Abstract
Background
Wall-associated kinases (WAK), one of the receptor-like kinases (RLK), function directly in the connection and communication between the plant cell wall and the cytoplasm. WAK genes are highly conserved and have been identified in plants, such as rice, but there is little research on the WAK gene family in cotton.
Results
In the present study, we identified 29 GhWAK genes in Gossypium hirsutum. Phylogenetic analysis showed that cotton WAK proteins can be divided into five clades. The results of synteny and Ka/Ks analysis showed that the GhWAK genes mainly originated from whole genome duplication (WGD) and were then mainly under purifying selection. Transcriptome data and real-time PCR showed that 97% of GhWAK genes highly expressed in cotton fibers and ovules. β-glucuronidase (GUS) staining assays showed that GhWAK5 and GhWAK16 expressed in Arabidopsis leaf trichomes. Fourteen GhWAK genes were found to possess putative gibberellin (GA) response elements in the promoter regions, 13 of which were significantly induced by GA treatment. Ten GhWAK genes contained auxin (IAA) response elements and the expression level of nine GhWAKs significantly increased under auxin treatment.
Conclusions
We provide a preliminary analysis of the WAK gene family in G. hirsutum, which sheds light on the potantial roles of GhWAK genes in cotton fiber cell development. Our data also provides a useful resource for future studies on the functional roles of GhWAK genes.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-021-07378-8.
Keywords: Gossypium hirsutum, WAK genes, Expression analysis
Background
The cell wall is a slightly elastic structure surrounding the cell membrane and is a complex network composed of cellulose, hemicellulose, pectin and a small amount of structural proteins [1]. The cell wall functions in cell morphological structure and is the first defense against pathogen invasion [2]. Receptor like kinases (RLKs), are a kind of important plant protein kinases. A typical RLK protein contains a putative extracellular domain (ECD), a hydrophobic transmembrane region and a cytoplasmic Ser/Thr kinase domain [3], which could be activated by external and internal stimulus and play important roles in plant growth and development. Studies have shown that the wall-associated kinases (WAKs) and the proline rich extensine-like receptor kinases (PERKs) function in signal transduction between the extracellular matrix and cytoplasm [4].
WAKs are linked with the cell wall and are important proteins in the connection between plant cell wall and cytoplasm. Typical WAK structures mainly consist of four domains, including ECD, transmembrane domain, epidermal growth factor-like (EGF) and intracellular kinase domain [5]. The intracellular domain of WAKs is composed of Ser/Thr kinase, which functions in intracellular signal transduction [6]. Protease treatments demonstrated that the ECD locates at the N-terminal, is a galacturonan-binding extracellular domain, which directly connected to the cell wall [7, 8]. Near the transmembrane domain, there are EGF-repeats containing 12 conserved cysteine residues [5]; these amino acid residues are thought to be directly involved in protein-protein interactions and are related to protein functions. The transmembrane structure of WAK indicates that WAK is essential in signaling between the cytoplasm and the cell wall [9]. Results of enzyme-linked immunosorbent assays (ELISAs) and pectinase treatment suggest that WAKs covalently bind to pectin [9, 10], which is the main component of the cell wall and is involved in cell elongation and pathogen resistance.
The first member of the WAK gene family, pro25, was found in Arabidopsis, and contains the Ser/Thr kinase domain. After analyzing its biochemical characteristics, pro25 was found to be closely linked to the cell wall, so they renamed it WAK1 (wall-associated kinase 1) and proposed the concept of cell wall-linked protein kinase [3]. Researchers then discovered a family of genes containing WAK1, WAK2, WAK3, WAK4 and WAK5 in Arabidopsis and they were highly conservative [7].
WAKs play important functions in cell elongation. In Arabidopsis, AtWAK4 antisense gene resulted in significantly decreased cell elongation and inhibited lateral root development [11]. Also, antisense plants of five members of the WAK family in Arabidopsis showed smaller leaves than control plants with shorter cells [9, 12]. The WAK2 null allele, wak2–1, caused a loss of cell expansion in roots. In addition, WAK2 affected cell elongation by regulating the expression level of a vacuolar invertase, which separates sucrose into glucose and fructose to increase the concentration of solute and promote cell water swelling, elevating intracellular turgor and resulting in a loose cell wall [1]. Expression analysis of the five AtWAK members also showed that WAKs expressed at organ junctions, shoot apical meristems, root apical meristems, and expanding leaves [9]. WAKs expressed throughout plant development and are required for cell expansion and elongation. Plant cell expansion and elongation mechanisms are very complicated and involve phytohormones, such as gibberellin (GA) and auxin (IAA) [13–16].
In recent years, studies have shown that WAKs are pectin receptors not only required for elongation but also for an induced stress response [17]. In Arabidopsis, WAK1 can be induced by pathogen infection or by exogenous salicylic acid (SA) [18]. RLK family genes AtWAK1–3 are involved in the activation of the secondary bile acid deoxycholic acid (DCA), which can activate the plant immune system to decrease two kinds of bacteria [19]. ZmWAK responds to the maize fungal diseases corn leaf blight and head smut disease [20, 21]. In rice, OsWAK14, OsWAK91 and OsWAK92 positively regulate quantitative resistance and OsWAK112d is a negative regulator of resistance to rice blast fungus Magnaporthe oryzae [22, 23]. Wheat TtWAK2 enhances wheat resistance to Fusarium graminearum by binding to pectin fragments [24], and OsWAK/Xa4 increases rice blight resistance by strengthening the cell wall through increasing cellulose synthesis [25]. WAK genes function in resistance to bacterial and fungal diseases in cereal plants with increasing biosynthesis of cellulose and phytoalexin to increase cell wall strength [20, 21, 25, 26]. WAK proteins are also essential in responding to plant abiotic stresses. OsWAK11 defends against excess copper by regulating the methylesterification of the cell wall [27]. Overexpression of AtWAK1 enhanced A. thaliana root tolerance to aluminum [28].
G. hirsutum is an allotetraploid species. The homeologous of chloroplast DNAs and the large subunit of ribulose bisphosphate carboxylase analysis among Gossypium species showed that the allotetraploid cotton species originated from the hybridization of female parent G. arboreum (AA) and male parent G. raimondii (DD) 1–2 million years ago [29, 30]. G. hirsutum is planted worldwide because its valuable fiber is used as raw material in the textile industry and the length and strength of its fibers are very important qualities for this industry [31, 32]. Cotton fibers start from ovule epidermal cells and rapidly elongate to form a single cell seed coat [33, 34]. Cotton fiber development is divided into four overlapping stages: initiation, elongation, secondary cell wall deposition, and maturation [35]. The fiber length is determined during its elongation stage, 0 to 26 days post-anthesis (DPA), to reach its final length [36]. The cellulose of the fiber cell wall is synthesized from 15 to 40 DPA, which determines the quality of fiber strength [37]. WAKs directly connected with the cell wall and functioned in cell elongation and cellulose synthesis, WAK genes is important for cotton fiber development and lays foundation for improving fiber qualities through over-expression or silence of GhWAKs by genetic engineering. In the present study, we performed whole-genome analysis on the WAK gene family in G. hirsutum. The potential function of the WAK gene family in cotton fiber development was further elucidated through phylogenetic relationships, chromosome positions, expression profiles, GUS staining and protein structure analysis.
Results
Identification of WAK genes in Gossypium species and chromosomal distribution analysis
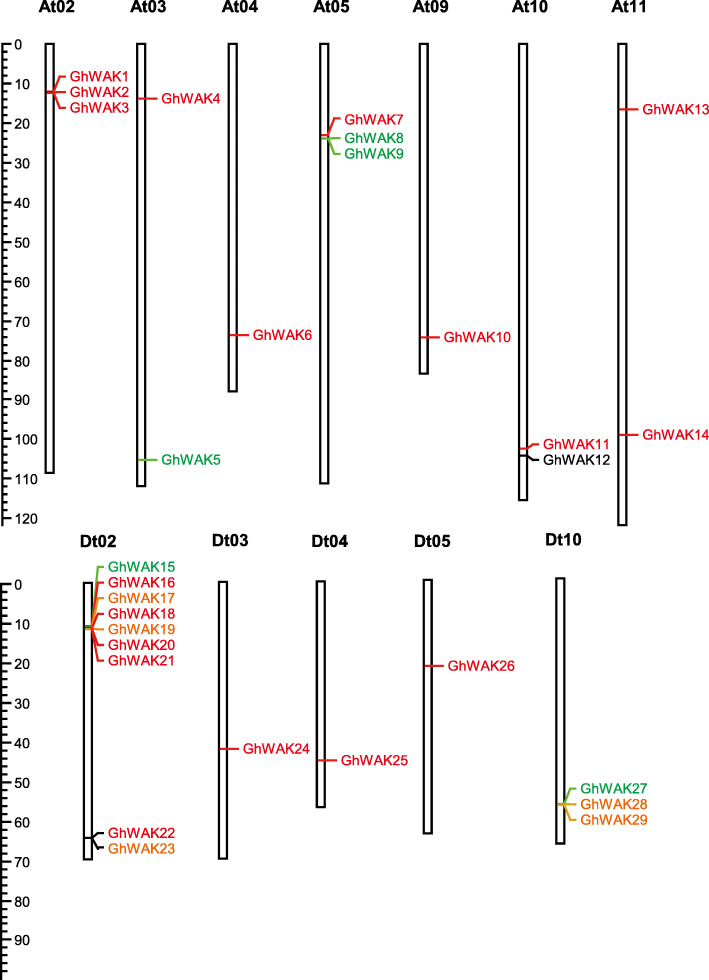
Whole genome sequences of three sequenced cotton species - G. hirsutum acc. TM-1 (ZJU_v2.1) [29], G. arboreum (CRI_v3.0) [38] and G. raimondii (JGI_v2.1) [39] - were used to identify WAK proteins. Arabidopsis AtWAK protein sequences were used as queries to search against the three reference genomes to screen out candidate WAK proteins in cotton. All the candidates (GhWAKs, GaWAKs and GrWAKs) with the four domains (signal peptide, 1–3 EGFs, transmembrane domain and protein kinase domain) were predicted to encode WAK proteins. In total, there were 29, 19 and 14 WAK genes in G. hirsutum, G. arboreum and G. raimondii, respectively. According to their relative chromosome locations, the 29 candidate GhWAK genes were named as GhWAK1 to GhWAK29 from chromosomes At01 to Dt13 (Fig. 1). Chromosome distribution analysis showed that the 29 GhWAKs unequally distributed on 12 chromosomes including seven At subgenomes and five Dt subgenomes. In chromosome Dt_02 nine GhWAK genes clustered together; At_02, At_05 and Dt_10 each had three GhWAK genes; At_10 had two GhWAK genes (Fig. 1); and all other chromosomes each had one GhWAK.
