Abstract
The outbreak of COVID-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been challenging human health worldwide. Loop-mediated isothermal amplification (LAMP) has been promptly applied to the detection of SARS-CoV-2 owing to its high amplification efficacy and less requirement of the thermal cycler. However, the vast majority of these LAMP-based assays depend on the non-specific detection of LAMP products, which can not discern the undesirable amplificons, likely to yield unreliable results. Herein, a sequence-specific LAMP assay was reported to detect SARS-CoV-2 using proofreading enzyme-mediated probe cleavage (named Proofman), which could realize real-time and visual detection without uncapping. This assay, introducing a proofreading enzyme and the fluorogenic probe to reverse-transcription LAMP (RT-Proofman-LAMP), can specifically detect the SARS-CoV-2 RNA with a detection limit of 100 copies. In addition to the real-time analysis, the assay is capable of endpoint visualization under a transilluminator within 50 min, providing a convenient reporting manner under the setting of point-of-care testing (POCT). In combination with different fluorophores, the one-pot multiplex assay was successfully achieved to detect multiple targets of SARS-CoV-2 and inner control simultaneously. In summary, the development of RT-Proofman-LAMP offers a versatile and highly-specific method for fast field screening and laboratory testing of SARS-CoV-2, making it a promising platform in COVID-19 diagnosis.
Keywords: LAMP, Proofman, Sequence-specific, SARS-CoV-2, COVID-19
1. Introduction
Over the past two decades, the world has faced several infectious disease outbreaks. Ebola, Influenza A (H1N1), SARS, MERS and Zika virus have had a massive global impact in terms of economic disruption, the strain on local and global public health resources and, above all, human health. Most recently, the pandemic of COVID-19, which caused more than one million deaths based on the Weekly Epidemiological Updates of WHO in October 2020, has been challenging the global public health system since December 2019 (Wang et al., 2020a). The high risk posed by the transmission of infectious agents like SARS-CoV-2 requires an efficient response mechanism in which rapid and accurate diagnostic methods are urgently demanded.
Nucleic acid detection techniques providing fast and reliable diagnosis could fulfill the requirement and play a critical role in the surveillance and prevention of infectious diseases. The reverse transcription polymerase chain reaction (RT-PCR) has been developed as a gold standard method to diagnose COVID-19 (Wang et al., 2020b). However, the strong reliance on thermal-cycler, as well as the time-consuming process, limit its broad use in some regions where the fast and on-site screening of potential infection cases are direly needed. Isothermal nucleic acid amplification (INAA) is widely used in detecting causative pathogens, which can provide a fast detection platform under constant temperature (Yan et al., 2014). Among the INAA methods, loop-mediated isothermal amplification (LAMP) is the most prevailing method as its high amplification efficiency and the good tolerance to crudely-processed biological samples (Kaneko et al., 2007; Notomi et al., 2000). Facing the challenge of COVID-19, many LAMP-based assays have been successfully applied to detect the SARS-CoV-2 virus, demonstrating LAMP is a robust and perspective method in COVID-19 detection (Lu et al., 2020; Park et al., 2020; Yan et al., 2020). These LAMP-based assays, however, mainly rely on the non-specific reporters, such as DNA intercalating dyes, metal ion indicators, reaction turbidity and labeled primers; that is to say, the non-specific analytic methods cannot discern the non-specific amplificon which would be problematic for some cases, especially in the case of detecting the samples sharing high homology in genetic sequence. Moreover, as a result of that LAMP is hard to perform multiplex detection in one-pot, many LAMP-based methods lack inner control to ensure sample collection procedure and avoid false positive. Most recently, two state-of-the-art techniques have been reported to solve the issues by using nanoparticle-based lateral flow biosensor (Zhu et al., 2020) or fluorogenic oligonucleotide strand exchange (OSD) probes (Bhadra et al., 2020). However, these two methods required uncapping operation or extra oligonucleotide strand, which enhances the risk of carry-over contamination or the complexity of probe design.
