Abstract
Autism spectrum disorder (ASD) is characterized by neurocognitive dysfunctions, such as impaired social interaction and language learning. Gene-environmental interactions play a pivotal role in ASD pathogenesis. Nuclear receptor corepressors (NCORs) are transcription co-regulators physically associated with histone deacetylases (HDACs) and many known players in ASD etiology such as transducin β-like 1 X-linked receptor 1 (TBL1XR1) and methyl-CpG binding protein 2 (MECP2). The epigenome-modifying NCOR complex is sensitive to many ASD risk factors, including HDAC inhibitor valproic acid (VPA) and a variety of endocrine factors, xenobiotic chemicals, or metabolites that can directly bind to multiple nuclear receptors. Here we review recent studies of NCORs in neurocognition using animal models and human genetics approaches. We discuss functional interplays between NCORs and other known players in ASD etiology. It is conceivable that the NCOR complex may bridge the in utero environmental risk factors of ASD with epigenetic remodeling and can serve as a converging point for many gene-environment interactions in the pathogenesis of ASD and intellectual disability.
Keywords: nuclear receptor corepressor (NCOR), silencing mediator for retinoid and thyroid hormone receptors (SMRT), histone deacetylase (HDAC), methyl-CpG binding protein 2 (MECP2), transducin β-like 1 X-linked receptor 1 (TBL1XR1), valproic acid, autism, memory, epigenetic, in utero environment
Introduction
Autism spectrum disorders (ASDs) are neurodevelopmental conditions characterized by abnormal social interaction, repetitive behaviors, impaired language learning, and other cognitive dysfunctions. ASD affects about 1% of the general global population [1], and 45% of ASD cases are associated with intellectual disability [2]. Syndromic ASDs include fragile X syndrome, Rett syndrome (RTT), Phelan–McDermid syndrome, and Timothy syndrome, which are caused by specific genetic alternations [3]. However, most ASD cases are of unknown causes and display highly heterogeneous clinical phenotypes.
The gene-environmental interaction and epigenetic mechanisms are believed to play a critical role in the pathogenesis of ASD [4]. Major environmental factors include maternal exposure to dietary, xenobiotic chemicals, and endocrine factors during pregnancy [5]. Many endocrine factors, metabolic intermediates derived from diet or xenobiotics can serve as ligands for nuclear receptors, a family of transcription factors. Nuclear receptors recruit nuclear receptor coactivators (NCOAs) or nuclear receptor repressors (NCORs) in a ligand-dependent manner, which further recruit epigenome-modifying enzymes such as histone acetyltransferases (HATs) and histone deacetylases (HDACs) [6]. Thus, nuclear receptor co-regulator complexes bridge the environmental risk factors of ASD with epigenetic remodeling, which offers a clue for general ASD etiology.
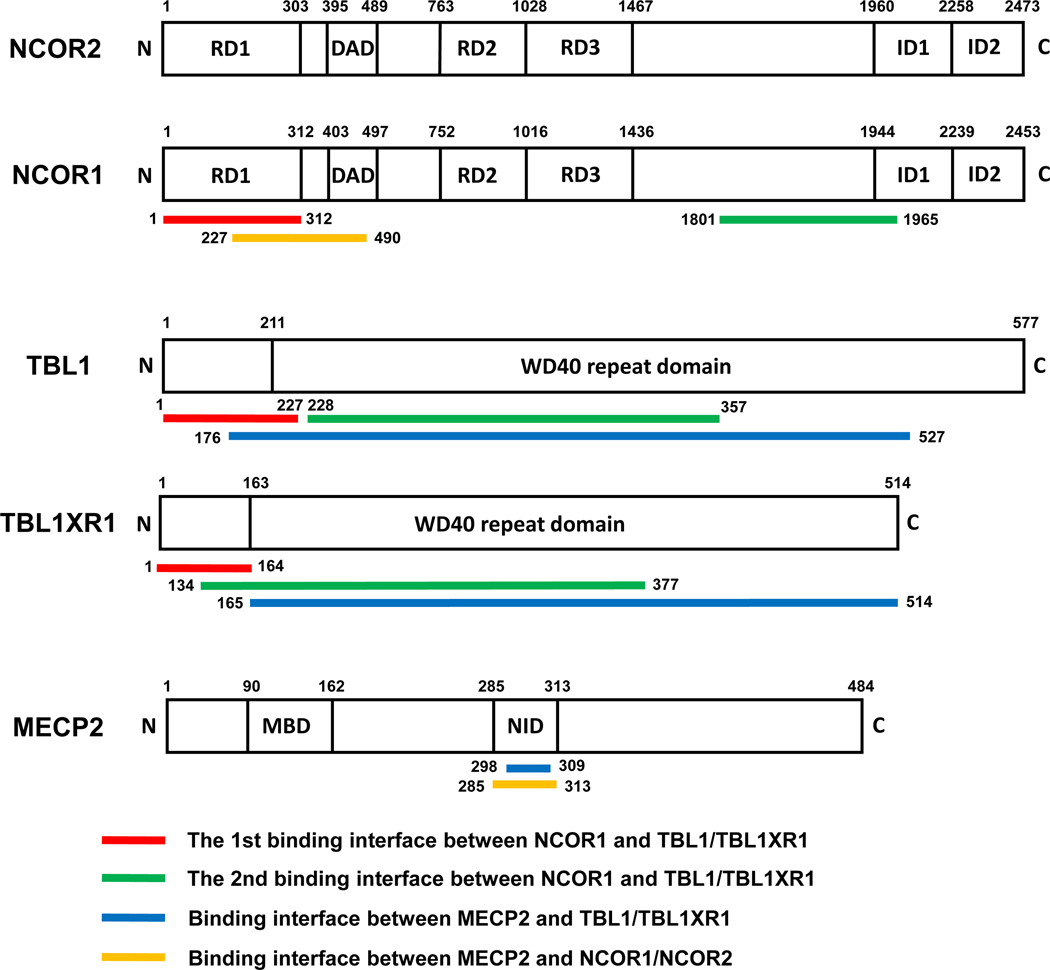
NCOR1 and its homolog NCOR2 (also known as silencing mediator for retinoid and thyroid hormone receptors, SMRT) are scaffolds for the multi-protein corepressor complex known as the NCOR complex [7]. Three repression domains (RD1, RD2, and RD3) at the N terminus can interact with epigenome-modifying enzymes and chromatin remodelers, while two interaction domains (ID1 and ID2) at the C terminus can interact with different nuclear receptors including thyroid hormone receptor (TR), estrogen receptor (ER), androgen receptor (AR), progesterone receptor (PR), glucocorticoid receptor (GR), retinoid X receptor (RXR) and orphan receptor Rev-erb [8] (Figure 1). HDAC3, a class I HDAC, is the major HDAC that confers the deacetylase enzyme activity to the NCOR complex. The enzyme activity of HDAC3 is dependent on binding to the deacetylase activating domain (DAD) on the N-terminus of NCOR1/2 [9] (Figure 1). Such binding causes a conformational change in HDAC3 protein, which opens its catalytic channel for substrate access [10]. Purified HDAC3 protein shows minimal enzyme activity in the absence of DAD [11]. In addition to DAD, a region near the C-terminus of NCOR1/2 can also interact with HDAC3 without affecting HDAC3 enzymatic activity [12]. Class IIa HDACs (HDAC4, HDAC5, HDAC7, and HDAC9) are also associated with the NCOR complex, but they have no intrinsic deacetylase enzymatic activity. As a result, their enzymatic activities are dependent on HDAC3[13]. The HDACs are implicated in neuronal functions in brain development, axon regeneration, memory and cognition, and neurodegeneration [14–18].
Figure 1. Illustration of protein-protein interactions between NCOR, TBL1XR1, and MECP2.
RD, repression domain; DAD, deacetylase activating domain; ID, interaction domain; MBD, methyl-CpG–binding domain; NID, NCOR/SMRT interaction domain. The amino acid numbers are based on human proteins.
NCORs in intellectual disability
The NCOR complex plays a vital role in neurocognition with implications in autism [19]. NCOR1−/− and NCOR2−/− mice are embryonic lethal, preventing the characterization of neurobehavioral phenotype [20, 21]. In a whole-body knock-in mouse model with mutations in the NCOR1/2 DADs that disrupt their binding to HDAC3 (NS-DADm mice), HDAC3 enzymatic activity is abolished in the brain [19]. These mice display social interaction deficits, impaired spatial learning and recognition memory, reduced anxiety, and enhanced locomotor coordination (Table 1). Gene expression profiling reveals altered expression of genes encoding GABAA receptor subunits, along with several ASD-related genes such as SH3 and multiple ankyrin repeat domains protein 3 (Shank3), Forkhead box protein P2 (Foxp2), and myocyte-specific enhancer factor 2c (Mef2c). Depletion of NCOR1/2 specifically in GABAergic neurons with Vgat-Cre (NS-V mice) is sufficient to cause learning and memory deficits [19]. The electrophysiological analysis identifies the hyperexcitability of GABAergic neurons in the lateral hypothalamus (LH) in NS-V mice. This hypothalamic excitatory/inhibitory (E/I) imbalance impairs synaptic plasticity in the hippocampus CA3 region through a monosynaptic LHGABA-to-CA3 neural projection, which accounts for the neurocognitive dysfunction in NS-V mice [19]. Injection of siRNA targeting Ncor1 into the amygdala of rats at 12 h or 28 h after birth reveals that Ncor1 knockdown increases juvenile social play behaviors in males and anxiety-like behaviors in both males and females. However, social interaction in the three-chamber assay is normal in these rats [22]. HDAC3 depletion specifically in mice hippocampal CA1 region does not impair memory but enhances long-term memory formation [23]. These results are in line with that the loss-of-function (LOF) for NCORs and HDAC3 in the hypothalamus account for cognitive dysfunction in NS-DADm and NS-V mice [19].
Table 1:
Genetic mouse models of the NCOR complex and related genes.
