Abstract
Background
UCEC is the most common gynecological malignancy in many countries, and its mechanism of occurrence and development is related to tumor mutation burden (TMB) and immune cell infiltration. Therefore, it is necessary to systematically explore the TMB-related gene profile in immune cells to improve the prognosis of UCEC.
Methods
We integrated TMB-related genes with basic clinical information of UCEC patients based on TCGA dataset. Differentially expressed genes (DEGs) were selected through differential expression screening, PPI, and enrichment analysis. Additionally, we analyzed the components of immune cell infiltration of the DEGs to obtain the differential immunity-related genes. A single factor and multifactor Cox regression analyses were conducted to establish new prognostic indicators of OS and DFS based on TMB-related immune genes. To further study the correlation between survival and immune cell infiltration, a Cox model based on these immune infiltration compositions was built. Using the clinical variables, we established nomograms for OS and DFS.
Results
393 DEGs were significantly associated with clinical outcomes and the immune component in patients with UCEC. Gene Ontology (GO) and Kyoto Encyclopedia of Genes, Genomes (KEGG) pathway and protein-protein interaction network (PPI) analyses revealed the role of these genes and information on related pathways. Then, two prognostic models were established based on the differential immune genes for OS (GFAP and MX2) and DFS (MX2, GFAP, IGHM, FGF20, and TRAV21). In DFS, the differential immune genes were related to CD4+ T cell, CD8+ T cell, macrophage, and neutrophil (all P < 0.05). B cell and CD8+ T cell were independent prognostic factors from among the immune cell elements in UCEC. Finally, the risk scores of these models were combined with the clinical elements-based nomogram models, and the AUC values were all over 0.7.
Conclusions
Our results identified several clinically significant differential immune genes and established relevant prognostic models, providing a basis for the molecular analysis of TMB and immune cells in UCEC, and identified potential prognostic and immune-related genes for UCEC. We added clinical related conditions for further analysis to confirm the identity of the genes and clinical elements-based models.
Keywords: Tumor mutation burden, Endometrial carcinoma, Immune-related survival, TCGA
Introduction
Uterine corpus endometrial carcinoma (UCEC) is a common type of gynecological tumor. It is estimated that there will be 65,620 new cases and 12,590 deaths caused by the disease in the United States in 2020, second only to ovarian cancer. At the same time, the mortality rate of UCEC has risen over the past 10 years [1]. The 5-year survival rate of stage I UCEC exceeds 90%, while stage IV can only reach 20% [2]. Traditional treatment methods mainly include surgery, radiotherapy, chemotherapy, and hormone therapy. For patients with metastases, surgery, and radiotherapy cannot achieve a satisfactory treatment level. These patients are treated using chemotherapy or hormone therapy, which also causes more significant damage to normal human cells and results in recurrence. Most importantly, options are often limited after traditional first-line treatment for advanced patients with metastases or recurrence, which has prompted many scholars to develop new treatment methods.
In recent years, immunotherapy has become an effective method for treating advanced cancer [3]. For example, typical clinical trials include lung cancer [4, 5] and melanoma [5–7]. Among these immunotherapies, the most important one is Immune Checkpoint Inhibitor (ICI), and its main inhibitory points include Cytotoxic T-lymphocyte antigen 4 (CTLA-4) and Programed cell death 1 receptor (PD-1) [7, 8]. More specifically, ICI blocks the checkpoint activity of T cells, which increases the sensitivity of immune cells to the recognition of cancer cells and triggers the immune system to attack and destroy cancer cells. Tumor mutation burden (TMB) refers to the total number of mutations per mega base in tumor tissue. There is increasing evidence that the burden of tumor mutations is related to immunotherapy in cancer patients [9]. Although most inhibitors are rarely clinically approved for humans, their immunotherapy has shown great potential in refractory tumors. Immunotherapy research has also been conducted on targeted therapy for UCEC. The level of TMB and microsatellite instability can predict whether patients with UCEC may benefit from immunotherapy [10, 11]. Uterine carcinosarcoma is a rare and aggressive histological variant of UCEC with a poor prognosis [10]. Bhangoo et al. [11] found that inactivation of DNA polymerase ɛ (POLE) mutation is associated with a high TMB and ICI in carcinosarcoma. In summary, the study of TMB on immune invasiveness of UCEC requires further research. Therefore, we aimed to study the relationship between the clinical prognosis of UCEC and the role of TMB and immunity.
Materials and methods
Data collection
Clinical data on a UCEC cohort, including age, stage, grade, histological type, together with transcriptome data, were obtained from the TCGA data portal (https://portal.gdc.cancer.gov/). Meanwhile, we downloaded the processed “mask somatic mutation” data using the VarScan algorithm in the TCGA database. The R package “maftools function” [12] was adopted to describe mutated genes in the UCEC samples. Since each dataset was retrieved from published reports, it was verified that written informed consent had been obtained.
TMB score and clinical characteristics
We used Perl script (JAVA8 platform) to remove silent mutations from the samples while calculating the number of base mutations and then corrected it to the number of base mutations per million bases, which was the TMB value. The samples were divided into a low group and high group based on the median value of TMB. Further, we used Kaplan-Meier statistics to analyze survival differences between these two groups. The Wilcoxon rank-sum test was used to evaluate the correlation between TMB and various clinical variables. Furthermore, X-tile software (Yale University, New Haven, Connecticut, USA) was utilized to select the optimal threshold value for age. We analyzed overall survival (OS) and disease-free survival (DFS) to determine the prognostic value of the two TMB groups of patients with UCEC.
Differentially expressed genes and enrichment analysis
The Limma package of R software was adopted to predict differentially expressed genes (DEGs) between the high-TMB group and low-TMB group. Genes that exhibited a |Fold Change|of > 1 with an adjusted P-value of < 0.05 were regarded as significant DEGs. Then, the “pheatmap” package of R software was used to assess the effect of the difference by constructing a heatmap plot. Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analyses were carried out to identify the primary biological attributes for the genes selected above using the “enrichplot,” “clusterProfiler,” “ggplot2” and “org.Hs.eg.db” functions of R software. Biological processes, cellular components, and molecular functions are the three primary functional components of GO enrichment analysis. KEGG analysis can identify the neighbor pathways of the DEGs.
Search tool for the retrieval of interacting genes database (STRING)
STRING (https://string-db.org/) is a database of known and predicted protein-protein interactions, and its latest version, 11.0, includes 24,584,628 proteins from 5090 organisms [13]. This tool was applied to explore the physical and functional associations between the DEGs and conduct the protein-protein interaction network (PPI) analysis of the DEGs. The results of the PPI analysis provided the number of adjacent nodes per gene, which was obtained through a histogram created using R software.
Gain differential immune genes and survival analysis
CIBERSOR is a general calculation method used to quantify cellular components from many tissue gene expression profiles (GEP), which could accurately estimate the immune composition of tumor biopsy [14]. We applied the immune cell expression profiles to the “CIBERSORT” algorithm to determine immune cell content in the UECE sample. The vioplot package was used to determine the expression differences of various immune cells in the high-TMB and low-TMB groups. After intersecting the immune cell-related genes derived from the immune database (https://www.immport.org/) and the DEGs, the differential immune genes were obtained using the “VennDiagram” package. Kaplan–Meier survival analysis was performed to assess the survival of the immune-related genes.
Establishment of differential immune genes-based prognostic index models
First, differential immune genes were extracted to be further study using a Cox single-factor and multifactor regression analysis for OS and DFS, which was performed using R software. The statistical indicators that were used were: hazard ratio (HR), 95% confidence interval (CIs), and log-rank P-value (a P-value of < 0.05 was considered statistically significant). The receiver operating characteristic (ROC) curve of each model was constructed. Furthermore, patients were divided into high-risk and low-risk groups based on the median risk score.
TIMER database
TIMER web server (https://cistrome.shinyapps.io/timer/) is a comprehensive resource for the systematical analysis of six immune infiltrates, including B cells, CD4+ T cells, CD8+ T cells, Neutrophils, Macrophages, and Dendritic cells, across diverse cancer types [15]. Moreover, the survival module in the TIMER database was also used to draw Kaplan–Meier plots for the immune infiltrates and hub immune genes to visualize survival differences. The P-value of the log-rank test used to compare the survival curves of the two groups is shown in each plot.
Outputs of the immune elements-based models
Surv (UCEC) ~ B_cell + CD8_Tcell + CD4_Tcell + Macrophage + Neutrophil + Dendritic was the formula of Cox’s regression model. This model was fitted using the “coxph” function of the R software package, “survival.” HR is presented as lower and upper 95% confidential intervals, and the threshold p-value was set at 0.05.
Construction of nomogram models for OS and DFS
The information on clinical characteristics obtained from public datasets and two risk groups were collected and divided into a modeling group and a verification group based on a 7:3 ratio. The “rms” package of R software was used to construct the nomogram and calibration plots. ROC curves were also plotted to determine the sensitivity and specificity of the nomogram models.
Statistical analysis
R 3.6.3 (https://www.r-project.org/) software was used for all statistical analyses. Additionally, the R package “survival” was utilized to conduct the univariate Cox regression analysis to examine the relationship between OS and DFS with gene expression. At the same time, a model was constructed based on the multivariate Cox regression analysis. X-tile software was used to group patient data based on age, with 69 years of age for OS and 59 years of age for DFS used the critical values (Figs. 3, 4). Then, nomogram plots were conducted for both clinical parameters and risk groups. The area under the curve (AUC) of the ROC curve was computed using the ROC function of the “survival” package in R software. A P value of < 0.05 was considered to indicate statistical significance.
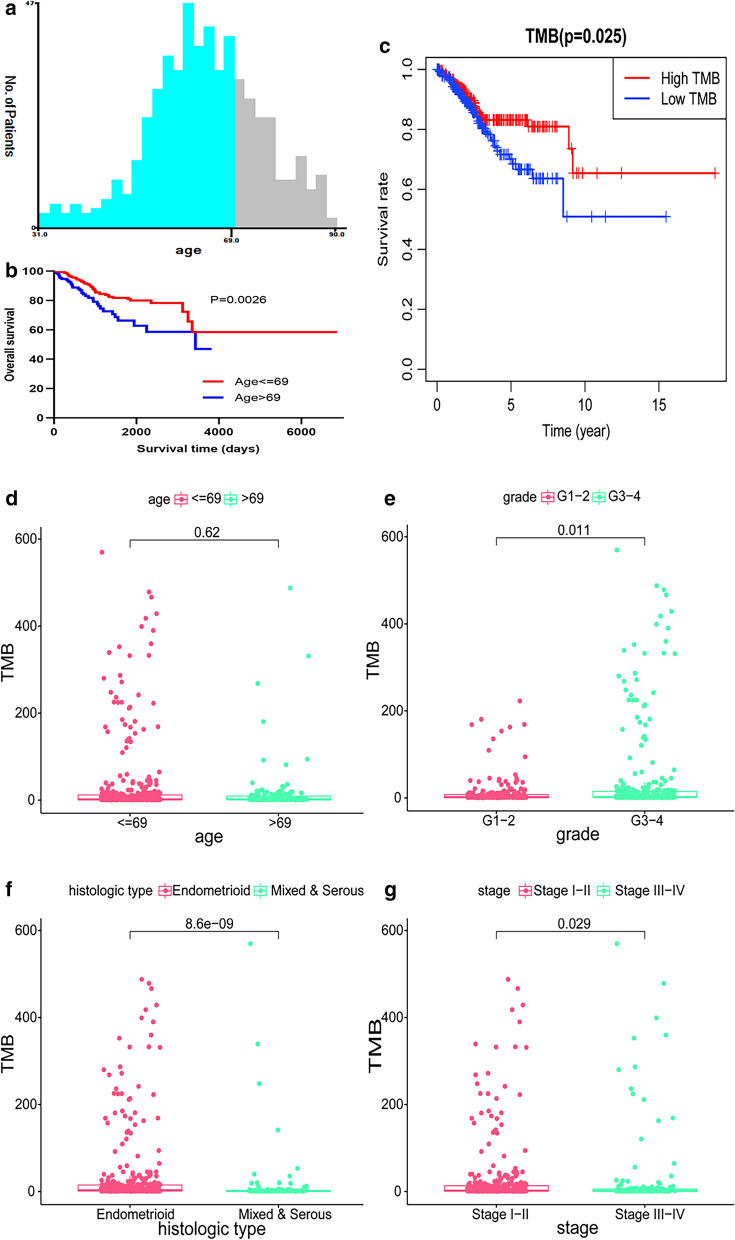
Fig. 3.