The enzymatic cleavage-based probes are highly popular in specific molecular diagnosis (Juskowiak 2011). Owing to the isothermal DNA polymerase lacks 5' → 3' exonuclease activity, enzymatic cleavage-mediated probes are hardly used in INAA. The inclusion of endonuclease IV to recombinase polymerase reaction (RPA) provided a strategy of sequence-specific detection based on enzymatic cleavage-mediated probes (Piepenburg et al., 2006). However, the probe requires the incorporation of tetrahydrofuran abasic–site mimic, complicating the probe preparation. In previous research, we briefly introduced the proofreading enzyme-mediated probe cleavage (termed Proofman) as developing recombinase assisted loop-mediated isothermal amplification (RALA) (Chen et al., 2020). When using Proofman probe, an additional DNA polymerase (Pfu) with proofreading activity was involved to cleave the probe from 3’ end, producing a fluorescent signal. In this study, we developed a sequence-specific detection platform for SARS-CoV-2 nucleic acid using LAMP coupled with the Proofman probe. In addition to real-time monitoring, visual detection could be realized under a transilluminator, providing a convenient amplification-to-result assay for rapid on-site detection. Furthermore, we evaluated the feasibility of multiplex detection, proving the Proofman-based LAMP could be an appealing choice for accurate diagnosis of COVID-19.
2. Material and methods
2.1. Nucleic acid preparation and in vitro transcription of RNA
The plasmid containing the SARS-CoV-2 gene N cDNA sequence was purchased from Sangon Biotech Company Limited (Shanghai, China). The SARS-CoV-2 gene Orf1ab cDNA was artificially synthesized through reverse transcription-PCR using pseudovirus (purchased from Yeasen Biotech Co., Ltd.) as the template, then conventional PCR was conducted followed by gel extraction and purification using commercial kit (Vazyme, China). The primers used were described below. To obtain the RNA sequence, the corresponding target sequences were amplified through PCR with the T7 promoter-tagged primers (as depicted in Figure S4). Then the PCR products were verified by sanger sequencing and thereafter, transcribed by RNA transcription kit (New England Biolabs, English). The transcribed RNA was purified and recycled using denaturing polyacrylamide gel electrophoresis. The final RNA quality and concentration were measured by the NanoDrop UV–Vis spectrophotometer (Thermo Fisher Scientific Corporation, USA). Then the transcribed RNA was verified via RT-PCR kit (TaKaRa Bio Inc, Japan).
2.2. Primer and probe design
The primers for LAMP (FIP, BIP, F3, B3, LF, LB) were designed according to the SARS-CoV-2 (GenBank accession number: MN985325.1) genome target sequences gene N (location: 28,274 to 29,533) and Orf1ab (location: 13,051 to 13,282). The Proofman probes were designed based on primer LB sequence, with the fluorophore and the quencher labeled at the end of 3′ end mismatch nucleotide and 5′ end nucleotide, respectively. The primers used for reverse transcription-PCR, conventional PCR were designed according to relative sequences. All sequences used in this study were presented in Tables S1.
2.3. PCR and RT-PCR reactions
For PCR reaction, it was performed in 50 μL condition containing 5 U EasyTaq® DNA Polymerase, 5 μL 10 × EasyTaq® buffer (TransGen Biotech, Beijing, China), 0.2 mM deoxyribonucleoside triphosphates (dNTP), and 0.4 μM each of forward and reverse primer. The PCR reaction was incubated in a C1000TM Thermal Cycler PCR (Bio-Rad Laboratories, USA) for a total of 30 cycles (94 °C for 1 min, followed by 30 cycles of 15 s at 94 °C, 10 s at 55 °C and 20 s at 72 °C. A final elongation step at 72 °C for 5 min). For RT-PCR, the One Step PrimeScript RT-PCR kit (TaKaRa Bio Inc, Japan) was used. The reaction was carried out in 25 μL condition according to the manufacturer's instructions with the exception that the concentration of forward and reverse primers was 0.64 μM and the 0.4 × SYBR Green I was used to monitor the real-time amplification. The RT-PCR reaction was incubated in PikoReal 24 RealTime PCR System (Thermo Fisher Scientific Corporation, USA) at 42 °C for 5 min, 95 °C for 10 s and then 50 cycles of 5 s at 95 °C, 30 s at 55 °C followed by a signal collection.
2.4. Proofman-LAMP reaction
For LAMP reaction, the reaction was performed in 25 μL mixture containing 2.5 μL 10 x reaction buffer (200 mM Tris-HCl (pH 8.8), 100 mM (NH4)2SO4, 50 mM MgSO4 and 600 mM KCl), 0.8 mM dNTP, 4 U Bsm DNA polymerase large fragment (Thermo Fisher Scientific), 0.8 μM each of inner primers (FIP, BIP), 0.4 μM of loop primer (LF), 0.2 μM each of outer primers (F3, B3), 1M betaine (Sigma-Aldrich), and 1 μL DNA template. All the LAMP reactions were conducted at 60 °C. For the multiplex assay, it should be noted that the Bsm DNA polymerase and AMV reverse transcriptase were doubled compared to singleplex assay.
