Zeng Q.-T.; Yang Y.-Z.; Cheng X.; Li C.; Yang R.; Wang C.-C.; Wu G.; Lu Q.-L.; Bai Y.; Huang Y.-F.; Yin D.; Yang Q.; Wang X.-J.; Dai D.-P.; Zhang R.-F.; Wan J.; Ren J.-H.; Li S.-S.; Zhao Y.-Y.; Fu F.-F.; Huang Y.; Li Q.-X.; Shi S.-W.; Lin N.; Pan Z.-W.; Li Y.; Yu B.; Wu Y.-X.; Ke Y.-H.; Lei J.; Wang N.; Luo C.-Y.; Ji L.-Y.; Gao L.-J.; Li L.; Liu H.; Huang E.-W.; Cui J.; Jia N.; Ren X.; Li H.; Ke T.; Zhang X.-Q.; Liu J.-Y.; Liu M.-G.; Xia H.; Yang B.; Shi L.-S.; Xia Y.-L.; Tu X.; Wang Q. K. Genome-Wide Association Identifies a Susceptibility Locus for Coronary Artery Disease in the Chinese Han Population. Nat. Genet. 2011, 43, 345–349. 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- Patti G. J.; Yanes O.; Siuzdak G. Metabolomics: The Apogee of the Omics Trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J. K.; Lindon J. C.; Holmes E. “Metabonomics”: Understanding the Metabolic Responses of Living Systems to Pathophysiological Stimuli via Multivariate Statistical Analysis of Biological NMR Spectroscopic Data. Xenobiotica 1999, 29, 1181–1189. 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- Cajka T.; Fiehn O. Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics. Anal. Chem. 2016, 88, 524–545. 10.1021/acs.analchem.5b04491. [DOI] [PubMed] [Google Scholar]
- Raterink R.-J.; Lindenburg P. W.; Vreeken R. J.; Ramautar R.; Hankemeier T. Recent Developments in Sample-Pretreatment Techniques for Mass Spectrometry-Based Metabolomics. TrAC, Trends Anal. Chem. 2014, 61, 157–167. 10.1016/j.trac.2014.06.003. [DOI] [Google Scholar]
- Wang X.; Chen S.; Jia W. Metabolomics in Cancer Biomarker Research. Curr. Pharmacol. Rep. 2016, 2, 293–298. 10.1007/s40495-016-0074-x. [DOI] [Google Scholar]
- Dinis-Oliveira R. J. Metabolomics of Methadone: Clinical and Forensic Toxicological Implications and Variability of Dose Response. Drug Metab. Rev. 2016, 48, 568–576. 10.1080/03602532.2016.1192642. [DOI] [PubMed] [Google Scholar]
- Khamis M. M.; Adamko D. J.; El-Aneed A. Mass Spectrometric Based Approaches in Urine Metabolomics and Biomarker Discovery. Mass Spectrom. Rev. 2017, 36, 115–134. 10.1002/mas.21455. [DOI] [PubMed] [Google Scholar]
- Qiu F.; Zhang Y.-Q. Metabolic Effects of Mulberry Branch Bark Powder on Diabetic Mice Based on GC-MS Metabolomics Approach. Nutr. Metab. 2019, 16, 10. 10.1186/s12986-019-0335-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer K.; Aronov P. A.; Hammock B. D. Mass Spectrometry-Based Metabolomics. Mass Spectrom. Rev. 2007, 26, 51–78. 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoridis G.; Gika H. G.; Wilson I. D. Mass Spectrometry-Based Holistic Analytical Approaches for Metabolite Profiling in Systems Biology Studies. Mass Spectrom. Rev. 2011, 30, 884–906. 10.1002/mas.20306. [DOI] [PubMed] [Google Scholar]
- Beale D. J.; Pinu F. R.; Kouremenos K. A.; Poojary M. M.; Narayana V. K.; Boughton B. A.; Kanojia K.; Dayalan S.; Jones O. A. H.; Dias D. A. Review of Recent Developments in GC–MS Approaches to Metabolomics-Based Research. Metabolomics 2018, 14, 152. 10.1007/s11306-018-1449-2. [DOI] [PubMed] [Google Scholar]