| Brain region/cell type targeted | Target gene | Mouse model | Life span | Body weight | Brain weight | Behavioral phenotype | Ref. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Breath regularity | Motility | Hindlimb clasping | Motor coord. | Anxiety | Social Interact. | Learning/memory | E/I balance | Hippo LTP | |||||||
| Whole body | Mecp2 | Mecp2 −/− or Mecp2 -/y (Mecp2-KO) | ↓ | ↓/← | ↓ | ↓ | ↓ | ↑ | ↓ | ↓ | ↑/↓ | ↓ | CA1↑ CA3↑ Bs↑Ctx↓ | – | [44–51] |
| Mecp2R306C/y | ↓ | – | ↓ | – | ↓ | ↑ | ↓ | – | – | – | – | – | [29] | ||
| Mecp2T308A/y | – | ← | ↓ | – | – | ↑ | ↓ | – | – | – | – | – | [28] | ||
| Ncor1/2 | Ncor1Y478A/Y478A; Ncor2Y470A/Y470A (NS-DADm) | ← * | ← | ← | ← | ↑ | ← | ↑ | ↓ | ↓ | ↓ | – | – | [19] | |
| Shank3 | Shank3−/− | – | ← | – | – | ←/↓ | – | ↓/← | ←/↑ | ↓ | ↓/← | CA1↑/← | ↓ | [132–136] | |
| Mef2c | Mef2c+/− | ↓ | ↓ | – | – | ← | ↑ | – | ↑ | ↓ | ↓ | Hippo↑ | – | [165] | |
| Foxp2 | Foxp2−/− | ↓ | ↓ | – | – | – | – | ↓ | – | ↓ | – | – | – | [152, 153] | |
| Foxp2+/− | ← | ←/↓ | – | – | – | - | ← | – | ↓ | ← | – | – | [152, 153] | ||
| GABA neuron | Mecp2 | Vgat-Cre; Mecp2loxP/y | ↓ | ↓ | – | ↓ | ↓ | ↑ | ↓ | ← | ↑ | ↓ | Ctx PN↑ Str GN↑ | ↓ | [52] |
| Ncor1/2 | Vgat-Cre; Ncor1loxP/loxP; Ncor2loxP/loxP (NS-V) | – | – | – | – | ← | – | – | ← | ↓ | ↓ | Hypo↑ CA3↓ | ↓ | [19] | |
| Shank3 | Vgat-Cre;Shank3loxP/loxP | – | – | – | – | ↓ | – | – | ↑ | ←/↓ | – | DL Str Neuron↓ | – | [140] | |
| Forebrain neuron | Mecp2 | Camk2-Cre;Mecp2loxP/loxP | ↓ | ↑ | ↓ | ↓ | ↓/← | ← | ↓ | ↑ | ↑ | ↓ | – | – | [53, 54] |
| Hdac3 | Camk2-Cre;Hdac3loxP/loxP | – | – | – | – | ↑ | ↑ | ↓ | – | ↓ | ↓ | – | – | [43] | |
| Mef2c | Camk2-Cre;Mef2cloxP/loxP | – | – | – | – | ↑ | ↑ | ↓ | ← | ← | ← | ← | ← | [169] | |
| Neural progenitor | Mef2c | Nestin-Cre;Mef2cloxP/- | ↓ | ↓ | ↓ | – | ← | ↑ | - | ↑ | – | ↓ | Ctx↓ | – | [166] |
| hGFAP-Cre;Mef2cloxP/- | – | ↓ | ← | – | ← | ↑ | ↓ | ← | – | ↓ | – | – | [167] | ||
| Ctx and Hippo excitatory neuron | Mef2c | Emx1-Cre;Mef2cloxP/loxP | – | – | – | – | ↑ | – | ← | – | ↓ | ↓ | Ctx↓ | – | [168] |
| Foxp2 | Nex-Cre;Foxp2lop/loxP | – | ← | – | – | – | – | – | ← | ↓ | – | – | – | [154] | |
| Hippo | Hdac3 | AAV-Cre into CA1 of Hdac3loxP/loxP | – | – | – | – | – | – | – | – | – | ↑ | – | – | [23] |
| AAV-Cre into CA3 of Hdac3loxP/loxP | – | – | – | – | – | – | – | – | – | ← | – | – | [19] | ||
| Mecp2 | AAV-shRNA at adult dorsal hippocampus | – | – | – | – | ← | – | – | ← | – | ↓ | – | – | [55] | |
| Hypo | Hdac3 | AAV-Cre into Hdac3loxP/loxP | – | – | – | – | – | – | – | – | – | ↓ | – | – | [19] |
| Mecp2 | Sim1-Cre; Mecp2loxP/y | ← | ↑ | – | – | ← | ↑ | ← | ↑ | ← | ← | – | – | [56] | |
no change
decrease
increase
not determined; Bs, brainstem; Ctx, Cortex; Hippo, hippocampus; Hypo, hypothalamus; Str, striatum; PN, pyramidal neuron; DL, dorsolateral.
survive for at least 1 year.
In addition to mouse models, several de novo genetic variants in NCOR1, NCOR2, and HDAC3 are found in pediatric patients with intellectual disabilities or ASD (Table 2). A heterozygous 152 kb deletion affecting the NCOR1 gene is found in a 10-year-old boy with ASD [19]. He shows no language expression, intellectual disability, poor motor coordination, stereotyped hand, and head movements, and auto-aggressive attitude. Another 8-year-old boy with ASD harbors a de novo NCOR1 heterozygous variant at canonical splice donor site (c.2182+ 2T>G) [19]. He displays global developmental delay, learning difficulties, epilepsy, absences and drop attacks, tip-toe walking, and double incontinence. Interestingly, an adjacent variant (c.2182 + 1G > T) affects the same splice site is identified in a 3-year-old girl with ASD [24]. The girl displays ASD with stereotypical and compulsive behaviors, narrowed interests, and communication defects such as poor eye contact. She also has mild developmental delay, thoracic scoliosis, slightly tented mouth, and bifid uvula. Both mutations are predicted to cause exon skipping and reading frameshift, which would produce a truncated NCOR1 protein or reduced protein production from the allele due to nonsense-mediated mRNA decay. If a truncated protein is produced, it is predicted to lack the RD3 and all ID domains on the C-terminus. Another study identifies de novo missense [c.3122C>T (P1025L)] and splice-acceptor (c.3449–1G>C) NCOR1 variants from two ASD probands, respectively [25]. In addition to NCOR1, two other pediatric patients with learning difficulties carry de novo variants in NCOR2 [c.6887G> A (R2296Q)] and HDAC3 [c.797T> C (L266S)] respectively [19]. A heterozygous NCOR2 missense variant [c.1940C>T (S647L)] is found in atypical RTT patients with no MECP2 mutation [26]. A patient with RTT-like microcephaly and severe intelligence disability carries compound heterozygous variants [c.3983A>G (E1328G)] and [c.1399G>A (V467I)] of NCOR2 [27].
Table 2.
NCOR complex genetic variants in human
| Gene | Mutation types | Changes of nucleotides or amino acids sequence | Phenotypes | Ref. | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ASD | Intellectual deficiency | Brain malformation | Learning disability | Development delay | Hearing loss | West syndrome | Pierpont syndrome | ||||
| TBL1XR1 | Deletion | 3q26.31q26.32 (175,507,453–177,095,072) | √ | [100] | |||||||
| 3q26.32 (176,025,379–177,377,006) | √ | √ | √ | [101] | |||||||
| Micro-deletion | 3q26.32 (176,221,801−176,929,584 ) | √ | √ | [102] | |||||||
| Micro-duplication | 3q26.32 (176,648,502–176,957,675) | √ | √ | √ | √ | √ | [103] | ||||
| 3q26.32 (176,627,832–177,149,304) | √ | √ | √ | [103] | |||||||
| Missense | G70D | √ | √ | √ | √ | [104] | |||||
| Y446C | √ | √ | √ | √ | [106, 107] | ||||||
| L282P | √ | [30] | |||||||||
| H441R | √ | [109] | |||||||||
| D370Y | √ | [109] | |||||||||
| D328G | √ | [109] | |||||||||
| P444R | √ | √ | [109] | ||||||||
| H213Q | √ | √ | [109] | ||||||||
| C325Y | √ | √ | √ | √ | √ | [105] | |||||
| Y446H | √ | √ | √ | √ | [105] | ||||||
| Y446S | √ | √ | [108] | ||||||||
| Frameshift | I269YfsTer8 | √ | √ | [109] | |||||||
| I397SfsX19 | √ | [30] | |||||||||
| NCOR1 | Splicing site | c.2182+2T>G | √ | √ | √ | [19] | |||||
| c.2182 +1G > T | √ | √ | √ | [24] | |||||||
| c.3449–1G>C | √ | [25] | |||||||||
| Deletion | 17:15869204–16021258 | √ | √ | [19] | |||||||
| Missense | P1025L | √ | [25] | ||||||||
| NCOR2 | Missense | R2296Q | √ | [19] | |||||||
| S647L | √ | [26] | |||||||||
| E1328G and V467I | √ | √ | √ | [27] | |||||||
| HDAC3 | Missense | L266S | √ | [19] | |||||||
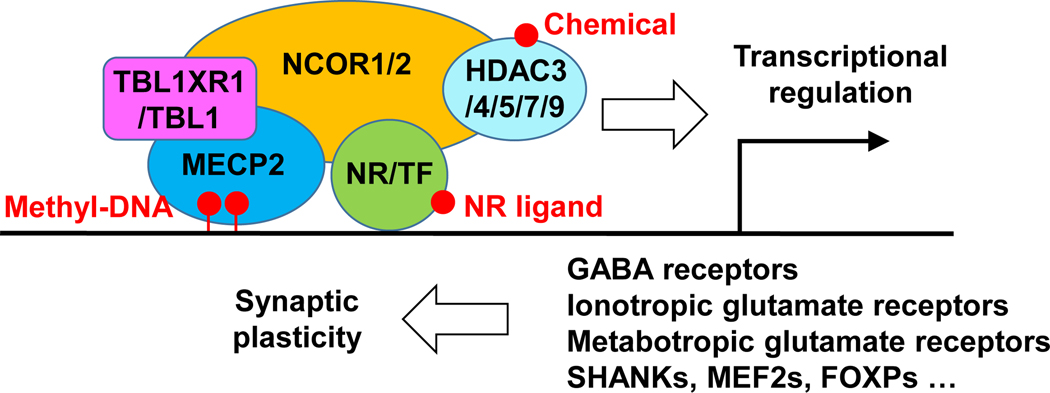
The NCOR complex is unique in many ways in the context of ASD (Figure 2). (1) The NCOR complex is directly involved in the action of steroid hormones that are critical in maintaining in utero environment during the prenatal neurodevelopment. (2) It directly interacts with multiple canonical epigenomic factors closely related to autism and, therefore, provides a converging point for different epigenomic factors to interplay. The NCOR-binding region of MECP2 is a hotspot for disease-causing mutations [28, 29]. TBL1XR1 (also known as TBLR1), a stable component of the NCOR complex, is among the top candidate genes associated with ASD identified by multiple studies [30, 31]. (3) The NCOR complex is druggable. Unlike most transcription factors, nuclear hormone receptors can directly interact with diverse small-molecule ligands and metabolites, which provides many opportunities for drug targeting [6, 32, 33]. Besides, HDACs are the crucial components of the NCOR complex and rely on NCORs for their deacetylase enzymatic activity [11]. Many small-molecule HDAC inhibitors are being tested for treating a variety of diseases, including ASD [34–36]. Below we discuss the relevance of NCORs-mediated cognitive regulation in the context of different ASD etiologies.
Figure 2. A working model for NCOR functions in the brain.
The NCOR complex is recruited to the genome either by DNA-binding nuclear receptors (NRs), other transcription factors (TFs), or MECP2 that recognizes methylated DNA regions. The NCOR complex is sensitive to hormones, metabolites, xenobiotics, and chemicals that can function as NR ligands or HDAC inhibitors. The NCOR complex regulates the transcription of multiple genes involved in neurotransmission and synaptic plasticity.
NCORs in MECP2-mediated cognitive regulation
RTT is traditionally an X-linked syndromic ASD caused by mutations in MECP2 [37]. MECP2 mutations are also found in non-RTT autistic patients. MECP2 can recruit the NCOR complex to repress gene expression [38]. Accumulating evidence suggests that disruption of the NCOR complex contributes to ASD pathogenesis caused by some MECP2 LOF mutations. (1) Trichostatin A (TSA), an HDAC inhibitor, inhibits transcription repression by MECP2 in a reporter gene assay in mouse fibroblasts expressing GAL4 DNA-binding domain fused with MECP2 amino acids 207–492 [39]. This finding indicates that the HDAC enzyme activity is required for MECP2-mediated transcriptional repression. (2) A subset of RTT-causing MECP2 LOF mutations disrupt the MECP2-NCOR interaction. A radically truncated MECP2 protein that only retains the methyl-CpG binding domain (MBD) and NCOR/SMRT interaction domain (NID), when expressed in the brain of Mecp2 knockout (KO) mice, is able to extend survival and improve behavioral abnormalities in tremor, gait, and activity [40]. This finding suggests that NID and MBD are critical regions for the function of MECP2. Missense mutations within NID of MECP2, including P302R, K304E, K305R, and R306C, disrupt the MECP2-NCOR interaction [29], leading to reduced transcriptional repression function of MECP2 (Figure 1). MECP2R306C accounts for 5% of classical RTT cases. Mecp2R306C knock-in mice demonstrate hindlimb clasping, compromised mobility, and impaired motor coordination [29]. Phosphorylation of MECP2 at T308 impairs the MECP2-NCOR interaction and transcriptional repression ability of MECP2 [28]. Mecp2T308A disrupts activity-dependent MeCP2 phosphorylation in neurons, and its knock-in mice have RTT-like phenotypes such as lower brain weight, seizures, hindlimb clasping, and motor abnormalities [28]. These findings suggest that the MECP2-NCOR interaction is delicately regulated since too much (Mecp2T308A) or too little interaction (Mecp2R306C) can lead to RTT-like phenotypes. In addition to RTT, MECP2 variants identified in idiopathic autistic disorder patients can also alter the MECP2-NCOR interaction. MECP2 R294X, a nonsense de novo mutation previously found in RTT, also exists in non-RTT autistic patients and leads to a truncated MECP2 protein lacking the NCOR-binding regions (Figure 1) [41]. (3) MECP2 depletion disrupts HDAC3 or NCOR chromatin binding. Chromatin immunoprecipitation (ChIP) assays reveals that HDAC3 binding at transcription start sites (TSS) of several lipogenic genes is decreased in the MECP2-depleted mouse liver compared to the control [42]. The elevated histone acetylation on H3K27 is also consistent with suppressed HDAC3 binding and activity. ChIP-seq demonstrates decreased HDAC3 binding at promoters of Arrdc2, Dusp4, Klf10, Tle1, Bdnf, and Nr4a1 in the hippocampus at postnatal day 45 in Mecp2-KO mice [43]. MECP2 enrichment at these promoter regions remains unchanged in hippocampal CA1 neurons in 3-month-old mice with HDAC3 depleted in forebrain excitatory neurons [43]. MECP2-dependent HDAC3 genomic recruitment in the hippocampus is abolished by R306C mutation [43], indicating that the MECP2-NCORs interaction is critical for HDAC3 genomic recruitment at these genes. (4) There are similarities in behavioral phenotype, gene expression changes, and neuronal activity changes between mouse models for MECP2 LOF and NCOR/HDAC3 LOF (Table 1). It is described in more detail below.
