Abstract
Early diagnosis and ultrahigh sample throughput screening are the need of the hour to control the geological spread of the COVID-19 pandemic. Traditional laboratory tests such as enzyme-linked immunosorbent assay (ELISA), reverse transcription polymerase chain reaction (RT-PCR) and computed tomography are implemented for the detection of COVID-19. However, they are limited by the laborious sample collection and processing procedures, longer wait time for test results and skilled technicians to operate sophisticated facilities. In this context, the point of care (PoC) diagnostic platform has proven to be the prospective approach in addressing the abovementioned challenges. This review emphasizes the mechanism of viral infection spread detailing the host-virus interaction, pathophysiology, and the recent advances in the development of affordable PoC diagnostic platforms for rapid and accurate diagnosis of COVID-19. First, the well-established optical and electrochemical biosensors are discussed. Subsequently, the recent advances in the development of PoC biosensors, including lateral flow immunoassays and other emerging techniques, are highlighted. Finally, a focus on integrating nanotechnology with wearables and smartphones to develop smart nanobiosensors is outlined, which could promote COVID-19 diagnosis accessible to both individuals and the mass population at patient care.
Keywords: COVID-19 diagnosis, Optical biosensors, Electrochemical biosensors, Wearable biosensors, Smart nanobiosensors, Nanoparticles
Graphical abstract
1. Introduction
The outbreak of novel emerging viral infectious diseases presents a significant threat to public health globally and constitutes public health emergencies of international concern (PHEIC). In this connection, the outset of highly contagious novel severe acute respiratory syndrome coronavirus (SARS-CoV-2) disease has affected the global economy, physical and mental wellness of human beings. The infectious disease caused by the SARS-CoV-2 virus is addressed as COVID-19. The history of human coronavirus infectious diseases started long back in 1965, reporting four subtypes of coronaviruses (two β-coronaviruses, namely HKU1 and OC43, two α-coronaviruses, namely NL63 and 229E) causing severe respiratory tract infections [1,2]. In the past two decades, the world has faced the emergence of lethal virulent infectious diseases, including severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002, Middle Eastern Respiratory Syndrome (MERS) in 2012, and the novel severe acute respiratory syndrome coronavirus (SARS-CoV 2/COVID -19) in 2019 [1,3,4]. However, the rapid spread and uncertain evolution of the COVID-19 has imposed strict restrictive measures, which eventually impacted the global economy [2].
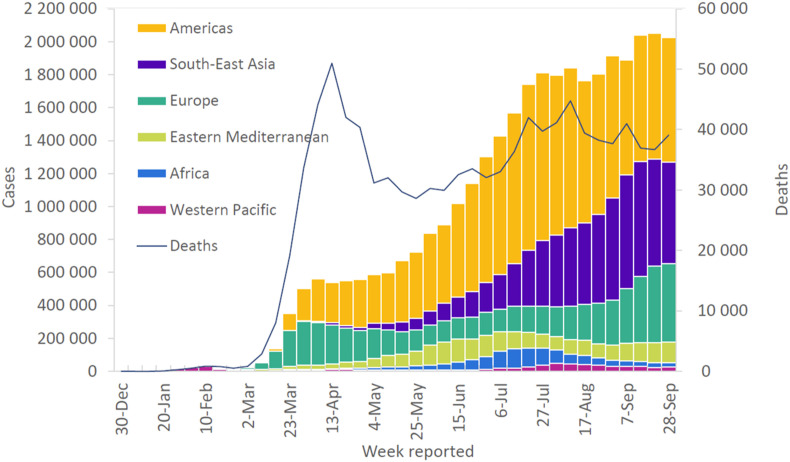
According to the World Health Organisation (WHO) report in April 2020, the COVID-19 pandemic has affected 213 countries, with the number of infected cases exceeding millions [5]. India is the third-worst COVID-19 infection affected country after the United States and Brazil, with the number of daily reported cases increases from 10% to 18.84% in July 2020. There are 35,848,254 cases of COVID-19 reported including 1, 048, 181 deaths globally from December 2019 till October 7, 2020 (Fig. 1 ) [2,[6], [7], [8], [9], [10]]. The global spread of this novel COVID-19 is highly contagious compared to the earlier reported coronavirus infections [11]. Despite the widespread of the COVID-19 disease at a faster pace, the global deaths reported account for 6% compared to SARS-CoV (10% mortality) and MERS-CoV (35% mortality) [12]. Further, COVID-19 infection creates more inconvenience to healthcare professionals, elders, and people with pre-existing health problems.
Fig. 1.
Number of COVID-19 cases and deaths reported weekly to the World Health Organisation (WHO). The number of affected cases globally reached 35 million, with over 1 million deaths till October 7, 2020. The majority of the deaths were reported in the Americas (55%) and Europe (23%). Reproduced with permission from Ref. [13].
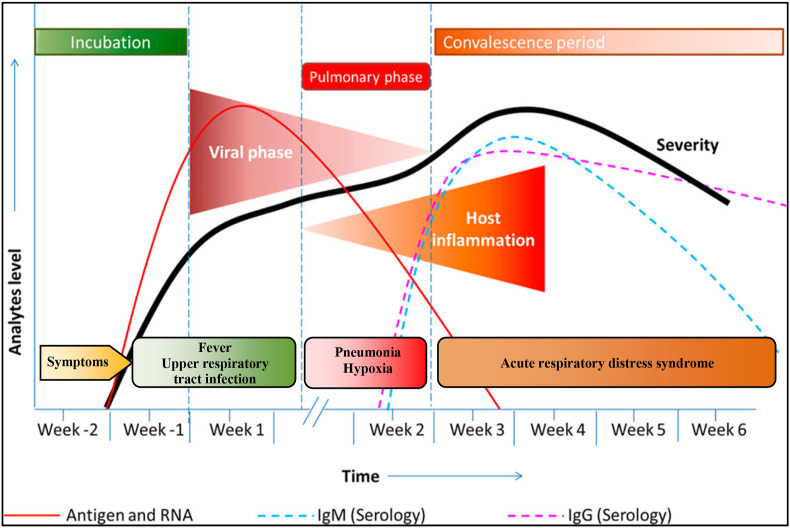
Coronaviruses are of zoonotic origin and are widely found in mammals such as bats and rodents, which are the high-risk viral reservoirs and source for diseases affecting human beings. However, the transmission of this infectious viral agent COVID-19 to humans is unknown [14]. Once this infectious agent is transmitted to humans, they are affected with viral pneumonia accompanied by symptoms of fever, cough and respiratory syndromes. Subsequently, a rapid spread within the local community might arise within a short period through respiration droplets [4,15]. Currently, clinicians rely upon conventional diagnostic tools such as reverse transcription-polymerase chain reaction (RT-PCR), enzyme-linked immunosorbent assay (ELISA), and chest X-ray radiography for COVID-19 diagnosis [16]. The RT-PCR is one of the molecular diagnostic techniques which is considered to be the gold standard as it involves amplification of viral nucleic acids of COVID-19, ensuring specific and sensitive diagnosis. In addition to this, radiological tests, including computed tomography (CT) imaging, are believed to be the assistive diagnostic method in which the presence of consolidation lesions in CT images indicates the onset of the COVID-19 disease [12,17,18]. However, these methods are entirely dependent on sophisticated laboratory facilities and skilled laboratory personnel, which render them inconsiderate at public health centres and remote countryside for mass screening during this pandemic [19,20]. Moreover, viral load started to decrease with the onset of the disease resulting in the unreliability of the molecular diagnostic test results (Fig. 2 ). Apart from the decrease in the viral load, there is a release of antibodies against the COVID-19 virus due to the immune response of the infected patients [21]. The foremost challenge besides the false-positive result is the long-time-gap between sampling and diagnosis. These key factors motivate the scientific community to develop a robust and affordable point of care (PoC) diagnostic systems for mass screening.