Connection between TMB and clinical variables for OS. a, b Age grouping using X-tile in OS; c survival analysis of the high-TMB and low-TMB groups; d–g the link between TMB and clinical features, including age, grade, histological type, and stage
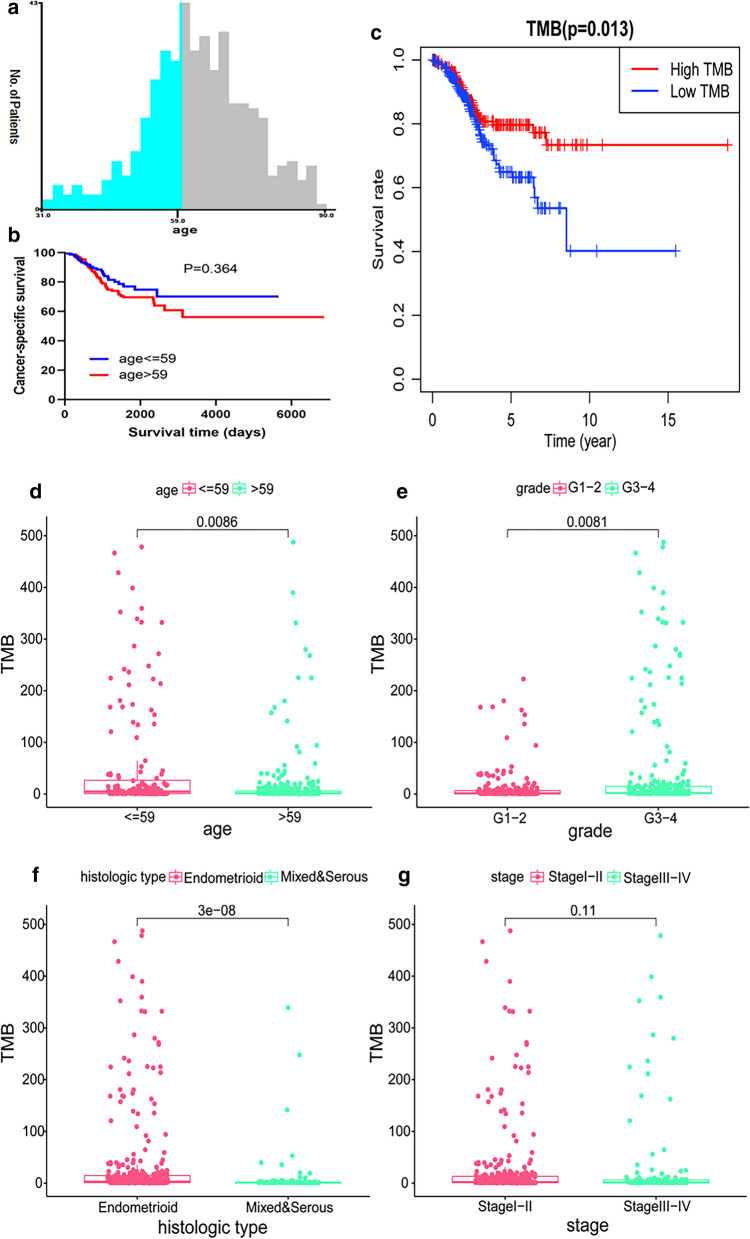
Fig. 4.
Relationship between TMB and clinical characteristics for DFS. a, b Age grouping using X-tile in DFS; c survival analysis of the high-TMB and low-TMB groups; d–g the link between TMB and clinical features, including age, grade, histological type, and stage
Results
Analyze mutation data in UCEC
The somatic mutation data of 375 UCEC patients were downloaded from the TCGA database, and these materials were visualized using the “maftools” R package. Diagrams a–c in Fig. 1 provides a summary of the types of mutations in the samples, in which missense mutations occupied the leading position (Fig. 1a), C>T was the most common single nucleotide variant (SNV) (Fig. 1c). Only one mutation type, Single Nucleotide Polymorphism (SNP), was found in UCEC (Fig. 1b). The mutation value and median of each sample are shown in Fig. 1d, e. Furthermore, we showed the top 10 mutated genes in the samples, which included TTN (35%), MUC16 (22%), PTEN (46%), RYR2 (20%), CSMD3 (20%), PIK3CA (46%), TP53 (31%), ARID1A (23%), CHD4 (19%) and CTNNB1 (22%) (Fig. 1f). The waterfall chart shown in Fig. 2a demonstrates the specific mutation types and proportions of the mutant genes in each sample. Figure 2b shows the connection between two mutant genes, in which the green color indicates a positive correlation, while a brown color indicates a negative correlation.
Fig. 1.
Summary description of gene mutations in UCEC. a Statistics of the number of variant classifications: missense mutation, nonsense mutation, splice site, translation start site, and nonstop mutation; b counting of variant types: only SNPs in UCEC; c summary of base mutations: T>G, T>A, T>C, C>T, C>G and C>A. d, e TMB in tissues; f top 10 mutated genes
Fig. 2.
Display of mutation information in UCEC. a The waterfall of mutation information: the ordinate on the left indicates the name of the gene, and the abscissa represents different samples. Six different colors were used to show the six different types of mutations: gray color indicated no mutation, and the icon shows the color that represents the remaining five types of mutations; b relationship between pairwise mutant genes: green color indicates a positive correlation and a brown color represents a negative correlation
Relationship between TMB and clinical features
The TMB of UCEC cases was calculated to determine its clinical significance. For OS, initially, we found that 69 years of age was the best age cut-off value using x-tile software, and the median value of TMB was used as the optimal cut-off value to divide these samples into two groups: low and high TMB groups. It could be detected that the low TMB group showed a more unsatisfactory survival outcome compared to the high TMB group, based on values in the log-rank test and Kaplan–Meier curve (P = 0.025, Fig. 3c). The high TMB group was closely related to the grade 3 or 4 (P = 0.011, Fig. 3e) endometroid pathological type (P < 0.001, Fig. 3f) and stage I or II (P = 0.029, Fig. 3g), while there was no significant difference between TMB and age (P = 0.62, Fig. 3d). For DFS, Fig. 4 shows that 59 years of age was the optimal threshold and that the association between TMB and survival were similar to the results for OS. Additionally, expression levels of TMB decreased due when age was ≤ 59, the grade was 3–4, and endometrioid histological type (all P < 0.05), and there were no significant associations between TMB and stage (P = 0.11).
Enrichment analyses and the PPI of genes in the different TMB group
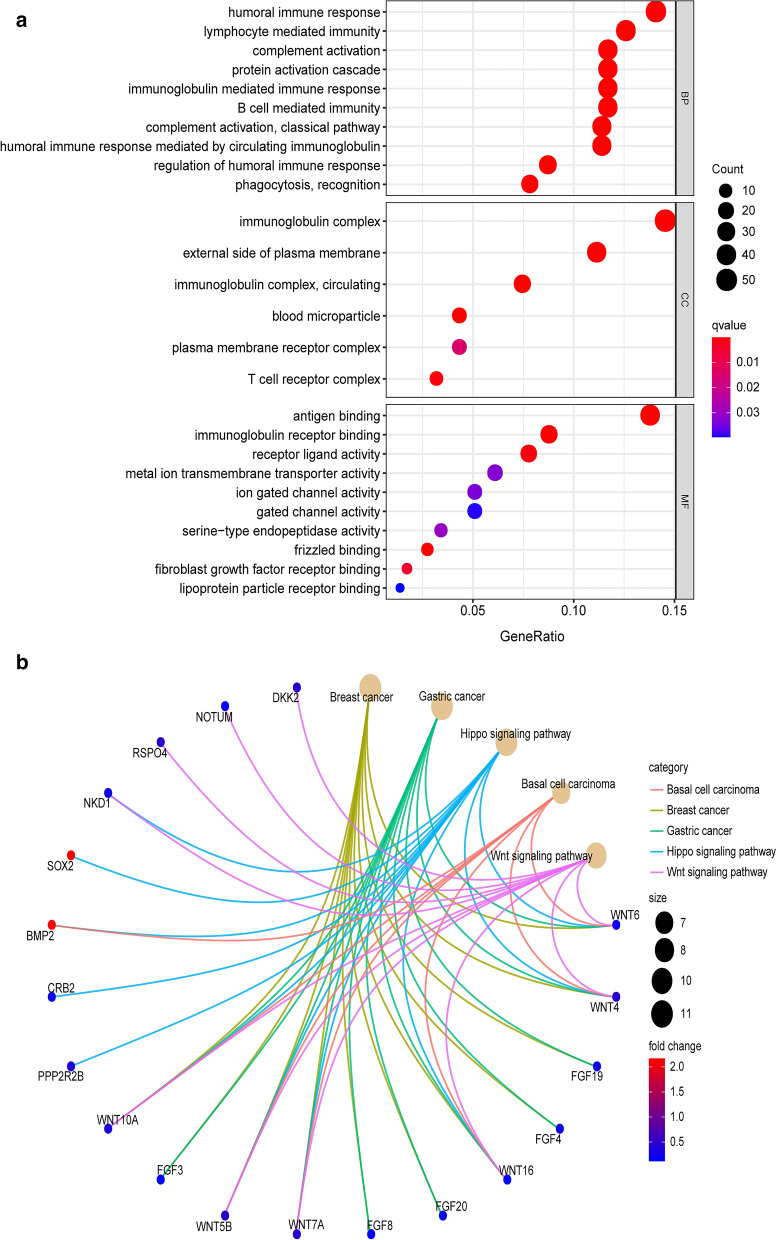
We found 393 differentially expressed genes (DEGs) between the high TMB and low TMB groups, and the top 19 DEGs are shown in Table 1. The heatmap constructed shows the expression levels of the DEGs (Fig. 5a). The Venn diagram was created to show 1713 immune-related genes and 393 DEGs to obtain 98 differential immune genes (Fig. 5b). After that, the KEGG pathway and GO term enrichment analyses of these DEGs were conducted. Figure 6a and Table 2 show that the GO terms primarily enriched for biological processes included humoral immune response and lymphocyte-mediated immunity. Simultaneously, the Immunoglobulin complex, external side of the plasma membrane, and immunoglobulin complex were the cellular components that were mainly enriched.
Table 1.
The top 19 DEGs in UCEC
| Gene | Low group | High group | logFC | P-value | FDR |
|---|---|---|---|---|---|
| ERVMER34-1 | 8.007284856 | 2.261724965 | − 1.823889628 | 0.000229986 | 0.001203484 |
| CRYGN | 0.421748475 | 0.208045874 | − 1.019481175 | 7.82E−11 | 4.35E−09 |
| IGKV2OR22-4 | 0.446894633 | 1.119245354 | 1.324519704 | 0.010006045 | 0.026333049 |
| MYT1 | 0.28577249 | 0.12531108 | − 1.189353053 | 3.48E−08 | 7.38E−07 |
| IGHV4-31 | 8.410528443 | 17.48325048 | 1.05570508 | 6.91E−05 | 0.00044633 |
| SIX3 | 2.3926374 | 0.838171842 | − 1.513283815 | 0.001384316 | 0.005254444 |
| PAGE2 | 4.185109335 | 2.030715895 | − 1.043276898 | 0.000968098 | 0.003926813 |
| AC011008.2 | 0.31787423 | 0.150634816 | − 1.0774008 | 1.59E−07 | 2.63E−06 |
| CYP4F3 | 0.403131219 | 0.823924444 | 1.031262531 | 0.00367986 | 0.011629492 |
| COL22A1 | 1.05899893 | 0.495353661 | − 1.096170312 | 0.00192648 | 0.006859294 |
| GRIK5 | 2.776782427 | 1.332647212 | − 1.05911923 | 7.20E−05 | 0.000460392 |
| TTYH1 | 0.84036637 | 0.407639698 | − 1.043723874 | 0.002854537 | 0.009465163 |
| TCL1A | 0.149056807 | 0.377160857 | 1.339317699 | 0.00311425 | 0.010161885 |
| IGLV5-45 | 13.93991507 | 28.93674286 | 1.053680768 | 0.000152768 | 0.000861658 |
| HMGN2P6 | 0.199381466 | 0.404795154 | 1.021660709 | 0.01678836 | 0.03972772 |
| CYTL1 | 5.128458024 | 1.579728719 | − 1.698848284 | 1.13E−12 | 1.26E−10 |
| HTR1E | 0.400763548 | 0.090015782 | − 2.154501428 | 0.000105609 | 0.000635234 |
| IGLL1 | 0.189352489 | 0.900723467 | 2.250009862 | 0.0085663 | 0.023218884 |
| HOXC8 | 0.283649843 | 0.703358842 | 1.310149851 | 5.47E−05 | 0.000365756 |
Fig. 5.