2.5. Amplification analysis
For conventional fluorescent reaction, 0.4 x SYBR Green I (Thermo Fisher Scientific, USA) was added to the reaction for real-time fluorescent monitoring. As for Proofman-LAMP, instead of SYBR Green I, 0.4 μM of Proofman probe was added. The fluorescent signal was collected with 1 min interval by the CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, USA) at corresponding channels (FAM, HEX and ROX channels). The endpoint visual image was taken by the camera under the Blue LED Light Transilluminator (Biofriend, Wuxi, China).
2.6. The sensitivity and specificity of the assay
The sensitivity was evaluated by using 10-fold serial dilutions of the RNA templates. The specificity of the assay was measured by using the extracted genomics from 5 common respiratory tract pathogens including Pseudomonas aeruginosa (ATCC 9027), Escherichia coli (ATCC 25922), Klebsiella pneumoniae (ATCC BAA-2146), Staphylococcus aureus (ATCC 25923), Candida tropicalis (ATCC 4563) and synthetic SARS-CoV (NCBI accession number: NC_004718.3) gene N/Orf1ab RNA sequences. All the genomic DNA of these pathogens used in the study were preserved in the laboratory. The relative sequence information was provided in Table S2.
2.7. Evaluation of the method using contrived samples
The saliva samples were collected from consent informed healthy persons through published saliva specimen collection procedure. Then the pseudovirus (purchased from Yeasen Biotech Co., Ltd.) were spiked into the collected samples and taken as contrived real-life samples. The contrived samples were processed either by viral RNA isolation kit (Foregene, Chengdu, China) or cellular direct lysis buffer (Sigma-Aldrich, USA).
3. Results and discussion
3.1. The principle of proofman coupled with LAMP
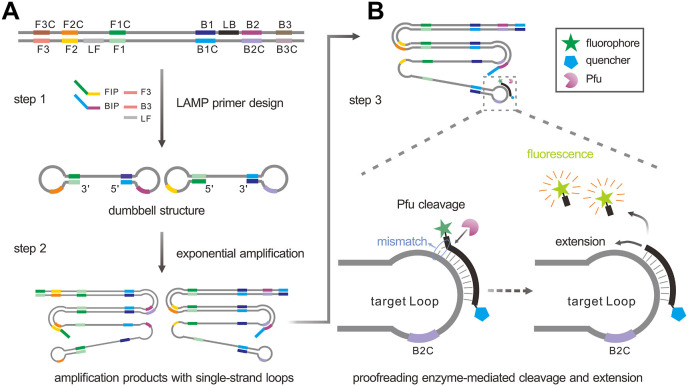
The schematic strategy was shown in Fig. 1 A, the primers used for LAMP amplification were designed according to the target sequence. LAMP could produce the dumbbell structure in the initial amplification phase (step 1). After the initial phase, the products containing single-strand loop were generated during the exponential amplification phase (step 2). Then the Proofman probe, respectively labeled with fluorophore and quencher at 3′ end and 5′ end, together with a thermostable proofreading DNA polymerase (Pfu) were introduced to realize the sequence-specific detection (Fig. 1B). Of note was that a deliberate mismatch at the 3’ end of the Proofman probe was necessary to trigger the 3' → 5′ exonuclease activity of the proofreading enzyme Pfu. Upon the binding of the Proofman probe to the target segment in the loop domain, the mismatched nucleotide would be cleaved by Pfu, releasing the fluorophore. Since then, the cleaved probe can further serve as an extendable primer which enhances the isothermal amplification efficiency (step 3). Based on the Proofman probe, the LAMP amplification products can be detected specifically through the increase of fluorescence intensity as the amplification reaction is proceeding. In our strategy, the sequence-specific detection of LAMP products was achieved by the binding and cleavage of the Proofman probe, thus enhancing the accuracy of LAMP reaction.
Fig. 1.
The schematic of specific detection of LAMP products using the Proofman probe. (A) LAMP reaction process including the initial amplification phase and exponential amplification phase. Primer design was based on the target sequence: FIP and BIP were the inner primers; F3 and B3 were the outer primers; LF was the loop primer. (B) The principle of sequence-specific detection using the Proofman probe. The Proofman probe was designed based on the target sequence and a deliberate mismatch at the 3′ end of the probe was needed to trigger the cleavage activity of the proofreading enzyme (Pfu). Once the Proofman probe binds to the target loop of LAMP products, the Pfu can cleave the probe at the mismatching nucleotide, releasing the fluorophore. Then, the cleaved probe serves as a loop primer to enhance the amplification efficiency.