Fig. 2.
Different stages involved in the COVID-19 course of infection. Stage 1: Incubation phase. The patient will be having cough, headaches, and fever with the multiplication of virus inside the host over time. Stage 2: Pulmonary phase. In this phase, the patient suffers from hypoxia, pneumonia and the host immune system starts to respond to the infection with the release of antibodies and other metabolites. Stage 3: Convalescence phase. In this phase, the patient suffers from fatal acute respiratory distress syndrome. Reproduced with permission from Ref. [22].
In the present scenario, due to the absence of a safe and effective vaccine against COVID-19, there is an increased demand for PoC diagnostic methods for screening individuals and the mass [[23], [24], [25]]. In the context of PoC diagnostics, immunoassay platforms have received attention over molecular diagnostics. They could accomplish better control by early on-site diagnosis with reduced false-positive cases promoting effective isolation of the infected cases. Besides, the in-situ detection of SARS-CoV-2 could be an easy, rapid, and cost-effective technique that requires no additional skills for device operability and offers enhanced sensitivity. More research work towards improving the development of PoC units to be deployed for practical usage in epidemiological research is currently in progress [[26], [27], [28], [29]].
This review outlines the mechanism of the spread of COVID-19 virus infection, host-virus interaction, pathophysiology, recent advances in the development of affordable PoC diagnostic platforms for rapid diagnosis of COVID-19. A focus on the role of nanotechnology in the development of these PoC diagnostic platforms, along with optical and electrochemical biosensors, is outlined. A comprehensive section on smart nano-biosensors, including wearable biosensors with smartphone interfaced applications to promote rapid detection of COVID-19, is presented. The problems related to sensitivity, specificity and the challenges associated with quantitative analysis for in-situ diagnosis are also discussed.
2. COVID-19 virus-host interaction mechanism
2.1. Structure of COVID-19 virus
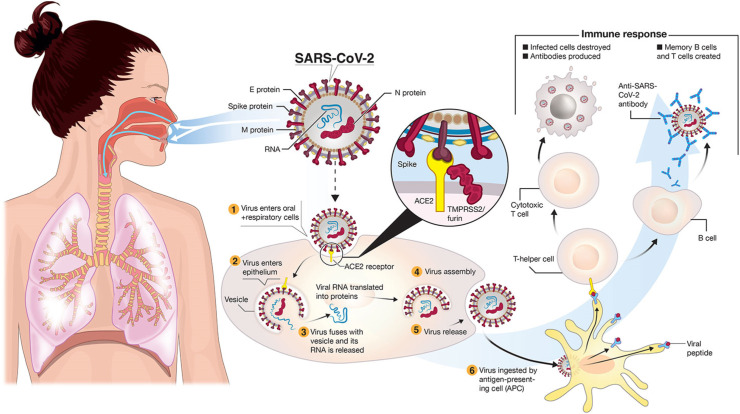
Structurally, the COVID-19 virus is composed of four main proteins, including spike (S) glycoprotein, small envelope (E) glycoprotein, membrane (M) glycoprotein and nucleocapsid (N) protein. It is a single-stranded ribonucleic acid (RNA) enveloped virus, as shown in Fig. 3 . The S glycoproteins assemble into trimers on the virion surface, forming peplomers embedded in an envelope to form a distinctive “corona” or crown-like appearance [30,31]. They bind to the host receptor via angiotensin-converting enzyme 2 (ACE2) and responsible for enabling viral entry into the cell [32]. This S glycoprotein has further two functional subunits S1 and S2, facilitating cell attachment and membrane binding, respectively [33]. The envelope or E protein is the smallest component and present in minor amounts in the COVID-19 virion envelope and plays an essential role in the virus replication and maturation [34]. Membrane or M protein is the most abundant component present in the virus structure, which defines the virus envelope [35]. Finally, the nucleocapsid or the N-protein is a structural protein that binds the single-stranded RNA genome to form a shell around the enclosed nucleic acid [36]. The N-protein plays a vital role in the viral replication cycle and host cell response to viral infection [34,37].
Fig. 3.
Structure and cell entry mechanism of COVID-19 virus and the associated immune response. COVID-19 is transmitted through respiratory droplets of infected subjects to mucosal cells in the oral and respiratory system. SARS-CoV-2 binds to the ACE2 receptor after activating the spike protein by transmembrane protease serine 2 (TMPRSS2). Reproduced with permission from Ref. [40].
2.2. Cell entry mechanism of COVID-19 viral infection
The COVID-19 virus has a genome sequence similar to its predecessor viruses responsible for severe acute respiratory syndrome [38]. COVID-19 spike protein S1 has a receptor-binding domain (RBD) that particularly identifies angiotensin-converting enzyme 2 (ACE2) as its receptor and binds to it for cellular uptake [13]. On binding, the virus is sacked inside an endosome and ingested into the cell. Inside the endosome cathepsin L, a lysosomal endopeptidase enzyme starts digesting the spike proteins of the COVID-19 virus, leaving the viral RNA intact for replication. ACE2 surface proteins are predominantly present in the nose, lungs, kidneys, blood vessels and gastrointestinal tract [39].
Furthermore, studies have demonstrated that the COVID-19 virus spike protein exhibits 10 to 20 times higher affinity for ACE2 compared to SARS-CoV. ACE2 receptors convert the angiotensin II peptides to molecules capable of vasodilation in increased blood pressure and vice versa. Also, it plays a vital role in physiological functions, which include regulating blood pressure, retaining sodium and potassium balance, maintaining fluid volume, and inflammatory response [41,42]. In addition to RBD, previous studies have shown that protease activators such as transmembrane protease/serine subfamily member 2 (TMPRSS2) play a crucial role in the COVID-19 virus fusing on to a cell surface [43]. TMPRSS2, a single-pass type 2 membrane protein, has been shown to proteolytically cleave the viral spike protein, thereby activating the virus and releasing the viral RNA into the host cell. For a virus to enter the cell, the spike protein must be primed through an enzyme called protease. COVID-19 uses TMPRSS2 to complete this process [44]. As a result, the viral RNA gets entry into the host cell environment for subsequent replication of the virus [13].
2.3. Pathophysiology of COVID-19 viral infection
Once the virus enters the cell, patients show signs of fever, dry cough, headache and myalgia at the incubation phase. Followed by seven days from the onset of the COVID-19 infection, the patient slowly develops pneumonia, which eventually gets worsened into acute respiratory distress syndrome and is fatal at the convalescence phase. At every stage of infection, there will be a release of significant biomarkers in the host system. Biomarkers play a crucial role in identifying the prognosis and pathogenesis of the disease. During the incubation phase, the clinically significant biomarkers reported in the literature for the COVID-19 diagnosis include 2019 new Coronavirus RNA (2019n-CoV) and SARS-CoV-2 antigen, which are collected from nasal and throat swabs. After seven days from the onset of the disease, patients produce IgG and IgM antibodies (developed in the patient's serum against the viral infection), which could be analyzed by sampling blood samples. These biomarkers are crucial in evaluating the epidemiological factors such as the extent of viral infection, age ranked seroprevalence and disease severity ratio [[45], [46], [47]]. In support of this statement, neutralization monoclonal antibodies such as CA1 and CB6 were identified from the convalescent SARS-CoV-2 affected individuals, and among which, CB6 is found to be the potential therapeutic agent for COVID-19. Also, metabolites, lipid molecules and other diagnostic factors including white blood cell (WBC) count, eosinophil count, serum ferritin and serum amyloid A (SAA) have the most clinical significance during infection [[48], [49], [50], [51]]. Moreover, there will be alterations in the levels of cytokines such as interleukin-6 (IL-6), IL-10, IL-2 and interferons-γ (IFN-γ) in the peripheral blood as COVID-19 infection is accompanied by cytokine storm [52,53] and could be analyzed by sampling sweat, tears and urine samples. All these biomarkers are present in very low concentrations, however, could be rapidly and accurately diagnosed with the help of biosensors.