Heatmap and enrichment analysis of the DEGs. a Distribution of genes in the high and low TMB groups: the red color represents a higher level of expression, and blue color represents a lower level of expression; b Venn diagram of the immune-related genes and DEGs
Fig. 6.
Enrichment analysis of the DEGs. a GO plots of the DEGs. b KEGG analysis results of the DEGss
Table 2.
Detailed GO profiles of the DEGs in UCEC
| ID | Description | GeneRatio | BgRatio | p.adjust | qvalue | geneID | Count | |
|---|---|---|---|---|---|---|---|---|
| BP | GO:0006958 | Complement activation, classical pathway | 38/337 | 137/18670 | 4.55E−31 | 4.27E−31 | IGHV4-31/IGLL1/IGHV4-34/IGLV1-44/IGHM/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGHV3-43/IGLV6-57/IGHV3-53/IGHV3-72/IGHV3-66/IGLV3-21/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGLV2-14/IGHV3-13/IGKV1-5/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLL5/IGLV7-43/IGHV3-30/IGHV3-64/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 38 |
| BP | GO:0002455 | Humoral immune response mediated by circulating immunoglobulin | 38/337 | 150/18670 | 9.97E−30 | 9.37E−30 | IGHV4-31/IGLL1/IGHV4-34/IGLV1-44/IGHM/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGHV3-43/IGLV6-57/IGHV3-53/IGHV3-72/IGHV3-66/IGLV3-21/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGLV2-14/IGHV3-13/IGKV1-5/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLL5/IGLV7-43/IGHV3-30/IGHV3-64/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 38 |
| BP | GO:0006956 | Complement activation | 39/337 | 175/18670 | 2.00E−28 | 1.88E−28 | IGHV4-31/IGLL1/IGHV4-34/IGLV1-44/IGHM/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGHV3-43/IGLV6-57/IGHV3-53/IGHV3-72/CFD/IGHV3-66/IGLV3-21/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGLV2-14/IGHV3-13/IGKV1-5/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLL5/IGLV7-43/IGHV3-30/IGHV3-64/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 39 |
| BP | GO:0072376 | Protein activation cascade | 39/337 | 198/18670 | 2.28E−26 | 2.14E−26 | IGHV4-31/IGLL1/IGHV4-34/IGLV1-44/IGHM/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGHV3-43/IGLV6-57/IGHV3-53/IGHV3-72/CFD/IGHV3-66/IGLV3-21/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGLV2-14/IGHV3-13/IGKV1-5/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLL5/IGLV7-43/IGHV3-30/IGHV3-64/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 39 |
| BP | GO:0016064 | Immunoglobulin mediated immune response | 39/337 | 218/18670 | 8.39E−25 | 7.88E−25 | IGHV4-31/IGLL1/IL13RA2/IGHV4-34/IGLV1-44/IGHM/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGHV3-43/IGLV6-57/IGHV3-53/IGHV3-72/IGHV3-66/IGLV3-21/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGLV2-14/IGHV3-13/IGKV1-5/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLL5/IGLV7-43/IGHV3-30/IGHV3-64/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 39 |
| BP | GO:0019724 | B cell mediated immunity | 39/337 | 221/18670 | 1.20E−24 | 1.12E−24 | IGHV4-31/IGLL1/IL13RA2/IGHV4-34/IGLV1-44/IGHM/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGHV3-43/IGLV6-57/IGHV3-53/IGHV3-72/IGHV3-66/IGLV3-21/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGLV2-14/IGHV3-13/IGKV1-5/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLL5/IGLV7-43/IGHV3-30/IGHV3-64/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 39 |
| BP | GO:0006959 | Humoral immune response | 47/337 | 356/18670 | 2.12E−24 | 1.99E−24 | IGHV4-31/IGLL1/IGHV4-34/IGLV1-44/IGHM/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGHV3-43/IGLV6-57/IGHV3-53/REG1A/IGHV3-72/CFD/IGHV3-66/IGLV3-21/IGHV4-59/IGHV3-20/IGHV3-23/KLK7/IGHV3-73/IGLV2-14/IGHV3-13/KLK3/IFNG/IGKV1-5/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/CAMP/IGLL5/IGLV7-43/IGHV3-30/KLK5/IGHV3-64/IGKV1-17/IGKV1D-12/IGKC/CXCL9/IGLC7/IGHV3-74/CXCL13/IGHG2/IGHV2-5 | 47 |
| BP | GO:0006910 | Phagocytosis, recognition | 26/337 | 84/18670 | 9.61E−23 | 9.03E−23 | IGHV4-31/IGLL1/IGHV4-34/IGHM/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 26 |
| BP | GO:0002920 | Regulation of humoral immune response | 29/337 | 134/18670 | 1.10E−20 | 1.04E−20 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGLV6-57/IGHV3-53/IGLV3-21/IGHV4-59/IGHV3-23/KLK7/IGLV2-14/IGHV3-13/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/KLK5/IGKV1-17/IGKV1D-12/IGKC/IGLC7/CXCL13/IGHG2/IGHV2-5 | 29 |
| BP | GO:0002449 | Lymphocyte mediated immunity | 42/337 | 352/18670 | 5.32E−20 | 5.00E−20 | IGHV4-31/IGLL1/IL13RA2/IGHV4-34/IGLV1-44/IGHM/IGKV2-30/IGKV5-2/ULBP2/LAG3/IGHG4/IGLV2-8/IGHV3-43/IGLV6-57/IGHV3-53/IGHV3-72/CD8A/IGHV3-66/IGLV3-21/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGLV2-14/IGHV3-13/IGKV1-5/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLL5/IGLV7-43/IGHV3-30/IGHV3-64/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 42 |
| BP | GO:0002460 | Adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains | 42/337 | 361/18670 | 1.29E−19 | 1.22E−19 | IGHV4-31/IGLL1/IL13RA2/IGHV4-34/IGLV1-44/IGHM/IGKV2-30/IGKV5-2/ULBP2/IGHG4/IGLV2-8/IGHV3-43/IGLV6-57/IGHV3-53/IGHV3-72/CD8A/IGHV3-66/IGLV3-21/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGLV2-14/IGHV3-13/IGKV1-5/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLL5/IGLV7-43/IGHV3-30/IGHV3-64/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHV3-74/CXCL13/IGHG2/IGHV2-5 | 42 |
| BP | GO:0030449 | Regulation of complement activation | 26/337 | 115/18670 | 4.87E−19 | 4.57E−19 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGLV6-57/IGHV3-53/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHG2/IGHV2-5 | 26 |
| BP | GO:2000257 | Regulation of protein activation cascade | 26/337 | 116/18670 | 5.70E−19 | 5.35E−19 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGLV6-57/IGHV3-53/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHG2/IGHV2-5 | 26 |
| BP | GO:0050853 | B cell receptor signaling pathway | 27/337 | 129/18670 | 6.11E−19 | 5.74E−19 | IGHV4-31/IGLL1/IGHV4-34/IGHM/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGLC7/CD22/IGHV3-74/IGHG2/IGHV2-5 | 27 |
| BP | GO:0006911 | Phagocytosis, engulfment | 26/337 | 118/18670 | 7.88E−19 | 7.41E−19 | IGHV4-31/IGLL1/IGHV4-34/IGHM/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 26 |
| BP | GO:0006909 | Phagocytosis | 41/337 | 369/18670 | 1.63E−18 | 1.53E−18 | IGHV4-31/IGLL1/IGHV4-34/IGLV1-44/IGHM/IGKV2-30/IGKV5-2/SIRPG/IGHG4/IGLV2-8/IGHV3-43/APOA1/IGLV6-57/IGHV3-53/IGHV3-72/IGHV3-66/IGLV3-21/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGLV2-14/IGHV3-13/IFNG/IGKV1-5/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLL5/IGLV7-43/IGHV3-30/IGHV3-64/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 41 |
| BP | GO:0099024 | Plasma membrane invagination | 26/337 | 127/18670 | 5.10E−18 | 4.79E−18 | IGHV4-31/IGLL1/IGHV4-34/IGHM/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 26 |
| BP | GO:0002377 | Immunoglobulin production | 30/337 | 193/18670 | 2.21E−17 | 2.08E−17 | IGLV5-45/IL13RA2/IGLV8-61/IGKV3D-15/IGLV1-44/IGKV2-30/IGKV5-2/IGLV2-8/IGLV6-57/IGKV1-27/IGLV3-21/IGLV1-36/IGLV2-14/IGLV4-69/IGLV3-16/IGKV1-5/IGKV1D-43/TRDV1/IGLV3-19/IGLV7-43/IGKV1D-13/IGKV1-17/IGKV1D-12/TRAV19/IGKV6D-21/IGKC/TRAV14DV4/CD22/IGLV7-46/IGLV4-60 | 30 |
| BP | GO:0010324 | Membrane invagination | 26/337 | 135/18670 | 2.34E−17 | 2.20E−17 | IGHV4-31/IGLL1/IGHV4-34/IGHM/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 26 |
| BP | GO:0002433 | Immune response-regulating cell surface receptor signaling pathway involved in phagocytosis | 26/337 | 139/18670 | 4.58E−17 | 4.31E−17 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGLV6-57/IGHV3-53/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHG2/IGHV2-5 | 26 |
| BP | GO:0038096 | Fc-gamma receptor signaling pathway involved in phagocytosis | 26/337 | 139/18670 | 4.58E−17 | 4.31E−17 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGLV6-57/IGHV3-53/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHG2/IGHV2-5 | 26 |
| BP | GO:0038094 | Fc-gamma receptor signaling pathway | 26/337 | 142/18670 | 7.34E−17 | 6.90E−17 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGLV6-57/IGHV3-53/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHG2/IGHV2-5 | 26 |