Whole-body Mecp2-KO mice [44–46] and mice with whole-body NCOR1/2 DAD mutation knock-in (NS-DADm mice) [19] exhibit similar deficits in anxiety, social interaction, and explicit memory. There are also some phenotypic differences. Unlike Mecp2-KO mice, NS-DADm mice have normal brain weight, hindlimb activity, and breathing patterns (Table 1). They can also survive for over a year. However, NS-DADm mice display hyperactivity, superior locomotor coordination, and better motor learning, while Mecp2-KO mice demonstrate defects in locomotor coordination [45, 47–51]. GABA neurons play an important role in the behavioral phenotype caused by both MECP2 LOF and NCOR1/2 LOF. Vgat (also known as Viaat) promoter can restrict Cre recombinase expression within GABAergic neurons. Vgat-Cre; Mecp2loxP/y mice display normal anxiety levels but impaired spatial memory [52]. This is similar to NS-V (Vgat-Cre;Ncor1loxP/loxP; Ncor2loxP/loxP) mice [19]. However, Vgat-Cre; Mecp2loxP/y mice have increased social interest, which is opposite to NS-V mice.
In addition to the neuron type, different brain regions also play different roles in MECP2 and NCOR/HDAC3 LOF. HDAC3 depletion in neural progenitor cells using the Nestin-Cre mouse line causes lethality within 16 hours after birth [14], while MECP2 depletion using the Nestin-Cre can survive for ten weeks [53]. Depletion of HDAC3 or MECP2 in forebrain neurons using the Camk2-Cre line causes similar phenotypes, including hindlimb clasping and deficits in motor coordination, social interactions, and cognition [14, 43, 53, 54]. Stereotaxic injection of adenovirus or adeno-associated virus (AAV) expressing Cre in Hdac3loxP/loxP mice leads to region-specific depletion of HDAC3 in the brain. Depletion of HDAC3 in the hippocampal CA1 region enhances long-term memory formation as measured by location-dependent object recognition memory [23]. In contrast, HDAC3 depletion in the hippocampal CA3 region does not influence learning or memory in novel object recognition or Morris water maze tests [19] (Table 1). Stereotaxic injection of AAV delivering shRNA targeting Mecp2 into the dorsal hippocampus leads to impaired long-term object location memory but normal short-term memory [55]. Hippocampus-specific knockdown of Mecp2 also impairs cued fear conditioning but not contextual fear conditioning [55]. Besides hippocampus, depletion of MECP2 in the hypothalamus in Sim1-Cre; Mecp2loxP/y mice demonstrates increased anxiety in open field test and increased aggression. Learning and memory is normal in these mice as measured by conditioned fear [56]. By comparison, HDAC3 depletion in the lateral hypothalamus through AAV-Cre leads to robust memory deficits [19]. These studies suggest that the phenotypic outcome of the altered MECP2-NCORs interaction is modulated by brain region-specific factors.
Gene expression changes show some similarities between MECP2 and NCOR LOF mouse models. Expression of γ-aminobutyric acid receptor subunit α−2 (Gabra2) and neuromedin B receptor (Nmbr), genes related to synapses and neurotransmission, is suppressed in the NS-DADm, NS-V, and Mecp2-KO hypothalamus compared to their respective control [19, 57, 58]. However, brain-derived neurotrophic factor (Bdnf), glutamate decarboxylase 1 (Gad1) and glutamate decarboxylase 2 (Gad2) are altered in the Mecp2-KO hypothalamus but keep unchanged in the NS-DADm hypothalamus. ChIP-qPCR shows that both MECP2 and NCOR1 bind to the promoter region of Mef2c in the hypothalamus. However, Mef2c expression is downregulated in the NS-DADm hypothalamus and upregulated in the Mecp2-KO hypothalamus [19, 57, 58]. The expression of genes related to synapses and neurotransmission (such as Chrna4), synapses and dendrites (such as Tanc1), cognition (such as Adcy1, Egr1, and Foxp2) and neurogenesis (such as Cit, Gdpd5, Mef2c, and Ntng1) are downregulated in Mecp2-KO hippocampus and NS-DADm hypothalamus as compared to their respective wild-type control [19, 59]. Forebrain-specific HDAC3 depletion in Camk2-Cre; Hdac3loxP/loxP mice leads to downregulation of hippocampal genes that are also downregulated in Mecp2-KO mice (37 overlapping genes) or hESC-derived MECP2-KO neurons (81 overlapping genes) [43].
Dysfunction of the GABA signaling is implicated in ASD. Dysregulation of GABAA receptor subunits is found in the cortex, hippocampus, and cerebellum of post-mortem brain tissues from ASD patients [60], which suggests reduced GABAergic signaling and the increased ratio between excitation and inhibition in ASD. Reduced GABA level is also found in the frontal lobe and perisylvian region of the left hemisphere by proton magnetic resonance spectroscopy in ASD patients [61]. Reduced expression of GABAA receptors in the frontal cortex and limbic areas is detected by single-photon emission computed tomography and positron emission tomography in the superior and medial frontal cortex, frontotemporal cortex, and limbic areas[60, 62–64]. Knockout of the ASD risk gene Engrailed-2 results in autism-like behaviors and reduced GABAergic neurons [65]. GABAergic system is also disturbed in fragile X syndrome and RTT mouse models and RTT patients. Pharmacological intervention on GABAergic system deficits could ameliorate fragile X syndrome and RTT-like phenotypes in Drosophila and mice models [66].
Electrophysiological changes and excitatory/inhibitory (E/I) imbalance are observed in MECP2 and NCORs LOF mouse models (Table 1). Multiple GABAA receptor subunits are downregulated in NS-DADm mice [19]. This downregulation plays a causative role in the behavioral phenotype because diazepam, a positive modulator of the GABAA receptor, rescues the behavior abnormalities of NS-DADm mice. The downregulation of GABAA receptor subunits leads to hyperexcitability in lateral hypothalamus GABA neurons. Such an E/I imbalance inhibits long-term potentiation (LTP) formation in the hippocampus through a monosynaptic LHGABA > CA3 projection [19]. In RTT mouse models, Mecp2-KO and Mecp2A140V mutation cause E/I imbalance and hyperactivity of hippocampal CA3 or CA1 pyramidal neurons [67, 68]. Increased E/I ratio is also observed in the brainstem of Mecp2-KO mice [69]. In contrast, reduction in miniature excitatory postsynaptic currents (mEPSC) amplitudes in the Mecp2-KO cortex leads to a decreased E/I ratio [70]. In Vgat-Cre; Mecp2loxP/y mice, the amplitude of miniature inhibitory postsynaptic currents (mIPSCs) of pyramidal neurons and striatal GABAergic neuron is dramatically reduced with no alterations in frequency [52]. In summary, some similarities exist in electrophysiological characters between MECP2 LOF and NCORs LOF mouse models. Disrupted E/I balance and altered GABA receptors are observed in some brain regions and neuron types in both cases.
In addition to GABA receptors, the expression of NMDA receptor Grin2c and glutamate receptor mGluR4 (Grm4) is also decreased in the hypothalamus of NCOR LOF NS-DADm mice. The frameshift mutation in GRIN2C has been identified in ASD cases [71]. Whole-body Grin2c knockout in mice leads to cognitive deficits [72]. Grm4 expression is decreased in the caudate putamen and nucleus accumbens of an ASD mouse model with mu opioid receptor deficiency (Oprm1−/−) [73]. The mGluR4 allosteric modulator VU0155041 alleviates behavioral changes of Oprm1−/− mice in social interaction, anxiety, stereotyped grooming and circling, and marble burying. However, mGluR4 knockout mice demonstrate improved short-term and long-term memory and long-term potentiation (LTP) of hippocampal CA1 [74]. MECP2 LOF also leads to dysregulation of glutamate receptors in the brain. Microarray analysis shows that the glutamate system-related genes such as Grin2d, Grin3a, Grik2, Grik5, and Grm2 are upregulated in the hypothalamus of Mecp2-transgenic (Tg) mice (a MECP2 gain-of-function model) and downregulated in Mecp2-KO mice as compared to the WT control [57]. The results of NMDA receptors Grin2d and mGluR2 (Grm2) are further confirmed by RNA-seq [58]. In the hippocampi of Mecp2-KO mice, the expression of glutamate receptors including Grin2d, Grin3a, Grm2, and Grm3 is significantly decreased [59]. Taken together, dysfunction of glutamate system may also contribute to the phenotypes in mouse models of NCOR LOF and MECP2 LOF.
Both NCORs and MECP2 are traditionally considered as transcriptional repressors. However, nonbiased transcriptome profiling has identified prominent gene downregulation in both NCORs LOF and MECP2 LOF conditions. While 225 genes are downregulated in the hypothalamus of NS-DADm mice compared to the control, only 50 are upregulated [19]. The downregulated genes are highly enriched in neurotransmission or synapse function, while the upregulated genes are enriched in seemingly unrelated biological processes such as chondrocyte differentiation or insulin secretion. ChIP analyses confirmed the binding of HDAC3 on downregulated genes such as Gabra2, Gabra4, and Gabrd, suggesting that the NCOR/HDAC3 complex may directly activate these genes, although an indirect or compensatory mechanism cannot be ruled out at this point [19]. These results are consistent with other studies about HDAC3 and MECP2 [43, 57, 75, 76]. Among the 300 genes differentially expressed in the hippocampus CA1 region of Camk2-Cre; Hdac3loxP/loxP mice compared to WT control, around 200 are downregulated while only 100 are upregulated. Gene ontology analysis reveals that downregulated genes are associated with brain functions including nervous system development, synaptic transmission, and neuron projection, which are not observed in upregulated genes [43]. For MECP2, over 2,000 genes are downregulated in the hypothalamus of Mecp2-KO mice or upregulated in Mecp2-Tg mice, as compared to their respective WT control mice [57]. In contrast, less than 400 genes are upregulated in Mecp2-KO mice or downregulated in Mecp2-Tg mice in the hypothalamus. Compared to MECP2-repressed genes, MECP2-activated genes in the hypothalamus are more enriched in neuron-related biological processes such as neurotransmitter biosynthetic process, neurotransmitter transport, fear, and memory. Similar results are obtained in the cerebellum, with nearly 800 genes upregulated but only a little over 400 genes downregulated in Mecp2-Tg mice compared to WT, and more than 800 genes downregulated but less than 300 genes upregulated in Mecp2-KO mice [75]. Cerebellar MECP2-activated genes are enriched in neuropeptide hormone activity and excitatory ion channel activity, while MECP2-repressed genes are enriched in protein dimerization activity, protein domain specific binding, olfactory receptor activity, and phospholipase C activity. In addition to mouse models, MECP2 KO in hESC-derived neurons results in downregulation of genes that are more than 10 times over the number of genes that are upregulated [76]. In summary, more genes are activated by the NCOR complex or MECP2 in the brain, directly or indirectly, as compared to genes that are repressed, especially for those genes associated with neuronal functions.