Fig. 1.
Chromosomal distribution of GhWAKs. Red lines show gene pairs involved in segmental duplication, green lines show gene pairs involved in tandem duplication, orange lines show gene pairs involved in proximal duplication
The number of amino acids (aa) in the GhWAK protein sequences ranged from 606 aa (GhWAK15) to 1200 aa (GhWAK29) with an average length of 755 aa. The molecular weights (MWs) of the predicted encoded proteins varied from 67.36 kDa (GhWAK15) to 134.02 kDa (GhWAK29) with an average of 84.32 kDa. According to the isoelectric point (pI) analysis, 20 GhWAKs had pI < 7.0 (with an average of 6.02) and were acidic in nature, whereas nine GhWAKs were predicted to encode proteins with pI > 7.0 (average of 8.34) and were basic in nature. Based on the instability index analysis, 23 GhWAK proteins have instability index values less than 40.0 and six GhWAK proteins have instability index values greater than 40.0 (GhWAK5, GhWAK15, GhWAK16, GhWAK20, GhWAK22 and GhWAK23). All the GhWAK proteins had negative grand average of hydropathicity (GRAVY) scores, indicating that all the GhWAK proteins were hydrophilic (Table 1). The detailed physicochemical parameters of GaWAK and GrWAK proteins were listed in Additional file 2: Table S1.
Table 1.
Physicochemical parameters of 29 GhWAK genes in G.hirsutum genome
| Gene ID | Gene Name | Chromosomes | Number of amino acids | Molecular weight | Theoretical pI | Instability index | Aliphatic index | Grand average of hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|---|---|
| GH_A02G0756 | GhWAK1 | A02 | 751 | 83,410.25 | 6.17 | 35.88 | 87.88 | −0.188 |
| GH_A02G0757 | GhWAK2 | A02 | 743 | 82,937.57 | 5.91 | 39.16 | 85.25 | −0.153 |
| GH_A02G0759 | GhWAK3 | A02 | 706 | 78,946.34 | 6.8 | 31.85 | 85.58 | −0.238 |
| GH_A03G0713 | GhWAK4 | A03 | 650 | 72,826.38 | 8.56 | 35.71 | 85 | −0.176 |
| GH_A03G1857 | GhWAK5 | A03 | 1014 | 113,495.2 | 5.89 | 42.75 | 72.38 | −0.371 |
| GH_A04G1037 | GhWAK6 | A04 | 636 | 70,572.08 | 8.62 | 33.77 | 94.26 | −0.051 |
| GH_A05G2392 | GhWAK7 | A05 | 752 | 83,052.99 | 5.11 | 32.64 | 84.88 | −0.216 |
| GH_A05G2396 | GhWAK8 | A05 | 709 | 79,129.46 | 8.35 | 36.54 | 82.61 | −0.201 |
| GH_A05G2397 | GhWAK9 | A05 | 746 | 82,336.37 | 6.52 | 31.05 | 84.68 | −0.213 |
| GH_A09G1705 | GhWAK10 | A09 | 637 | 70,724.12 | 8.79 | 34.59 | 93.36 | −0.103 |
| GH_A10G1999 | GhWAK11 | A10 | 684 | 76,591.69 | 6.38 | 36.21 | 84.06 | −0.214 |
| GH_A10G2055 | GhWAK12 | A10 | 745 | 83,174.38 | 5.99 | 39.57 | 79.93 | −0.236 |
| GH_A11G1483 | GhWAK13 | A11 | 654 | 73,748.88 | 6.4 | 39.24 | 88.04 | −0.193 |
| GH_A11G2681 | GhWAK14 | A11 | 636 | 71,097.94 | 8.7 | 38.26 | 87.04 | −0.148 |
| GH_D02G0763 | GhWAK15 | D02 | 606 | 67,363.5 | 7.09 | 40.44 | 81.85 | −0.23 |
| GH_D02G0764 | GhWAK16 | D02 | 745 | 82,717.14 | 5.2 | 40.76 | 88.97 | −0.154 |
| GH_D02G0769 | GhWAK17 | D02 | 745 | 82,795.27 | 5.33 | 38.37 | 88.05 | −0.157 |
| GH_D02G0772 | GhWAK18 | D02 | 747 | 83,034.53 | 5.63 | 35.96 | 84.95 | −0.207 |
| GH_D02G0775 | GhWAK19 | D02 | 751 | 83,325.86 | 5.83 | 37.04 | 85.43 | −0.222 |
| GH_D02G0777 | GhWAK20 | D02 | 743 | 83,260.15 | 6.2 | 40.28 | 85.9 | −0.156 |
| GH_D02G0778 | GhWAK21 | D02 | 716 | 80,229 | 6.54 | 35.66 | 86.7 | −0.209 |
| GH_D02G2021 | GhWAK22 | D02 | 993 | 111,861.5 | 7.96 | 44.65 | 78.89 | −0.337 |
| GH_D02G2022 | GhWAK23 | D02 | 1037 | 116,248.6 | 6.14 | 45.02 | 74.06 | −0.368 |
| GH_D03G1229 | GhWAK24 | D03 | 691 | 76,829.83 | 8.55 | 33.01 | 84.91 | −0.176 |
| GH_D04G1370 | GhWAK25 | D04 | 636 | 70,615.13 | 8.45 | 33.79 | 95.33 | −0.039 |
| GH_D05G2414 | GhWAK26 | D05 | 753 | 83,958.06 | 5.23 | 35.97 | 83.73 | −0.254 |
| GH_D10G2159 | GhWAK27 | D10 | 725 | 80,672.86 | 6.73 | 34.01 | 80.54 | −0.218 |
| GH_D10G2163 | GhWAK28 | D10 | 769 | 86,285.9 | 6.24 | 34.86 | 77.44 | −0.253 |
| GH_D10G2171 | GhWAK29 | D10 | 1200 | 134,018.2 | 6.22 | 35.24 | 74.99 | −0.255 |
Gene structure and phylogenetic relationships among GhWAK proteins
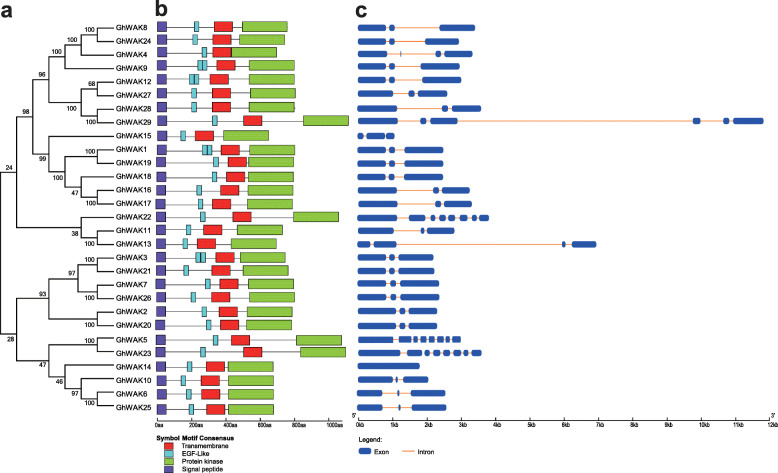
To fully display the protein domains and gene structures, a phylogenetic tree was constructed with the coding sequences (CDS) of GhWAKs (Fig. 2a). According to previous studies, WAK proteins contain highly conserved domains. All the GhWAK proteins have a signal peptide at the N-terminal end, the EGF-like domain is located near the transmembrane domain and the protein kinase domain is located at the C-terminal end (Fig. 2b). The intron and exon structure diagrams of GhWAK genes were constructed according to the CDS of the genes. GhWAK14 gene has no intron structure, GhWAK13 has four exons, and most of the GhWAKs have three exons (Fig. 2c).
Fig. 2.