| BP | GO:0050871 | Positive regulation of B cell activation | 26/337 | 142/18670 | 7.34E−17 | 6.90E−17 | IGHV4-31/IGLL1/IGHV4-34/IGHM/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 26 |
| BP | GO:0002431 | Fc receptor mediated stimulatory signaling pathway | 26/337 | 145/18670 | 1.22E−16 | 1.14E−16 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGLV6-57/IGHV3-53/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHG2/IGHV2-5 | 26 |
| BP | GO:0070613 | Regulation of protein processing | 28/337 | 180/18670 | 2.69E−16 | 2.52E−16 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGLV6-57/IGHV3-53/SPON1/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/CCBE1/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHG2/IGHV2-5 | 28 |
| BP | GO:0002440 | Production of molecular mediator of immune response | 34/337 | 286/18670 | 3.04E−16 | 2.86E−16 | IGLV5-45/IL13RA2/IGLV8-61/IGKV3D-15/IGLV1-44/IGKV2-30/IGKV5-2/IGLV2-8/APOA1/IGLV6-57/IGKV1-27/IGLV3-21/IGLV1-36/KLK7/IGLV2-14/IGLV4-69/KLK3/IGLV3-16/IGKV1-5/IGKV1D-43/TRDV1/IGLV3-19/IGLV7-43/IGKV1D-13/KLK5/IGKV1-17/IGKV1D-12/TRAV19/IGKV6D-21/IGKC/TRAV14DV4/CD22/IGLV7-46/IGLV4-60 | 34 |
| BP | GO:1903317 | Regulation of protein maturation | 28/337 | 182/18670 | 3.37E−16 | 3.16E−16 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGLV6-57/IGHV3-53/SPON1/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/CCBE1/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHG2/IGHV2-5 | 28 |
| BP | GO:0002673 | Regulation of acute inflammatory response | 26/337 | 159/18670 | 1.14E−15 | 1.07E−15 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGLV6-57/IGHV3-53/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHG2/IGHV2-5 | 26 |
| BP | GO:0002429 | Immune response-activating cell surface receptor signaling pathway | 42/337 | 473/18670 | 1.29E−15 | 1.22E−15 | IGHV4-31/IGLL1/IGHV4-34/IGLV1-44/IGHM/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGHV3-43/IGLV6-57/IGHV3-53/IGHV3-72/IGHV3-66/IGLV3-21/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGLV2-14/IGHV3-13/IGKV1-5/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLL5/MUCL1/TRBV7-9/IGLV7-43/IGHV3-30/IGHV3-64/IGKV1-17/IGKV1D-12/TRAV19/IGKC/IGLC7/CD22/IGHV3-74/IGHG2/IGHV2-5 | 42 |
| BP | GO:0016485 | Protein processing | 35/337 | 328/18670 | 2.60E−15 | 2.44E−15 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGLV6-57/IGHV3-53/SPON1/KLK6/IGLV3-21/IGHV4-59/IGHV3-23/PCSK1N/IGLV2-14/IGHV3-13/KLK3/CCBE1/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/KLK1/IGHV3-30/PCSK9/SCG5/IGKV1-17/IGKV1D-12/IGKC/IGLC7/CPXM2/IGHG2/IGHV2-5 | 35 |
| BP | GO:0050864 | Regulation of B cell activation | 27/337 | 184/18670 | 4.15E−15 | 3.90E−15 | IGHV4-31/IGLL1/IGHV4-34/IGHM/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGLC7/CD22/IGHV3-74/IGHG2/IGHV2-5 | 27 |
| BP | GO:0008037 | Cell recognition | 27/337 | 215/18670 | 2.20E−13 | 2.07E−13 | IGHV4-31/IGLL1/IGHV4-34/IGHM/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/CRTAC1/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 27 |
| BP | GO:0002526 | Acute inflammatory response | 27/337 | 220/18670 | 3.81E−13 | 3.58E−13 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGLV6-57/IGHV3-53/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/IGKV1-17/IGKV1D-12/IGKC/IL1A/IGLC7/IGHG2/IGHV2-5 | 27 |
| BP | GO:0051604 | Protein maturation | 35/337 | 397/18670 | 8.46E−13 | 7.95E−13 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGLV6-57/IGHV3-53/SPON1/KLK6/IGLV3-21/IGHV4-59/IGHV3-23/PCSK1N/IGLV2-14/IGHV3-13/KLK3/CCBE1/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/KLK1/IGHV3-30/PCSK9/SCG5/IGKV1-17/IGKV1D-12/IGKC/IGLC7/CPXM2/IGHG2/IGHV2-5 | 35 |
| BP | GO:0038095 | Fc-epsilon receptor signaling pathway | 23/337 | 169/18670 | 4.43E−12 | 4.16E−12 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGLV2-8/IGLV6-57/IGHV3-53/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/IGKV1-5/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHV2-5 | 23 |
| BP | GO:0042742 | Defense response to bacterium | 31/337 | 330/18670 | 5.32E−12 | 5.00E−12 | IGHV4-31/IGLL1/IGHV4-34/IGHM/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/KLK7/IGHV3-73/IGHV3-13/KLK3/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/CAMP/IGLL5/IGHV3-30/KLK5/IGHV3-64/IGKC/IGLC7/IGHV3-74/CXCL13/IGHG2/IGHV2-5 | 31 |
| BP | GO:0051251 | Positive regulation of lymphocyte activation | 31/337 | 334/18670 | 7.18E−12 | 6.74E−12 | IGHV4-31/IGLL1/IGHV4-34/IGHM/SIRPG/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/CCL5/TNFSF9/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IFNG/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGF2/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 31 |
| BP | GO:0038093 | Fc receptor signaling pathway | 26/337 | 241/18670 | 2.36E−11 | 2.22E−11 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGLV6-57/IGHV3-53/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/IGKV1-17/IGKV1D-12/IGKC/IGLC7/IGHG2/IGHV2-5 | 26 |
| BP | GO:0051249 | Regulation of lymphocyte activation | 36/337 | 485/18670 | 5.50E−11 | 5.17E−11 | IGHV4-31/IGLL1/IGHV4-34/IGHM/SOX11/SIRPG/LAG3/IGHG4/FOXN1/IGHV3-43/IGHV3-53/TIGIT/IGHV3-72/CCL5/TNFSF9/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IFNG/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGF2/IGLC7/CD22/IGHV3-74/IGHG2/IGHV2-5 | 36 |
| BP | GO:0050851 | Antigen receptor-mediated signaling pathway | 29/337 | 316/18670 | 5.80E−11 | 5.45E−11 | IGHV4-31/IGLL1/IGHV4-34/IGHM/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/TRBV7-9/IGHV3-30/IGHV3-64/TRAV19/IGKC/IGLC7/CD22/IGHV3-74/IGHG2/IGHV2-5 | 29 |
| BP | GO:0002696 | Positive regulation of leukocyte activation | 31/337 | 380/18670 | 2.01E−10 | 1.89E−10 | IGHV4-31/IGLL1/IGHV4-34/IGHM/SIRPG/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/CCL5/TNFSF9/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IFNG/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGF2/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 31 |
| BP | GO:0002697 | Regulation of immune effector process | 34/337 | 458/18670 | 2.32E−10 | 2.18E−10 | IL13RA2/IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/ULBP2/LAG3/IGHG4/IGLV2-8/APOA1/IGLV6-57/IGHV3-53/IGLV3-21/IGHV4-59/IGHV3-23/KLK7/IGLV2-14/IGHV3-13/IFNG/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/KLK5/IGKV1-17/IGKV1D-12/IGKC/IGLC7/CD22/IGHG2/IGHV2-5 | 34 |
| BP | GO:0050867 | Positive regulation of cell activation | 31/337 | 394/18670 | 4.90E−10 | 4.60E−10 | IGHV4-31/IGLL1/IGHV4-34/IGHM/SIRPG/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/CCL5/TNFSF9/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IFNG/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGF2/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 31 |
| BP | GO:0042113 | B cell activation | 27/337 | 310/18670 | 1.14E−09 | 1.07E−09 | IGHV4-31/IGLL1/IGHV4-34/IGHM/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGLC7/CD22/IGHV3-74/IGHG2/IGHV2-5 | 27 |
| BP | GO:0006898 | Receptor-mediated endocytosis | 27/337 | 316/18670 | 1.74E−09 | 1.63E−09 | APOB/IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGLV2-8/APOA1/IGLV6-57/IGHV3-53/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/IGKV1-5/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/PCSK9/IGKV1-17/IGKV1D-12/IGKC/AMBP/IGLC7/IGHV2-5 | 27 |
| BP | GO:0050900 | Leukocyte migration | 34/337 | 499/18670 | 2.23E−09 | 2.10E−09 | IGLL1/APOB/IGHV4-34/IGLV1-44/IGHM/IGKV2-30/IGKV5-2/SIRPG/IGLV2-8/IGLV6-57/IGHV3-53/CCL5/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/IGKV1-5/IGHV2-70/IGHV1-69/IGLV3-19/MAG/SLC7A10/IGLV7-43/IGHV3-30/IGKV1-17/IGKV1D-12/IGKC/CXCL9/IGLC7/ATP1B2/EDN3/CXCL13/IGHV2-5 | 34 |
| BP | GO:0050727 | Regulation of inflammatory response | 30/337 | 485/18670 | 3.25E−07 | 3.05E−07 | IGHV4-34/IGLV1-44/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/APOA1/IGLV6-57/IGHV3-53/CCL5/GBP5/IGLV3-21/IGHV4-59/IGHV3-23/IGLV2-14/IGHV3-13/IGKV1-5/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLV7-43/IGHV3-30/IGKV1-17/IGKV1D-12/IGKC/LRFN5/IGLC7/IGHG2/IGHV2-5 | 30 |
| BP | GO:0042475 | Odontogenesis of dentin-containing tooth | 10/337 | 90/18670 | 0.000340129 | 0.000319658 | WNT6/KLK4/SCN5A/FGF4/DLX3/BMP2/KLK5/CA2/FGF8/WNT10A | 10 |