3.2. Establishment of Proofman-based LAMP assay for SARS-CoV-2 detection
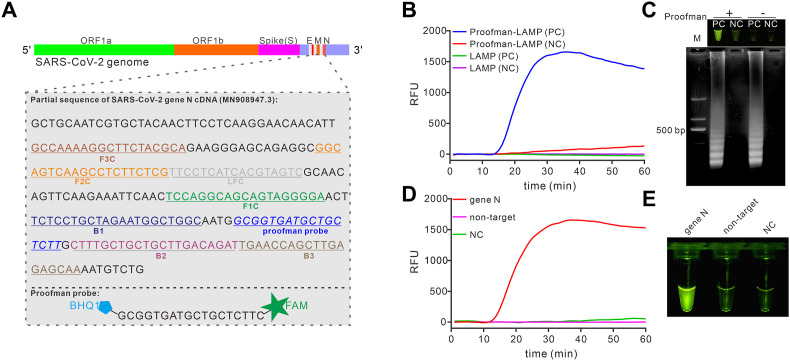
According to the genome information of SARS-CoV-2 from Global Initiative on Sharing All Influenza Data (GISAID, https://www.gisaid.org) and WHO guidance for laboratory testing of COVID-19 (https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117), SARS-CoV-2 gene N (nucleocapsid phosphoprotein) was a conservative target in COVID-19 diagnosis (Corman et al., 2020). To develop a specific detection platform for COVID-19 using the Proofman probe combined with LAMP, the same region of gene N was chosen to design LAMP primer sets (Fig. 2 A). The plasmid containing the SARS-CoV-2 gene N cDNA sequence was successfully amplified by the conventional LAMP reaction in the absence of the probe (Figure S1A and S1B). Then the Proofman probe (labeled with FAM and BHQ1, Fig. 2A) was designed based on the loop domain of LAMP products and verified to be cleavable by proofreading enzyme Pfu under LAMP condition (Figure S1C). Next, the probe was introduced to the developed LAMP assay. As shown in Fig. 2B, the reaction tube showed an increase in fluorescence intensity with 0.4 μM of Proofman probe, while reactions without the probe or template remained no change in fluorescence intensity. When verified with gel electrophoresis, as expected, all of the reactions except negative controls (without target template) presented characterized product bands on the gel electrophoresis (Fig. 2C, bottom). These results confirmed that the Proofman system was compatible with the established LAMP reactions and able to monitor the LAMP reaction real-timely. Moreover, we found that visualized detection could be realized under the 475 nm blue light (Fig. 2C, upper), indicating that the assay was capable of end-point detection without uncapping operation or adding extra dyes. Due to the fluorescence intensity derives from the cleavage of the Proofman probe, we further investigated the concentration of the probe and Pfu to obtain the optimal condition. As shown in Figure S2A, increasing the concentration of the probe from 0.2 μM to 1.2 μM produced increasing fluorescence intensity, while the reaction rate remained unchanged. However, changing the concentration of Pfu did not influence the fluorescence intensity (Figure S2B). Thus, the final concentration of the probe and Pfu used for the following study were 0.4 μM and 0.1 U, respectively. To investigate the feasibility of Proofman-LAMP in the detection of SARS-CoV-2 RNA, we artificially synthesized the RNA template of gene N by in vitro transcription and verified it with RT-PCR (Figure S4). Avian Myeloblastosis Virus (AMV) reverse transcriptase was chosen to amplify the RNA template because it keeps active even under the LAMP temperature (60 °C, Figure S5A). We explored the effect of AMV reverse transcriptase on the Proofman-LAMP reaction and found that the 1 U of AMV reverse transcriptase was enough to develop the reverse transcription Proofman-LAMP, namely RT-Proofman-LAMP (Figure S5B). Then, the RT-Proofman-LAMP reactions were carried out with the target template and non-target template parallelly. As illustrated in Fig. 2D, only the reaction containing the target template displayed an increase in fluorescence intensity. In contrast, the signal change in reactions containing non-target template or negative control was undetectable. Additionally, the end-point detection showed a clear visual difference between positive reactions and other reactions (Fig. 2E). These results demonstrated that the Proofman-coupled LAMP was successfully applied to the detection of the targeted sequence of SARS-CoV-2.