Phylogenetic relationships of protein architecture and gene structure. a Phylogenetic relationships of GhWAK genes. The CDS of GhWAKs were used to construct the phylogenetic tree in MEGA 7.0, using the NJ-method and tested with 1000 bootstrap iterations. b Conserved domains of GhWAK proteins. Four motifs of GhWAK proteins determined using MEME. The red rectangular box indicates the transmembrane domain, the blue rectangular box indicates the EGF-Like domain, the green rectangular box indicates proteins kinase, and the purple rectangular box indicates signal peptide. c Gene structure analysis of GhWAKs. The exons and introns are indicated with blue filled boxes and orange lines, respectively
Phylogenetic analysis of GhWAK proteins
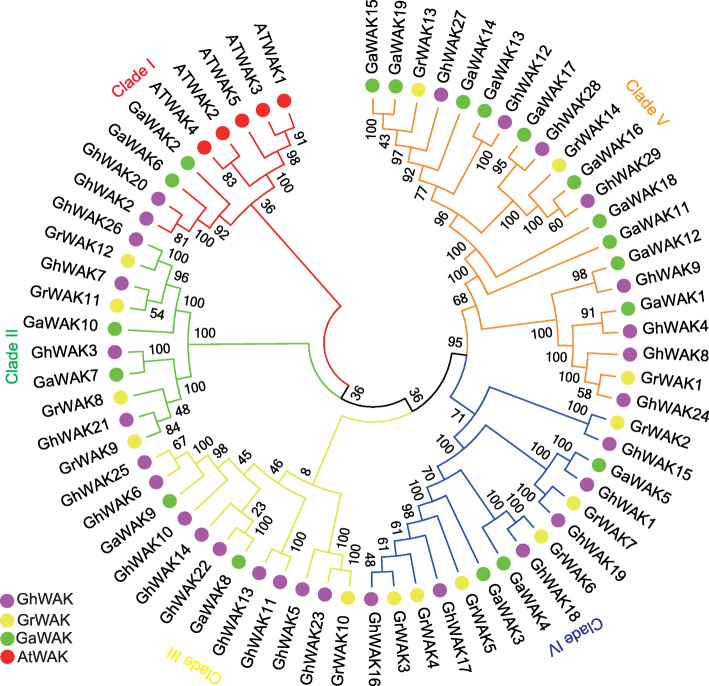
The phylogenetic tree of the WAK gene family contains WAK gene members from four species: G. hirsutum, G. arboreum, G. raimondii and A. thaliana. The phylogenetic tree was constructed using the neighbor-joining (NJ) method and tested by 1000 bootstrap replicates. The tree was resolved into five subgroups: Clade I, Clade II, Clade III, Clade IV and Clade V (Fig. 3). The GhWAK genes that clustered together in Fig. 1 were also preferentially clustered into the same subgroup in the phylogenetic tree. For example, GhWAK2 and GhWAK3 were clustered on chromosome At_02 and they were also clustered together into Clade II in the phylogenetic tree in Fig. 3. This phenomenon indicated that GhWAK genes may originate from gene duplication.
Fig. 3.
Phylogenetic tree of GhWAK GaWAK, GrWAK and AtWAK proteins. The phylogenetic tree was constructed using the full length of 29 GhWAKs, 19 GaWAKs, 14 GrWAKs and five AtWAKs, AtWAK1 (AT1G21210.1), AtWAK2 (AT1G21230.1), AtWAK3 (AT1G21240.1), AtWAK4 (AT1G21250.1) and AtWAK5 (AT1G21270.1) protein amino acid sequences. The phylogenetic tree was constructed using the neighbor-joining method with bootstrapping with 1000 iterations by MEGA 7.0
Duplication events of GhWAKs
Gene duplication events are usually the main reason for expansion of gene family members. Therefore, we further analyzed the gene duplication types of the WAK gene family in G. hirsutum, G. arboreum and G. raimondii using MCScanX. In total, 18 GhWAKs (accounting for 62.07%, ten GhWAKs from subgenome_At and eight GhWAKs from subgenome_Dt) were produced by whole genome duplication (WGD), five GhWAKs (17.24%) were produced by tandem duplication and five GhWAKs (17.24%) were produced by proximal duplication event (Additional file 2: Table S2). These results indicate that WGD events play a predominant role in the expansion of GhWAK genes, while GaWAKs and GrWAKs were mainly produced from tandem duplication events, accounting for 57.89 and 42.86%, respectively (Additional file 2: Table S2).
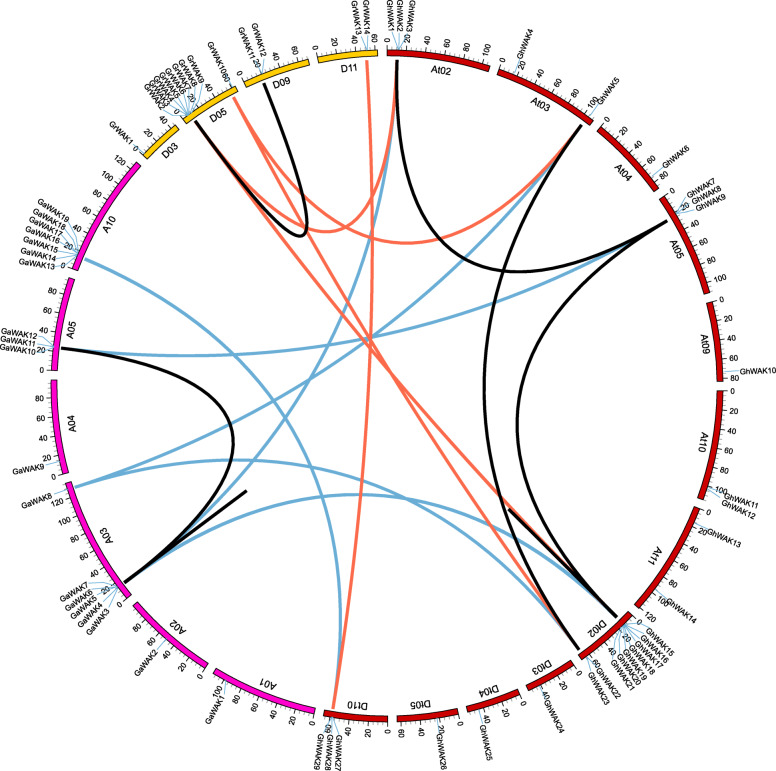
To compare the synteny and collinearity relationships of WAKs among G. arboreum, G. raimondii and G. hirsutum, we identified the orthologous and paralogous genes among the three released cotton genomes (Additional file 2: Table S3). Twelve GhWAKs had orthologous genes in G. arboreum, showing an AA genome origin, and seven GhWAKs genes had orthologous genes in G. raimondii, showing a DD genome origin (Fig. 4 and Additional file 1: Figure S1). To further analyze the gene duplication events of GhWAKs, we characterized four paralogous gene pairs (GhWAK15/GhWAK16, GhWAK9/GhWAK2, GhWAK8/GhWAK20, and GhWAK5/GhWAK22) in the G. hirsutum genome, two paralogous gene pairs (GaWAK7/GaWAK6 and GaWAK3/GaWAK12) in G. arboreum and one paralogous gene pair (GrWAK7/GrWAK11) in G. raimondii. Homologous exchange was observed among cotton genomes, which may lead to the orthologous gene pairs in different numbered chromosome among G. raimondii, G. arboreum and G. hirsutum.
Fig. 4.
The synteny relationships of WAK genes among three sequenced cotton species. G. hirsutum, G. raimondii and G. arboreum chromosomes are indicated in red, orange and pink, respectively. The putative orthologous WAKs between G. arboreum and G. hirsutum, and between G. raimondii and G. hirsutum are connected by blue and orange lines, respectively. Black lines connect the putative paralogous gene pairs (GhWAK15/GhWAK16, GhWAK2/GhWAK9, GhWAK8/GhWAK20, GhWAK5/GhWAK22, GaWAK7/GaWAK6, GaWAK3/GaWAK12 and GrWAK7/GrWAK11)
To further investigate the selective pressure after a gene duplication event, the non-synonymous (Ka) and synonymous (Ks) substitution rates were calculated. All the GhWAK gene pairs had Ka/Ks < 1.0 (Additional file 2: Table S4), indicating that the GhWAK genes had experienced purifying selection pressure after gene duplication events. Furthermore, the Ks ratio is less affected by selection and is usually used to estimate the evoluting divergence time [40]. The Ks ratios of six GhWAK gene pairs ranged from 0.0755 to 0.946, and duplication events occurred from approximately 14.51 million years ago (MYA) to 181.90 MYA (Additional file 2: Table S4). The Ks ratios of four GaWAK gene pairs ranged from 0.204 to 0.680, and the tandem duplication events occurred 39.20 MYA to 130.81 MYA. The Ks ratios of three GrWAK gene pairs ranged from 0.0362 to 0.272, and the tandem and proximal duplication events occurred 6.97 MYA to 52.23 MYA. These results suggest that the duplication events occurred before the polyploidization event because the G. hirsutum polyploidization occurred about 1–2 MYA.
Expression patterns of GhWAKs in different tissues
To investigate the tissue-specific expression patterns of GhWAK gene family members, transcriptome data from the roots, stems, leaves and 0 dpa ovules were used. As shown in Additional file 1: Figure S2, GhWAK6 highly expressed in young roots; GhWAK4, GhWAK5 and GhWAK16 showed stem-specific expression; GhWAK1, GhWAK3, GhWAK10, GhWAK17 and GhWAK19 dramatically expressed in young leaves; GhWAK2, GhWAK13, GhWAK18 and GhWAK29 expressed in flower-specific manner; GhWAK8, GhWAK9, GhWAK14, GhWAK15, GhWAK20, GhWAK21, GhWAK22, GhWAK23, GhWAK24, GhWAK27, GhWAK28 highly expressed in ovule-preferential manner. These results imply that GhWAKs expressed in tissue-specific manner in cotton.