| BP | GO:0031128 | Developmental induction | 6/337 | 34/18670 | 0.001891025 | 0.001777212 | SIX3/WNT4/HOXC11/BMP2/FGF8/GDNF | 6 |
| BP | GO:0009952 | Anterior/posterior pattern specification | 14/337 | 219/18670 | 0.002987205 | 0.002807418 | SIX3/HOXC8/HOXC6/ALX1/CRB2/FOXC2/HOXC10/HOXC11/BMP2/FGF8/HOXC9/CDX1/BARX1/NKD1 | 14 |
| BP | GO:0021984 | Adenohypophysis development | 4/337 | 13/18670 | 0.004101352 | 0.003854509 | WNT4/BMP2/FGF8/SOX2 | 4 |
| BP | GO:0003002 | Regionalization | 18/337 | 351/18670 | 0.004611317 | 0.004333781 | SIX3/HOXC8/HOXC6/ALX1/NKX2-2/CRB2/FOXC2/HOXC10/HOXC11/BMP2/FGF8/WNT7A/GDNF/HOXC9/CDX1/BARX1/DMRTA2/NKD1 | 18 |
| BP | GO:0045165 | Cell fate commitment | 15/337 | 270/18670 | 0.007471561 | 0.00702188 | WNT6/WNT4/FOXN1/NKX2-2/WNT16/FOXC2/HOXC10/BMP2/FGF8/TENM4/WNT7A/WNT5B/SOX2/WNT10A/DMRTA2 | 15 |
| BP | GO:0098742 | Cell–cell adhesion via plasma-membrane adhesion molecules | 15/337 | 273/18670 | 0.008281927 | 0.007783474 | PTPRT/APOA1/CLDN9/CRB2/CDH22/BMP2/MAG/LRRC4C/CLDN19/PCDH10/TENM4/CLDN6/TRO/PCDHB5/LRFN5 | 15 |
| BP | GO:0042476 | Odontogenesis | 10/337 | 132/18670 | 0.008307838 | 0.007807825 | WNT6/KLK4/SCN5A/FGF4/DLX3/BMP2/KLK5/CA2/FGF8/WNT10A | 10 |
| BP | GO:0061844 | Antimicrobial humoral immune response mediated by antimicrobial peptide | 7/337 | 73/18670 | 0.01961805 | 0.018437324 | REG1A/KLK7/KLK3/CAMP/KLK5/CXCL9/CXCL13 | 7 |
| BP | GO:0007389 | Pattern specification process | 19/337 | 446/18670 | 0.028662281 | 0.026937221 | SIX3/HOXC8/WNT6/HOXC6/ALX1/NKX2-2/CRB2/FOXC2/HOXC10/HOXC11/BMP2/FGF8/WNT7A/GDNF/HOXC9/CDX1/BARX1/DMRTA2/NKD1 | 19 |
| BP | GO:0001759 | Organ induction | 4/337 | 22/18670 | 0.032449794 | 0.03049678 | HOXC11/BMP2/FGF8/GDNF | 4 |
| BP | GO:0048762 | Mesenchymal cell differentiation | 12/337 | 219/18670 | 0.035439505 | 0.033306553 | ALX1/SOX11/WNT4/FGF19/CRB2/WNT16/FOXC2/BMP2/FGF8/GDNF/EDN3/WNT10A | 12 |
| BP | GO:0001658 | Branching involved in ureteric bud morphogenesis | 6/337 | 59/18670 | 0.035439505 | 0.033306553 | WNT6/ADAMTS16/WNT4/BMP2/FGF8/GDNF | 6 |
| BP | GO:0016338 | Calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules | 4/337 | 23/18670 | 0.036228036 | 0.034047626 | CLDN9/BMP2/CLDN19/CLDN6 | 4 |
| BP | GO:0045109 | Intermediate filament organization | 4/337 | 23/18670 | 0.036228036 | 0.034047626 | TCHH/NEFH/GFAP/KRT71 | 4 |
| BP | GO:0021871 | Forebrain regionalization | 4/337 | 24/18670 | 0.042179675 | 0.039641061 | SIX3/BMP2/FGF8/DMRTA2 | 4 |
| BP | GO:0046886 | Positive regulation of hormone biosynthetic process | 3/337 | 11/18670 | 0.042595197 | 0.040031574 | WNT4/IFNG/PPARGC1A | 3 |
| BP | GO:0048864 | Stem cell development | 7/337 | 85/18670 | 0.042595197 | 0.040031574 | ALX1/SOX11/FGF19/FOXC2/WNT7A/GDNF/EDN3 | 7 |
| BP | GO:0021983 | Pituitary gland development | 5/337 | 42/18670 | 0.044221556 | 0.04156005 | SIX3/WNT4/BMP2/FGF8/SOX2 | 5 |
| BP | GO:0070268 | Cornification | 8/337 | 112/18670 | 0.04586542 | 0.043104977 | TCHH/FLG/KRT71/KRT36/KRT5/KLK5/KRT31/KRT34 | 8 |
| BP | GO:0008211 | Glucocorticoid metabolic process | 4/337 | 25/18670 | 0.04586542 | 0.043104977 | WNT4/APOA1/BMP2/SERPINA6 | 4 |
| CC | GO:0019814 | Immunoglobulin complex | 51/354 | 159/19717 | 1.95E−47 | 1.78E−47 | IGHV4-31/IGLV5-45/IGLL1/IGLV8-61/IGHV4-34/IGKV3D-15/IGLV1-44/IGHM/IGKV2-30/IGKV5-2/IGHG4/IGLV2-8/IGHV3-43/IGLV6-57/IGHV3-53/IGHV3-72/IGHJ1/IGKV1-27/IGHV3-66/IGLV3-21/IGHV4-59/IGHV3-20/IGLV1-36/IGHV3-23/IGHV3-73/IGLV2-14/IGLV4-69/IGHV3-13/IGLV3-16/IGKV1-5/IGKV1D-43/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLL5/IGLV7-43/IGKV1D-13/IGHV3-30/IGHV3-64/IGKV1-17/IGKV1D-12/IGKV6D-21/IGKC/IGLC7/IGHV3-74/IGLV7-46/IGLV4-60/IGHG2/IGHV2-5 | 51 |
| CC | GO:0042571 | Immunoglobulin complex, circulating | 26/354 | 72/19717 | 3.37E−-25 | 3.07E−25 | IGHV4-31/IGLL1/IGHV4-34/IGHM/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 26 |
| CC | GO:0009897 | External side of plasma membrane | 39/354 | 393/19717 | 3.99E−16 | 3.63E−16 | IGHV4-31/IGLL1/IL13RA2/IGHV4-34/IGHM/CNTFR/ULBP2/CCR10/LAG3/LY6G6C/IGHG4/IGHV3-43/GPIHBP1/IGHV3-53/IGHV3-72/CD8A/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/GFRA2/IGHV3-13/GFRA1/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/PCSK9/IGHV3-64/FGF8/IGKC/CXCL9/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 39 |
| CC | GO:0072562 | Blood microparticle | 15/354 | 147/19717 | 5.34E−06 | 4.86E−06 | IGHM/IGKV2-30/IGHG4/APOA1/IGLV3-21/IGHV3-23/IGHV3-13/IGKV1-5/IGHG3/IGKV1-17/IGKV1D-12/IGKC/AMBP/PON1/IGHG2 | 15 |
| CC | GO:0042101 | T cell receptor complex | 11/354 | 127/19717 | 0.001186365 | 0.001080471 | TRBV7-6/CD8A/TRBV4-2/TRAV21/TRDV1/TRBV7-9/TRAV19/TRAV12-1/TRAV14DV4/TRBV4-1/TRAV26-2 | 11 |
| CC | GO:0098802 | Plasma membrane receptor complex | 15/354 | 295/19717 | 0.016378041 | 0.01491615 | GRIK5/CNTFR/TRBV7-6/CD8A/TRBV4-2/TRAV21/TRDV1/BMP2/TRBV7-9/CHRNA3/TRAV19/TRAV12-1/TRAV14DV4/TRBV4-1/TRAV26-2 | 15 |
| MF | GO:0003823 | Antigen binding | 41/300 | 160/17697 | 3.33E−34 | 2.97E−34 | IGHV4-31/IGLL1/IGHV4-34/IGLV1-44/IGHM/IGKV2-30/IGKV5-2/LAG3/IGHG4/IGLV2-8/IGHV3-43/IGLV6-57/IGHV3-53/IGHV3-72/IGHV3-66/IGLV3-21/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGLV2-14/IGHV3-13/IGKV1-5/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLV3-19/IGLL5/TRBV7-9/IGLV7-43/IGHV3-30/IGHV3-64/IGKV1-17/IGKV1D-12/TRAV19/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 41 |
| MF | GO:0034987 | Immunoglobulin receptor binding | 26/300 | 76/17697 | 5.43E−25 | 4.85E−25 | IGHV4-31/IGLL1/IGHV4-34/IGHM/IGHG4/IGHV3-43/IGHV3-53/IGHV3-72/IGHV3-66/IGHV4-59/IGHV3-20/IGHV3-23/IGHV3-73/IGHV3-13/IGHV2-26/IGHG3/IGHV2-70/IGHV1-69/IGLL5/IGHV3-30/IGHV3-64/IGKC/IGLC7/IGHV3-74/IGHG2/IGHV2-5 | 26 |
| MF | GO:0005109 | Frizzled binding | 8/300 | 39/17697 | 3.98E−05 | 3.55E−05 | WNT6/MYOC/WNT4/RSPO4/WNT16/WNT7A/WNT5B/WNT10A | 8 |
| MF | GO:0048018 | Receptor ligand activity | 23/300 | 482/17697 | 0.001065684 | 0.000951118 | WNT4/FGF19/APOA1/REG1A/CCL5/TNFSF9/FGF4/INSL4/SLURP1/IFNG/TRH/FGF20/BMP2/FGF8/CXCL9/WNT7A/GDNF/IL1A/FGF3/IGF2/EDN3/WNT10A/CXCL13 | 23 |
| MF | GO:0005104 | Fibroblast growth factor receptor binding | 5/300 | 25/17697 | 0.00536912 | 0.004791912 | FGF19/FGF4/FGF20/FGF8/FGF3 | 5 |
| MF | GO:0004252 | Serine-type endopeptidase activity | 10/300 | 160/17697 | 0.033673709 | 0.030053615 | KLK4/KLK6/CFD/KLK7/KLK3/GZMH/KLK8/KLK1/KLK5/PCSK9 | 10 |
| MF | GO:0046873 | Metal ion transmembrane transporter activity | 18/300 | 438/17697 | 0.036391745 | 0.032479448 | GRIK5/TTYH1/GPM6A/CACNA1S/SCN5A/KCNE5/LRRC55/KCNJ15/SLC5A8/JPH3/CACNG6/CACNA1I/ATP2A3/SLC6A15/SLC13A5/SCN1A/SLC13A2/ATP1B2 | 18 |
| MF | GO:0022839 | Ion gated channel activity | 15/300 | 334/17697 | 0.037581202 | 0.033541033 | GRIK5/TTYH1/CACNA1S/SCN5A/ANO2/KCNE5/LRRC55/KCNJ15/P2RX2/GABRA3/JPH3/CACNG6/CACNA1I/CHRNA3/SCN1A | 15 |
| MF | GO:0022836 | Gated channel activity | 15/300 | 343/17697 | 0.043831647 | 0.039119522 | GRIK5/TTYH1/CACNA1S/SCN5A/ANO2/KCNE5/LRRC55/KCNJ15/P2RX2/GABRA3/JPH3/CACNG6/CACNA1I/CHRNA3/SCN1A | 15 |
| MF | GO:0070325 | Lipoprotein particle receptor binding | 4/300 | 26/17697 | 0.044441069 | 0.039663429 | APOB/APOA1/PCSK9/SYT1 | 4 |
| MF | GO:0008236 | Serine-type peptidase activity | 10/300 | 182/17697 | 0.049388397 | 0.044078894 | KLK4/KLK6/CFD/KLK7/KLK3/GZMH/KLK8/KLK1/KLK5/PCSK9 | 10 |
| MF | GO:0005343 | Organic acid:sodium symporter activity | 4/300 | 28/17697 | 0.049388397 | 0.044078894 | SLC5A8/SLC6A15/SLC13A5/SLC13A2 | 4 |
| MF | GO:0017171 | Serine hydrolase activity | 10/300 | 186/17697 | 0.049706953 | 0.044363203 | KLK4/KLK6/CFD/KLK7/KLK3/GZMH/KLK8/KLK1/KLK5/PCSK9 | 10 |
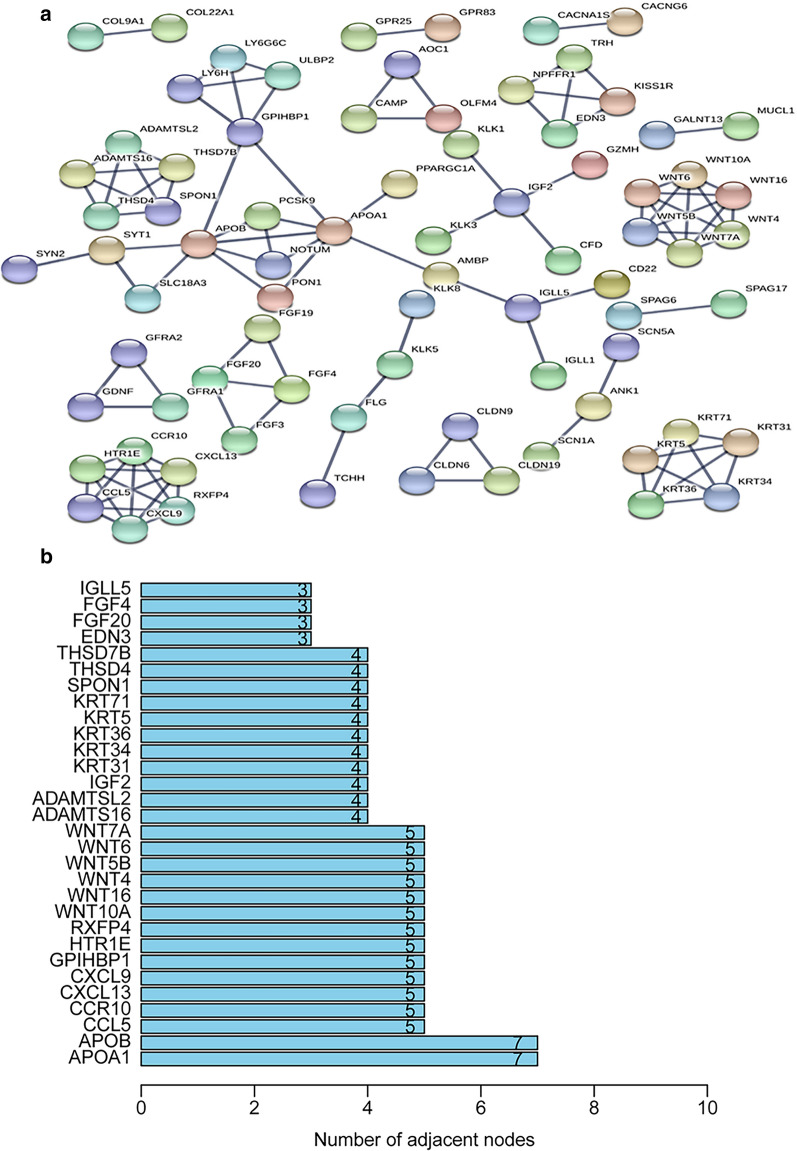
Regarding molecular functions, most genes were enriched for antigen binding, immunoglobulin receptor binding, and receptor-ligand activity. In the KEGG pathway enrichment analysis of the DEGs, these genes showed notable associations with breast cancer, basal cell carcinoma, gastric cancer, hippo signaling pathway, and the Wnt signaling pathway (Fig. 6b and Table 3). String was utilized to confirm the relationship between DEGs and then construct networks describing their interactions (Fig. 7a). The most common neighbors in the PPI diagram were APOA1 and APOB (Fig. 7b).
Table 3.