Fig. 2.
Establishment of Proofman-LAMP for SARS-CoV-2 detection. (A) Design of LAMP primer sets and the Proofman probe based on the conservative sequence of SARS-CoV-2 gene N. (B) LAMP assay coupled with or without the Proofman to amplify the pUC57-N plasmid containing SARS-CoV-2 gene N cDNA. (C) Agarose gel image of the LAMP reaction (with/without Proofman probe) products (bottom) and endpoint image under 475 nm blue light (upper). PC, positive reaction with the template; NC, negative control reaction without the template; M, DNA marker. (D) Detection of SARS-CoV-2 gene N RNA using RT-Proofman-LAMP. (E) Endpoint image of RT-Proofman-LAMP after 50 min incubation. The reactions contained 1 × 106 copies of DNA/RNA templates except for negative control (NC). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Analysis of sensitivity and specificity
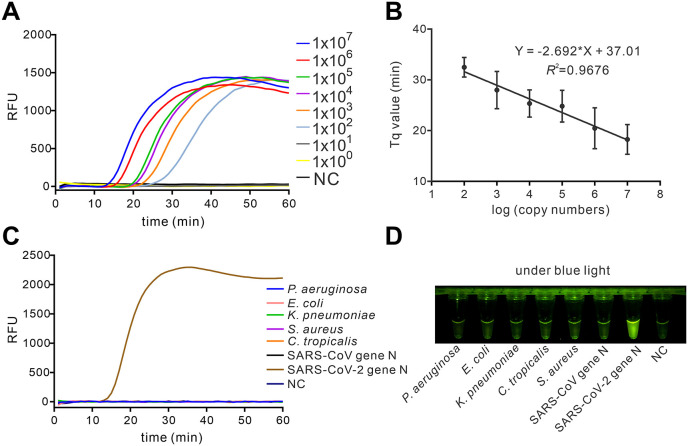
To determine the sensitivity of the assay, the serially diluted synthetic SARS-CoV-2 RNA templates were taken as input samples for RT-Proofman-LAMP. As depicted in Fig. 3 A, the RT-Proofman-LAMP could efficiently detect as low as 100 copies of gene N RNA in less than 1 h. A quantification time Tq, which is similar to the Cq in quantitative PCR, was introduced to the real-time RT-Proofman-LAMP assay as the reaction time when fluorescence intensity exceeds an arbitrary threshold. As shown in Fig. 3B, the Tq values of the assay were in linear correlation to the concentration of the target, suggesting the assay has the potential for quantitative analysis. The end-point visual outcome also presented unambiguous distinguishment between positive and negative reactions after 50 min incubation, confirming the assay was a sensitive method (Figure S6). The reported viral load of infected patients at the onset of symptom could be 105–107 copies per milliliter of sputum specimen (Kim et al., 2020), which indicates that the limit of detection for the RT-Proofman-LAMP was sufficient to realize the detection at the early phase of COVID-19. Next, we sought to verify the specificity of the RT-Proofman-LAMP. We conducted an assay using RT-Proofman-LAMP to discern the SARS-CoV-2 from 5 different pathogens, which are closely related to pathogenic infection. The results illustrated in Fig. 3C that the RT-Proofman-LAMP correctly discerned different pathogens and showed no cross-reactivity. The visualized results under the blue light also confirmed the high specificity of the assay (Fig. 3D). It is worth mentioning that the SARS-CoV-2 is extremely high homology to SARS-CoV at the nucleotide level17. The results demonstrated that RT-Proofman-LAMP could efficiently distinguish these two homologous sequences (Fig. 3C and D). Therefore, the RT-Proofman-LAMP assay could be a sensitive method for SARS-CoV-2 detection with high specificity.
Fig. 3.
Sensitivity and specificity of RT-Proofman-LAMP. (A) The real-time RT-Proofman-LAMP using serially diluted SARS-CoV-2 gene N synthetic RNA (107 copies to 100 copies). (B) The linear relationship between the Tq value and the logarithm of the copy number in the range of 107 copies to 102 copies of SARS-CoV-2 gene N synthetic RNA. The error bars are the standard deviation of three repetitive measurements. (C) The specificity of RT-Proofman-LAMP in the detection of SARS-CoV-2 gene N synthetic RNA. (D) The visual image of specific detection of SARS-CoV-2 gene N synthetic RNA after 50 min incubation. The amount of Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus, Candida tropicalis and SARS-CoV-gene N was 26 ng, 14 ng, 42 ng, 12 ng, 20 ng and 1 × 106 copies, respectively. The SARS-CoV gene N homologous domain RNA was obtained by in vitro transcription (Figure S7).