In contrast to the canonical view on transcription repressors and activators, accumulating evidence suggests that transcription regulators are versatile and the repressor/activator function is highly dependent on the chromatin context [77–84]. The molecular mechanisms underlying the switch between repressor and activator functions are diverse and not completely understood. For example, NCOR/HDAC3 LOF can repress gene expression by reducing DNA-binding activities of FOXO3 through increasing the acetylation on FOXO3 [43]. Rpd3, the yeast ortholog of HDAC3, can function as a histone chaperone and prevent histone eviction from the nucleosome in a deacetylase-independent manner, leading to chromatin stabilization and gene repression [85]. Rpd3-mediated histone deacetylation also facilitates nucleosome displacement, which enhances transcription factors binding to the DNA and promotes gene activation in response to stress [86, 87].
In addition to MECP2 LOF as described above, MECP2 gain-of-function (GOF) also causes diseases. Excess MECP2 protein expression causes MECP2 duplication syndrome characterized by intellectual disability and impaired motor function [88]. Homozygous Mecp2 overexpression from the Mapt (Tau) locus (Tau-MeCP2) is lethal. Heterozygous Tau-MeCP2 mice display reduced body weight and irregular breathing without changes in anxiety, motor coordination, or lethality [89]. MECP2R306C, a mutant unable to bind NCOR, does not induce lethality and other abnormalities when overexpressed at the same level as wild-type MECP2 [89]. Meanwhile, overexpression of MECP2T158M or MECP2R133C, mutants with disrupted DNA binding activities and reduced MECP2 protein stability, causes hindlimb clasping but do not affect survival. This study suggests that interaction with NCORs seems more critical for MECP2 GOF toxicity compared to its DNA binding activity. However, overexpression of MECP2 in the NS-DADm mice lacking HDAC3 enzymatic activity causes similar phenotype as its overexpression on the wild-type mice [89], suggesting that the toxic effect of MECP2 GOF does not rely on HDAC3 enzymatic activities. NCORs may have HDAC3-independent functions in the context of MECP2 GOF.
NCORs and TBL1XR1 in neurodevelopmental disorders
TBL1XR1 has about 86% identity with Transducin β-Like 1 (TBL1) based on the amino acid sequence. Both TBL1 and TBL1XR1 have six WD-40 domains and are essential components of the NCOR complex [90]. They interact with NCORs through the N terminal region and C terminal WD-40 repeats[91, 92] (Figure 1). Conversely, NCORs interact with TBL1/TBL1XR1 through its N-terminal RD1 region and a C terminal region (amino acid 1801–1965 in NCOR1) [51]. GST pull-down assay shows that TBL1/TBL1XR1 interacts with core histones H2B and H4 [92]. Simultaneous knockdown of TBL1 and TBL1XR1 attenuates thyroid hormone receptor (TR)-mediated repression of a luciferase reporter gene in HeLa cells [58]. ChIP assay reveals that TBL1/TBL1XR1 deficiency leads to decreased NCORs chromatin binding on the promoter of deiodinase 1 (D1), a gene repressed by unliganded TR [93]. TBL1XR1 selectively mediates the exchange of NCOR for NCOA upon nuclear receptors (NRs) binding to the ligand, which is required for ligand-induced transcriptional activation [94]. In addition to the NCOR/NCOA switch, TBL1/TBL1XR1 also targets NCORs for ubiquitination and degradation after NRs ligand binding [94]. Therefore, TBL1/TBLXR1 regulates the chromatin occupancy of NCORs both positively and negatively, depending on the absence or presence of the ligand for the NRs that recruit the NCOR complex. Pull-down assay using biotin tagged NCOR interaction domain (NID) peptide of MECP2 demonstrates its efficient binding with NCOR1 fragment (residues 227–490), which is abolished by NID of MECP2 K305R mutation. Co-immunoprecipitation (co-IP) assay shows that NCORs use similar regions for interaction with TBL1/TBL1XR1 and MECP2. The correlation of pull-down efficiency of NCOR1 fragments with MECP2 and TBL1/TBL1XR1 suggests that MECP2 and NCORs interaction can be bridged by TBL1/TBLXR1 [90]. Consistent with this notion, MECP2 interacts with the WD40 domain of TBL1/TBL1XR1 directly through NID, and this interaction is abolished by RTT-causing mutations (K304E, K305R, R306C, and R209X) as shown by pull-down assay [90]. In summary, TBL1/TBL1XR1 are stable components of the NCOR complex and are essential for its transcriptional function. TBL1/TBL1XR1 are also involved in the physical interaction between NCORs and MECP2.
TBL1/TBL1XR1 expression and activity are tightly regulated by several signaling pathways. Toll-like receptor 2 (TLR2) activation recruits calcium/calmodulin-dependent protein kinase 2 γ (CaMK2γ) to the NCOR complex, leading to TBL1XR1 phosphorylation and NCOR clearance from the promoters of inflammatory genes [95]. SUMOylation of TBL1/TBLR1 upon Wnt activation leads to their dissociation from the NCOR complex and increased binding of β-catenin to the promoter of Wnt target genes to facilitate their transcription [96].
TBL1 deficiency was originally found in cases with sensorineural deafness related to ocular albinism [97]. In the nervous system, Drosophila TBL1 homolog ebi forms a complex with transcriptional factor AP-1 and SMRTER (Drosophila counterpart of NCOR1/2) and is required for photoreceptor neuron survival by regulating anti-apoptotic gene hid expression [98]. TBL1XR1 is critical for brain development, and its de novo mutations have been identified in neurological disorders (Table 2). Targeted sequencing of over 200 genes in >11,730 cases of ASD, intellectual disability, seizure, microcephaly, or macrocephaly and >2,867 controls identifies 13 cases carrying disruptive mutations in TBL1XR1 [31]. Targeted sequencing of 2,446 ASD probands suggests that TBL1XR1 is one of the six genes with recurrent disruptive mutations that collectively contribute to 1% of sporadic ASDs [30]. Another study did not find TBL1XR1 among ASD risk genes [99]. However, many individual cases suggest a causative role of TBL1XR1 mutations in ASD (Table 2). Deletion of 1.6Mb (175,507,453–177,095,072) in the 3q26.31q26.32 region or 1.3 Mb deletion in the 3q26.32 (176,025,379–177,377,006) region that includes TBL1XR1 gene causes intellectual deficiency [100, 101]. A girl with 708Kb microdeletion on chromosome 3q26.32 (176,221,801−176,929,584 ) encompassing only TBL1XR1 has an intellectual disability and brain malformation [102]. 309Kb and 521Kb microduplications of TBL1XR1 leads to developmental delay, intellectual disability, ASD, and hearing loss[103]. De novo missense TBL1XR1 mutation [c.209 G>A (G70D)] is identified in a Japanese girl with West syndrome and ASD features [104]. Whole-exome sequencing of Pierpont syndrome patients identifies Y446C, C325Y and Y446H variants in TBL1XR1 as potentially disease-causing [105–107]. TBL1XR1 variant Y446S is also detected in autosomal dominant mental retardation 41 [108]. A de novo missense mutation (L282P) and a frameshift mutation (I397SfsX19) are associated with ASD [30]. Six de novo mutations of TBL1XR1 (H441R, D370Y, D328G, P444R, H213Q, and I269YfsTer8) lead to global developmental delay in the Deciphering Developmental Disorders (DDD) study [109]. Among them, three variants (P444R, H213Q, and I269YfsTer8) cause ASD phenotypes. All six mutations are located within the first 3 WD-40 repeats (amino acids 200–500) of TBL1XR1, a region essential for its direct interaction with the NCOR complex (Figure 1). Therefore, TBL1XR1 and NCORs variants found in ASD patients likely cause neurological and behavioral abnormalities by abolishing their direct physical interactions. It has not been tested experimentally in mammals whether TBL1/TBL1XR1 mutations indeed cause neurological disorders and, if so, whether it is mediated by disrupting the function of NCOR complex.
Valproic acid, histone deacetylase inhibitor, and NCORs
Valproate is an antiepileptic drug. Population-based study reveals that prenatal exposure VPA increases the risk of ASD and attention-deficit/hyperactivity disorder (ADHD) [110–112]. Studies in rodent models and non-human primates show that VPA in utero treated animals show behavior and electrophysiological deficits analogous to ASD children, supporting the causative role of prenatal VPA in ASD [113–116]. Although only a fraction of ASD cases in human are related to prenatal exposure to VPA [117], the VPA animal model remains a valuable research tool to probe gene-environmental interactions in ASD etiology, especially in the context of epigenomic modifications. Exposure to VPA at the embryonic stage by single intraperitoneal injection at a dose of 500 mg/kg leads to autism-like behavioral phenotypes in rats and mice [118]. Social interactions deficits are identified by three-chambered social approach task and ultrasonic vocalizations. Evaluation of behaviors like self-grooming, marble-burying, digging, and the Y-maze test demonstrates increased repetitive behaviors [117, 118]. Maternal exposure to VPA in non-human primates also impairs social interaction and stereotypies phenotype in juvenile offspring, indicating an evolutionary conserved pathogenic mechanism for VPA-induced ASD [119].
VPA can act as an HDAC inhibitor (HDI) and is known to inhibit HDAC3 [120]. Prenatal exposure to VPA causes hyperacetylation of histone H3 and H4 of the embryonic brain [120]. Valpromide, a VPA analog lacking HDI activity, does not affect histones acetylation and mice behaviors, indicating that in utero HDAC inhibition contributes to ASD pathogenesis. Treatment with another HDI TSA in utero also elevates histone H3K9 and H4K8 acetylation in embryonic brain and similarly causes abnormal social and repetitive behaviors, as measured by ultrasonic vocalization, olfactory motivation, three-chamber sociability, grooming, and digging [121]. These studies suggest that HDAC inhibition may underlie the neurological abnormalities upon in utero VPA treatment.
It is conceivable that inhibition of HDAC3 could contribute to VPA-induced autism behaviors, considering the following findings. (1) There are similarities in behavioral phenotypes between NCOR/HDAC3 LOF mouse models and prenatal VPA treatment (Table 1). HDAC3 enzymatic activity is suppressed in all tested brain regions in NS-DADm mice, which mimics what VPA does [19]. In line with this, neuronal depletion of HDAC3 using Camk2-Cre in mice impairs locomotor coordination, sociability, and cognition [43]. (2) The E/I imbalance is detected in NS-V mice and VPA rodent models. NCORs depletion causes hyperexcitability of lateral hypothalamus (LH) GABA neurons, with a decreased amplitude of mIPSCs and increased E/I ratio. In the hippocampal CA3 region of the NS-V mice, the E/I ratio is decreased. Rats prenatally exposed to VPA show enhanced local recurrent connectivity and decreased parvalbumin (PV)-positive GABAergic inhibitory interneurons in the cortex [122, 123]. The frequency and amplitude of mEPSCs increase significantly in the lateral nucleus (LA) neurons from the amygdala in rats prenatally exposed to VPA [124]. (3) There are similarities in dysregulated gene expression between the VPA animal model and the NS-DADm mouse model. Shank3 and Mef2c are two ASD-related genes downregulated in NS-DADm mice in the hypothalamus [19]. Similarly, TSA treatment at E12.5 decreases Shank3 expression in cortical neurons [125]. Treated with VPA at E12.5 decreases protein level of MEF2C in the prefrontal cortex in rats [126].
In addition to acting as an HDI, other mechanisms can also contribute to VPA-induced autism-like phenotypes. For example, VPA can activate Wnt and Ras-ERK-p21 signaling, as well as elevate GABA levels [127]. The role of HDACs or HDI in autism is complex and not completely understood. It is unknown whether histone hyperacetylation exists during the juvenile or adult stage with embryonic exposure to VPA in most animal models. As a matter of fact, VPA intraperitoneal injection of cynomolgus monkeys (Macaca fascicularis) on gestational day 26 and 29 induces decreases, rather than increases, H3K27 acetylation in the prefrontal cortex of the offspring [119]. In VPA-induced autism-like rodents, HDIs such as pentyl-4-yn-VPA, SAHA, and MS-275 can ameliorate social cognition deficits [128, 129]. Pharmacological inhibition of class I HDAC or RNAi knockdown of Hdac2 rescues social deficits in Shank3-deficient mouse models [35]. Of note, HDIs are used on animals after birth as therapeutics in these studies, while VPA is used at the embryonic stage to induce ASD. It is possible that HDACs play opposite roles in neurocognition at different developmental stages. Future studies with accurate profiling of the expression and activities of HDACs in different developmental stages in the VPA animal model will help clarify the interrelationships among VPA, HDACs, and ASD.