The top KEGG exhaustive items of the DEGs in UCEC
| ID | Description | GeneRatio | BgRatio | p.adjust | qvalue | Count |
|---|---|---|---|---|---|---|
| hsa05224 | Breast cancer | 11/115 | 147/8058 | 0.000788178 | 0.000687802 | 11 |
| hsa05226 | Gastric cancer | 11/115 | 149/8058 | 0.000788178 | 0.000687802 | 11 |
| hsa04390 | Hippo signaling pathway | 11/115 | 157/8058 | 0.000865824 | 0.00075556 | 11 |
| hsa04310 | Wnt signaling pathway | 10/115 | 160/8058 | 0.003453597 | 0.003013774 | 10 |
| hsa05217 | Basal cell carcinoma | 7/115 | 63/8058 | 0.001385335 | 0.00120891 | 7 |
Fig. 7.
Protein interactions between the DEGs. a PPI network of the DEGs. b Histogram of the number of adjacent nodes
Association between differential immune gene expressions and immune infiltration
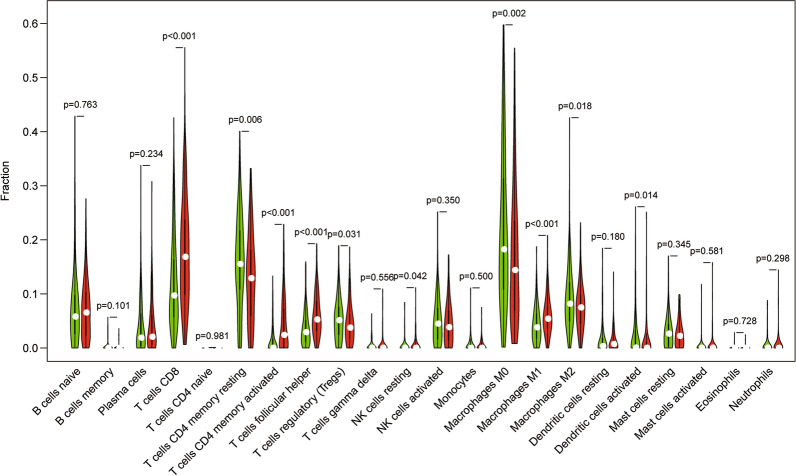
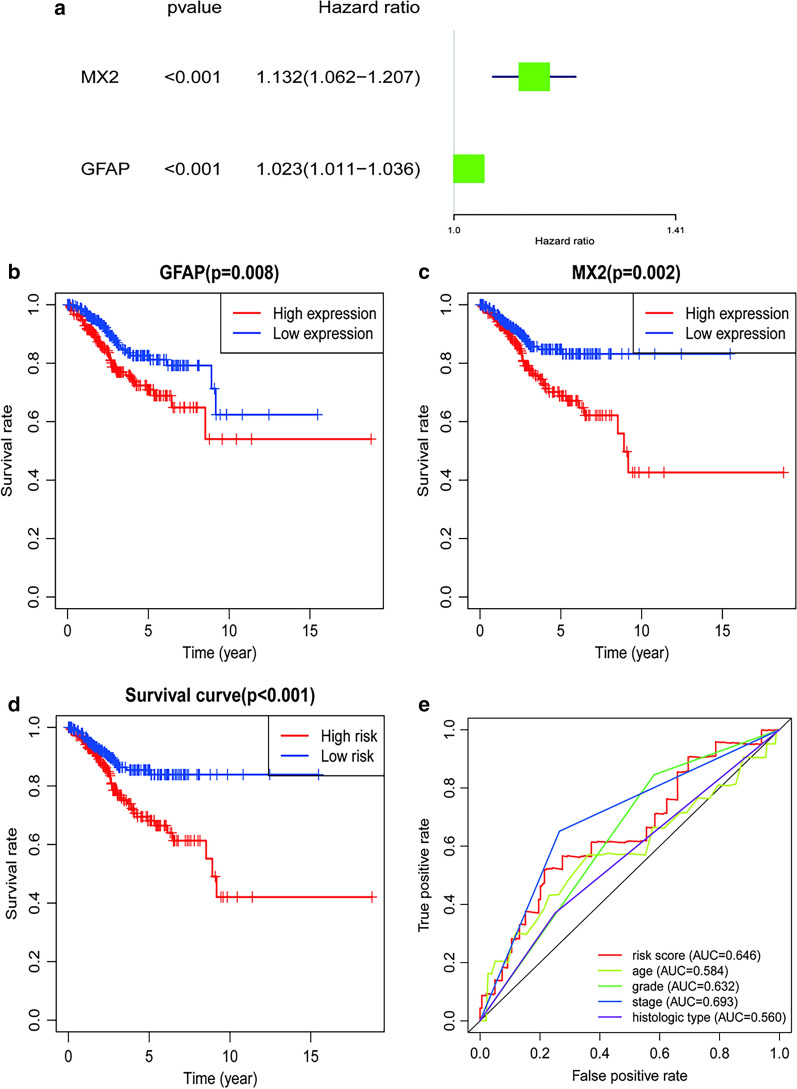
The violin plot shows the difference between various immune cell types in the high and low TMB groups. As suggested by the results, T cells CD8 (P < 0.001), T cells CD4 memory resting (P = 0.006), T cells CD4 memory activated (P < 0.001), T cells follicular helper (P < 0.001), T cells regulatory (P = 0.031), NK cells resting (P = 0.042), Macrophages M0 (P = 0.002), Macrophages M1 (P < 0.001), Macrophages M2 (P = 0.018) and Dendritic cells activated (P = 0.014) showed significant differences between the high and low TMB groups (Fig. 8). Differential immune genes showed significant associations between their expression patterns, and the OS and DFS for UCEC patients were used to conduct a univariate Cox regression analysis. Eventually, we found GFAP (HR 1.023, 95%CI 1.011,1.036, P < 0.001) and MX2 (HR 1.132, 95%CI 1.062,1.207, P < 0.001) to be related to overall survival after multivariate Cox regression analysis was used to remove genes that were not independent indicators of prognosis (Fig. 9a). It could be detected that high expression of GFAP and MX2 were related to poorer OS in endometrial cancer patients, based on the values of the long-rank test and Kaplan-Meier curve (Fig. 9b, c). MX2 (HR 1.130, 95%CI 1.058, 1.207, P < 0.001), GFAP (HR 1.025, 95%CI 1.012, 1.038, P < 0.001), IGHM (HR 0.999, 95%CI 0.998, 1.000, P = 0.039), FGF20 (HR 0.939, 95%CI 0.883, 0.998, P = 0.041) and TRAV21 (HR 0.611, 95%CI 0.407, 0.917, P = 0.017) were analyzed for DFS (Fig. 10a). Regarding DFS, UECE patients with a higher expression level of MX2 and GFAP had a worse prognosis, while patients with higher mRNA levels of IGHM, FGF20, and TRAV21 showed a favorable outcome (Fig. 8c–h). These genes and clinical factors, including age, grade, stage, histological type, which are related to prognosis, were selected for model construction. The prognosis index formula multiplied the gene expression level and clinical characteristics in each case with the corresponding Cox regression coefficient. All selected genes were classified into either a high-risk or low-risk group based on the median prognosis index value, and survival curves were plotted. Moreover, Kaplan-Meier plots were drawn to analyze differences in survival time between both groups, indicating that cases in the low-risk group were significantly associated with increased survival probability than those in the high-risk group, for both OS and DFS (Figs. 9d and 10h). As shown in Figs. 8e and 9b, the largest AUC areas were found for the risk score of OS (0.646) and DFS (0.743), respectively.
Fig. 8.
The violin diagram depicts the expression analysis of multiple immune cells in the high and low TMB groups. The green color is used for the high TMB group, and the red color is used for the low TMB group
Fig. 9.
Differential immune genes in OS. a Forest plot of the differential immune genes; b, c survival analysis of the GFAP and MX2 genes; d survival analysis of the risk score; e the ROC curves of the models
Fig. 10.

Differential immune genes in DFS. a Forest plot of the differential immune genes; b the ROC curves of the models; c–g Survival analysis of GFAP, MX2, FGF20, IGHM, and TRAV21 genes; h survival analysis of the risk score
Association between risk score and immune infiltrates
The relationship between risk score and different types of immune cells were obtained from the Timer website using R software. The risk score of DFS showed a negative correlation with CD4+ T cell, CD8+ T cell, macrophage, and neutrophil (all P < 0.05, Fig. 11a). However, the risk score of OS was not associated with immune cells (all P < 0.05, Fig. 11b).
Fig. 11.
Correlation analysis of the risk score and immune cells in OS (a) and DFS (b)
High B cell and CD8 + T cell infiltration indicates a better outcome
To understand the association between immune infiltrating cells and survival in UCEC, we used the Cox regression equation to calculate the expression levels of CD4+ T cell (HR 0.001, 95%CI 0, 0.052, P = 0.005), CD8+ T cell (HR 0.001, 95%CI 0, 0.052, P = 0.001) and Neutrophil (HR 2314.933, 95%CI 1.836, 2919028.574, P = 0.033), which had declined due to cancer progression (Table 4). Additionally, Kaplan-Meier plots were applied to determine that UECE patients with a higher expression level of B cell and CD8+ T cell had a better prognosis (Fig. 12).
Table 4.
Multi-factor analysis of six immune infiltrating cells in UCEC
| HR | 95%CI_low | 95%CI_up | p.value | |
|---|---|---|---|---|
| B cell | 0.102 | 0 | 47.135 | 0.466 |
| CD8+ Tcell | 0.001 | 0 | 0.052 | 0.001** |
| CD4+ Tcell | 0 | 0 | 0.068 | 0.005** |
| Macrophage | 24.031 | 0.178 | 3252.991 | 0.204 |
| Neutrophil | 2314.933 | 1.836 | 2919028.574 | 0.033* |
| Dendritic | 4.928 | 0.183 | 132.438 | 0.342 |
*P < 0.05; **P < 0.01
Fig. 12.
The Kaplan–Meier plot of the six immune cells: B cell, CD8+ T cell, CD4+ T cell, macrophage, neutrophil, and dendritic cell
Nomogram of the differential immune genes and clinical variables for OS and DFS
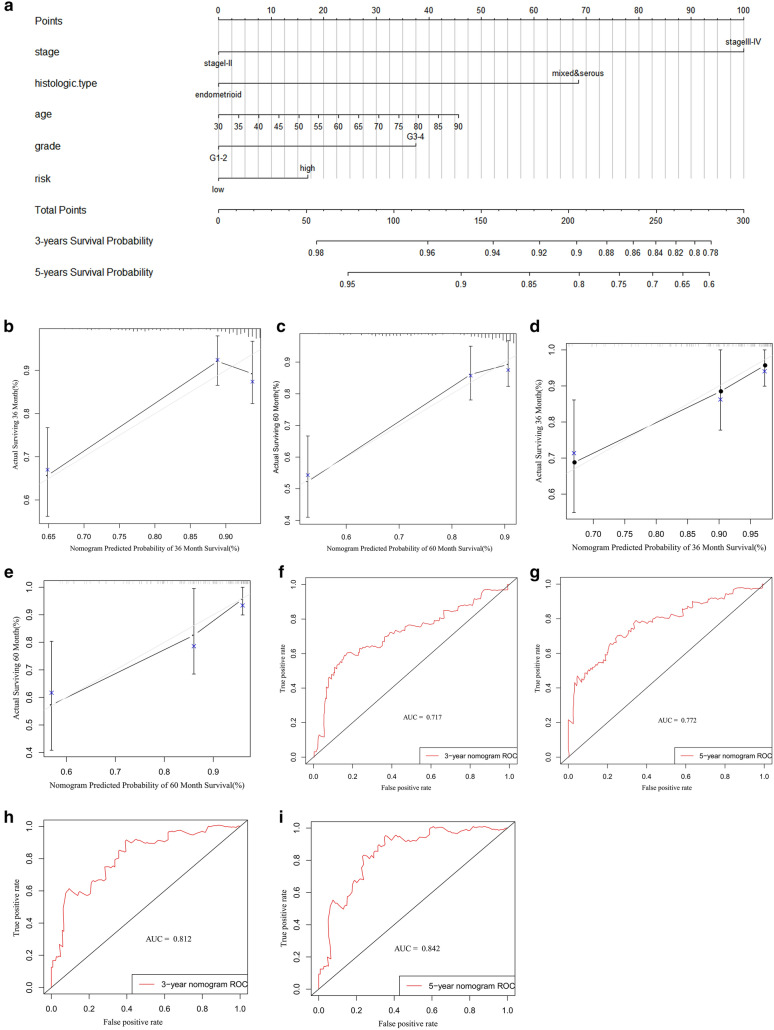
The basic clinical characteristics of the patients are shown in Table 5 for OS and in Table 6 for DFS. Through univariate and multivariate Cox analyses of the modeling group and the entire cohort of patients, we found that the risk score (especially for DFS) may be an independent risk factor for UCEC patients (Tables 7, 8, 9, 10). Additionally, we used risk group, age, grade, stage, and histological type to construct nomogram models for OS (Fig. 13a) and DFS (Fig. 14a). To verify the predictive value of the models, 3-year and 5-year calibration charts were drawn for the modeling group and the verification group for OS (Fig. 13b–e) and DFS (Fig. 14b–e), which suggested the two models produced consistent results. ROC curves were constructed, and their AUC areas were greater than 0.7, indicating that these two models showed a high level of diagnostic accuracy.