Transcriptome analysis of GhWAK genes during fiber cell development
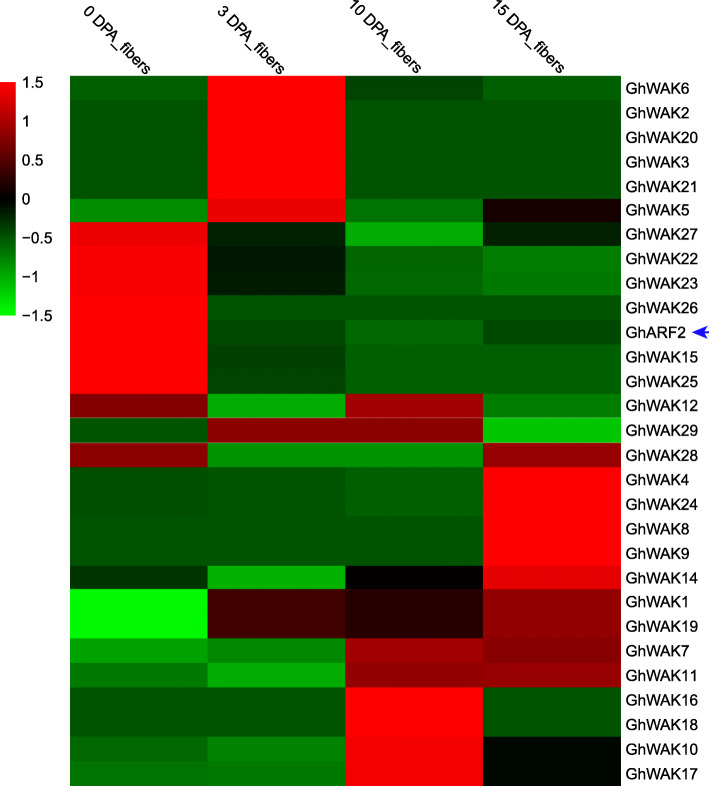
To further explore whether GhWAK genes potentially contribute to fiber cell development, the expression profiles of each gene were investigated using transcriptome data from different developmental stages (0 DPA, 3 DPA, 10 DPA and 15 DPA). To better show the expression values, we introduced GhARF2 gene (GH_D12G2130) [41], which highly expressed at the cotton fiber cell initiation stage, into the profile analysis of GhWAK genes. Our result showed that GhARF2 highly expressed in 0 DPA, which is consistent with the published result [41]. Furthermore, we investigated the expression level of six GhWAK genes using qRT-PCR experiment. Our data showed that these GhWAK genes were up-regulated in 3 DPA samples (Additional file 1: Figure S3), which is similar to the results from the transcriptome data, suggesting that the transcriptome data is credible. Twenty-eight out of Twenty-nine GhWAKs showed expression values in cotton fiber development process. The GhWAKs highly expressed during the fiber cell development process (Fig. 5). GhWAK2, 3, 5, 6, 12, 15, 20, 21, 22, 23, 25, 26, 27 and GhARF2 showed high expression levels at fiber cell initiation stage (0 and 3 DPA). The other GhWAK members highly expressed at fiber cell elongation stage (10 and 15 DPA). Arabidopsis trichomes are organs similar to fiber cells in cotton [42].
Fig. 5.
Heatmap of RNA-seq data of GhWAK gene expression levels during fiber cell development stages. The transcriptome data were normalized and visiualized by the pheatmap package with default parameters in R language. The colors from green to red indicate the expression levels from low to high, respectively. GhARF2 is involved in fiber initiation development and used as a marker gene
Atfer the initiation, fiber cells undergo the fast elongation stage (about 3 DPA to 10 DPA), in which fiber cells elongate about 2 mm per day. Therefore, we selected GhWAK5 with the highest expression level at 3 DPA and GhWAK16 with the peak value in 10 DPA fiber cells as the examples to further investigate the potential expression patterns of GhWAKs by GUS staining assays. The confused vectors of PGhWAK5::GUS and PGhWAK16::GUS were constructed and introduced into Arabidopsis. Various tissues at different developmental stages of the transgenic Arabidopsis were collected and stained by GUS solution. As shown in Fig. 6a-f, GhWAK5 promoter-driven GUS expression was mainly observed in veins, trichomes, nodes, primary roots, hypocotyls and nodes. GhWAK16 promoter-driven GUS gene expression mainly present in leaf veins and hypocotyls, trichomes, nodes, primary roots and nodes (Fig. 6g-m). These results suggest that GhWAK5 and GhWAK16 may contribute to fiber development because trichomes are the similar organs to cotton fibers.
Fig. 6.

The phenotype of GUS staining of PGhWAK5:GUS and PGhWAK16:GUS transgenic A. thaliana plants. a and g Five-day-old transgenic seedlings. d eight-day-old transgenic plant. h 15-day-old transgenic plant. c and k Root. f and l flower. m stem. Scale bars = 2.5 mm in (a, b, c, d, f, g, h, i, k, l, m), 50 μm (e, j)
Analysis of cis-acting elements related to phytohormones in putative GhWAK promoter regions
Plant hormones play important roles in cotton fiber initiation, elongation, secondary cell wall deposition and maturation stages, and any slight change in its content will cause obvious changes in fiber qualities [35]. Cotton fibers are single cells and start from ovule epidermis. In order to investigate the potential regulatory mechanism of GhWAK genes, 2000 bp promoter sequences upstream from inition codons (ATG) were scanned in the PLACE database to obtain the cis-acting elements related to phytohormones. The results showed that fourteen GhWAK promoters harbor at least one salicylic acid (SA) related cis-acting elements (Additional file 2: Table S5); twenty, fourteen and ten GhWAKs contain jasmonic acid (JA), gibberellin (GA) and auxin related cis-acting elements, respectively, indicating that GhWAKs may be induced by various plant hormones.
GhWAK genes were stimulated by phytohormones
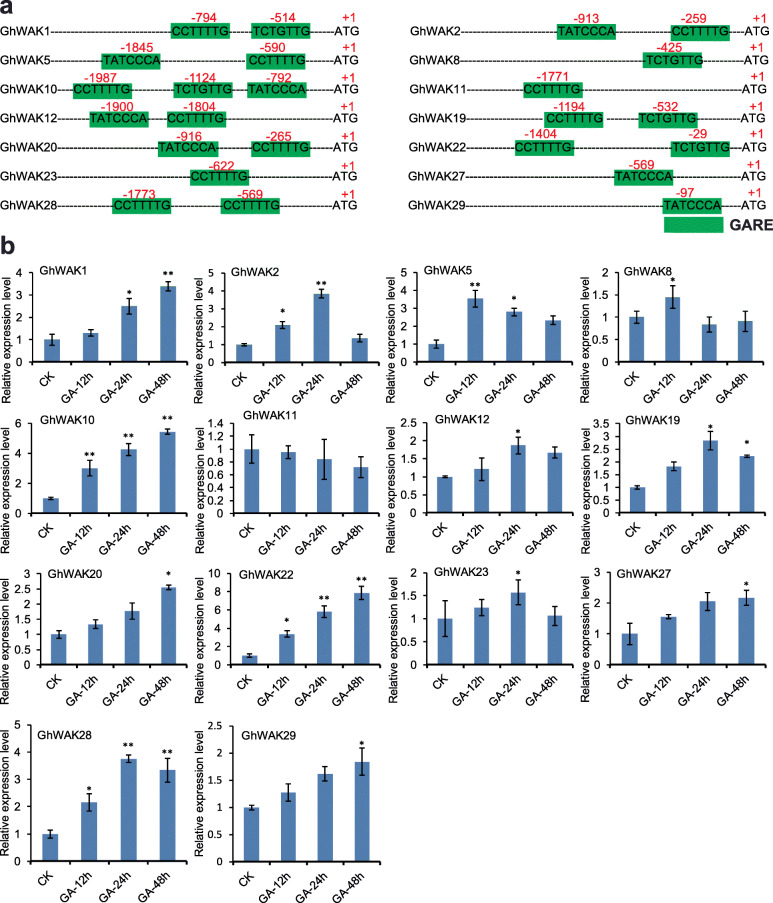
Phytohormones, especially gibberellin and auxin, are reported to regulate cotton fibers cell development [43, 44]. In order to explore the relationship between GhWAK genes and gibberellin acid (GA), we analyzed the cis-elements in the GhWAK promoter regions. A number of gibberellin-responsive cis-elements (GAREs), including TCTGTTG, CCTTTTG and TATCCCA, were observed within GhWAK promoter regions (Fig. 7a). Fourteen out of 29 GhWAK genes possessed at least one cis-acting element involved in gibberellin-responsiveness. These results suggest that the expression level of GhWAK genes may be regulated by GA. To confirm this finding, expression analysis of GhWAK genes was carried out after treatment with GA. Our results showed that a total of 13 GhWAK genes were induced by GA treatment (Fig. 7b), except for GhWAK11.
Fig. 7.
GhWAK gene transcription is induced by gibberellic acid (GA). a Identification of putative gibberellic acid response elements (GARE) in the promoter regions of the GhWAK gene family members. b GA-induced transcription in the GhWAK genes
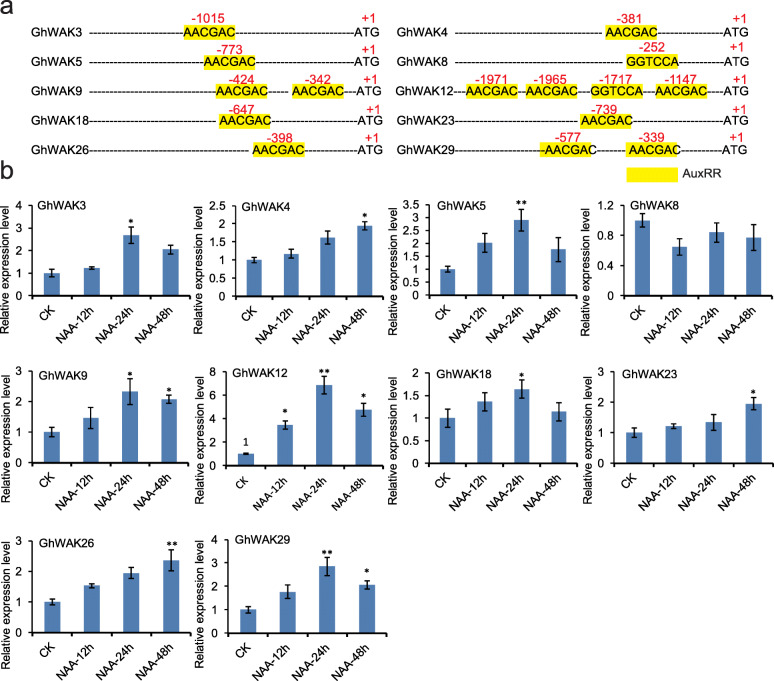
Auxin is also reported to promote cotton fiber cell development [43]. In order to explore the relationship between GhWAK genes and auxin (IAA), we also searched the cis-acting regulatory element involved in auxin responsiveness. As a result, AuxRR-core (GGTCCAT and AACGAC), and two kinds of auxin-responsive cis-elements were successfully identified in the GhWAKs promoter regions. Our data showed that a total of ten GhWAK genes contained an auxin-responsive cis-element (Fig. 8a). The qRT-PCR analysis showed that the expression levels of nine GhWAK genes were significantly increased after treatment with auxin (Fig. 8b). These results suggest that GhWAK genes may be involved in phytohormone-mediated fiber cell development in cotton.