2.4. State of currently developed methods for COVID-19 diagnosis
Early identification of SARS-CoV-2 is critical for timely control and isolation of reported cases to deter further transmission of the virus. Currently, reverse transcriptase-polymerase chain reaction (RT-PCR) is considered as a “gold standard” for the detection of SARS-CoV-2 [54]. RT-PCR is a nucleic acid amplification technique wherein the ribonucleic acid (RNA) is translated into complementary deoxyribonucleic acids (cDNA) and repeatedly multiplied for detection [55]. It detects the presence of viral nucleic acids in nasopharyngeal swab samples. However, the technique has certain caveats demanding complicated and expensive devices for testing [56]. The PCR framework to handle high sample throughput is inadequate in community hospitals outside metropolitan cities. Though it is a highly-sensitive technique, there is still a risk of RT-PCR eliciting false-negative results [38].
Loop-mediated isothermal amplification (LAMP) is another unique technique that can quickly amplify specific nucleic acid under isothermal conditions with increased specificity [57]. The method is performed using 4 to 6 specially designed primers and a DNA polymerase with chain displacement activity [58]. RNA is first translated into cDNA by usual reverse transcriptase implemented in RT-PCR. The LAMP system requires a constant temperature of approximately 60–65 °C [59], and the device can be more straightforward and economical than the thermos-cycler used in conventional RT-PCR [60]. One of the limitations of the LAMP technique is the intricate primer design, which could impede target site selection [61].
Serological tests rely on blood samples instead of swab samples used in molecular diagnostic tests and are employed for detecting antibodies generated in the human body in response to viral infection [62]. Once the patient is affected with COVID-19 infection, Immunoglobulin M (IgM) will be produced after three to six days of COVID-19 viral infection, and immunoglobulin G (IgG) will be detectable after eight days of infection [63]. Some of the serological tests include lateral flow immunoassay (LFIA), chemiluminescence immunoassay (CLIA) and enzyme-linked immunosorbent assay (ELISA) are in practice for COVID-19 diagnosis [64]. However, such types of tests are not useful for diagnosing a viral infection in individual subjects.
Several low cost immunoassays and molecular diagnostic kits, including Abbott's ID NOWTM, Lumex Instruments' Microchip RT-PCR and Roche's Cobas, etc., have been developed [[65], [66], [67], [68]]. However, serological tests are used as a surveillance technique to understand better the epidemiology of the population-wide immunity environment of COVID-19 [69]. As mentioned earlier, RT-PCR results in low sensitivity which eventually led to missed diagnosis and rapid spread of this disease [70]. In this context, chest computed tomography (CT) has been considered an assistive tool for the accurate diagnosis of COVID-19 [71]. In CT, a narrow beam of X-ray measurements is taken at different angles across the patient's chest to generate cross-sectional images [72,73]. The chest CT features associated with COVID-19 demonstrated ground-glass opacities [74], reticular pattern [75], reverse halo sign [76], pleural effusion [77], paraseptal emphysema [78] and bronchial wall thickening [79]. Meanwhile, CT scans should be performed with strict safety measures to avoid unnecessary radiation and COVID-19 exposure to patients and the healthcare workforce [80]. Since COVID-9 is classified as a class B infectious disease, a high level of disinfection and sterilization processes need to be followed in the equipment operating room [81].
3. Point of care diagnostic platforms for COVID-19 diagnosis
Biosensors are the appropriate PoC diagnostic platforms and defined as portable analytical tools developed to deliver care near the patient [82]. Biosensors involve the utilization of biological molecules such as antibodies, proteins, enzymes, aptamers, cells, and nucleic acids as receptor probes. They are generally immobilized on the transducer surface utilizing physisorption or chemisorption. At the transducer surface, the biochemical interaction events such as affinity reaction between the antigen and antibody, nucleic acid hybridization, protein-protein interactions are measured employing various transduction modalities such as optical, electrochemical, piezoelectric, mechanical, magnetic and thermal [23,46,83]. The three critical components of the biosensor include the selection of (i) bioreceptors for specific detection of biomarkers (ii) transduction mechanism to interpret the biochemical events as readable signal (iii) detector for quantification and further analysis [84]. These have evolved considerably and are generally categorized into handheld diagnostic units and compact benchtop systems [85]. The commercial market of PoC diagnostic platforms are governed by optical and electrochemical biosensors, which include glucometers and pregnancy test strips. In both optical and electrochemical biosensors, nanoparticles are utilized to promote ultrasensitive detection of biomarkers present in the biofluidic samples.
3.1. Optical biosensors
Generally, optical biosensors are defined as the analytical devices in which the analyte-bioreceptor interaction event is interpreted in terms of changes in optical measurements such as luminescence, fluorescence, absorbance and reflectance, which correlates to the concentration of the analyte [86]. From the viewpoint of PoC based optical biosensing platforms, lateral flow immunoassay (LFIA) test strips are considered the economical alternative for the instant diagnosis of COVID-19 in remote locations and public health centres [87] (Fig. 4 ). Although LFAs are proven to be the gold standard, more research is focused on promoting high-throughput immune-analyzers for mass screening. The sensing principle behind LFAs involves detecting analytes (could be an antigen or antibody) with the help of secondary antibodies conjugated with labels such as gold nanoparticles, fluorescent molecules and quantum dots, promoting visual sensing of color changes. The utilization of nanoparticles as labels has gained attention in developing rapid diagnostic test kits for improved diagnosis and treatment [88,89].
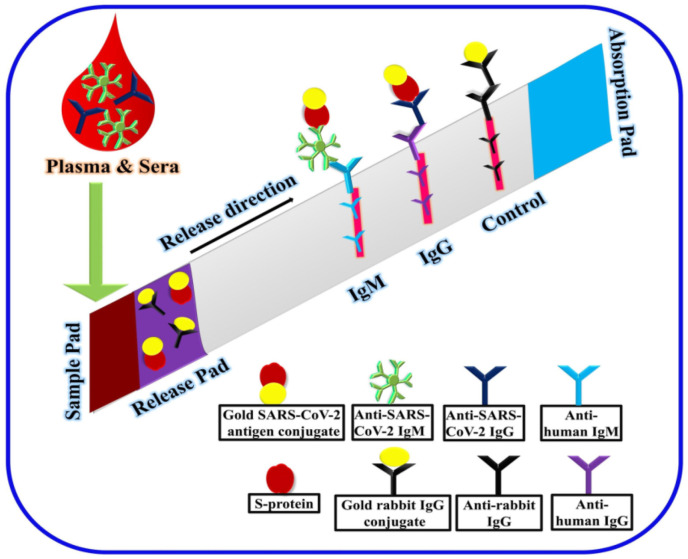
Fig. 4.
Lateral flow immunoassay based sensing of COVID-19 viral infection. The patients' serum sample containing IgG and IgM antibodies is introduced in the sample pad and flows through the strip via capillary action. The release pad contains the gold SARS-CoV-2 antigen conjugate and gold rabbit IgG conjugate. The two test lines are immobilized with anti-human IgG and anti-human IgM, and the control line is immobilized with anti-rabbit IgG. The respective antigen and antibody interactions are measured as red lines with the help of gold bioconjugates. Reproduced with permission from Ref. [98].
Nanoparticles gain much significance in developing PoC diagnostic platforms as they offer biocompatibility, ease in bioconjugation, robust, rapid and ultrasensitive diagnosis with less sample volume [90]. The property of nanoparticles offering enhanced sensitivity with minimal sample volume is considerable for COVID-19 detection. A limited amount of biosamples such as throat swab, nose swab, saliva sputum and blood of the patient is collected for analysis. Also, nanoparticles exhibit excellent optical and electronic properties such as high chemical reactivity, excellent stability, enhanced sensitivity and large surface area to volume ratio for better interaction [91]. Some of the widely used nanoparticles as biosensor labels include noble metal nanoparticles (gold and silver) [92], magnetic nanoparticles (iron oxide and magnetic beads) and fluorescent nanoparticles (fluorescent molecules and quantum dots) [93].