SHANKs and NCORs
SH3 and multiple ankyrin repeat domains protein (SHANK) family have several members in mammals (SHANK1, SHANK2, and SHANK3) and are major scaffold proteins interacting with glutamate receptor complexes, anchoring proteins, and actin cytoskeleton at the excitatory synapse [130]. They play crucial roles in regulating synaptic development, function, and plasticity. Deletions or de novo single nucleotide variant (SNV) mutations can lead to SHANK haploinsufficiency and contribute to approximately 1% of all ASD cases (SHANK1 mutation 0.04%, SHANK2 mutation 0.17%, SHANK3 mutation 0.69%) [131]. Shank3 deficiency or knock-in mouse models demonstrate defective social interaction, repetitive behaviors, and impaired synapse function [130, 132–136] (Table. 1). Restoring Shank3 expression from adulthood in Shank3-knockout mice ameliorates autism-like phenotypes [137]. The autism-like morphological, behavioral and electrophysiological abnormalities in Shank3 deficient mice can also be rescued by manipulation of the mGluR5 activity or targeting actin regulators such as cofilin, Rac1, and PAK [138, 139]. Impaired novel object recognition and performance in Morris water maze are observed in two Shank3 mutant lines, exons 4–9B/ANK repeat deletion (Δex4–9B) and exons 4–9J/ANK repeat deletion (Δex4–9J) [132]. The electrophysiological analysis reveals that Shank3 deficiency leads to decreased mEPSC amplitude in the hippocampal CA1 region and striatum. SHANK3 depletion in GABAergic neurons leads to enhanced anxiety and repetitive behaviors, decreased courtship ultrasonic vocalizations, and increased direct social interactions while three-chamber social interaction is indistinguishable from the wild-type control [140]. E/I balance in dorsolateral striatal neurons is disrupted as the frequency and amplitude of mEPSC are decreased and mIPSCs keep unchanged[140]. Shank1 mutant (Δex14–15) mice demonstrate altered synaptic protein expression, decreased mEPSC frequency in the hippocampal CA1 region, and impaired social interaction [132]. Impaired memory and social interaction are also observed in Shank2 mutant (Δex6–7 of Shank2a) mice [141].
SHANK gene expression is regulated by epigenetic mechanisms including histone acetylation. Prenatal exposure to HDAC inhibitor TSA decreases Shank2 and Shank3 expression in cortical neurons [125]. The Shank3 expression is reduced in the hypothalamus of NS-DADm mice [19]. Conversely, SHANK3 can also modulate HDAC expression and activities. The expression of Hdac2 is upregulated in the prefrontal cortex of Shank3 deficient mice [35]. HDAC inhibitors MS-275 and romidepsin or RNAi-mediated knockdown of Hdac2 rescue social deficits in Shank3 deficient mice [35, 36]. Therefore, HDAC inhibition could be a therapeutic strategy for ASD induced by SHANK3 mutations. Interestingly, both SHANK3 GOF and LOF can activate HDAC activity, which seems to contribute to the neurotoxic effect. Shank3 overexpression transgenic mice demonstrate manic-like behavior, decreased sociability, and defective ultrasonic vocalization [142]. The manic-like behaviors can be rescued after VPA treatment. These studies suggest that the expression of SHANK3 should be fine-tuned to maintain proper HDAC activity for normal neurodevelopment. It is not clear whether and how SHANK1 and SHANK2 can modulate HDACs. It is conceivable that SHANKs may contribute to the NCOR/HDAC3 complex-mediated cognitive regulation and vice versa, considering the phenotypic similarities between some SHANK LOF mouse models and the NCOR/HDAC3 LOF mouse models in terms of impaired social interaction, cognition dysfunction, and electrophysiological abnormalities [19, 132, 137–139, 141].
FOXP2 and NCORs
Forkhead box protein P2 (FOXP2) is a transcription factor associated with inherited language or speech disorder [143]. FOXP2 genetic variants are associated with verbal, linguistic and grammatical difficulties [144–146]. Studies on Foxp2 KO (Foxp2−/−) mice demonstrate that FOXP2 controls synaptogenesis and vocalization [147]. FOXP2 is vital for the function of the mirror neuron system (MNS), a class of visuomotor neurons that fire in monkeys when they do object-directed actions or observe others performing similar behaviors [148]. Mirror neurons are crucial for action understanding, imitation, language, mentalization, social communication, empathy, and social cognition. MNS dysfunction is found in children with ASD [149]. Although there are some controversial reports, large-scale analysis using a novel approach of homozygous haplotype mapping of genetic heterogeneous ASD cases identifies FOXP2 as an ASD candidate gene [150]. FOXP2 SNP rs1456031 is found to be significantly associated with ASD in the Chinese population [151].
Whole-body knockout of Foxp2 in mice causes motor impairment, premature death, and impaired social communication as indicated by the absence of ultrasonic vocalizations [152] (Table 1). Foxp2−/− mice die at postnatal day 21 while Foxp2+/− mice are viable [152, 153]. Reduced body weight is observed in Foxp2 homozygous and heterozygous KO mice [152]. However, Foxp2+/− mice perform similarly as wild-type mice in the Morris water maze test [152]. Nex-Cre;Foxp2loxP/loxP mice with cortical Foxp2 deletion show behavior abnormalities in social interaction [154]. In addition to FOXP2, two additional FOXP family members including FOXP1 and FOXP4, are expressed in the brain, which can form heterodimers to regulate gene transcription [155]. FOXP1 variants are found in ASD patients [156]. Foxp1 and Foxp4 knockout in mice leads to embryonic lethality [155].
FOXP2 regulates the expression of many ASD-related genes, including CNTNAP2, MET, and SRPX2 [157–159]. In the VPA-induced ASD model, the number of FOXP2 positive neuron increases in the cerebral cortex layer V, but decreases in layer VI, suggesting that FOXP2 expression is regulated by HDACs and histone acetylation [160]. FOXP2 expression is decreased in the hypothalamus of the NS-DADm mice [19], suggesting that the NCOR complex is an upstream regulator of FOXP2. It is possible that FOXP2 LOF contributes to some of the behavioral phenotypes in the NCOR LOF mouse models.
MEF2C and NCORs
Four myocyte-specific enhancer factor 2 (MEF2) members (MEF2A-D) are expressed in various brain regions, including cortex, hippocampus, thalamus, and striatum [161]. MEF2s share highly conserved N-terminal DNA binding/dimerization domains and C-terminal transcription regulation regions. MEF2s can associate with class IIa HDACs or HATs to regulate neuronal gene expression in an activity-dependent manner. MEF2s are important for neuronal survival, differentiation, migration, axon guidance, dendrite formation, and synapse plasticity. MEF2A and MEF2D regulate the expression of ASD-related genes, including Ube3A, Slc9A6, Pcdh10, and C3orf58 [162]. Knockdown of Mef2a and Mef2d in cultured rat hippocampal neuron increased excitatory synapses number and mEPSC frequency [163]. Patients carrying 318 kb or 1.1 Mb deletions flanking MEF2C have severe mental retardation, ASD, and epilepsy [164]. Mef2c+/− mice demonstrate neurobehavioral deficits, E/I imbalance, neuronal death, and decreased neurogenesis due to haploinsufficiency effects [165]. MEF2C depletion in Nestin-Cre; Mef2cloxP/- mice leads to deficits in neuronal differentiation, decreased E/I ratio, altered anxiety, impaired memory and paw clasping [166] (Table 1). hGFAP-Cre; Mef2cloxP/- mice demonstrate efficient MEF2C depletion in neurons and glial cells derived from radial glial cells [167]. They are slightly smaller than wild-type and display hind/forelimb clasping behaviors, but with normal brain architecture, locomotion, and anxiety. Contextual dependent learning is impaired while cue-dependent learning is normal in these mice, as measured in the fear conditioning paradigm. The frequency of mEPSCs in the dentate gyrus granule neurons is increased while the amplitude keeps unchanged. MEF2C depletion in differentiated forebrain excitatory neurons of Emx1-Cre; Mef2loxP/loxP mice leads to E/I imbalance and downregulation of several ASD-related genes such as Ntng1, Nlgn1, Nrxn1, Nrxn3, Pcdh19, Shank2, Shank3, Pten, and Htr1b. They demonstrate autism-like behaviors including fewer ultrasonic vocalizations, impaired social interaction, increased repetitive jumping behavior and repetitive fine motor movements, and deficits in learning and memory [168]. Camk2-Cre; Mef2cloxP/loxP mice with Mef2c deletion in postnatal forebrain regions show increased spine numbers in the hippocampus, hyperactivity, abnormal motor coordination, hindlimb clasping [169]. However, there is no change in learning and memory, long-term potentiation in the hippocampus CA1 region induced by theta-burst stimulation, and social interactions.
The expression of MEF2C is potentially regulated by NCORs/HDAC complex. In the hypothalamus of NS-DADm mice with abolished HDAC3 enzymatic activity, the Mef2c expression level is decreased [19]. In line with this, MEF2C transcription is induced in rat striatum by cocaine treatment, which requires phosphorylation of HDAC5 by salt-inducible kinase SIK1 [170]. HDAC5 is class IIa HDAC and gains its enzyme activity by interacting with the NCOR complex and recruiting HDAC3 [13]. HDAC4, HDAC5, HDAC7, and HDAC9 stimulate SUMOylation of MEF2C and further inhibit its transcriptional activity [171]. In addition to being regulated by HDACs, MEF2C can also bind to HDAC4 and HDAC5 to suppress miR-9 expression, which is critical for neurogenesis, neuronal differentiation, and neurite elongation [172, 173]. In addition to MEF2C, MEF2D can be deacetylated by the NCOR/HDAC3 complex, which contributes to MEF2D-mediated repression of downstream target genes in HEK293 cells [174]. Thus, NCORs regulates MEF2 function at both transcriptional and post-translational levels.
This raises the possibility that MEF2s dysregulation may contribute to the cognitive dysfunction in the NCOR/HDAC3 LOF scenario. In line with this thought, NS-DADm and NS-V showed observed E/I imbalance, reduced anxiety, hyperactivity, deficits in cognition and social interactions, which resembled multiple Mef2c LOF mouse models including Mef2c+/−, Nestin-Cre;Mef2cloxP/-, hGFAP-Cre;Mef2cloxP/-, Emx1-Cre;Mef2cloxPloxP, and Camk2-Cre;Mef2cloxP/loxP mice [19, 164–168] (Table 1). However, hindlimb clasping, motor coordination is normal in NS-DADm mice. It is conceivable that MEF2C functionally interplays with other NCORs-target genes, which collectively contributes to NCOR-mediated cognitive regulation. It is also possible that MEF2A and MEF2D can compensate for MEF2C dysregulation in the context of NCORs LOF.
Conclusion
The critical role of the NCOR complex in intellectual disability and ASD starts to gain recognition recently. (1) The NCOR complex physically interacts with MECP2 and TBL1XR1, proteins closely related to ASD and intellectual disability from human genetics studies. Dysfunction of the NCOR complex may contribute to ASD pathogenesis in the presence of disease-associated MECP2 or TBL1XR1 genetic variants. (2) Multiple de novo genetic variants in NCOR1, NCOR2, and HDAC3 are associated with ASD and intellectual disability. (3) Studies on mouse models of NCORs and HDAC3 support a role of the NCOR complex in neurocognitive functions and ASD-like behaviors. (4) The NCOR-associated HDAC activity is sensitive to VPA, an environmental risk factor of ASD when exposed prenatally. NCOR-associated HDAC activity is also amenable for pharmaceutic manipulation by HDAC inhibitors, which show some therapeutic values against a variety of neurological diseases under certain conditions. (5) The NCOR complex is directly involved in the epigenetic effects of many hormones, metabolites, and xenobiotic chemical that can serve as ligands or antagonists for a variety of nuclear receptors, which are critical in maintaining an in utero environment during the prenatal neurodevelopment. Further investigations are warranted to dissect the role of the NCOR complex in neurocognition, which will not only provide mechanistic insights into the ASD etiologies but also lay intellectual foundations for treatment or prevention of many neurological diseases.