Fig. 8.
GhWAK gene transcription is induced by auxin (NAA). a Identification of putative cis-acting regulatory element involved in auxin responsiveness (AuxRRs) in the promoter regions of the GhWAK gene family members. b NAA-induced transcription in the GhWAK genes
Discussion
Wall-associated kinase (WAK), one of the receptor-like kinases (RLKs), is essential in signal transduction between the cell wall and the cytoplasm. WAKs are highly conservative and contains four typical domains (signal peptide, transmembrane, EGF and protein kinase). In Arabidopsis, there are five WAK proteins, which function directly in cellular cell elongation and cell wall formation [5]. However, the WAK gene family has not been investigated in G. hirsutum, which is famous for its high quality fiber [45].
GhWAK family members amplified by gene duplication events
G. hirsutum is an allotetraploid species that is originated from hybridization of the female parent G. arboreum (AA, 2n = 2x = 26) and the male parent G. raimondii (DD, 2n = 2x = 26) 1–2 million years ago [30, 46]. In this study, 29 GhWAKs, 19 GaWAKs and 14 GrWAKs were identified from G. hirsutum, G. arboretum and G. raimondii, respectively. The number of WAKs in G. hirsutum was nearly six times that in Arabidopsis, indicating that gene expansion occurred in WAK gene family during the evolution of cotton genomes. Analysis of gene duplication events showed that GrWAK and GaWAK family members were mainly produced from tandem duplication and GhWAK family members were mainly produced from WGD. According to our divergence time analysis, the divergence time of GhWAKs, GaWAKs and GrWAKs were earlier than 6.97 MYA, which indicated that the gene duplication events occurred before the polyploidization event.
Previous phylogenetic analysis of WAK proteins among Arabidopsis, rice and Populus showed that WAK proteins form species-specific clusters [6, 47, 48], which indicates that WAK proteins are highly conserved during evolution. In this study, we identified a total of 29 GhWAKs in G. hirsutum, including 15 from the Dt subgenome and 14 genes from the At subgenome (Additional file 2: Table S1). Notably, 14 GrWAKs genes were found in the G. raimondii genome and 19 GaWAKs were found in the G. arboreum genome, suggesting that duplication events and gene loss occurred in the GhWAK gene family after the polyploidization in Gossypium, which further confirmed previous work showing that a large number of genes were lost in allotetraploid cotton [30]. Further chromosome localization analysis of GhWAKs shows that 29 GhWAKs are distributed unequally on 12 chromosomes and there are a number of genes clustered closely on one chromosome with high sequence similarity, which can prevent the loss of functions during evolution [49].
Roles of GhWAKs in plant growth and hormone treatment
Plant cell wall is a complicated network structure that is involved in physiological processes such as plant cell growth and signal transduction. Studies in Arabidopsis, corn, rice and wheat showed that WAKs play functions in loosing cell wall to elongate cell and increasing cell wall strength to enhance pathogen resistance by promoting biosynthesis of cellulose and phytoalexin [9–12, 20–24]. Cotton fibers are single cells, and the cell length as well as cell wall strength determine fiber qualities [33, 34]. Considering the important functions of WAKs in plant cell walls, the promoters and expression patterns of GhWAKs were analyzed in G. hirsutum.
Salicylic acid (SA) and jasmonic acid (JA) are important signal molecules in plant pathgenon defense responses by promoting cell wall synthesis and integrity [50]. Previous studies have shown that TtWAK2, OsWAK/Xa4, and ZmWAK-RLK1 can increase pathogen resistance by regulating cellulose synthesis to strength the cell wall [20, 24, 25]. In the promoter regions analysis, we found SA and JA responsive cis-elements in GhWAK promoters (Additional file 2: Table S5). Furthermore, these GhWAKs highly expressed at 15 DPA, which was the secondary cell wall development stage, the important for fiber strength [51].
Both auxin and GA are phytohormones that promote cell elongation [52–54]. In this study, we identified auxin- and GA-responsive cis-elements harbored in most of the GhWAK promoter regions and these GhWAKs can also respond to GA and auxin. Coincidently, studies have shown that WAKs were involved in the regulation of cell elongation. Expression of an inducible antisense AtWAK2 led to a 50% reduction in WAK protein levels, with a subsequent loss of cell elongation, and hence dwarf in Arabidopsis [8]. On the basis of cis-element analysis, 14 GhWAKs possessed at least one cis-acting element involved in gibberellin-responsiveness and 13 of them were significantly induced by gibberellin treatment. A total of ten GhWAKs were found to contain an auxin-responsive cis-element. Among them, nine GhWAKs were significantly induced after treatment with auxin (Figs. 7 and 8). GUS staining indicated that the GhWAK5 and GhWAK16 expressed in Arabidopsis trichomes, stems, roots and nodes. Also, previous studies demonstrated that trichomes are organs similar to cotton fibers [54]. Therefore, we speculated that WAKs may play functions in promoting the elongation of cotton cells.
Conclusions
In the present study, we identified 29 GhWAKs from G. hirsutum. Phylogenetic analysis showed that cotton WAK proteins can be divided into five clades. The results of synteny and Ka/Ks analysis showed that the GhWAKs mainly originated from whole genome duplication (WGD) and were then mainly under purifying selection. Transcriptome data and qRT-PCR showed that 28 out of 29 GhWAKs highly expressed in cotton fibers. Both RNA-seq data and promoter GUS stainning analysis showed that GhWAKs expressed with tissue-specific manner. We also identified putative gibberellin (GA), auxin (IAA), jasmonate acid (JA) and salicylic acid (SA) response elements in the promoter regions. Furthermore, qRT-PCR data showed that the GhWAKs with transcripts significantly induced by GA and IAA. Fourteen GhWAKs possessed at least one cis-acting element involved in gibberellin-responsiveness and 13 of them were significantly indcued by gibberellin treatment. A total of ten GhWAKs were found to contain an auxin-responsive cis-element. Among them, nine GhWAKs were significantly induced after treatment with auxin. Phytohormones auxin and GA positively regulate cell elongation. Combining the high expression of GhWAK genes in fiber cells with the increased GhWAK transcripts induced by auxin and GA, we think that GhWAK genes might invovle in auxin- and GA-regulating cotton fiber growth.
Methods
Identification of WAKs in G. hirsutum, G. arboreum and G. raimondii
Genome sequences of G. hirsutum acc. TM-1 (ZJU_v2.1), G. raimondii (JGI_v2.1) and G. arboreum (CRI_v3.0) were downloaded from the CottonGen database (https://cottonfgd.org/about/download.html). The five AtWAKs of A. thaliana were downloaded from the Arabidopsis Information Resource (TAIR, Additional file 2: Table S6) [55] The WAKs of G. hirsutum, G. raimondii and G. arboreum were retrieved by BLASTp with threshold value 1e-5 [56]. Based on previous studies of WAK proteins in Arabidopsis, Oryza sativa and Malus domestica [3, 5, 27, 47], WAK proteins contain four typical conserved domains, signal peptide, 1–3 EGFs, a transmembrane domain and a protein kinase domain, therefore, all Blastp results were confirmed by identification of the four typical domains.
Gene locations, structures and physicochemical properties analysis
The position information for all GhWAKs was obtained from the gene annotations in gene feature format (GFF) files were downloaded from the CottonGen database and visualized using MapChart [57]. The exon-intron structure of GhWAKs were analyzed according to the GFF and visualized by the online software Gene Structure Display Server Program (GSDS) [58].
The isoelectric point (pI) and molecular weight (MW) were predicted with the online software ExPASy [59]. The subcellular localization of GhWAK proteins was predicted using WoLF PSORT [60] online software. An instability index greater than 40 indicates that the in vivo half-life is less than 5 h and an index less than 40 indicate that the in vivo half-life is more than 16 h [61]. A grand average of hydropathicity (GRAVY) less than zero indicates that proteins are hydropathic [62].
GhWAK protein alignment, phylogenetic analysis and conserved domain analysis
The multiple sequences of GhWAKs and five AtWAKs (AT1G21210.1, AT1G21230.1, AT1G21240.1, AT1G21250.1 and AT1G21270.1) were aligned using ClustalX 2.0 [63], and an unrooted phylogenetic tree was generated using the NJ method in MEGA 7.0 [64].
The conserved domains of GhWAKs were predicted using online software as described below. The signal peptides were scanned using SignalP-5.0 Server [65]; transmembrane domains were predicted using the Prediction of transmembrane helices in proteins (TMHMM Server v. 2.0) [66]; and the EGF and protein kinase domain were predicted using the ScanProsite tool [67].