3.4. Multiplex detection using RT-proofman-LAMP
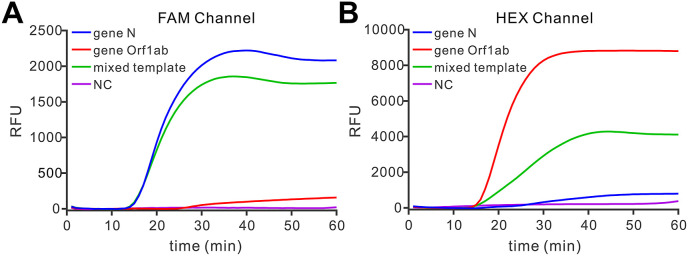
As a probe-based method, the Proofman-coupled LAMP assay was theoretically accessible to realize multiplex detection which could be helpful to improve the accuracy of the COVID-19 detection. Gene Orf1ab (ORF1ab polyprotein), another conservative domain in SARS-CoV-2 genome, was also recommended as a target for COVID-19 diagnosis according to the standard testing guideline published by WHO and Chinese Center for Disease Control and Prevention (Jung et al., 2020; Prevention 2020). Thus, we also sought to develop a Proofman-based assay for the detection of gene Orf1ab. In this case, the Proofman probe used for Orf1ab was labeled by another fluorophore HEX. After the establishment of singleplex detection of gene Orf1ab using RT-Proofman-LAMP (Figure S9), we investigated the feasibility of multiplex detection of RT-Proofman-LAMP by combining two sets of primers and probes in one tube to detect SARS-CoV-2 gene N and gene Orf1ab concurrently. As shown in Fig. 4 A and B, only the reaction tube containing the mixed template (equal mixing of SARS-CoV-2 gene N and Orf1ab RNA) presented the fluorescence curves both in FAM and HEX channels, while the reaction tube containing one kind of templates (SARS-CoV-2 gene N or Orf1ab) presented fluorescence curve just in one fluorescence collection channel, demonstrating the duplex was available using RT-Proofman-LAMP. Given that inner control plays an important role in clinical detection, we aimed to develop a triplex assay in which the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene mRNA was taken as inner control. As illustrated in Figure S10, when adding triple-mixed template (containing SARS-CoV-2 gene N, gene Orf1ab and human GAPDH mRNA) to the reaction comprising three sets of corresponding primers and probes, the fluorescence curves appeared both in three fluorescence collection channels (FAM, HEX and ROX), suggesting the triplex assay was successfully conducted. Although multiplex detection is a favorable method in molecular detection, few LAMP-based methods are capable of multiplex detection in one pot due to the lack of suitable reporters. The Proofman-based LAMP assay provided a good alternative to achieve multiplex detection in a single step, which could further strengthen the specificity of the assay. Moreover, the inclusion of the inner control in one pot could guarantee the quality of specimen processing, decreasing the false-positive results.
Fig. 4.
Duplex RT-Proofman-LAMP. (A) The real-time fluorescence in the FAM channel of duplex assay using different templates. (B) The real-time fluorescence in the HEX channel of duplex assay using different templates. The mixed template comprised an equal amount (1 × 106 copies) of SARS-CoV-2 gene N and gene Orf1ab RNA. The gene Orf1ab RNA template was obtained by in vitro transcription and verified through RT-PCR (Figure S8).