Acknowledgement
The authors are thankful for the following funds that support the laboratories, although none of them directly supports the work described in this review. The funds are NIH grant CA215591, DK111436, and ES027544 (Z.S.), American Diabetes Association grant 1–19-PDF-012 (W.Z.), National Natural Science Foundation of China 31200804 (Y.K.), and the Fundamental Research Funds for the Central Universities 2242019K40122 (Y.K.). We thank John S. Dunn Foundation and Mrs. Clifford Elder White Graham Endowed Research Fund. We are also thankful for the Cardiovascular Research Institute, Dan L Duncan Comprehensive Cancer Center (P30CA125123), Texas Medical Center Digestive Diseases Center (P30DK056338), and the SPORE program in lymphoma (P50 CA126752) at Baylor College of Medicine, and Gulf Coast Center for Precision Environmental Health (P30ES030285).
Footnotes
Declaration of Interests
The authors declare no actual or potential competing financial interests.
References
- 1.Lord C, Elsabbagh M, Baird G, Veenstra-Vanderweele J. Autism spectrum disorder. Lancet. 2018;392:508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai M-C, Lombardo MV, Baron-Cohen S. Autism. Lancet. 2014;383:896–910. [DOI] [PubMed] [Google Scholar]
- 3.Ayhan F, Konopka G. Genomics of autism spectrum disorder: approach to therapy. F1000Res. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Telese F, Gamliel A, Skowronska-Krawczyk D, Garcia-Bassets I, Rosenfeld MG. ‘Seq-ing’ insights into the epigenetics of neuronal gene regulation. Neuron. 2013;77:606–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forsberg SL, Ilieva M, Maria Michel T. Epigenetics and cerebral organoids: promising directions in autism spectrum disorders. Transl Psychiatry. 2018;8:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lonard DM, O’Malley BW. Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat Rev Endocrinol. 2012;8:598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emmett MJ, Lazar MA. Integrative regulation of physiology by histone deacetylase 3. Nat Rev Mol Cell Biol. 2019;20:102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu X, Lazar MA. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metab. 2000;11:6–10. [DOI] [PubMed] [Google Scholar]
- 9.You S-H, Lim H-W, Sun Z, Broache M, Won K-J, Lazar MA. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat Struct Mol Biol. 2013;20:182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Codina A, Love JD, Li Y, Lazar MA, Neuhaus D, Schwabe JWR. Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors. Proc Natl Acad Sci USA. 2005;102:6009–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oberoi J, Fairall L, Watson PJ, Yang J-C, Czimmerer Z, Kampmann T, et al. Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery. Nat Struct Mol Biol. 2011;18:177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bottomley MJ, Lo Surdo P, Di Giovine P, Cirillo A, Scarpelli R, Ferrigno F, et al. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J Biol Chem. 2008;283:26694–26704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norwood J, Franklin JM, Sharma D, D’Mello SR. Histone deacetylase 3 is necessary for proper brain development. J Biol Chem. 2014;289:34569–34582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hervera A, Zhou L, Palmisano I, McLachlan E, Kong G, Hutson TH, et al. PP4-dependent HDAC3 dephosphorylation discriminates between axonal regeneration and regenerative failure. EMBO J. 2019;38:e101032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwapis JL, Alaghband Y, López AJ, Long JM, Li X, Shu G, et al. HDAC3-Mediated Repression of the Nr4a Family Contributes to Age-Related Impairments in Long-Term Memory. J Neurosci. 2019;39:4999–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alaghband Y, Kwapis JL, López AJ, White AO, Aimiuwu OV, Al-Kachak A, et al. Distinct roles for the deacetylase domain of HDAC3 in the hippocampus and medial prefrontal cortex in the formation and extinction of memory. Neurobiol Learn Mem. 2017;145:94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penney J, Tsai L-H. Histone deacetylases in memory and cognition. Sci Signal. 2014;7:re12. [DOI] [PubMed] [Google Scholar]
- 19.Zhou W, He Y, Rehman AU, Kong Y, Hong S, Ding G, et al. Loss of function of NCOR1 and NCOR2 impairs memory through a novel GABAergic hypothalamus-CA3 projection. Nat Neurosci. 2019;22:205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jepsen K, Hermanson O, Onami TM, Gleiberman AS, Lunyak V, McEvilly RJ, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. [DOI] [PubMed] [Google Scholar]
- 21.Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim H-J, Glass CK, et al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. [DOI] [PubMed] [Google Scholar]
- 22.Jessen HM, Kolodkin MH, Bychowski ME, Auger CJ, Auger AP. The nuclear receptor corepressor has organizational effects within the developing amygdala on juvenile social play and anxiety-like behavior. Endocrinology. 2010;151:1212–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, et al. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31:764–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakaguchi Y, Uehara T, Suzuki H, Sakamoto Y, Fujiwara M, Kosaki K, et al. Haploinsufficiency of NCOR1 associated with autism spectrum disorder, scoliosis, and abnormal palatogenesis. Am J Med Genet A. 2018;176:2466–2469. [DOI] [PubMed] [Google Scholar]
- 25.Wang T, Guo H, Xiong B, Stessman HAF, Wu H, Coe BP, et al. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat Commun. 2016;7:13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sajan SA, Jhangiani SN, Muzny DM, Gibbs RA, Lupski JR, Glaze DG, et al. Enrichment of mutations in chromatin regulators in people with Rett syndrome lacking mutations in MECP2. Genet Med. 2017;19:13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwama K, Mizuguchi T, Takeshita E, Nakagawa E, Okazaki T, Nomura Y, et al. Genetic landscape of Rett syndrome-like phenotypes revealed by whole exome sequencing. J Med Genet. 2019;56:396–407. [DOI] [PubMed] [Google Scholar]
- 28.Ebert DH, Gabel HW, Robinson ND, Kastan NR, Hu LS, Cohen S, et al. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature. 2013;499:341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyst MJ, Ekiert R, Ebert DH, Merusi C, Nowak J, Selfridge J, et al. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat Neurosci. 2013;16:898–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stessman HAF, Xiong B, Coe BP, Wang T, Hoekzema K, Fenckova M, et al. Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental-disability biases. Nat Genet. 2017;49:515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. [DOI] [PubMed] [Google Scholar]
- 33.Mottis A, Mouchiroud L, Auwerx J. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes Dev. 2013;27:819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kratsman N, Getselter D, Elliott E. Sodium butyrate attenuates social behavior deficits and modifies the transcription of inhibitory/excitatory genes in the frontal cortex of an autism model. Neuropharmacology. 2016;102:136–145. [DOI] [PubMed] [Google Scholar]
- 35.Qin L, Ma K, Wang Z-J, Hu Z, Matas E, Wei J, et al. Social deficits in Shank3-deficient mouse models of autism are rescued by histone deacetylase (HDAC) inhibition. Nat Neurosci. 2018;21:564–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma K, Qin L, Matas E, Duffney LJ, Liu A, Yan Z. Histone deacetylase inhibitor MS-275 restores social and synaptic function in a Shank3-deficient mouse model of autism. Neuropsychopharmacology. 2018;43:1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. [DOI] [PubMed] [Google Scholar]
- 38.Lyst MJ, Bird A. Rett syndrome: a complex disorder with simple roots. Nat Rev Genet. 2015;16:261–275. [DOI] [PubMed] [Google Scholar]
- 39.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. [DOI] [PubMed] [Google Scholar]
- 40.Tillotson R, Selfridge J, Koerner MV, Gadalla KKE, Guy J, De Sousa D, et al. Radically truncated MeCP2 rescues Rett syndrome-like neurological defects. Nature. 2017;550:398–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carney RM, Wolpert CM, Ravan SA, Shahbazian M, Ashley-Koch A, Cuccaro ML, et al. Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatr Neurol. 2003;28:205–211. [DOI] [PubMed] [Google Scholar]
- 42.Kyle SM, Saha PK, Brown HM, Chan LC, Justice MJ. MeCP2 co-ordinates liver lipid metabolism with the NCoR1/HDAC3 corepressor complex. Hum Mol Genet. 2016;25:3029–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nott A, Cheng J, Gao F, Lin Y-T, Gjoneska E, Ko T, et al. Histone deacetylase 3 associates with MeCP2 to regulate FOXO and social behavior. Nat Neurosci. 2016;19:1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. [DOI] [PubMed] [Google Scholar]
- 45.Belichenko NP, Belichenko PV, Li HH, Mobley WC, Francke U. Comparative study of brain morphology in Mecp2 mutant mouse models of Rett syndrome. J Comp Neurol. 2008;508:184–195. [DOI] [PubMed] [Google Scholar]
- 46.Li W, Pozzo-Miller L. Beyond Widespread Mecp2 Deletions to Model Rett Syndrome: Conditional Spatio-Temporal Knockout, Single-Point Mutations and Transgenic Rescue Mice. Autism Open Access. 2012;2012:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chao H-T, Zoghbi HY, Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007;56:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stearns NA, Schaevitz LR, Bowling H, Nag N, Berger UV, Berger-Sweeney J. Behavioral and anatomical abnormalities in Mecp2 mutant mice: a model for Rett syndrome. Neuroscience. 2007;146:907–921. [DOI] [PubMed] [Google Scholar]
- 50.Pelka GJ, Watson CM, Radziewic T, Hayward M, Lahooti H, Christodoulou J, et al. Mecp2 deficiency is associated with learning and cognitive deficits and altered gene activity in the hippocampal region of mice. Brain. 2006;129:887–898. [DOI] [PubMed] [Google Scholar]
- 51.Weng S-M, McLeod F, Bailey MES, Cobb SR. Synaptic plasticity deficits in an experimental model of rett syndrome: long-term potentiation saturation and its pharmacological reversal. Neuroscience. 2011;180:314–321. [DOI] [PubMed] [Google Scholar]
- 52.Chao H-T, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. [DOI] [PubMed] [Google Scholar]
- 54.Gemelli T, Berton O, Nelson ED, Perrotti LI, Jaenisch R, Monteggia LM. Postnatal loss of methyl-CpG binding protein 2 in the forebrain is sufficient to mediate behavioral aspects of Rett syndrome in mice. Biol Psychiatry. 2006;59:468–476. [DOI] [PubMed] [Google Scholar]
- 55.Gulmez Karaca K, Brito DVC, Zeuch B, Oliveira AMM. Adult hippocampal MeCP2 preserves the genomic responsiveness to learning required for long-term memory formation. Neurobiol Learn Mem. 2018;149:84–97. [DOI] [PubMed] [Google Scholar]
- 56.Fyffe SL, Neul JL, Samaco RC, Chao H-T, Ben-Shachar S, Moretti P, et al. Deletion of Mecp2 in Sim1-expressing neurons reveals a critical role for MeCP2 in feeding behavior, aggression, and the response to stress. Neuron. 2008;59:947–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chahrour M, Jung SY, Shaw C, Zhou X, Wong STC, Qin J, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen L, Chen K, Lavery LA, Baker SA, Shaw CA, Li W, et al. MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc Natl Acad Sci USA. 2015;112:5509–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baker SA, Chen L, Wilkins AD, Yu P, Lichtarge O, Zoghbi HY. An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell. 2013;152:984–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cellot G, Cherubini E. GABAergic signaling as therapeutic target for autism spectrum disorders. Front Pediatr. 2014;2:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harada M, Taki MM, Nose A, Kubo H, Mori K, Nishitani H, et al. Non-invasive evaluation of the GABAergic/glutamatergic system in autistic patients observed by MEGA-editing proton MR spectroscopy using a clinical 3 tesla instrument. J Autism Dev Disord. 2011;41:447–454. [DOI] [PubMed] [Google Scholar]