Duplication and Synteny analysis of GhWAK genes
The duplication types of GhWAK, GrWAK and GaWAK were classified using the MCScanX software with the default parameters [68], the synteny and collinearity were determined and analyzed using the Multiple Collinearity Scan toolkit (MCScanX) software [68]. The synteny relationships of GhWAK, GaWAK and GrWAK genes were visualized with Circos software [69]. The gene sequences of each paralogous gene pairs were used to calculate the synonym substitution (Ks) and non-synonymous substitution (Ka) by using the PAML package [70].
Plant growth conditions and plant hormone treatments
G. hirsutum cultivar Xuzhou 142 was grown with a 16 h light and 8 h dark cycle at 30 °C under controlled climate conditions as previously reported [54, 71]. The fresh cotton fibers were separated from the ovules at 0, 3, 10 and 15 DPA and immediately frozen in liquid nitrogen. Experiments of phytohormone treatment were performed with 30 cotton ovules at 1 day post anthesis (1 DPA) and each treatment had three biological replicates [72]. The 1 DPA ovules were carefully dissected, sterilized and cultured on liquid medium [71, 72], containing 1 μM auxin 1-Naphthylacetic acid (NAA, Sigma), 1 μM gibberellin acid (GA3, Sigma), or the treatment without any phytohormone as the control (CK), respectively. After the treatments, the ovules were collected and immediately frozen in liquid nitrogen, and stored at − 80 °C for qRT-PCR experiments.
Identification of plant growth regulator-related cis-elements
The upstream 2000 bp genomic DNA sequences of the initiation codon (ATG) were proposed as promoter regions for GhWAKs [72, 73]. The cis-elements of GhWAK promoter regions were detected using the plant cis-acting regulatory element (Plant CARE) database with the default parameters [74]. Phytohormones-related cis-elements were summarized and tabulated in Additional file 2: Table S5.
GUS staining analysis of GhWAK5 promoter
The putative genomic promoter sequences of GhWAK5 and GhWAK16 were cloned from Xuzhou 142, respectively. Then, the cloned putative promoters were assembled into the vector pCAMBIA1300 by In-Fusion® HD Cloning Kit (Vazyme Biotech, China) to drive the GUS gene expression. The fused vector was introduced into Arabidopsis using transformation method as previous work [75]. All the primers and cutting-sites used in vector construction are listed in Additional file 2: Table S7.
For GUS staining, various tissues at different developmental stages of the transgenic Arabidopsis were collected, stained with GUS staining solution according to the instruction and then washed with 70% ethanol for several times to remove background [76]. Stained samples were observed with DMRX microscope (Leica, Germany) and photographed by digital camera (Nikon, Japan).
RNA isolation and expression profiling analysis
The expression levels of GhWAKs in different tissues (stem, root, leaf and ovules) and fibers at various developmental stages (0 DPA, 3 DPA, 10 DPA and 15 DPA) were obtained from previously reported transcriptome data [77]. The datas were available in the National Center for Biotechnology Information (NCBI) under accession number SRA180756. The expression analysis of each gene was conducted and normalized using the fragments per kilobase of transcript per million mapped reads (FPKM) method. To show GhWAK expression patterns during fiber development, an auxin responsive factor gene (GhARF2, GH_D12G2130) was used as a marker gene, which is one of the key regulators of cotton fiber initiation [41]. The transcriptome data were normalized and visualized using the pheatmap package with default parameters in an R environment (https://stackoverflow.com/questions/33292067/pheatmap-annotation-colors-and-border).
Total RNA from ovules treated with plant hormone as described above was isolated using the PureLink™ RNA mini kit (Invitrogen, Lot no. 1687455) and the cDNA was reverse-transcribed from 1.0 μg total RNA using PrimeScript RT Regent kit (Takara, Japan). OLIGO 7 was used to design gene-specific primers [78] for quantitative real-time PCR (qRT-PCR) (Additional file 2: Table S7).
The qRT-PCR assays were performed using a Bio-RadReal Time PCR detection system (Bio-Rad CFX96Touch, USA). The SYBR Green qRT-PCR reactions contained 10 μl SYBR® Premix Ex Taq™ II (Takara, Japan), 0.5 μl of 10 μM primers, and 2 ng cDNA template. The final volume was adjusted to 20 μl with ddH2O. The PCR cycling conditions were 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. The cotton ubiquitin gene GhUBQ7 was used as the internal control for normalization of gene expression in each qRT-PCR experiment [79]. The experiment was performed with three biological replicates and each biological replicate was performed with three technical replicates. The expression levels were compared with the control treatments and relative gene expression was calculated using the 2−ΔΔCt method [80]. SPSS 16.0 software was used for one-way statistical variance to analyze relative expression levels [81].
Supplementary Information
Additional file 1: Figure S1. Synteny comparison of WAK regions from the homeologous in three cotton genomes. Black and orange ovals indicate WAK genes from dispersed and proximal duplication, respectively. The red lines indicate WAK gene from WGD. Green dots indicate WAK gene from tandem duplication. The blue lines indicate orthologous gene pairs. Figure S2. Heatmap of RNA-seq data of GhWAK gene expression levels in five different tissues of G. hirsutum (Xuzhou 142). The transcriptome data were normalized and visualized by the pheatmap package in R language. The colorful bars from green to red indicate the expression levels from low to high, respectively. Figure S3. Expression profile of six GhWAK genes during fiber cell development stages.
Additional file 2: Table S1. Physicochemical parameters of 14 GrWAK and 19 GaWAK genes in G. raimondii and G. arboreum genomes. Table S2. Analysis of duplication events of GhWAK, GaWAK and GrWAK in G. hirsutum, G. arboreum and G. raimondii, respectively. Table S3. Orthologous and paralogous relationships among WAK genes in G. hirsutum, G. arboreum and G. raimondii. Table S4. Ka/Ks analysis and divergence times of duplicated GhWAK gene pairs of G. hirsutum, G.raimondii and G. arboreum. Table S5. cis-elements related to plant growth regulators in the promoters of GhWAK genes. Table S6. Physicochemical parameters of five AtWAK genes in the A. thaliana genome. Table S7. GhWAK gene primer pairs used for qRT-PCR and GUS staining.
Acknowledgements
The authors thank the College of Life Sciences, Shaanxi Normal University for providing experimental support. The authors are very grateful to Yang Yan from Wuhan University for his help in the revision of the article.
Abbreviations
- RLK
Receptor like kinase
- ECD
Extracellular domain (ECD
- WAK
Wall-associated kinase
- PERK
Proline rich extensine-like receptor kinases
- EGF
Epidermal growth factor-like
- ELISA
Enzyme-linked immunosorbent assay
- GA
Gibberellin
- IAA
Auxin
- DCA
Deoxycholic acid
- SA
Salicylic acid
- DPA
Day post-anthesis
- NJ
Neighbor-joining
- WGD
Whole genome duplication
- MYA
Million years ago
- Ka
Non-synonymous
- Ks
Synonymous
- GAREs
Gibberellin-responsive cis-elements
- GRAVY
Grand average of hydropathicity
- NAA
1-Naphthylacetic acid
- JA
Jasmonic acid
Authors’ contributions
GHX, HBL and HHS designed the experiment; LLD and ZFL performed experiments and data analysis; QS, HRS and CSZ analyzed data, GHX wrote the manuscript. WBW, HZL, HBL and HHS revised the manuscript. All authors contributed to the research and read and approved the final manuscript.
Funding
This work was sponsored by the State Key Laboratory of Cotton Biology Open Fund (CB2019A03, 2019A09 and CB2020A12), the National Natural Science Foundation of China (31872175), the Fundamental Research Funds for the Central Universities (GK202001004), Natural Science Basic Research Plan in Shaanxi Province of China (2018JZ3006 and 2019JQ-062), Shaanxi Youth Entrusted Talents Program (20190205), Shaanxi Postdoctoral Project (2018BSHYDZZ76), Construction project of selenium-rich crop planting demonstration base in Shaanxi province ([2017]-01–09), Comprehensive Scientific research Fund project of Xianyang Normal University (XSYK20002) and Young Elite Scientists Sponsorship Program by CAST (2019-2021QNRC001). The funding bodies provided the financial support to the research projects, but didn’t involve in study design, data collection, analysis, or preparation of the manuscript.
Availability of data and materials
The genome sequences of G. hirsutum acc. TM-1 (ZJU_v2.1), G. raimondii (JGI_v2.1) and G. arboreum (CRI_v3.0) are available in the CottonGen website (https://cottonfgd.org/about/download.html). The Arabidopsis genome was available from the TAIR 10 (https://www.arabidopsis.org/). The transcriptome data of G. hirsutum is available from NCBI Sequence Read Archive (SRA) with the accession number SRA180756.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing of interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lingling Dou, Zhifang Li and Qian Shen contributed equally to this work.
Contributor Information
Lingling Dou, Email: xjyldll@126.com.
Zhifang Li, Email: lizhifang2009@163.com.
Qian Shen, Email: shenqian429@126.com.
Huiran Shi, Email: s15829765138@163.com.
Huaizhu Li, Email: lihuaizhu121@126.com.
Wenbo Wang, Email: wenbowang@163.com.
Changsong Zou, Email: cs.zou@163.com.
Haihong Shang, Email: shh9119@163.com.
Hongbin Li, Email: lihbabc@163.com.
Guanghui Xiao, Email: guanghuix@snnu.edu.cn.