3.5. Evaluation of the assay using contrived samples
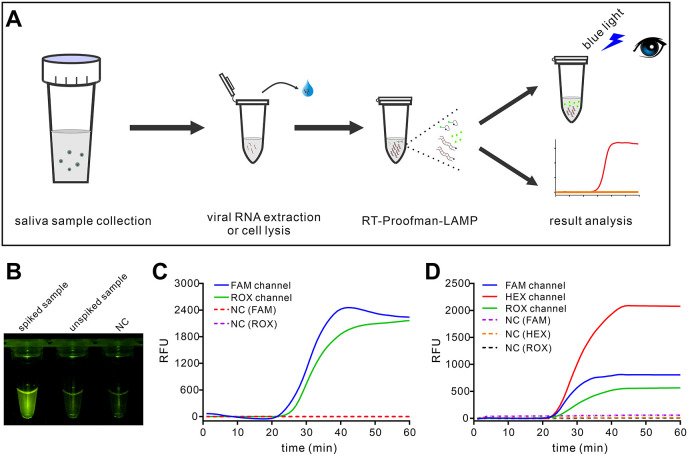
After the development of the assay, we sought to evaluate the performance of the RT-Proofman-LAMP to detect real-life samples. The saliva specimen has been proposed as a reliable tool for the diagnostic of SARS-CoV-2 (Azzi et al., 2020), which is easy to collect and highly appreciated in POCT. Based on the RT-Proofman-LAMP, a general workflow was proposed to realize the rapid detection of SARS-CoV-2 (Fig. 5 A). Due to the lacking of clinical samples from infected patients, the pseudovirus containing the SARS-CoV-2 virus partial genome and the retrovirus envelope was spiked to saliva samples from informed individuals to simulate real-life samples. The spiked saliva samples were processed either by viral RNA extraction kit or commercial direct lysis buffer. Then 1 μL of processed samples were inputted to the RT-Proofman-LAMP assays. We first checked the ability of the assay to detect the pseudovirus in the presence of genomic RNA (RNA extraction Kit) or cellular lysate (lysis buffer). The results demonstrated that RT-Proofman-LAMP was able to detect the target viral RNA without cross-reaction with genomic RNA or cellular lysate (Figure S11). Then the spiked samples were used to evaluate the singleplex and multiplex RT-Proofman-LAMP assay. For the singleplex assay, as shown in Fig. 5B, after 50 min incubation only the tube with spiked samples showed an increased fluorescence under the blue light, indicating the RT-Proofman-LAMP could be a qualitative method through visual results. Additionally, the method was compared to conventional RT-PCR and showed comparable analytical performance (Figure S12A). Afterward, the duplex assay was examined. When using a duplex assay that simultaneously detects SARS-CoV-2 gene N and the inner control, both the fluorescence signals in FAM and ROX channel were detected in the tube containing spiked samples (Fig. 5C). Another duplex detection targeting gene Orf1ab and inner control was also successfully achieved (Figure S12B). Next, the triplex assay that includes both two targets of the viral genome and the inner control was tested. As can be seen from Fig. 5D, three corresponding fluorescence signals of FAM (blue curve), HEX (red curve) and ROX (green curve) were concurrently appeared in one tube containing spiked samples, verifying the accessibility of the triplex assay. These results validated that the RT-Proofman-LAMP was capable of detecting the contrived samples in the way of the singleplex or multiplex assay. Facing the challenge of COVID-19, varied diagnostic requirements are needed under different settings. Based on the above results, we provided three solutions to meet diverse diagnostic demands. The simplest singleplex assay could be readily used to fast pre-screening of suspected cases in some resource-limited regions where the sophisticated instrument is out of reach. To improve the diagnostic accuracy, the duplex assay established above was a convenient one that can guarantee the sureness of a test and avoid the uncertain test results caused by improper sample processing. As an evolving virus, SARS-CoV-2 may generate some latent mutants in its genome, thus the triplex assay that detects two targets of SARS-CoV-2 genome could be an efficient way to prevent the situation. Taken together, the RT-Proofman-LAMP was a versatile method that embraces the potential to realize accurate POCT.
Fig. 5.
Evaluation of RT-Proofman-LAMP using spiked saliva samples. (A) The general workflow to detect saliva samples using Proofman-coupled LAMP. (B) Visual detection of singleplex RT-Proofman-LAMP. The spiked sample contained 50 copies of pseudovirus. (C) Duplex assay of RT-Proofman-LAMP using spiked saliva sample (containing 50 copies of pseudovirus). (D) Triplex assay of RT-Proofman-LAMP using spiked saliva sample (containing 50 copies of pseudovirus).
4. Conclusions
In summary, this work combined the proofreading-based detection method with the LAMP condition for the first time, which realizes the sequence-specific detection of LAMP amplicon. With the Proofman probe, the multiplex detection can be achieved in one pot, which bypasses the uncapping operation. In addition to real-time monitoring, colorimetric detection was also realized under blue light, which can give fast qualitative results. Unlike many reported LAMP-based methods that achieved the fast colorimetric SARS-CoV-2 detection with non-specific reporters, the RT-Proofman-LAMP assay could improve the diagnostic accuracy by providing a sequence-specific colorimetric result. Based on the visualized singleplex assay, the fast pre-screening of suspected individuals could be finished in 50 min using direct lysed samples, which is highly suitable for POCT. Using multiple fluorogenic probes, the one-pot multiplex assay could provide a robust laboratory testing platform for accurate diagnosis of COVID-19. All in all, the Proofman-based LAMP offers a highly specific platform for the detection of SARS-CoV-2, promising in enhancing the accuracy of the COVID-19 diagnosis.