- Wu Z.; Huang Z.; Lehmann R.; Zhao C.; Xu G. The Application of Chromatography-Mass Spectrometry: Methods to Metabonomics. Chromatographia 2009, 69, 23–32. 10.1365/s10337-009-0956-8. [DOI] [Google Scholar]
- Rathahao-Paris E.; Alves S.; Junot C.; Tabet J.-C. High Resolution Mass Spectrometry for Structural Identification of Metabolites in Metabolomics. Metabolomics 2016, 12, 10. 10.1007/s11306-015-0882-8. [DOI] [Google Scholar]
- Rubert J.; Zachariasova M.; Hajslova J. Advances in High-Resolution Mass Spectrometry Based on Metabolomics Studies for Food – a Review. Food Addit. Contam., Part A 2015, 32, 1685–1708. 10.1080/19440049.2015.1084539. [DOI] [PubMed] [Google Scholar]
- Madsen R.; Lundstedt T.; Trygg J. Chemometrics in Metabolomics—A Review in Human Disease Diagnosis. Anal. Chim. Acta 2010, 659, 23–33. 10.1016/j.aca.2009.11.042. [DOI] [PubMed] [Google Scholar]
- Castillo-Peinado L. S.; Luque de Castro M. D. Present and Foreseeable Future of Metabolomics in Forensic Analysis. Anal. Chim. Acta 2016, 925, 1–15. 10.1016/j.aca.2016.04.040. [DOI] [PubMed] [Google Scholar]
- Hu X.; Wu G.; Zhang M.; Pan S.; Wang R.; Ouyang J.; Liu J.; Chen Z.; Tian H.; Liu D. GC–MS-Based Metabolic Profiling Reveals Metabolic Changes in Anaphylaxis Animal Models. Anal. Bioanal. Chem. 2012, 404, 887–893. 10.1007/s00216-012-6129-x. [DOI] [PubMed] [Google Scholar]
- Rousseau G.; Chao de la Barca J. M.; Rougé-Maillart C.; Teresiński G.; Jousset N.; Dieu X.; Chabrun F.; Prunier-Mirabeau D.; Simard G.; Reynier P.; Palmiere C. A Serum Metabolomics Signature of Hypothermia Fatalities Involving Arginase Activity, Tryptophan Content, and Phosphatidylcholine Saturation. Int. J. Legal Med. 2019, 133, 889–898. 10.1007/s00414-018-1937-y. [DOI] [PubMed] [Google Scholar]
- Wang D.; Wang X.; Wu J.; Su R.; Kong J.; Yu X. Metabolic Risk Factors Associated with Sudden Cardiac Death (SCD) during Acute Myocardial Ischemia. Forensic Sci. Res. 2017, 2, 126–131. 10.1080/20961790.2017.1343269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio D.; Consonni V. Classification Tools in Chemistry. Part 1: Linear Models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. 10.1039/C3AY40582F. [DOI] [Google Scholar]
- Brown S. G. A. The Pathophysiology of Shock in Anaphylaxis. Immunol. Allergy Clin. North Am. 2007, 27, 165–175. 10.1016/j.iac.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Lin H.; Su X.; He B. Protein Lysine Acylation and Cysteine Succination by Intermediates of Energy Metabolism. ACS Chem. Biol. 2012, 7, 947–960. 10.1021/cb3001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo C.; Hwang Y.-H.; Lee H.-S.; Kim Y.; Shin T. H.; Lee G.; Son Y.-J.; Kim H.; Yee S.-T.; Park A. K.; Paik M.-J. Metabolomic Study for Monitoring of Biomarkers in Mouse Plasma with Asthma by Gas Chromatography–Mass Spectrometry. J. Chromatogr., B 2017, 1063, 156–162. 10.1016/j.jchromb.2017.08.039. [DOI] [PubMed] [Google Scholar]
- Davies P. M.; Fairbanks L. D.; Dulby J. A.; Simmonds H. A. Urinary Uracil Concentrations Are a Useful Guide to Genetic Disorders Associated with Neurological Deficits and Abnormal Pyrimidine Metabolism. J. Inherit. Metab. Dis. 1997, 20, 328–330. 10.1023/A:1005332606213. [DOI] [PubMed] [Google Scholar]