References
- 1.Kohorn BD, Kobayashi M, Johansen S, Riese J, Huang LF, Koch K, Fu S, Dotson A, Byers N. An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant J. 2006;46(2):307–316. doi: 10.1111/j.1365-313X.2006.02695.x. [DOI] [PubMed] [Google Scholar]
- 2.Underwood W. The plant cell wall: a dynamic barrier against pathogen invasion. Front Plant Sci. 2012;3:85. doi: 10.3389/fpls.2012.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cunwu Z, Yulian L, Zhigang G, Juan M, Mingyu C, Baihong C. Genome-wide annotation and expression responses to biotic stresses of the wall-associated kinase-Rcecptor-like kinase (WAK-RLK) gene family in apple (Malus domestica) Eur J Plant Pathol. 2019;153:771–785. doi: 10.1007/s10658-018-1591-8. [DOI] [Google Scholar]
- 4.Nakhamchik A, Zhao Z, Provart NJ, Shin-Han S, Keatley SK, Cameron RK, Goring DR. A comprehensive expression analysis of the Arabidopsis proline-rich extensin-like receptor kinase gene family using bioinformatic and experimental approaches. Plant Cell Physiol. 2004;45(12):1875–1881. doi: 10.1093/pcp/pch206. [DOI] [PubMed] [Google Scholar]
- 5.He ZH, Cheeseman I, He D, Kohorn BD. A cluster of five cell wall-associated receptor kinase genes, Wak1–5, are expressed in specific organs of Arabidopsis. Plant Mol Biol. 1999;39(6):1189–1196. doi: 10.1023/A:1006197318246. [DOI] [PubMed] [Google Scholar]
- 6.Tocquard K, Lafon-Placette C, Auguin D, Muries B, Bronner G, Lopez D, Fumanal B, Franchel J, Bourgerie S, Maury S. In silico study of wall-associated kinase family reveals large-scale genomic expansion potentially connected with functional diversification in Populus. Tree Genet Genomes. 2014;10(5):1135–1147. doi: 10.1007/s11295-014-0748-7. [DOI] [Google Scholar]
- 7.Wagner TA, Kohorn BD. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell. 2001;13(2):303–318. doi: 10.1105/tpc.13.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He ZH, Fujiki M, Kohorn BD. A cell wall-associated, receptor-like protein kinase. J Biol Chem. 1996;271:19789–19793. doi: 10.1074/jbc.271.33.19789. [DOI] [PubMed] [Google Scholar]
- 9.Kanneganti V, Gupta AK. Wall associated kinases from plants-an overview. Physiol Mol Biol Plants. 2008;14(1–2):109–118. doi: 10.1007/s12298-008-0010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decreux PAJ, Physiology C. Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 2005;46(2):268–278. doi: 10.1093/pcp/pci026. [DOI] [PubMed] [Google Scholar]
- 11.Lally D, Ingmire P, Tong HY, He ZH. Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell. 2001;13(6):1317–1331. doi: 10.1105/tpc.13.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson CM, Wagner TA, Perret M, He ZH, He D, Kohorn BD. WAKs: cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol Biol. 2001;47(1–2):197–206. doi: 10.1023/A:1010691701578. [DOI] [PubMed] [Google Scholar]
- 13.Li C, Zheng L, Wang X, Hu Z, Zheng Y, Chen Q, Hao X, Xiao X, Wang X, Wang G, et al. Comprehensive expression analysis of Arabidopsis GA2-oxidase genes and their functional insights. Plant Sci. 2019;285:1–13. doi: 10.1016/j.plantsci.2019.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Han S, Jia MZ, Yang JF, Jiang J. The integration of ACS2-generated ACC with GH3-mediated IAA homeostasis in NaCl-stressed primary root elongation of Arabidopsis seedlings. Plant Growth Regul. 2019;88:151–158. doi: 10.1007/s10725-019-00495-1. [DOI] [Google Scholar]
- 15.Yongqi P, Jintong L, Bishu Q, Mi T, Lirong S, Xuechen W, Biology H. Aquaporin AtTIP5;1 as an essential target of gibberellins promotes hypocotyl cell elongation in Arabidopsis thaliana under excess boron stress. Funct Plant Biol. 2018;45(3):305–314. doi: 10.1071/FP16444. [DOI] [PubMed] [Google Scholar]
- 16.Lv S, Yu D, Sun Q, Jiang J. Activation of gibberellin 20-oxidase 2 undermines auxin-dependent root and root hair growth in NaCl-stressed Arabidopsis seedlings. Plant Growth Regul. 2018;84:225–236. doi: 10.1007/s10725-017-0333-9. [DOI] [Google Scholar]
- 17.Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. PNAS. 2020;107(20):9452–9457. doi: 10.1073/pnas.1000675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Z, He D, Kohorn BD. Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 1998;14(1):55–63. doi: 10.1046/j.1365-313X.1998.00092.x. [DOI] [PubMed] [Google Scholar]
- 19.Zarattini M, Launay A, Farjad M, Wénès E, Taconnat L, Boutet S, Bernacchia G, Fagard M. The bile acid eoxycholate elicits defenses in Arabidopsis and reduces bacterial infection. Mol Plant Pathol. 2017;18(4):540–554. doi: 10.1111/mpp.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang P, Praz C, Li B, Singla J, Robert CAM, Kessel B, Scheuermann D, Lüthi L, Ouzunova M, Erb M, et al. Fungal resistance mediated by maize wall-associated kinase ZmWAK-RLK1 correlates with reduced benzoxazinoid content. New Phytol. 2019;221(2):976–987. doi: 10.1111/nph.15419. [DOI] [PubMed] [Google Scholar]
- 21.Zuo W, Chao Q, Zhang N, Ye J, Tan G, Li B, Xing Y, Zhang B, Liu H, Fengler K. A maize wall-associated kinase confers quantitative resistance to head smut. Nat Genet. 2015;47:151–157. doi: 10.1038/ng.3170. [DOI] [PubMed] [Google Scholar]
- 22.Delteil A, Gobbato E, Cayrol B, Estevan J, Michel-Romiti C, Dievart A, Kroj T, Morel JB. Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 2016;16:17. doi: 10.1186/s12870-016-0711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cayrol B, Delteil A, Gobbato E, Kroj T, Morel JB. Three wall-associated kinases required for rice basal immunity form protein complexes in the plasma membrane. Plant Signal Behav. 2016;11(4):e1149676. doi: 10.1080/15592324.2016.1149676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gadaleta A, Colasuonno P, Giove SL, Blanco A, Giancaspro A. Map-based cloning of QFhb.mgb-2A identifies a WAK2 gene responsible for Fusarium Head Blight resistance in wheat. Sci Rep. 2019;9(1):6929. doi: 10.1038/s41598-019-43334-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu K, Cao J, Zhang J, Xia F, Ke Y, Zhang H, Xie W, Liu H, Cui Y, Cao Y. Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat Plants. 2017;3:17009. doi: 10.1038/nplants.2017.9. [DOI] [PubMed] [Google Scholar]
- 26.Hurni S, Scheuermann D, Krattinger SG, Kessel B, Wicker T, Herren G, Fitze MN, Breen J, Presterl T, Ouzunova M. The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc Natl Acad Sci U S A. 2015;112(28):8780–8785. doi: 10.1073/pnas.1502522112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan X, Yin S, Zhang K, Shi X, Lian C, Zhang H, Hu Z, Shen Z. OsWAK11, a rice wall-associated kinase, regulates cu detoxification by alteration the immobilization of cu in cell walls. Environ Exp Bot. 2018;150:99–105. doi: 10.1016/j.envexpbot.2018.03.005. [DOI] [Google Scholar]
- 28.Sivaguru M, Ezaki B, He ZH, Tong H, Matsumoto H. Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol. 2003;132(4):2256–2266. doi: 10.1104/pp.103.022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y, Chen J, Fang L, Zhang Z, Ma W, Niu Y, Ju L, Deng J, Zhao T, Lian J, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet. 2019;51(4):739–748. doi: 10.1038/s41588-019-0371-5. [DOI] [PubMed] [Google Scholar]
- 30.Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, Ma Z, Shang H, Ma X, Wu J. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nature Biotechnol. 2015;33(5):524–530. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- 31.Du X, Huang G, He S, Yang Z, Sun G, Ma X, Li N, Zhang X, Sun J, Liu M. Resequencing of 243 diploid cotton accessions based on an updated a genome identifies the genetic basis of key agronomic traits. Nat Genet. 2018;50(6):796–802. doi: 10.1038/s41588-018-0116-x. [DOI] [PubMed] [Google Scholar]
- 32.Zou C, Lu C, Zhang Y, Song G. Distribution and characterization of simple sequence repeats in Gossypium raimondii genome. Bioinformation. 2012;8(17):801–806. doi: 10.6026/97320630008801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao C, Jin X, Zhu L, Li H. Two-dimensional gel electrophoresis-based proteomic analysis reveals N-terminal truncation of the Hsc70 protein in cotton fibers in vivo. Sci Rep. 2016;6:36961. doi: 10.1038/srep36961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji SJ, Lu YC, Feng JX, Wei G, Li J, Shi YH, Fu Q, Liu D, Luo JC, Zhu YX. Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Res. 2003;31(10):2534–2543. doi: 10.1093/nar/gkg358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuttle JR, Nah G, Duke MV, Alexander DC, Guan X, Song Q, Chen ZJ, Scheffler BE, Haigler CH. Metabolomic and transcriptomic insights into how cotton fiber transitions to secondary wall synthesis, represses lignification, and prolongs elongation. BMC Genomics. 2015;16(1):477. doi: 10.1186/s12864-015-1708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao W, Dong H, Zahoor R, Zhou Z, Snider JL, Chen Y, Siddique KH, Wang Y. Ameliorative effects of potassium on drought-induced decreases in fiber length of cotton (Gossypium hirsutum L.) are associated with osmolyte dynamics during fiber development. Crop J. 2019;7(5):619–634. doi: 10.1016/j.cj.2019.03.008. [DOI] [Google Scholar]
- 37.Song Z, Chen Y, Zhang C, Zhang J, Huo X, Gao Y, Pan A, Du Z, Zhou J, Zhao Y, et al. RNA-seq reveals hormone-regulated synthesis of non-cellulose polysaccharides associated with fiber strength in a single-chromosomal-fragment-substituted upland cotton line. Crop J. 2019;8(2):1–14. [Google Scholar]