Noble metal nanoparticle-based biosensing platforms involve applying gold and silver nanoparticles with various geometrical dimensions as labels. These noble metal nanoparticles exhibit excellent optical absorption and emission characteristics, which are entirely dependent on the dielectric properties of the metal, surrounding medium, particle size and shape [[94], [95], [96]]. Generally, gold nanofilms and gold nanoparticles (GNPs), along with receptor binding domains, are employed to develop plasmonic biosensors such as surface plasmon resonance (SPR) and localized surface plasmon resonance (LSPR) sensors. In localized surface plasmon resonance (LSPR) based sensors, metallic nanoparticles with a size smaller than the wavelength of light show strong dipolar excitations. LSPRs are non-propagating excitations of the conduction electrons of metallic nanoparticles coupled to the electromagnetic field, vital for ultrasensitive detection of biomolecules [97].
A literature report utilizing gold nanoparticles as labels were presented to develop an LFIA test strip to detect SARS-CoV-2 IgG antibody present in the patient serum. The development of the LFIA test strip involves the utilization of (i) nitrocellulose membrane, (ii) labels such as gold, carbon, magnetic, colored latex and polystyrene nanoparticles, and (iii) antibodies. LFIA test strips consist of a sample pad where the sample can enter and interact with the immobilized antibodies and conjugates, followed by removing excess sample using the absorption pad. The SARS-CoV-2 nucleocapsid protein is immobilized onto the LFA strip and functions as capture receptors, whereas anti-human IgG conjugated gold nanoparticles are utilized as signal reporters. Upon interaction of the analyte with the gold nanoparticles conjugated antibodies, a red-colored line will be produced, indicating the presence of the analyte in the sample. The developed LFA strips achieved rapid detection (10–15 min) in a less sample volume (10 μL of the patient serum) with improved sensitivity of 69.1% [99]. Furthermore, immunochromatographic test strips for the simultaneous detection of IgG and IgM viral antibodies instead of single viral antibodies were investigated to facilitate better sensitivity [100,101]. The study revealed that the sensitivity of the assay was low in the early stage (<7 days) and increased from intermediate (8–14 days) to a later stage (>15 days), thus confirming the release of antibodies after 7 days from the onset of the disease. The efficiency of the strips for COVID-19 diagnosis was blindly proofed with the RT-PCR results.
In the timeline of COVID-19 viral infections, some of the metabolites are upregulated, and few others are downregulated. In this context, teslin paper substrate-based mass spectrometry (PS-MS) was reported to detect these metabolomic biomarkers. It could be considered as a rapid assistive technique for PCR in the early diagnosis and prognosis of the COVID-19 infection [102]. Teslin, a microporous polyolefin silica matrix, offers more prolonged sample activation and retention, thereby minimizing the interferences and provide a better response compared to conventional cellulose substrates. The statistical analysis of Teslin PS-MS showed a 93.3% correlation with the PCR results. Targeting glycoprotein of COVID-19 enables a faster diagnosis and therapeutics with high sensitivity. A recent study showed that the glycoprotein spikes have a higher affinity to angiotensin-converting enzyme-2 compared to other antibodies [103].
Besides LFA and paper-based substrates, several PoC diagnostic platforms were reported in the literature for COVID-19 diagnosis, which includes optical fiber probes [104,105] and microfluidic chips. PoC diagnosis using optical fiber probes relies on the principle of evanescence [106]. Generally, light propagates through the optical waveguide employing total internal reflection (TIR). When TIR takes place at the core-clad interface, reflected photons generate an electric field in the direction opposite to that of the interface [107]. This decaying electric field generated at the interface is called evanescence, which is exploited for sensing, stating that the analytes should present in the vicinity of the evanescent field. This decaying electric field could be enhanced with various factors, including fiber geometry and (utilization of plasmonic nanoparticles/nanofilms. Towards this end, a plasmonic fiber optic absorbance biosensor (P-FAB) using both labeled and label free immunosensing approaches was proposed for the detection of nucleocapsid (N) proteins in the COVID-19 virus [108]. Herein, a U-bent decladded optical fiber surface is considered to be the sensing region. The immunoassay is realized on the surface of the U-bent portion and the binding of the analyte with the bioreceptor is realized in the form of a decrease in the optical intensity.
In addition to proteins, genetic biomarkers are also employed to detect the viruses through a hybridization reaction [109]. Generally, GNPs absorbs light at the plasmonic region and emits non-radiative relaxation (heat). This dual property is used to develop an LSPR biosensor based on the plasmonic photothermal effect (PPT). Gold nanoislands were fabricated and conjugated with complementary DNA receptors to detect the SARS-CoV-2 sequence through hybridization of nucleic acid sequences. The heat generated when exposed at the plasmonic region assist in the hybridization reaction [110]. The proposed LSPR biosensor achieved a limit of detection down to 0.22 pM and encouraged selective detection of the specific target sequence in a multi-gene sample. An SPR sensor with gold nanometal films produced utilizing vacuum evaporation was reported for multiplex detection of respiratory viruses. In this study, the gene probes are printed onto the thin gold film and reported enhanced sensitivity through hybridization detection [111].
Other nanomaterials such as fluorescent nanoclusters and quantum dots also exhibit excellent optical properties, thereby improving sensing and disease diagnosis [112,113]. The optical behavior of fluorescent nanomaterials from group II and VI is due to the quantum confinement and increased surface to volume ratio. Furthermore, they demonstrate color-tunable emission, large stoke shift and higher quantum yield [113]. Fluorescent labels, in conjunction with the optical fibers prove to be a simpler means of developing sensor probes. A localized surface plasmon coupled fluorescence-based fiber optic biosensor (LFI-GNPs) was reported for the enhanced detection of COVID-19 nucleocapsid protein (N protein) [114]. In this report, PMMA optical fiber was utilized in which the decladded portion is subjected to immobilization of biorecognition antibodies and labels conjugated with fluorophores. The sensor showed 0.1 pg/ml sensitivity, which is 104 fold higher than the conventional ELISA test and proved to be a suitable candidate for high throughput sensing. A low-cost microfluidic platform utilizing polycarbonate was developed for multiplexed detection of IgG/IgM/SARS-CoV-2 antigen using fluorescent molecules as labels. The salient features of the microchip are in-built centrifugation, fluorescence immunodetection, display for quantitative analysis and European CE certification [115].
Various other PoC platforms developed using magnetic nanoparticles are discussed in detail as follows: Magnetic nanoparticle-based sensors have shown promising potential in biosensing applications over the decade [116]. Magnetic materials such as iron, cobalt, nickel, and manganese are used to fabricate the nanosized particles. This involves the application of magnetic nanoparticles and magnetic beads as labels to conjugate with the bioreceptors. These magnetic nanoparticles could substitute the tedious filtration process of biomolecular separation thereby replacing the heavy systems such as centrifugation and filtration units. Besides, they offer low background noise as the biological molecules are non-magnetic. The fast response and relaxation of magnetic nanoparticles open a new paradigm for biomedical diagnostics to improve the separation and purification of biomolecules, enhanced signal amplification, and ultrasensitive signal read-out. Opto-magnetic sensors, magneto-resistive sensors, giant magnetoresistance, magnetic reed switch are the currently used methodologies for magnetic biosensing applications [113,117]. Magnetic biosensors have been reported for various applications which include detecting cancer biomarker, stem cell, pathogens (bacteria and virus) and toxic-metal ions.