- Liebler D. C.; Kling D. S.; Reed D. J. Antioxidant Protection of Phospholipid Bilayers by Alpha-Tocopherol. Control of Alpha-Tocopherol Status and Lipid Peroxidation by Ascorbic Acid and Glutathione. J. Biol. Chem. 1986, 261, 12114–12119. 10.1016/S0021-9258(18)67210-2. [DOI] [PubMed] [Google Scholar]
- Chen H.; Yau K.; Tsai K.-S. Urinary Uric Acid/Creatinine Ratio as an Additional Marker of Perinatal Asphyxia. J. formosan Med. Assoc. 2000, 99, 771–774. [PubMed] [Google Scholar]
- Fenves A. Z.; Kirkpatrick H. M.; Patel V. V.; Sweetman L.; Emmett M. Increased Anion Gap Metabolic Acidosis as a Result of 5-Oxoproline (Pyroglutamic Acid): A Role for Acetaminophen. Clin. J. Am. Soc. Nephrol. 2006, 1, 441 LP–447. 10.2215/CJN.01411005. [DOI] [PubMed] [Google Scholar]
- Liss D. B.; Paden M. S.; Schwarz E. S.; Mullins M. E. What Is the Clinical Significance of 5-Oxoproline (Pyroglutamic Acid) in High Anion Gap Metabolic Acidosis Following Paracetamol (Acetaminophen) Exposure?. Clin. Toxicol. 2013, 51, 817–827. 10.3109/15563650.2013.844822. [DOI] [PubMed] [Google Scholar]
- Luyasu S.; Wamelink M. M. C.; Galanti L.; Dive A. Pyroglutamic Acid-Induced Metabolic Acidosis: A Case Report. Acta Clin. Belg. 2014, 69, 221–223. 10.1179/2295333714Y.0000000022. [DOI] [PubMed] [Google Scholar]
- Goyal V.; Jassal D. S.; Dhalla N. S. Pathophysiology and Prevention of Sudden Cardiac Death. Can. J. Physiol. Pharmacol. 2016, 94, 237–244. 10.1139/cjpp-2015-0366. [DOI] [PubMed] [Google Scholar]
- Xie G.; Zhou B.; Zhao A.; Qiu Y.; Zhao X.; Garmire L.; Shvetsov Y. B.; Yu H.; Yen Y.; Jia W. Lowered Circulating Aspartate Is a Metabolic Feature of Human Breast Cancer. Oncotarget 2015, 6, 33369–33381. 10.18632/oncotarget.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabson R.; Tolbert N. E.; Kearney P. C. Formotion of Serine and Glyceric Acid by the Glycolate Pathway. Arch. Biochem. Biophys. 1962, 98, 154–163. 10.1016/0003-9861(62)90161-3. [DOI] [PubMed] [Google Scholar]
- Pesko B. K.; Weidt S.; McLaughlin M.; Wescott D. J.; Torrance H.; Burgess K.; Burchmore R. Postmortomics: The Potential of Untargeted Metabolomics to Highlight Markers for Time Since Death. OMICS: J. Integr. Biol. 2020, 24, 649–659. 10.1089/omi.2020.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluskal T.; Castillo S.; Villar-Briones A.; Orešič M. MZmine 2: Modular Framework for Processing, Visualizing, and Analyzing Mass Spectrometry-Based Molecular Profile Data. BMC Bioinformatics 2010, 11, 395. 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J.; Wishart D. S.; Xia J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinf. 2019, 68, e86 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]
- Kind T.; Wohlgemuth G.; Lee D. Y.; Lu Y.; Palazoglu M.; Shahbaz S.; Fiehn O. FiehnLib: Mass Spectral and Retention Index Libraries for Metabolomics Based on Quadrupole and Time-of-Flight Gas Chromatography/Mass Spectrometry. Anal. Chem. 2009, 81, 10038–10048. 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J. D.; Tibshirani R. Statistical Significance for Genomewide Studies. Proc. Natl. Acad. Sci. 2003, 100, 9440 LP–9445. 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.