Table 5.
Clinical information of UCEC patients for OS obtained from TCGA database
| Variables | UCEC |
|---|---|
| N = 521(%) | |
| Age | 64(57,71) |
| Stage | |
| Stage I–II | 374(71.8%) |
| Stage III–IV | 147(28.2%) |
| Grade | |
| G1–2 | 212(40.7%) |
| G3–4 | 309(59.3%) |
| Histologic type | |
| Endometrioid | 388(74.5%) |
| Mixed & serous | 133(25.5%) |
| Risk | |
| Low | 261(50.1%) |
| High | 260(49.9%) |
Table 6.
Clinical baseline of UCEC patients for DFS obtained from TCGA database
| Variables | UCEC |
|---|---|
| N = 450(%) | |
| Age | 63(56.8,70) |
| Stage | |
| Stage I–II | 328(72.9%) |
| Stage III–IV | 122(27.1%) |
| Grade | |
| G1–2 | 195(43.3%) |
| G3–4 | 255(56.7%) |
| Histologic type | |
| Endometrioid | 339(75.3%) |
| Mixed & serous | 111(24.7%) |
| Risk | |
| Low | 224(49.8%) |
| High | 226(50.2%) |
Table 7.
Univariate and multivariate Cox analysis of UCEC patients for OS in the training set
| Variables | Univariate analysis | Multivariable analysis | ||
|---|---|---|---|---|
| HR(95%CI) | P-value | HR(95%CI) | P-value | |
| Age | 1.020(0.995–1.046) | 0.177 | 1.009(0.981–1.037) | 0.546 |
| Stage | ||||
| Stage I–II | Reference | Reference | ||
| Stage III–IV | 4.175(2.506–6.957) | < 0.001 | 3.045(1.769–5.241) | < 0.001 |
| Grade | ||||
| G1–2 | Reference | Reference | ||
| G3–4 | 3.264(1.697–6.279) | < 0.001 | 1.518(0.707–3.261) | 0.284 |
| Histologic type | ||||
| Endometrioid | Reference | Reference | ||
| Mixed & serous | 3.939(2.374–6.537) | < 0.001 | 2.146(1.101–4.181) | 0.025 |
| Risk | ||||
| Low | Reference | Reference | ||
| High | 2.435(1.403–4.226) | 0.002 | 1.209(0.628–2.325) | 0.57 |
Table 8.
Univariate and multivariate Cox analysis of all UCEC patients for OS
| Variables | Univariate analysis | Multivariable analysis | ||
|---|---|---|---|---|
| HR(95%CI) | P-value | HR(95%CI) | P-value | |
| Age | 1.031(1.009–1.053) | 0.005 | 1.023(1.000-1.046) | 0.047 |
| Stage | ||||
| Stage I–II | Reference | Reference | ||
| Stage III–IV | 3.819(2.471–5.901) | < 0.001 | 3.091(1.957–4.882) | < 0.001 |
| Grade | ||||
| G1–2 | Reference | Reference | ||
| G3–4 | 3.589(2.018–6.382) | < 0.001 | 2.135(2.122–4.064) | 0.021 |
| Histologic type | ||||
| Endometrioid | Reference | Reference | ||
| Mixed & serous | 2.856(1.851–4.406) | < 0.001 | 1.241(0.734–2.097) | 0.421 |
| Risk | ||||
| Low | Reference | Reference | ||
| High | 2.166(1.368–3.431) | 0.001 | 1.271(0.758–2.130) | 0.363 |
Table 9.
Univariate and multivariate Cox analysis of UCEC patients for DFS in the modeling group
| Variables | Univariate analysis | Multivariable analysis | ||
|---|---|---|---|---|
| HR(95%CI) | P-value | HR(95%CI) | P-value | |
| Age | 1.005(0.980–1.031) | 0.692 | 0.992(0.967–1.019) | 0.572 |
| Stage | ||||
| Stage I–II | Reference | Reference | ||
| Stage III–IV | 2.430(1.423–4.149) | 0.001 | 2.026(1.143–3.593) | 0.016 |
| Grade | ||||
| G1–2 | Reference | Reference | ||
| G3–4 | 1.989(1.096–3.609) | 0.024 | 1.330(0.655-2.700) | 0.429 |
| Histologic type | ||||
| Endometrioid | Reference | Reference | ||
| Mixed & serous | 2.082(1.213–3.573) | 0.008 | 1.281(0.661–2.481) | 0.463 |
| Risk | ||||
| Low | Reference | Reference | ||
| High | 3.159(1.715–5.818) | 0.002 | 3.000(1.606–5.606) | 0.001 |
Table 10.
Univariate and multivariate Cox analysis of all UCEC patients for DFS
| Variables | Univariate analysis | Multivariable analysis | ||
|---|---|---|---|---|
| HR(95%CI) | P-value | HR(95%CI) | P-value | |
| Age | 1.009(0.989–1.030) | 0.395 | 0.995(0.974–1.017) | 0.676 |
| Stage | ||||
| Stage I–II | Reference | Reference | ||
| Stage III–IV | 2.125(1.389–3.251) | 0.001 | 1.746(1.101–2.769) | 0.018 |
| Grade | ||||
| G1–2 | Reference | Reference | ||
| G3–4 | 2.103(1.312–3.372) | 0.002 | 1.478(0.852–2.564) | 0.164 |
| Histologic type | ||||
| Endometrioid | Reference | Reference | ||
| Mixed & serous | 2.212(1.437–3.404) | < 0.001 | 1.251(0.734–2.133) | 0.411 |
| Risk | ||||
| Low | Reference | Reference | ||
| High | 3.015(1.868–4.866) | 0.001 | 2.799(1.707–4.589) | < 0.001 |
Fig. 13.
The model of risk score and clinical factors in OS. a 3-year and 5-year survival nomogram; b–e 3-year and 5-year calibration charts for the training and verification groups; f–i 3-year and 5-year ROC graphs for the training and verification groups
Fig. 14.
The model of risk score and clinical factors in DFS. a 3-year and 5-year survival nomogram; b–e 3-year and 5-year verification plots in testing and verification cohort; f–i 3-year and 5-year AUC areas in testing and verification cohort
Discussion
Immunotherapy has been used to treat recurring or metastatic tumors in clinical settings, but it is only effective for certain tumors, such as breast cancer [16] and prostate cancer [17]. In recent years, an increasing number of research studies have focused on immune molecular features and immunotherapeutic intervention for UCEC [18], owing to the lower survival rates of advanced UCEC patients. It was found that TMB can be beneficial to understand gene mutations in cancer cells, which is closely related to the efficacy of immunotherapy [19]. During the further analysis of the correlation between TMB and UCEC, we first explored the association between the high and low TMB groups with clinical survival. We found that for both OS and DFS, high TMB indicated a better prognosis. High TMB indicates that cancer cells have a high level of mutations, suggesting that cancer cells are more different from normal cells. They can be easily discovered by the human immune system, allowing the tumor cells to be killed.
The more effective the immunotherapy, the longer the survival [20, 21]. Of course, the detail mechanisms of that are unclear. A leading research demands that the process of mutations may create neoantigens which play an important role in the response of patients to immune checkpoint inhibitors. On the other hand, the expense of immunotherapy was still high, and many studies have paid an increasing amount of attention to determine whether patients were suitable for immunotherapy. TMB assessment can be used to predict the efficacy of immunotherapy further. Wu et al. [22] found that progression-free survival (PFS) (combined HR 0.59, 95%CI 0.49, 0.71, P < 0.001) and OS (combined HR 0.68, 95%CI 0.53, 0.89, P = 0.004) of the high TMB group significantly improved through the study of TMB levels and the effect of immunotherapy on various tumors. In the predictive analysis, TMB was found to be an independent predictor of immune checkpoint inhibitor treatment.
In contrast, group testing found that patients with high TMB showed an excellent response to immunotherapy [23], indicating that a higher TMB is expected to improve the efficacy of immune checkpoint inhibitors in cancers. Then, we used the CIBERSORT algorithm to calculate the proportion of immune cell infiltration in each patient sample. Additionally, the two TMB groups were strongly associated with specific immune infiltrating cells, which further proved that TMB was associated with the immune response. At the same time, high TMB was related to immunotherapy, and this phenomenon may involve immune infiltrating cells.
Using the median value of TMB as the critical value, we identified 393 DEGs between the low and high expression groups of TMB. Functional enrichment analyses of those DEGs provided an understanding of their biological roles. In the GO analysis, immune cells and receptors were found to be associated with the DEGs. Sonoda et al. [24] suggested that RCAS1 was a ligand for immune cell receptors, which was significantly associated with the stage of UCEC, the degree of myometrial invasion, positive peritoneal cytology, and overall survival. In the KEGG analysis, they were associated with various tumor pathways or genes, such as basal cell, breast, and gastric carcinoma. This implied that they had specific tumor pathways in common. For example, there were some similarities between UCEC and breast cancer at the molecular level. IDO1 was involved in the anti-tumor immune process of both tumors and was related to TMB [18]. After the protein network map of the DEGs was constructed, we detected the core expression levels of these genes in the PPI network, which may play an essential role in UCEC. APOA1 could promote the increase of macrophage infiltration, decrease TMB and metastasis, and improve the survival rate, similar to its effects in colorectal cancer [25, 26]. APOB caused a high mutation burden of cancer genes and tumorigenesis [27].
We utilized differential immune genes to establish Cox risk models for OS and DFS. Two genes (GFAP and MX2) were identified in the OS model, while five genes (GFAP, MX2, FGF20, IGHM, and TRAV21) were identified in the DFS model. In this study, we further explored the role of the above-mentioned differential immune genes and immune infiltration in the high and low TMB groups in UCEC. These genes were the core differential immune genes, and the risk scores obtained from the Cox multivariable analysis were grouped. For DFS, the risk score was related to some immune cells. The higher the risk score for DFS, the lower the level of CD4+ T cell, CD8+ T cell, macrophage, and neutrophil infiltration.
Additionally, through the survival analysis, we found that high-risk scores indicated better survival. MX2 is a viral interferon, which is the key to block human immunodeficiency virus type 1 [28]. GFAP can delay the development of type 1 diabetes by regulating T cell differentiation [29]. Inhibition of the FGFR family of genes can prevent the development of tumors by blocking paracrine signaling, which was related to immune escape in the tumor microenvironment [30].
We found that decreases in the expression of B cell and CD8+ T cell showed an apparent association with a poorer prognosis. In breast cancer, the presence of CD8+ T cells decreased the risk of breast cancer death by about 20% [31]. CD8+ T cells induced prolonged survival for patients with various types of tumors, including liver cancer [32] and rectal cancer [33]. Previous research [34] has proven that CD8+ T lymphocytes are an independent risk factor and that UCEC patients with high expression levels show better survival rates, which is consistent with our results. The role of B cells on tumors is unclear. B cells promote squamous cell carcinoma (SCC) development by depositing immunoglobulin- containing immune complexes [35]. In our study, high B cell expression was associated with a good outcome. Although the role of B cells in tumors is uncertain, the prognostic value of B-cell gene expression signatures in tumors has been demonstrated [36]. From the above the mechanisms that B cell and CD8+ T cell were linked to immunosuppression in tumor microenvironment are incompletely clear. Advanced research implied that these immunotherapy are potentially related to the expression of PD1 and CTLA4 from CD8+ T cell.