- 38.Li F, Fan G, Wang K, Sun F, Yuan Y, Song G, Li Q, Ma Z, Lu C, Zou C, et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet. 2014;46(6):567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- 39.Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L, Shang H, Zhu S, et al. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet. 2012;44(10):1098–1103. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- 40.Fang X, Wang L, Deng X, Wang P, Ma Q, Nian H, Wang Y, Yang C. Genome-wide characterization of soybean P1B-ATPases gene family provides functional implications in cadmium responses. BMC Genomics. 2016;17(1):376. doi: 10.1186/s12864-016-2730-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao G, Peng H, Peng Z, Hao L, Li Z, Pang C, Yu J. Genome-wide identification of GhARF gene family reveals GhARF2 and GhARF18 are involved in cotton fibre cells initiation. J Exp Bot. 2018;69(18):4323–4337. doi: 10.1093/jxb/ery219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Huang GQ, Zou D, Yan JQ, Li Y, et al. The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 2018;217:625–640. doi: 10.1111/nph.14864. [DOI] [PubMed] [Google Scholar]
- 43.Zhang M, Zheng X, Song S, Zeng Q, Hou L, Li D, Zhao J, Wei Y, Li X, Luo M. Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol. 2011;29(5):453–458. doi: 10.1038/nbt.1843. [DOI] [PubMed] [Google Scholar]
- 44.Nml S, Kusakina J, Fernández-López Á, Chembath A, Belbin FE, Dodd AN. The energy-signaling hub SnRK1 is important for sucrose-induced hypocotyl elongation. Plant Physiol. 2018;176(2):1299–1310. doi: 10.1104/pp.17.01395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang M, Tu L, Yuan D, Zhu D, Shen C, Li J, Liu F, Pei L, Wang P, Zhao G, et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat Genet. 2019;51(2):224–229. doi: 10.1038/s41588-018-0282-x. [DOI] [PubMed] [Google Scholar]
- 46.Sunilkumar G, Campbell LAM, Puckhaber L, Stipanovic RD, Rathore KS. Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc Natl Acad Sci U S A. 2006;103(48):18054–18059. doi: 10.1073/pnas.0605389103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang S, Chen C, Li L, Meng L, Singh J, Jiang N, Deng XW, He ZH, Lemaux PG. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 2005;139(3):1107–1124. doi: 10.1104/pp.105.069005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohorn BD. WAKs; cell wall associated kinases. Curr Opin Cell Biol. 2001;13(5):529–533. doi: 10.1016/S0955-0674(00)00247-7. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka K, Nakamura Y, Asami T, Yoshida S, Matsuo T, Okamoto S. Physiological roles of brassinosteroids in early growth of Arabidopsis: brassinosteroids have a synergistic relationship with gibberellin as well as auxin in light-grown hypocotyl elongation. J Plant Growth Regul. 2003;22(3):259–271. doi: 10.1007/s00344-003-0119-3. [DOI] [Google Scholar]
- 50.Takahashi H, Kanayama Y, Ming SZ, Kusano T, Shah J. Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene-for-gene resistance to cucumber mosaic virus. Plant Cell Physiol. 2004;45(6):803–809. doi: 10.1093/pcp/pch085. [DOI] [PubMed] [Google Scholar]
- 51.Qin YM, Zhu Y. A brief summary of major advances in cotton functional genomics and molecular breeding studies in China. Chin Sci Bull. 2007;23:8–12. [Google Scholar]
- 52.Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Matsui M. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 2001;25(2):213–221. doi: 10.1046/j.1365-313x.2001.00957.x. [DOI] [PubMed] [Google Scholar]
- 53.De Lucas M, Davière J-M, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451(7177):480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, He P, Yang Z, Huang G, Wang L, Pang C, Xiao H, Zhao P, Yu J, Xiao G. A genome-scale analysis of the PIN gene family reveals its functions in cotton fiber development. Front Plant Sci. 2017;8:461. doi: 10.3389/fpls.2017.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, et al. The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis. 2015;53:474–485. doi: 10.1002/dvg.22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang W, Yu M, Zou C, Lu C, Yu D, Cheng H, Jiang P, Feng X, Zhang Y, Wang Q, et al. Genome-wide comparative analysis of RNA-binding glycine-rich protein family genes between Gossypium arboreum and Gossypium raimondii. PLoS One. 2019;14(6):e0218938. doi: 10.1371/journal.pone.0218938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93(1):77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 58.Hu B, Jin J, Guo AY, Zhang H, Luo J, et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Banerjee A, Jana A, Pati BR, Mondal KC, Das Mohapatra PK. Characterization of tannase protein sequences of bacteria and fungi: an in silico study. Protein J. 2012;31(4):306–327. doi: 10.1007/s10930-012-9405-x. [DOI] [PubMed] [Google Scholar]
- 62.Lan T, Gao J, Zeng QY. Genome-wide analysis of the LEA (late embryogenesis abundant) protein gene family in Populus trichocarpa. Tree Genet Genomes. 2013;9(1):253–264. doi: 10.1007/s11295-012-0551-2. [DOI] [Google Scholar]
- 63.Sun H, Hao P, Ma Q, Zhang M, Qin Y, Wei H, Su J, Wang H, Gu L, Wang N, et al. Genome-wide identification and expression analyses of the pectate lyase (PEL) gene family in cotton (Gossypium hirsutum L.) BMC Genomics. 2018;19(1):661. doi: 10.1186/s12864-018-5047-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 66.Chen Y, Yu P, Luo J, Jiang Y. Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm Genome. 2003;14:859–865. doi: 10.1007/s00335-003-2296-6. [DOI] [PubMed] [Google Scholar]
- 67.Gattiker A, Gasteiger E, Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl Bioinforma. 2002;1:107–108. [PubMed] [Google Scholar]
- 68.Yupeng W, Haibao T, Debarry JD, Xu T, Jingping L, Xiyin W, Tae-Ho L, Huizhe J, Barry M, Hui G. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13(5):555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 71.Shi YH, Zhu SW, Mao XZ, Feng JX, Zhu YX. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell. 2006;18(3):651–664. doi: 10.1105/tpc.105.040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He P, Yang Y, Wang Z, Zhao P, Xiao G. Comprehensive analyses of ZFP gene family and characterization of expression profiles during plant hormone response in cotton. BMC Plant Biol. 2019;19(1):329. doi: 10.1186/s12870-019-1932-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y, He P, Ma X, Yang Z, Pang C, Yu J, Wang G, Friml J, Xiao G. Auxin-mediated statolith production for root gravitropism. New Phytol. 2019;224(2):761–774. doi: 10.1111/nph.15932. [DOI] [PubMed] [Google Scholar]
- 74.Rombauts S, Dehais P, Van Montagu M, Rouze P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999;27(1):295–296. doi: 10.1093/nar/27.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313X.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 76.Huang J, Ge X, Sun M. Modified CTAB protocol using a silica matrix for isolation of plant genomic DNA. Biotechniques. 2000;28:432–434. doi: 10.2144/00283bm08. [DOI] [PubMed] [Google Scholar]
- 77.Zhang T, Hu Y, Jiang W, Fang L, Guan X, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 78.Rychlik W. OLIGO 7 primer analysis software. Methods Mol Biol. 2007;402:35–60. doi: 10.1007/978-1-59745-528-2_2. [DOI] [PubMed] [Google Scholar]
- 79.Xiao GH, Wang K, Huang G, Zhu YX. Genome-scale analysis of the cotton KCS gene family revealed a binary mode of action for gibberellin a regulated fiber growth. J Integr Plant Biol. 2016;58(6):577–589. doi: 10.1111/jipb.12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 81.George D, Mallery P. SPSS for Windows step by step: A simple guide and reference; Boston: Allyn & Bacon, Inc.; 2008.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Synteny comparison of WAK regions from the homeologous in three cotton genomes. Black and orange ovals indicate WAK genes from dispersed and proximal duplication, respectively. The red lines indicate WAK gene from WGD. Green dots indicate WAK gene from tandem duplication. The blue lines indicate orthologous gene pairs. Figure S2. Heatmap of RNA-seq data of GhWAK gene expression levels in five different tissues of G. hirsutum (Xuzhou 142). The transcriptome data were normalized and visualized by the pheatmap package in R language. The colorful bars from green to red indicate the expression levels from low to high, respectively. Figure S3. Expression profile of six GhWAK genes during fiber cell development stages.
Additional file 2: Table S1. Physicochemical parameters of 14 GrWAK and 19 GaWAK genes in G. raimondii and G. arboreum genomes. Table S2. Analysis of duplication events of GhWAK, GaWAK and GrWAK in G. hirsutum, G. arboreum and G. raimondii, respectively. Table S3. Orthologous and paralogous relationships among WAK genes in G. hirsutum, G. arboreum and G. raimondii. Table S4. Ka/Ks analysis and divergence times of duplicated GhWAK gene pairs of G. hirsutum, G.raimondii and G. arboreum. Table S5. cis-elements related to plant growth regulators in the promoters of GhWAK genes. Table S6. Physicochemical parameters of five AtWAK genes in the A. thaliana genome. Table S7. GhWAK gene primer pairs used for qRT-PCR and GUS staining.
Data Availability Statement
The genome sequences of G. hirsutum acc. TM-1 (ZJU_v2.1), G. raimondii (JGI_v2.1) and G. arboreum (CRI_v3.0) are available in the CottonGen website (https://cottonfgd.org/about/download.html). The Arabidopsis genome was available from the TAIR 10 (https://www.arabidopsis.org/). The transcriptome data of G. hirsutum is available from NCBI Sequence Read Archive (SRA) with the accession number SRA180756.