- 62.Mori T, Mori K, Fujii E, Toda Y, Miyazaki M, Harada M, et al. Evaluation of the GABAergic nervous system in autistic brain: (123)I-iomazenil SPECT study. Brain Dev. 2012;34:648–654. [DOI] [PubMed] [Google Scholar]
- 63.Yamashita Y, Matsuishi T, Ishibashi M, Kimura A, Onishi Y, Yonekura Y, et al. Decrease in benzodiazepine receptor binding in the brains of adult patients with Rett syndrome. J Neurol Sci. 1998;154:146–150. [DOI] [PubMed] [Google Scholar]
- 64.Mendez MA, Horder J, Myers J, Coghlan S, Stokes P, Erritzoe D, et al. The brain GABA-benzodiazepine receptor alpha-5 subtype in autism spectrum disorder: a pilot [(11)C]Ro15–4513 positron emission tomography study. Neuropharmacology. 2013;68:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sgadò P, Genovesi S, Kalinovsky A, Zunino G, Macchi F, Allegra M, et al. Loss of GABAergic neurons in the hippocampus and cerebral cortex of Engrailed-2 null mutant mice: implications for autism spectrum disorders. Exp Neurol. 2013;247:496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Braat S, Kooy RF. The GABAA Receptor as a Therapeutic Target for Neurodevelopmental Disorders. Neuron. 2015;86:1119–1130. [DOI] [PubMed] [Google Scholar]
- 67.Calfa G, Li W, Rutherford JM, Pozzo-Miller L. Excitation/inhibition imbalance and impaired synaptic inhibition in hippocampal area CA3 of Mecp2 knockout mice. Hippocampus. 2015;25:159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma L-Y, Wu C, Jin Y, Gao M, Li G-H, Turner D, et al. Electrophysiological phenotypes of MeCP2 A140V mutant mouse model. CNS Neurosci Ther. 2014;20:420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kline DD, Ogier M, Kunze DL, Katz DM. Exogenous brain-derived neurotrophic factor rescues synaptic dysfunction in Mecp2-null mice. J Neurosci. 2010;30:5303–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dani VS, Chang Q, Maffei A, Turrigiano GG, Jaenisch R, Nelson SB. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc Natl Acad Sci USA. 2005;102:12560–12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu Y, Lin Y, Takasaki Y, Wang C, Kimura H, Xing J, et al. Rare loss of function mutations in N-methyl-D-aspartate glutamate receptors and their contributions to schizophrenia susceptibility. Transl Psychiatry. 2018;8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hillman BG, Gupta SC, Stairs DJ, Buonanno A, Dravid SM. Behavioral analysis of NR2C knockout mouse reveals deficit in acquisition of conditioned fear and working memory. Neurobiol Learn Mem. 2011;95:404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Becker JAJ, Clesse D, Spiegelhalter C, Schwab Y, Le Merrer J, Kieffer BL. Autistic-like syndrome in mu opioid receptor null mice is relieved by facilitated mGluR4 activity. Neuropsychopharmacology. 2014;39:2049–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iscru E, Goddyn H, Ahmed T, Callaerts-Vegh Z, D’Hooge R, Balschun D. Improved spatial learning is associated with increased hippocampal but not prefrontal long-term potentiation in mGluR4 knockout mice. Genes Brain Behav. 2013;12:615–625. [DOI] [PubMed] [Google Scholar]
- 75.Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009;18:2431–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Y, Wang H, Muffat J, Cheng AW, Orlando DA, Lovén J, et al. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell. 2013;13:446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Soshnev AA, Li X, Wehling MD, Geyer PK. Context differences reveal insulator and activator functions of a Su(Hw) binding region. PLoS Genet. 2008;4:e1000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu Z, Syu L-J, Mellerick DM. Contextual interactions determine whether the Drosophila homeodomain protein, Vnd, acts as a repressor or activator. Nucleic Acids Res. 2005;33:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ruiz i Altaba A. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development. 1999;126:3205–3216. [DOI] [PubMed] [Google Scholar]
- 80.Adams GE, Chandru A, Cowley SM. Co-repressor, co-activator and general transcription factor: the many faces of the Sin3 histone deacetylase (HDAC) complex. Biochem J. 2018;475:3921–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kang J, Kang Y, Kim YW, You J, Kang J, Kim A. LRF acts as an activator and repressor of the human β-like globin gene transcription in a developmental stage dependent manner. Biochem Cell Biol. 2019;97:380–386. [DOI] [PubMed] [Google Scholar]
- 82.Abraham S, Paknikar R, Bhumbra S, Luan D, Garg R, Dressler GR, et al. The Groucho-associated phosphatase PPM1B displaces Pax transactivation domain interacting protein (PTIP) to switch the transcription factor Pax2 from a transcriptional activator to a repressor. J Biol Chem. 2015;290:7185–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sakabe NJ, Aneas I, Shen T, Shokri L, Park S-Y, Bulyk ML, et al. Dual transcriptional activator and repressor roles of TBX20 regulate adult cardiac structure and function. Hum Mol Genet. 2012;21:2194–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21:5979–5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen X-F, Kuryan B, Kitada T, Tran N, Li J-Y, Kurdistani S, et al. The Rpd3 core complex is a chromatin stabilization module. Curr Biol. 2012;22:56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alejandro-Osorio AL, Huebert DJ, Porcaro DT, Sonntag ME, Nillasithanukroh S, Will JL, et al. The histone deacetylase Rpd3p is required for transient changes in genomic expression in response to stress. Genome Biol. 2009;10:R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sertil O, Vemula A, Salmon SL, Morse RH, Lowry CV. Direct role for the Rpd3 complex in transcriptional induction of the anaerobic DAN/TIR genes in yeast. Mol Cell Biol. 2007;27:2037–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Van Esch H, Bauters M, Ignatius J, Jansen M, Raynaud M, Hollanders K, et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am J Hum Genet. 2005;77:442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koerner MV, FitzPatrick L, Selfridge J, Guy J, De Sousa D, Tillotson R, et al. Toxicity of overexpressed MeCP2 is independent of HDAC3 activity. Genes Dev. 2018;32:1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kruusvee V, Lyst MJ, Taylor C, Tarnauskaitė Ž, Bird AP, Cook AG. Structure of the MeCP2–TBLR1 complex reveals a molecular basis for Rett syndrome and related disorders. PNAS. 2017;114:E3243–E3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, et al. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci USA. 2000;97:7202–7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoon H-G, Chan DW, Huang Z-Q, Li J, Fondell JD, Qin J, et al. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yoon H-G, Choi Y, Cole PA, Wong J. Reading and Function of a Histone Code Involved in Targeting Corepressor Complexes for Repression. Mol Cell Biol. 2005;25:324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A Corepressor/Coactivator Exchange Complex Required for Transcriptional Activation by Nuclear Receptors and Other Regulated Transcription Factors. Cell. 2004;116:511–526. [DOI] [PubMed] [Google Scholar]
- 95.Huang W, Ghisletti S, Perissi V, Rosenfeld MG, Glass CK. Transcriptional integration of TLR2 and TLR4 signaling at the NCoR de-repression checkpoint. Mol Cell. 2009;35:48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Choi H-K, Choi K-C, Yoo J-Y, Song M, Ko SJ, Kim CH, et al. Reversible SUMOylation of TBL1-TBLR1 regulates β-catenin-mediated Wnt signaling. Mol Cell. 2011;43:203–216. [DOI] [PubMed] [Google Scholar]
- 97.Bassi MT, Ramesar RS, Caciotti B, Winship IM, De Grandi A, Riboni M, et al. X-linked late-onset sensorineural deafness caused by a deletion involving OA1 and a novel gene containing WD-40 repeats. Am J Hum Genet. 1999;64:1604–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lim Y-M, Hayashi S, Tsuda L. Ebi/AP-1 suppresses pro-apoptotic genes expression and permits long-term survival of Drosophila sensory neurons. PLoS ONE. 2012;7:e37028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tabet A-C, Leroy C, Dupont C, Serrano E, Hernandez K, Gallard J, et al. De novo deletion of TBL1XR1 in a child with non-specific developmental delay supports its implication in intellectual disability. Am J Med Genet A. 2014;164A:2335–2337. [DOI] [PubMed] [Google Scholar]
- 101.Vaqueiro AC, de Oliveira CP, Cordoba MS, Versiani BR, de Carvalho CX, Alves Rodrigues PG, et al. Expanding the spectrum of TBL1XR1 deletion: Report of a patient with brain and cardiac malformations. Eur J Med Genet. 2018;61:29–33. [DOI] [PubMed] [Google Scholar]
- 102.Pons L, Cordier MP, Labalme A, Till M, Louvrier C, Schluth-Bolard C, et al. A new syndrome of intellectual disability with dysmorphism due to TBL1XR1 deletion. Am J Med Genet A. 2015;167A:164–168. [DOI] [PubMed] [Google Scholar]
- 103.Riehmer V, Erger F, Herkenrath P, Seland S, Jackels M, Wiater A, et al. A heritable microduplication encompassing TBL1XR1 causes a genomic sister-disorder for the 3q26.32 microdeletion syndrome. Am J Med Genet A. 2017;173:2132–2138. [DOI] [PubMed] [Google Scholar]
- 104.Saitsu H, Tohyama J, Walsh T, Kato M, Kobayashi Y, Lee M, et al. A girl with West syndrome and autistic features harboring a de novo TBL1XR1 mutation. J Hum Genet. 2014;59:581–583. [DOI] [PubMed] [Google Scholar]
- 105.Lemattre C, Thevenon J, Duffourd Y, Nambot S, Haquet E, Vuadelle B, et al. TBL1XR1 mutations in Pierpont syndrome are not restricted to the recurrent p.Tyr446Cys mutation. Am J Med Genet A. 2018;176:2813–2818. [DOI] [PubMed] [Google Scholar]
- 106.Slavotinek A, Pua H, Hodoglugil U, Abadie J, Shieh J, Van Ziffle J, et al. Pierpont syndrome associated with the p.Tyr446Cys missense mutation in TBL1XR1. Eur J Med Genet. 2017;60:504–508. [DOI] [PubMed] [Google Scholar]
- 107.Heinen CA, Jongejan A, Watson PJ, Redeker B, Boelen A, Boudzovitch-Surovtseva O, et al. A specific mutation in TBL1XR1 causes Pierpont syndrome. J Med Genet. 2016;53:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yamamoto T, Imaizumi T, Yamamoto-Shimojima K, Lu Y, Yanagishita T, Shimada S, et al. Genomic backgrounds of Japanese patients with undiagnosed neurodevelopmental disorders. Brain Dev. 2019;41:776–782. [DOI] [PubMed] [Google Scholar]
- 109.Laskowski RA, Tyagi N, Johnson D, Joss S, Kinning E, McWilliam C, et al. Integrating population variation and protein structural analysis to improve clinical interpretation of missense variation: application to the WD40 domain. Hum Mol Genet. 2016;25:927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Christensen J, Grønborg TK, Sørensen MJ, Schendel D, Parner ET, Pedersen LH, et al. Prenatal valproate exposure and risk of autism spectrum disorders and childhood autism. JAMA. 2013;309:1696–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bromley RL, Mawer G, Clayton-Smith J, Baker GA, Liverpool and Manchester Neurodevelopment Group. Autism spectrum disorders following in utero exposure to antiepileptic drugs. Neurology. 2008;71:1923–1924. [DOI] [PubMed] [Google Scholar]
- 112.Christensen J, Pedersen L, Sun Y, Dreier JW, Brikell I, Dalsgaard S. Association of Prenatal Exposure to Valproate and Other Antiepileptic Drugs With Risk for Attention-Deficit/Hyperactivity Disorder in Offspring. JAMA Netw Open. 2019;2:e186606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wagner GC, Reuhl KR, Cheh M, McRae P, Halladay AK. A new neurobehavioral model of autism in mice: pre- and postnatal exposure to sodium valproate. J Autism Dev Disord. 2006;36:779–793. [DOI] [PubMed] [Google Scholar]