Finally, the risk stratification in the models mentioned above and other clinical factors was used to conduct single-factor and multifactor Cox analyses to construct the corresponding Nomogram model. The ROC curve showed that the Nomogram model was more reliable than the other models. The nomogram could comprehensively evaluate the survival rate of patients with genetic and clinical factors, and they were more intuitive and performed well. Furthermore, the risk score in DFS showed a significantly greater impact on diagnosis than in OS. At present, an increasing number of genes have been used as models to predict the prognosis and improve the prognosis of UCEC [37].
However, this study also contains certain limitations: (i) One limitation of this study is that it was a retrospective study. The relationship between differential TMB-related immune genes and immune cell infiltration still needs to be confirmed using primary experimental evidence. For example, we can analyse the differential immune cell infiltration expression between TMBHigh and TMBLow goup of EC cells and further explore the expressed level of immune checkpoint inhibitors such as PD-1 or CTL-4. (ii) The lack of many clinical samples to verify the prognostic effect of TMB and its potential relationship with immune infiltration. Therefore, the relationship between the occurrence and development of EC needs to be further confirmed using many more studies. Due to cost and technological limitations, the application of polygenic models is restricted.
Conclusions
High TMB is related to prolonged survival and may promote immune infiltration. The immune gene models related to TMB levels were established. Clinical factors related to the models were determined to evaluate the prognosis of UCEC further and provide a base to predict immunotherapeutic outcomes. Modeling of immune infiltrating cells in endometrial cancer also showed that B cell and CD8+ T cell are the most important types of immune infiltrating cells. The function and mechanism of involvement of these hub immune genes, included in the models, are still unclear. Many more experiments need to be conducted to confirm their function in the immune system (Additional files 1, 2, 3, 4, 5, 6, 7).
Supplementary Information
Additional file 1. The disease-free survival of six immune cells.
Additional file 2. Clinical features of uterine corpus endometrial carcinoma patients in disease-free survival.
Additional file 3. Related genes in immune cells.
Additional file 4. The overall survival of six immune cells.
Additional file 5. Clinical features of uterine corpus endometrial carcinoma patients in overall survival.
Additional file 6. The tumor mutation burden of uterine corpus endometrial carcinoma samples.
Additional file 7. The mRNA data of uterine corpus endometrial carcinoma samples.
Acknowledgements
None.
Abbreviations
- AUC
Area under the curve
- CIs
Confidence interval
- CTLA-4
Cytotoxic T-lymphocyte antigen 4
- DEGs
Differentially expressed genes
- DFS
Disease-free survival
- GEP
Gene expression profiles
- GO
Gene Ontology
- HR
Hazard ratio
- ICI
Immune Checkpoint Inhibitor
- KEGG
Kyoto Encyclopedia of Genes, Genomes
- OS
Overall survival
- PD-1
Programed cell death 1 receptor
- PFS
Progression-free survival
- POLE
polymerase ɛ
- PPI
Protein–protein interaction network
- ROC
Receiver operating characteristic
- SCC
Squamous cell carcinoma
- SNP
Single Nucleotide Polymorphism
- SNV
Single nucleotide variant
- STRING
Search Tool for the Retrieval of Interacting Genes database
- TMB
Tumor mutation burden
- UCEC
Uterine corpus endometrial carcinoma
Authors’ contributions
LZ: project administration, data curation, manuscript writing. XF: methodology, investigation, manuscript writing. XH: formal analysis, methodology, software. YY: investigation, software, conceptualization. YY: formal analysis, investigation. JG: conceptualization, supervision, manuscript reviewing and editing. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
None.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-021-01774-6.
References
- 1.Siegel RL, MFiller KD, Jemal A. Cancer statistics. 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Braun MM, Overbeek-Wager EA, Grumbo RJ. Diagnosis and management of endometrial cancer. Am Family Phys. 2016;93(6):468–74. [PubMed] [Google Scholar]
- 3.Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, et al. Cancer treatment and survivorship statistics, 2019. Cancer J Clin. 2019;69(5):363–85. doi: 10.3322/caac.21565. [DOI] [PubMed] [Google Scholar]
- 4.Memon H, Patel BM. Immune checkpoint inhibitors in non-small cell lung cancer: a bird's eye view. Life Sci. 2019;233:116713. doi: 10.1016/j.lfs.2019.116713. [DOI] [PubMed] [Google Scholar]
- 5.Heeke S, Benzaquen J, Long-Mira E, Audelan B, Lespinet V, Bordone O, et al. In-house Implementation of tumor mutational burden testing to predict durable clinical benefit in non-small cell lung cancer and melanoma patients. Cancers. 2019;11(9):1271. doi: 10.3390/cancers11091271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang B, Yan X, Sheng X, Si L, Cui C, Kong Y, et al. Safety and clinical activity with an anti-PD-1 antibody JS001 in advanced melanoma or urologic cancer patients. J Hematol Oncol. 2019;12(1):7. doi: 10.1186/s13045-018-0693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forschner A, Battke F, Hadaschik D, Schulze M, Weissgraeber S, Han CT, et al. Tumor mutation burden and circulating tumor DNA in combined CTLA-4 and PD-1 antibody therapy in metastatic melanoma—results of a prospective biomarker study. J immunother Cancer. 2019;7(1):180. doi: 10.1186/s40425-019-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackburn SD, Shin H, Haining WN, Zou T, Workman CJ, Polley A, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10(1):29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–6. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones NL, Xiu J, Rocconi RP, Herzog TJ, Winer IS. Immune checkpoint expression, microsatellite instability, and mutational burden: Identifying immune biomarker phenotypes in uterine cancer. Gynecol Oncol. 2020;156(2):393–9. doi: 10.1016/j.ygyno.2019.11.035. [DOI] [PubMed] [Google Scholar]
- 11.Bhangoo MS, Boasberg P, Mehta P, Elvin JA, Ali SM, Wu W, et al. Tumor mutational burden guides therapy in a treatment refractory POLE-mutant uterine carcinosarcoma. Oncologist. 2018;23(5):518–23. doi: 10.1634/theoncologist.2017-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–56. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607-D13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–59. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108-e10. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emens LA. Breast Cancer Immunother Facts Hopes. Clinical Cancer Res. 2018;24(3):511–20. doi: 10.1158/1078-0432.CCR-16-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu X, Horner JW, Paul E, Shang X, Troncoso P, Deng P, et al. Effective combinatorial immunotherapy for castration-resistant prostate cancer. Nature. 2017;543(7647):728–32. doi: 10.1038/nature21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng X, Tang R, Zhang R, Wang H, Ji Z, Shao Y, et al. A comprehensive analysis of IDO1 expression with tumour-infiltrating immune cells and mutation burden in gynaecologic and breast cancers. J Cell Mol Med. 2020;24(9):5238–48. doi: 10.1111/jcmm.15176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budczies J, Seidel A, Christopoulos P, Endris V, Kloor M, Gyorffy B, et al. Integrated analysis of the immunological and genetic status in and across cancer types: impact of mutational signatures beyond tumor mutational burden. Oncoimmunology. 2018;7(12):e1526613. doi: 10.1080/2162402X.2018.1526613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNamara MG, Jacobs T, Lamarca A, Hubner RA, Valle JW, Amir E. Impact of high tumor mutational burden in solid tumors and challenges for biomarker application. Cancer Treat Rev. 2020;89:102084. doi: 10.1016/j.ctrv.2020.102084. [DOI] [PubMed] [Google Scholar]
- 21.Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30(1):44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Y, Xu J, Du C, Xia D, Lv W, Hu J. The predictive value of tumor mutation burden on efficacy of immune checkpoint inhibitors in cancers: a systematic review and meta-analysis. Front Oncol. 2019;9:1161. doi: 10.3389/fonc.2019.01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, Bai X, Wang J, Tang XR, Wu DH, Du SS, et al. Combination of TMB and CNA stratifies prognostic and predictive responses to immunotherapy across metastatic cancer. Clin Cancer Res. 2019;25(24):7413–23. doi: 10.1158/1078-0432.CCR-19-0558. [DOI] [PubMed] [Google Scholar]
- 24.Sonoda K, Miyamoto S, Hirakawa T, Kaku T, Nakashima M, Watanabe T, et al. Association between RCAS1 expression and clinical outcome in uterine endometrial cancer. Br J Cancer. 2003;89(3):546–51. doi: 10.1038/sj.bjc.6601126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo G, Wang Y, Zhou Y, Quan Q, Zhang Y, Wang H, et al. Immune cell concentrations among the primary tumor microenvironment in colorectal cancer patients predicted by clinicopathologic characteristics and blood indexes. J Immunother Cancer. 2019;7(1):179. doi: 10.1186/s40425-019-0656-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zamanian-Daryoush M, Lindner D, Tallant TC, Wang Z, Buffa J, Klipfell E, et al. The cardioprotective protein apolipoprotein A1 promotes potent anti-tumorigenic effects. J Biol Chem. 2013;288(29):21237–52. doi: 10.1074/jbc.M113.468967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J, Choudhry H, Cao W. Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like family genes activation and regulation during tumorigenesis. Cancer Sci. 2018;109(8):2375–82. doi: 10.1111/cas.13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dicks MD, Goujon C, Pollpeter D, Betancor G, Apolonia L, Bergeron JR, et al. Oligomerization requirements for MX2-mediated suppression of HIV-1 infection. J Virol. 2016;90(1):22–32. doi: 10.1128/JVI.02247-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jayasimhan A, Marino E, Dietary, SCFAs IL-22, and GFAP: the three musketeers in the gut-neuro-immune network in type 1 diabetes. Front Immunol. 2019;10:2429. doi: 10.3389/fimmu.2019.02429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh M. FGFR inhibitors: effects on cancer cells, tumor microenvironment and whole-body homeostasis (Review) Int J Mol Med. 2016;38(1):3–15. doi: 10.3892/ijmm.2016.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali HR, Provenzano E, Dawson SJ, Blows FM, Liu B, Shah M, et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol. 2014;25(8):1536–43. doi: 10.1093/annonc/mdu191. [DOI] [PubMed] [Google Scholar]
- 32.Kalathil SG, Hutson A, Barbi J, Iyer R, Thanavala Y. Augmentation of IFN-gamma + CD8 + T cell responses correlates with survival of HCC patients on sorafenib therapy. JCI insight. 2019;4(15):e130116. doi: 10.1172/jci.insight.130116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogura A, Akiyoshi T, Yamamoto N, Kawachi H, Ishikawa Y, Mori S, et al. Pattern of programmed cell death-ligand 1 expression and CD8-positive T-cell infiltration before and after chemoradiotherapy in rectal cancer. Eur J Cancer. 2018;91:11–20. doi: 10.1016/j.ejca.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Kondratiev S, Sabo E, Yakirevich E, Lavie O, Resnick MB. Intratumoral CD8 + T lymphocytes as a prognostic factor of survival in endometrial carcinoma. Clin Cancer Res. 2004;10(13):4450–6. doi: 10.1158/1078-0432.CCR-0732-3. [DOI] [PubMed] [Google Scholar]
- 35.Affara NI, Ruffell B, Medler TR, Gunderson AJ, Johansson M, Bornstein S, et al. B cells regulate macrophage phenotype and response to chemotherapy in squamous carcinomas. Cancer Cell. 2014;25(6):809–21. doi: 10.1016/j.ccr.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iglesia MD, Vincent BG, Parker JS, Hoadley KA, Carey LA, Perou CM, et al. Prognostic B-cell signatures using mRNA-seq in patients with subtype-specific breast and ovarian cancer. Clin Cancer Res. 2014;20(14):3818–29. doi: 10.1158/1078-0432.CCR-13-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying J, Wang Q, Xu T, Lyu J. Establishment of a nine-gene prognostic model for predicting overall survival of patients with endometrial carcinoma. Cancer Med. 2018;7(6):2601–11. doi: 10.1002/cam4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The disease-free survival of six immune cells.
Additional file 2. Clinical features of uterine corpus endometrial carcinoma patients in disease-free survival.
Additional file 3. Related genes in immune cells.
Additional file 4. The overall survival of six immune cells.
Additional file 5. Clinical features of uterine corpus endometrial carcinoma patients in overall survival.
Additional file 6. The tumor mutation burden of uterine corpus endometrial carcinoma samples.
Additional file 7. The mRNA data of uterine corpus endometrial carcinoma samples.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.