CRediT authorship contribution statement
Sheng Ding: conceived and designed this study, performed the experiments, Formal analysis, analyze the data, contributed the materials, Writing - original draft, drafted the manuscript. Gangyi Chen: conceived and designed this study, performed the experiments, contributed the materials, revised the manuscript. Yinghua Wei: performed the experiments, contributed the materials. Juan Dong: performed the experiments, contributed the materials. Feng Du: performed the experiments, contributed the materials. Xin Cui: Formal analysis, analyze the data, contributed the materials. Xin Huang: Formal analysis, analyze the data, contributed the materials. Zhuo Tang: conceived and designed this study, contributed the materials, revised the manuscript, Writing - original draft, drafted the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant numbers 21907090, 21708037, and 21877108); the Innovative Team of Sichuan Province (grant number 2017TD0021); Sichuan Science and Technology Program (grant numbers 2018TJPT0044, 2019ZDZX0048, 2020YFS0370 and 2019YFS0331); and the Youth Innovation Promotion Association CAS (No. 2020365).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113041.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A., Fasano M., Sessa F., Tettamanti L., Carinci F., Maurino V., Rossi A., Tagliabue A., Baj A. Saliva is a reliable tool to detect SARS-CoV-2. J. Infect. 2020;81(1):e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadra S., Riedel T., Lakhotia S., Tran N., Ellington A. 2020. High-surety Isothermal Amplification and Detection of SARS-CoV-2, Including with Crude Enzymes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Chen R., Ding S., Li M., Wang J., Zou J., Du F., Dong J., Cui X., Huang X., Deng Y., Tang Z. Recombinase assisted loop-mediated isothermal DNA amplification. Analyst. 2020;145(2):440–444. doi: 10.1039/c9an01701a. [DOI] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y., Park G.S., Moon J.H., Ku K., Beak S.H., Lee C.S., Kim S., Park E.C., Park D., Lee J.H., Byeon C.W., Lee J.J., Maeng J.S., Kim S.J., Kim S.I., Kim B.T., Lee M.J., Kim H.G. Comparative analysis of primer-probe sets for RT-qPCR of COVID-19 causative virus (SARS-CoV-2) ACS Infect. Dis. 2020;6(9):2513–2523. doi: 10.1021/acsinfecdis.0c00464. [DOI] [PubMed] [Google Scholar]
- Juskowiak B. Nucleic acid-based fluorescent probes and their analytical potential. Anal. Bioanal. Chem. 2011;399(9):3157–3176. doi: 10.1007/s00216-010-4304-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H., Kawana T., Fukushima E., Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods. 2007;70(3):499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Ko J.H., Kim Y., Kim Y.J., Kim J.M., Chung Y.S., Kim H.M., Han M.G., Kim S.Y., Chin B.S. Viral load kinetics of SARS-CoV-2 infection in first two patients in korea. J. Kor. Med. Sci. 2020;35(7):e86. doi: 10.3346/jkms.2020.35.e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Wu X., Wan Z., Li Y., Jin X., Zhang C. A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int. J. Mol. Sci. 2020;21(8) doi: 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G.S., Ku K., Baek S.H., Kim S.J., Kim S.I., Kim B.T., Maeng J.S. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2020;22(6):729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006;4(7) doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevention C. Technical guidelines for COVID-19 laboratory testing. China CDC Weekly. 2020;2:332–336. doi: 10.46234/ccdcw2020.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yao H., Xu X., Zhang P., Zhang M., Shao J., Xiao Y., Wang H. Limits of detection of 6 approved RT-PCR kits for the novel SARS-coronavirus-2 (SARS-CoV-2) Clin. Chem. 2020;66(7):977–979. doi: 10.1093/clinchem/hvaa099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26(6):773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Zhou J., Zheng Y., Gamson A.S., Roembke B.T., Nakayama S., Sintim H.O. Isothermal amplified detection of DNA and RNA. Mol. Biosyst. 2014;10(5):970–1003. doi: 10.1039/c3mb70304e. [DOI] [PubMed] [Google Scholar]
- Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S., Xing M., Chen H., Wang Y. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020;166:112437. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.