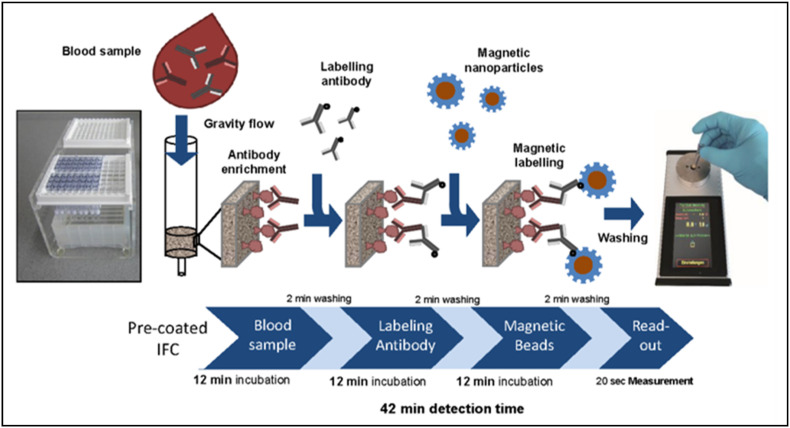
Extraction of the RNA from the virus is the crucial step involved in diagnosing COVID-19 using RT-PCR; however, nucleic acid extraction using the conventional column technique is laborious and time-consuming. In this perspective, amino-modified magnetic nanoparticles with multiple carboxyl groups (MNPs) were reported to promote automated nucleic extraction as there is a strong interaction between nucleic acid and carboxyl groups [118]. The synthesized MNPs were exploited to extract the RNA from pseudovirus samples to detect ORF1ab region through RT-PCR. Interestingly, the MNPs exhibited 100% RNA extraction, easy extraction process in less time (<30 min), reduced the false-negative result by avoiding RNA loss during elution, and compatible with various amplification procedures. A magnetic chemiluminescence immunoassay was reported to detect target specific COVID-19 antibodies (IgG and IgM)using synthetic biotinylated peptides and streptavidin-coated with magnetic nanobeads [118]. Herein, a luminescence is produced upon the interaction of the antibodies with the corresponding peptides coated magnetic nanobeads. The test results proved the excellent specificity offered by the assay and the detection rate for IgG (71.4%) is higher compared to IgM (57.2%). Compared to the commercial ELISA, magnetic chemiluminescence immunoassay showed higher specificity and sensitivity. Pietschmann and his team developed a novel magnetic immunodetection to promote PoC detection of COVID-19 by employing antigen-coated immunofiltration columns (IFC)as shown in Fig. 5 [119]. In the magnetic immunodetection technique, antibodies from the sample is allowed to flow through the precoated IFC, followed by enabling the secondary antibodies to flow. Then, functionalized magnetic nanobead labels are applied, and the IFC is washed to remove any unbound beads. Different concentration of antibodies were retained within the coloumn and further fed into a portable magnetic sensing device. The sensing of antibodies is made possible by exciting the coloumn at 49 KHz of magnetic field using the faraday coils. This magnetic immunoassasy detection provides higher sensitivity, portable, low cost and four fold less time than ELISAtest. This technique has high sensing ability and could be extended for multiplex detection thereby promoting PoC sensing of COVID-19.
Fig. 5.
Magnetic immunodetection technique using immunofiltration columns. The sample containing antibodies is made to flow through IFC followed by the injection of secondary antibodies. Magnetic nanoparticles labels are utilized to retain the antibodies, and the unbound molecules are washed. PoC sensing of COVID-19 is possible with portable magnetic read-out units. Reproduced with permission from Ref. [119].
3.2. Electrochemical biosensors
Generally, electrochemical biosensors transcribe biochemical events such as the interaction between the analyte and bioreceptor into a readable electrical signal using electrodes as sensing probes [120]. Various electrochemical biosensors for detecting vital biomarkers such as blood glucose, uric acid, ketones, lactate and deoxyribose nucleic acid (DNA) have been reported [121]. From the viewpoint of application and measurable signal at the output end, electrochemical biosensors are categorized into potentiometric, amperometric and conductometric sensors [122]. In a potentiometric sensor, a possible potential difference is measured between the working and reference electrodes due to the changes in ion concentration produced as a result of biochemical interactions. Amperometric biosensor generates an electric current, which is associated with the electron flow resulting from a redox reaction. The conductometric biosensor measures electrical conductivity, which is influenced by the variations in the ionic strength of a solution. Nanoparticles based on electrochemical biosensors have been developed and implemented for virus detection as they offer portability, cost-effectiveness and high sensitivity [123]. In this context, Tripathy and co-workers [124] developed an electrochemical biosensing technique in which gold nanoparticles deposited on a titanium (Ti) surface through electrodeposition acts as a sensing electrode as it offers stability, simplicity and inertness to severe chemical treatments. Platinum was used as reference and counter electrodes and a polydimethylsiloxane (PDMS) reservoir, which establishes the area of reaction and confinement of the electrodes. The hybridization of the viral RNA or cDNA with the complementary probe introduced into the reaction chamber results in an electrical signal, including voltage, current or impedance. In another study, a screen-printed carbon electrode was developed for COVID-19 diagnosis based on the potentiostat, and the efficiency of the electrode is compared with fluorine-doped tin oxide (FTO) electrode cast with gold nanoparticles [125]. Here, the monoclonal antibodies were immobilized onto the gold nanoparticles, and the change in electrical conductivity is measured. In this study, gold nanoparticles serve as a catalyst and amplify the electrochemical signal, thereby enhancing the electrical conductivity.
One of the significant variables influencing the replication of the SARS-CoV-2 infection within the host lungs is triggering mitochondrial reactive oxygen species (ROS) [[126], [127], [128], [129]]. In this direction, an electrochemical sensor utilizing multi-wall carbon nanotubes (MWCNTs) was proposed to detect ROS in the sputum sample, which is a vital source of lung epithelium. Earlier literature reports demonstrated that the MWCNTs are sensitive to super-oxidants, such as hydrogen peroxide (H2O2)/ROS [130]. Herein, the MWCNTs are immobilized on the tip of steel needles in three-electrode conformation consisting of working, reference, and counter electrodes [131]. In this three-electrode configuration, the reference electrode should have a stable potential and is used for measuring the working electrode potential, whereas, the counter electrode is used to complete the cell circuit in the electrochemical cell. With the help of cyclic voltammetry, the interaction of the ROS with the MWCNTs was monitored. The interaction resulted in the generation of cathodic ionic peak current, and in turn, the electric charges are released through the counter electrode. Moreover, the results correlate with the CT scan results of patients with 92% sensitivity and 94% specificity. In contrast, it presented 97% sensitivity and 91% specificity in correlation with the RT-PCR assay, the gold standard for detecting SARS-CoV-2 infections. Hence, this electrochemical sensor could be promoted as a PoC assistive diagnostic technique for early screening and rapid diagnosis in less than 30 s.
A biosensor based on bioelectric recognition assay (BERA) refers to the estimation of biochemical substances based on the precise and selective interaction of these substances with cells immobilized on a matrix presented [132]. A change in electric potential arises upon contact with the target molecule suspended in the gel matrix. A membrane engineered mammalian cell is attached with SARS-CoV-2 antibodies generate by electro-insertion [133]. A biosensor based on BERA was developed to identify the S1 functional subunit of spike protein present on the surface of the SARS-CoV-2 virus. This S1 subunit is responsible for binding the virus to the angiotensin-converting enzyme-2 (ACE-2) receptor in the host [13]. The interaction of the antibody with the S1 functional subunit results in a change in bioelectric properties. The device was custom fabricated on screen-printed electrodes covered by the PDMS layer attached to the electrode employing an adhesive. The technique provides rapid response, with a better detection limit of 1 fg/ml. Besides, no cross-reactivity was perceived against the SARS-CoV-2 nucleocapsid protein.