- 114.Gandal MJ, Edgar JC, Ehrlichman RS, Mehta M, Roberts TPL, Siegel SJ. Validating γ oscillations and delayed auditory responses as translational biomarkers of autism. Biol Psychiatry. 2010;68:1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mehta MV, Gandal MJ, Siegel SJ. mGluR5-antagonist mediated reversal of elevated stereotyped, repetitive behaviors in the VPA model of autism. PLoS ONE. 2011;6:e26077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schneider T, Przewłocki R. Behavioral alterations in rats prenatally exposed to valproic acid: animal model of autism. Neuropsychopharmacology. 2005;30:80–89. [DOI] [PubMed] [Google Scholar]
- 117.Nicolini C, Fahnestock M. The valproic acid-induced rodent model of autism. Exp Neurol. 2018;299:217–227. [DOI] [PubMed] [Google Scholar]
- 118.Rasalam AD, Hailey H, Williams JHG, Moore SJ, Turnpenny PD, Lloyd DJ, et al. Characteristics of fetal anticonvulsant syndrome associated autistic disorder. Dev Med Child Neurol. 2005;47:551–555. [DOI] [PubMed] [Google Scholar]
- 119.Zhao H, Wang Q, Yan T, Zhang Y, Xu H-J, Yu H-P, et al. Maternal valproic acid exposure leads to neurogenesis defects and autism-like behaviors in non-human primates. Transl Psychiatry. 2019;9:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kataoka S, Takuma K, Hara Y, Maeda Y, Ago Y, Matsuda T. Autism-like behaviours with transient histone hyperacetylation in mice treated prenatally with valproic acid. Int J Neuropsychopharmacol. 2013;16:91–103. [DOI] [PubMed] [Google Scholar]
- 121.Moldrich RX, Leanage G, She D, Dolan-Evans E, Nelson M, Reza N, et al. Inhibition of histone deacetylase in utero causes sociability deficits in postnatal mice. Behav Brain Res. 2013;257:253–264. [DOI] [PubMed] [Google Scholar]
- 122.Gogolla N, Leblanc JJ, Quast KB, Südhof TC, Fagiolini M, Hensch TK. Common circuit defect of excitatory-inhibitory balance in mouse models of autism. J Neurodev Disord. 2009;1:172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rinaldi T, Silberberg G, Markram H. Hyperconnectivity of local neocortical microcircuitry induced by prenatal exposure to valproic acid. Cereb Cortex. 2008;18:763–770. [DOI] [PubMed] [Google Scholar]
- 124.Lin H-C, Gean P-W, Wang C-C, Chan Y-H, Chen PS. The amygdala excitatory/inhibitory balance in a valproate-induced rat autism model. PLoS ONE. 2013;8:e55248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kawanai T, Ago Y, Watanabe R, Inoue A, Taruta A, Onaka Y, et al. Prenatal Exposure to Histone Deacetylase Inhibitors Affects Gene Expression of Autism-Related Molecules and Delays Neuronal Maturation. Neurochem Res. 2016;41:2574–2584. [DOI] [PubMed] [Google Scholar]
- 126.Luo Y, Zhou B, Liu F, Ai R, Wen M, Tong X. [Enhanced autophagy activates p38/MEF2C pathway to regulate the expression of synapse-associated proteins and improve the symptoms of autistic rats]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2019;35:236–242. [PubMed] [Google Scholar]
- 127.Kuo H-Y, Liu F-C. Molecular Pathology and Pharmacological Treatment of Autism Spectrum Disorder-Like Phenotypes Using Rodent Models. Front Cell Neurosci. 2018;12:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Foley AG, Cassidy AW, Regan CM. Pentyl-4-yn-VPA, a histone deacetylase inhibitor, ameliorates deficits in social behavior and cognition in a rodent model of autism spectrum disorders. Eur J Pharmacol. 2014;727:80–86. [DOI] [PubMed] [Google Scholar]
- 129.Foley AG, Gannon S, Rombach-Mullan N, Prendergast A, Barry C, Cassidy AW, et al. Class I histone deacetylase inhibition ameliorates social cognition and cell adhesion molecule plasticity deficits in a rodent model of autism spectrum disorder. Neuropharmacology. 2012;63:750–760. [DOI] [PubMed] [Google Scholar]
- 130.Monteiro P, Feng G. SHANK proteins: roles at the synapse and in autism spectrum disorder. Nat Rev Neurosci. 2017;18:147–157. [DOI] [PubMed] [Google Scholar]
- 131.Leblond CS, Nava C, Polge A, Gauthier J, Huguet G, Lumbroso S, et al. Meta-analysis of SHANK Mutations in Autism Spectrum Disorders: A Gradient of Severity in Cognitive Impairments. PLoS Genet. 2014;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jiang Y, Ehlers MD. Modeling Autism by SHANK Gene Mutations in Mice. Neuron. 2013;78:8–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang M, Bozdagi O, Scattoni ML, Wöhr M, Roullet FI, Katz AM, et al. Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. J Neurosci. 2012;32:6525–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20:3093–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Peça J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mei Y, Monteiro P, Zhou Y, Kim J-A, Gao X, Fu Z, et al. Adult restoration of Shank3 expression rescues selective autistic-like phenotypes. Nature. 2016;530:481–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Duffney LJ, Zhong P, Wei J, Matas E, Cheng J, Qin L, et al. Autism-like Deficits in Shank3-Deficient Mice Are Rescued by Targeting Actin Regulators. Cell Reports. 2015;11:1400–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang X, Bey AL, Katz BM, Badea A, Kim N, David LK, et al. Altered mGluR5-Homer scaffolds and corticostriatal connectivity in a Shank3 complete knockout model of autism. Nature Communications. 2016;7:11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yoo T, Cho H, Lee J, Park H, Yoo Y-E, Yang E, et al. GABA Neuronal Deletion of Shank3 Exons 14–16 in Mice Suppresses Striatal Excitatory Synaptic Input and Induces Social and Locomotor Abnormalities. Front Cell Neurosci. 2018;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Won H, Lee H-R, Gee HY, Mah W, Kim J-I, Lee J, et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486:261–265. [DOI] [PubMed] [Google Scholar]
- 142.Han K, Holder JL, Schaaf CP, Lu H, Chen H, Kang H, et al. SHANK3 overexpression causes manic-like behaviour with unique pharmacogenetic properties. Nature. 2013;503:72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Deriziotis P, Fisher SE. Speech and Language: Translating the Genome. Trends Genet. 2017;33:642–656. [DOI] [PubMed] [Google Scholar]
- 144.MacDermot KD, Bonora E, Sykes N, Coupe A-M, Lai CSL, Vernes SC, et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet. 2005;76:1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schulze K, Vargha-Khadem F, Mishkin M. Phonological working memory and FOXP2. Neuropsychologia. 2018;108:147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Argyropoulos GPD, Watkins KE, Belton-Pagnamenta E, Liégeois F, Saleem KS, Mishkin M, et al. Neocerebellar Crus I Abnormalities Associated with a Speech and Language Disorder Due to a Mutation in FOXP2. Cerebellum. 2019;18:309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen Y-C, Kuo H-Y, Bornschein U, Takahashi H, Chen S-Y, Lu K-M, et al. Foxp2 controls synaptic wiring of corticostriatal circuits and vocal communication by opposing Mef2c. Nat Neurosci. 2016;19:1513–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Corballis MC. FOXP2 and the mirror system. Trends Cogn Sci (Regul Ed). 2004;8:95–96. [DOI] [PubMed] [Google Scholar]
- 149.Khalil R, Tindle R, Boraud T, Moustafa AA, Karim AA. Social decision making in autism: On the impact of mirror neurons, motor control, and imitative behaviors. CNS Neurosci Ther. 2018;24:669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Casey JP, Magalhaes T, Conroy JM, Regan R, Shah N, Anney R, et al. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum Genet. 2012;131:565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gong X, Jia M, Ruan Y, Shuang M, Liu J, Wu S, et al. Association between the FOXP2 gene and autistic disorder in Chinese population. Am J Med Genet B Neuropsychiatr Genet. 2004;127B:113–116. [DOI] [PubMed] [Google Scholar]
- 152.Shu W, Cho JY, Jiang Y, Zhang M, Weisz D, Elder GA, et al. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc Natl Acad Sci USA. 2005;102:9643–9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.French CA, Groszer M, Preece C, Coupe A-M, Rajewsky K, Fisher SE. Generation of mice with a conditional Foxp2 null allele. Genesis. 2007;45:440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Medvedeva VP, Rieger MA, Vieth B, Mombereau C, Ziegenhain C, Ghosh T, et al. Altered social behavior in mice carrying a cortical Foxp2 deletion. Hum Mol Genet. 2019;28:701–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bowers JM, Konopka G. ASD-relevant Animal Models of the Foxp Family of Transcription Factors. Autism Open Access. 2012;Suppl 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mukamel Z, Konopka G, Wexler E, Osborn GE, Dong H, Bergman MY, et al. Regulation of MET by FOXP2, genes implicated in higher cognitive dysfunction and autism risk. J Neurosci. 2011;31:11437–11442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Toma C, Hervás A, Torrico B, Balmaña N, Salgado M, Maristany M, et al. Analysis of two language-related genes in autism: a case-control association study of FOXP2 and CNTNAP2. Psychiatr Genet. 2013;23:82–85. [DOI] [PubMed] [Google Scholar]
- 159.Bruneau N, Szepetowski P. The role of the urokinase receptor in epilepsy, in disorders of language, cognition, communication and behavior, and in the central nervous system. Curr Pharm Des. 2011;17:1914–1923. [DOI] [PubMed] [Google Scholar]
- 160.Kuo H-Y, Liu F-C. Valproic acid induces aberrant development of striatal compartments and corticostriatal pathways in a mouse model of autism spectrum disorder. FASEB J. 2017;31:4458–4471. [DOI] [PubMed] [Google Scholar]
- 161.Assali A, Harrington AJ, Cowan CW. Emerging roles for MEF2 in brain development and mental disorders. Curr Opin Neurobiol. 2019;59:49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Flavell SW, Kim T-K, Gray JM, Harmin DA, Hemberg M, Hong EJ, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Flavell SW, Cowan CW, Kim T-K, Greer PL, Lin Y, Paradis S, et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. [DOI] [PubMed] [Google Scholar]
- 164.Novara F, Beri S, Giorda R, Ortibus E, Nageshappa S, Darra F, et al. Refining the phenotype associated with MEF2C haploinsufficiency. Clin Genet. 2010;78:471–477. [DOI] [PubMed] [Google Scholar]
- 165.Tu S, Akhtar MW, Escorihuela RM, Amador-Arjona A, Swarup V, Parker J, et al. NitroSynapsin therapy for a mouse MEF2C haploinsufficiency model of human autism. Nat Commun. 2017;8:1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci USA. 2008;105:9397–9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Barbosa AC, Kim M-S, Ertunc M, Adachi M, Nelson ED, McAnally J, et al. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci USA. 2008;105:9391–9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Harrington AJ, Raissi A, Rajkovich K, Berto S, Kumar J, Molinaro G, et al. MEF2C regulates cortical inhibitory and excitatory synapses and behaviors relevant to neurodevelopmental disorders. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Adachi M, Lin P-Y, Pranav H, Monteggia LM. Postnatal Loss of Mef2c Results in Dissociation of Effects on Synapse Number and Learning and Memory. Biol Psychiatry. 2016;80:140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dietrich J-B, Takemori H, Grosch-Dirrig S, Bertorello A, Zwiller J. Cocaine induces the expression of MEF2C transcription factor in rat striatum through activation of SIK1 and phosphorylation of the histone deacetylase HDAC5. Synapse. 2012;66:61–70. [DOI] [PubMed] [Google Scholar]
- 171.Grégoire S, Yang X-J. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol Cell Biol. 2005;25:2273–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gu X, Fu C, Lin L, Liu S, Su X, Li A, et al. miR-124 and miR-9 mediated downregulation of HDAC5 promotes neurite development through activating MEF2C-GPM6A pathway. J Cell Physiol. 2018;233:673–687. [DOI] [PubMed] [Google Scholar]
- 173.Davila JL, Goff LA, Ricupero CL, Camarillo C, Oni EN, Swerdel MR, et al. A positive feedback mechanism that regulates expression of miR-9 during neurogenesis. PLoS ONE. 2014;9:e94348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Grégoire S, Xiao L, Nie J, Zhang X, Xu M, Li J, et al. Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol Cell Biol. 2007;27:1280–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]