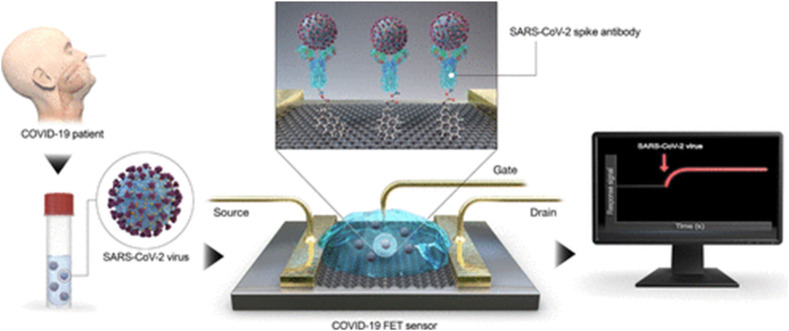
Recently, graphene is gaining significance as potential substrates for biosensing applications, mainly due to its excellent electronic conductivity, large surface area and robust in handling. A graphene-based Field Effect Transistor (FET) electrochemical biosensor was reported to detect the SARS-CoV-2 spike protein by immobilizing spike antibody onto the graphene surface using a crosslinker, and this offers excellent specificity over other coronaviruses due to the amino acid sequence diversity of the spike protein [134]. The basic principle in a FET type sensor is based on the potentiometric detection of changes in the charge-density induced at the gate insulator/solution interface. A schematic of a FET-based sensor is shown in Fig. 6 . The changes in surface charge bindings lead to changes in source-drain current measurements. The reported electrochemical biosensor offers excellent sensitivity (~1 fg/ml), the possibility of detection in a wide range of samples including buffer, clinical samples, cultured medium and nasopharyngeal swabs with no cross-reactivity. A graphene FET based on antibody-antigen interaction for real-time detection of spike protein S1 was investigated [135]. The sensor was fabricated by immobilizing an antibody of SARS-CoV-2 spike S1 subunit protein or ACE-2 on the surface of graphene. There is a change in conductance/resistance associated with graphene FET, which arises due to the binding of S1protein has a slight positive charge with SARS-CoV-2 spike S1 subunit protein or ACE2. This sensor has the advantages of providing rapid results, SARS-CoV-2 spike S1 subunit protein modified FET sensor showed higher sensitivity with a limit of detection of 0.2 pM.
Fig. 6.
Field Effect Transistor (FET) based electrochemical biosensor for COVID-19 detection. Here, graphene is used as a sensing medium and COVID-19 antibody is immobilized on a graphene sheet using 1-pyrenebutyric acid N-hydroxysuccinimide ester as a crosslinker. Reproduced with permission from Ref. [134].
3.3. Wearable and smart nanobiosensors
Despite the extensive research, most of the commercialized PoC biosensors are presented as the supportive diagnostic tool due to the challenges in automation, quantitative analysis and interference from complex biological samples. Another major limitation is remote health monitoring, real-time analysis and fall detection of the physiological parameters of the patients. This could be addressed with the advancements in the field of wearable sensors, nanotechnology and smartphone technology [136,137]. Wearable sensors are receiving attention mainly to promote continuous monitoring, non-invasive measurements and to overcome the laborious sample processing procedures involved in traditional laboratory diagnostic tests [[138], [139], [140]]. Moreover, biological samples such as sweat and tears offer selective detection compared to blood. Thus, wearable sensors offer PoC diagnosis and are efficient in addressing the mass level screening, which is vital in controlling the widespread of the disease.
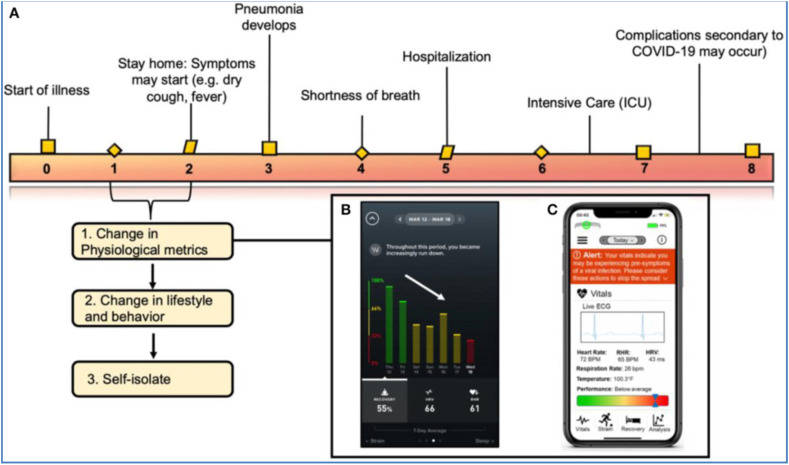
Seshadri et al., 2020 discussed the integration of wearables and android applications to predict and remotely monitor the alterations in the physiological status of the COVID-19 affected patients before the onset of clinical symptoms [141] (Fig. 7 ). Nowadays, there has been an increasing trend in the usage of smartwatches, including Fitbit, Amazefit, WHOOP and VivaLNK, for assessing the physical wellbeing of the individual. These wearables employ accelerometers and optical sensors to measure blood pressure, temperature and heart rate [142]. In this context, the usage of smartwatches and wearable devices for measuring clinically significant physiological parameters for COVID-19 diagnosis is proposed. During a viral infection, some of the physiological responses are elevated, including heart rate, core body temperature and sleep duration. With the increase in the severity of the infection, the person may exhibit lower SpO2 and arrhythmias. Herein, the abovementioned biological parameters such as heart rate, respiration rate, pulse rate, sleep activity, temperature are monitored using the commercial wearables to remotely track COVID-19 infection. This could ensure early identification and promote patient isolation before the onset of the COVID-19 illness [141]. Thus, these healthcare smartwatches are found to be viable for alerting individuals and identifying COVID-19 affected regions remotely with increasing efforts towards data security and handling.
Fig. 7.
Role of wearable technology and smartphone in the continuous monitoring of variations in the physiological parameters during the COVID-19 course of infection (A) Clinical symptoms and timeline of COVID-19 infection and correlating physiological parameters during the course of infection. (B) WHOOP application showing the decrease in physiological metrics and recovery of an individual diagnosed with COVID-19 (C) iPhone screen showing the physiological data measured from a wearable sensor and alerting the individual on his/her health status. Reproduced with permission from Ref. [141].
Apart from existing wearables measuring physiological parameters such as heart rate and other physical activity measurements, more research towards the development of wearables for identifying the vital parameters to detect COVID-19 infection, including respiratory rate, body temperature and respiratory activity in terms of cough frequency, is in progress. John A. Rogers and his group, in collaboration with the US Department of Health and Human Service's Biomedical Advanced Research and Development (BARDA) and Sonica Health, developed a chest mount patch sensor (Fig. 8 ) for measuring complete respiration related features, heart activity related vital parameters (heart sound, heart rate, and cardiac amplitude) and body temperature. The patch sensor consists of an accelerometer and a temperature sensor placed in direct contact with the skin at the base of the neck. The initial phase testing of the patch sensor was carried out with 50 subjects, and the results revealed the changes in the respiratory parameters are in correlation, which helped to understand the prognosis of COVID-19 viral infection [143]. Also, the patch sensor is robust, thereby achieving less discomfort to the patient and promoting the application for mass level screening.
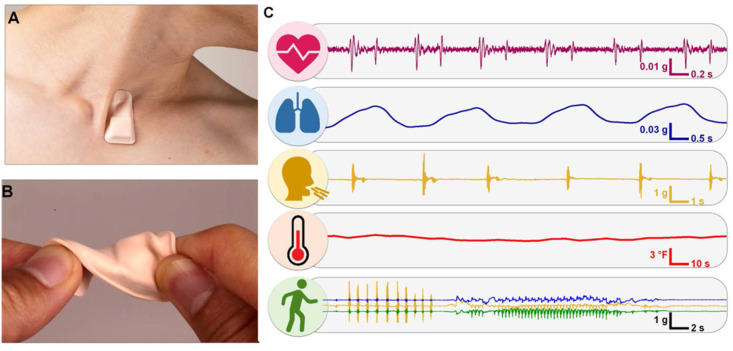
Fig. 8.
Chest mount wearable patch sensor for COVID-19 diagnosis along with the vital parameters measured (A) Wireless sensor placed at the neck position in such a way that the sensor is not affected by any movements/disturbances (B) Testing the robustness of the sensor (C) Physiological parameters including heart rate, respiration rate, coughing, temperature and patterns of activity measured using the sensor from a COVID-19 patient. Reproduced with permission from Ref. [143].
During this pandemic, face masks are found to be used by everyone as the foremost protective kit to control the widespread of COVID-19 infection [144]. There is a huge demand for face masks resulting in a global shortage of face masks. However, more prolonged usage of the facemask will have high risk and are more likely to affect themselves due to prolonged COVID-19 exposure. Predicting such risks with a color change in the facemask could be a better solution to control the spread. Such innovative high demanding face masks in combination with nanotechnology could pave the way for the development of wearable masks for diagnosis and self-protection [145,146]. Nanocoatings with polymer matrix could improve the filtration efficiency and hydrophobicity of the face mask, which could overcome the present concerns of disposable masks, including contamination, wetting, breathability and anti-microbial resistance.
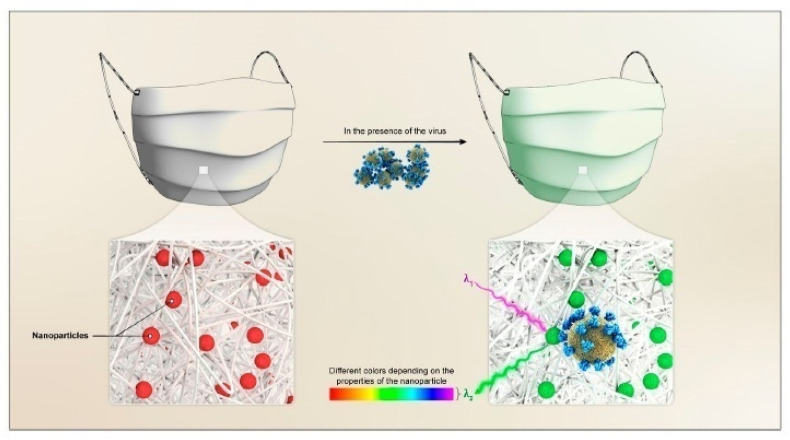
In addition to self-protection, a face mask for sensing application could be vital for early diagnosis. The metal-organic framework coated wearable mask was developed for real-time diagnosis of COVID-19 virus from the simple visual color changes. Herein, the changes in the optical properties of the noble metal nanoparticles are utilized to detect the presence of the virus. The nanoparticles are doped in a nanoporous matrix such as metal-organic frameworks (MOF) utilizing physisorption or chemisorption. This is followed by coating the nanoparticles doped MOF onto the surface of the mask (Fig. 9 ). When the doped nanoparticles interact with the virus, there will be alterations in the optical properties of the doped nanoparticles, which results in a visible color change.
Fig. 9.
Wearable mask for real-time detection of COVID-19. The mask is coated with nanoparticles doped metal-organic frameworks which undergo visual color changes upon interaction with the virus. Reproduced with permission from Ref. [135].
Moreover, smartphones could be utilized for color analysis and imaging to promote selective and quantitative detection [135]. There are studies reported on the application of MOF based biosensors for the detection of viruses including HIV-1, H1N1 and Zika virus [147]. Despite the ease in detection, more clinical trials are required to commercialize the MOF treated mask for COVID-19 diagnosis.
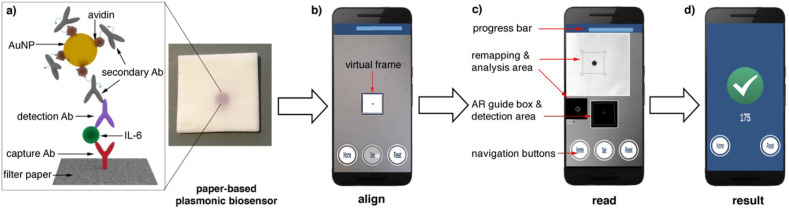
The integration of smartphones and nanotechnology has opened up smart nanobiosensors which could assist the public in making use of a smartphone as colorimetric, fluorimetric and electrochemical sensors [135,148]. Smartphones are used in two ways for biosensing applications (i) as a detector in interpreting the changes of the biochemical events [149] (ii) as a data analytics platform in creating a database, thereby facilitating the eHealth [150]. Herein, we discuss employing smartphones as a detector, ensuring better quantitative analysis in the following section. The progress in the smartphone PoC diagnostic platforms has been reviewed extensively which includes customized android application development, optoelectronics integration, and instrumental interface for imaging and color analysis, thereby ensuring colorimetric, electrochemical, and fluorimetric detection of an analyte in various biofluid samples such as saliva, urine, blood, sweat, and tears [151,152]. In this context, a paper-based plasmonic biosensor combined with a smartphone (Fig. 10 ) was developed for the automatic detection of interleukin −16 using augmented reality which is vital to diagnose COVID-19 [153].
Fig. 10.
Schematic showing the steps involved in the detection of Interleukin-6 (IL-6) using the paper-based smart plasmonic biosensor (a) Immunosensing scheme in which capture Ab (antibody) is immobilized onto the filter paper, and binding of IL-6 to capture Ab is detected using gold nanoparticles conjugated with avidin and biotinylated antibodies (b) Imaging of the colored spot on the filter paper with the help of virtual frame using smartphone (c) Detection screen displaying the automatic identification of the region of interest using augmented reality (d) The result screen indicating the pixel intensity value. Reproduced with permission from Ref. [154].
The smartphone app is utilized for the quantitative colorimetric analysis of the red spots created on the paper surface due to the interaction of gold nanoparticle labels with the cytokines from the sample. Various smartphone interfaced biosensor applications in conjunction with the microfluidic channels and bioassay cartridges to detect viral infections are tabulated in Table 1 . The smartphones are utilized to detect (i) colorimetric and fluorimetric changes in which smartphone camera and standalone android application are utilized for imaging and color analysis (ii) electrochemical alterations are monitored using a smartphone camera and CMOS detectors for further data interpretation and communication [148,151,152]. This is an easy to use technique and finds more scope towards PoC diagnostic platforms for controlling the COVID-19 and future pandemics.
Table 1.
Smartphone-based diagnostic platforms for detection of various viruses.
| Signal transduction | Analyte | Sensing substrates | Bioreceptors | Labels | Sensitivity | Ref |
|---|---|---|---|---|---|---|
| Electrochemical | COVID-19 RNA | Screen printed carbon electrode | Calixarene functionalized graphene | Au@Fe3O4 nanocomposites | 3 aM | [155] |
| Zika virus (ZIKV) | Functionalized interdigitated gold micro electrode | ZIKV specific envelope protein antibody | – | 10 pM | [156] | |
| Hepatitis C Virus | Three electrode system using PCB | – | – | 1 ng/μL | [157] | |
| Colorimetry | Zika virus (ZIKV) | Vial immunosensor | – | Platinum/gold core shell nanoparticles | 1 pg/ml | [158] |
| Avian influenza | PDMS microfluidic channel | H5 monoclonal antibodies | Gold nanoparticles with silver shells | 8 × 103 EID50/mLa | [159] | |
| Influenza A | Paper based microfluidic system | 14B11 antibody | Horse radish peroxidase enzyme conjugated 14F10 antibody | 32 × 10−4HAb | [160] | |
| Fluorimetry | Zika virus non-structural protein 1 | Nitrocellulose membrane | – | Quantum dot microspheres | 0.15 ng/ml | [161] |
| Influenza virus | CYTOP-on-coverslip | – | – | 3.0 × 104 PFU ml−1c | [162] | |
| Equine herpes virus 1 (EHV 1) | Silicon microfluidic chips | – | Evagreen dye | 5.5 × 104 copies/mLd | [163] |
Note: a-The amount of infectious virus present in the sample is expressed in terms of 50% embryo infectious dose per millilitre. b- HA refers to the measure of haemagglutinin protein content in the virus. c-The number of infective particles in the sample is expressed in terms of plaque-forming units per millilitre. d-The viral load in the sample is expressed as the number of copies per millilitre.
On the whole, the prospects of deploying wearable sensors for the real-time diagnosis of COVID-19 can aid in controlling the spread of the pandemic. The combination of nanotechnology with the smartphone-based detectors could serve for both qualitative and quantitative detection of COVID-19. However, this could offer only an initial screening for viral detection and followed by the recommendation of standard diagnostic protocols for further diagnosis and treatment.
4. Conclusions and summary
The global crisis of the COVID-19 pandemic has instigated more research towards developing affordable point of care diagnostic platforms for mass population screening and rapid diagnosis. In this perspective, this review highlighted the pathophysiology of COVID-19, the application of commercial existing lateral flow immunoassay strips for COVID-19 diagnosis, and other emerging diagnostic modalities, including optical, electrochemical and wearable biosensors. Highlighting the importance of quantification and PoC diagnosis, the integration of smartphones with nanotechnology and wearable sensors to develop smart nano-biosensors are discussed, which is crucial for the developing countries to control the pandemic.
Despite the rapid development of PoC immunoassays and molecular diagnostic kits for early screening and rapid diagnosis of COVID-19, there involves more optimization towards many factors to enter the commercial market for real-time deployment. Firstly, it involves a thorough clinical validation with a large number of clinical samples for mass sample screening. Secondly, a more accurate diagnosis is expected for early screening to minimize the false negatives and meet the universal standards. Finally, a robust database has to be formulated from the pilot studies to understand the unknown COVID-19 transmission, replication and immunity from the diverse clinical samples. Many low-cost COVID-19 diagnostic kits have been developed and are streamlined in the process of validation for FDA approval. However, there exist several hurdles, including limited sensitivity, multiplexing, reproducibility, real-time operability, miniaturization, sampling and patient willingness. These hurdles have to be addressed before they enter into the commercial market to operate in decentralized settings. Thus, the PoC diagnostic platforms in conjunction with advanced electronics, communication and data analytics, could strongly confront and combat the future pandemics.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Docea A.O., Tsatsakis A., Albulescu D., Cristea O., Zlatian O., Vinceti M., Moschos S.A., Tsoukalas D., Goumenou M., Drakoulis N., Dumanov J.M., Tutelyan V.A., Onischenko G.G., Aschner M., Spandidos D.A., Calina D. Int. J. Mol. Med. 2020;45:1631. doi: 10.3892/ijmm.2020.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singhal T. Indian J. Pediatr. 2020;87:281. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ezhilan M., Suresh I., Nesakumar N. Measurement. 2021;168:108335. doi: 10.1016/j.measurement.2020.108335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gralinski L.E., Menachery V.D. Viruses. 2020;12 doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coronavirus Disease 2019 (COVID-19) Situation Report – 88. World Health Organization; USA: 2020. [Google Scholar]
- 6.WHO Coronavirus Disease (COVID-19) Dashboard. 2020/9/13.
- 7.Chen B., Tian E.K., He B., Tian L., Han R., Wang S., Xiang Q., Zhang S., El Arnaout T., Cheng W. Signal Transduct. Target. Ther. 2020;5:89. doi: 10.1038/s41392-020-0190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mainardes R.M., Diedrich C. Ther. Deliv. 2020;11:411. doi: 10.4155/tde-2020-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh A.K., Singh A., Shaikh A., Singh R., Misra A. Diabetes Metab. Syndr. 2020;14:241. doi: 10.1016/j.dsx.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konwar A.N., Borse V. Sensors Int. 2020;1:100015. doi: 10.1016/j.sintl.2020.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascella M R.M., Cuomo A., Dulebohn S.C., Napoli R.D. StatPearls Publishing; 2020. Features, Evaluation, and Treatment of Coronavirus (COVID-19) [PubMed] [Google Scholar]
- 12.Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M., Zhang L.J. Radiology. 2020;296:E15. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Cell. 2020;181:281. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoeman D., Fielding B.C. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheikhi K., Shirzadfar H., Sheikhi M. A review on novel coronavirus (Covid-19): symptoms, transmission and diagnosis tests. Infect. Dis. Trop. Med. 2020;2:1. [Google Scholar]
- 16.Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R., Jervey S.R., Liu C. ACS Cent. Sci. 2020;6:591. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song F., Shi N., Shan F., Zhang Z., Shen J., Lu H., Ling Y., Jiang Y., Shi Y. Radiology. 2020;295:210. doi: 10.1148/radiol.2020200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Younes N., Al-Sadeq D.W., Al-Jighefee H., Younes S., Al-Jamal O., Daas H.I., Yassine H.M., Nasrallah G.K. Viruses. 2020;12 doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huff H.V., Singh A. Clin. Infect. Dis. 2020;71:2752. doi: 10.1093/cid/ciaa654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. ACS Nano. 2020;14:3822. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 21.Sethuraman N., Jeremiah S.S., Ryo A. JAMA. 2020;323:2249. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 22.Chauhan D.S., Prasad R., Srivastava R., Jaggi M., Chauhan S.C., Yallapu M.M. Bioconjugate Chem. 2020;31:2021. doi: 10.1021/acs.bioconjchem.0c00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morales-Narvaez E., Dincer C. Biosens. Bioelectron. 2020;163:112274. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravi N., Cortade D.L., Ng E., Wang S.X. Biosens. Bioelectron. 2020;165:112454. doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zainol Rashid Z O.S., Abdul Samat M.N., Ali U.K., Wong K.K. Malays. J. Pathol. 2020;42:13. [PubMed] [Google Scholar]
- 26.Kumar R., Nagpal S., Kaushik S., Mendiratta S. Virusdisease. 2020;31:97. doi: 10.1007/s13337-020-00599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhalla N., Pan Y., Yang Z., Payam A.F. ACS Nano. 2020;14:7783. doi: 10.1021/acsnano.0c04421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheikhzadeh E., Eissa S., Ismail A., Zourob M. Talanta. 2020;220:121392. doi: 10.1016/j.talanta.2020.121392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pokhrel P., Hu C., Mao H. ACS Sens. 2020;5:2283. doi: 10.1021/acssensors.0c01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pal M., Berhanu G., Desalegn C., Kandi V. Cureus. 2020;12 doi: 10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Viruses. 2012;4 doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letko M., Marzi A., Munster V. Nat. Microbiol. 2020;5:562. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schoeman D., Fielding B.C. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., Siddell S.G., Stamou D.G., Wilson I.A., Kuhn P., Buchmeier M.J. J. Struct. Biol. 2011;174:11. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McBride R., Van Zyl M., Fielding B.C. Viruses. 2014;6 doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Cell. Mol. Immunol. 2020;17:613. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H., Li X., Li T., Zhang S., Wang L., Wu X., Liu J. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1629. doi: 10.1007/s10096-020-03899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ni W., Yang X., Yang D., Bao J., Li R., Xiao Y., Hou C., Wang H., Liu J., Yang D., Xu Y., Cao Z., Gao Z. Crit. Care. 2020;24:422. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Funk C.D., Laferrière C., Ardakani A. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahmudpour M., Roozbeh J., Keshavarz M., Farrokhi S., Nabipour I. Cytokine. 2020;133:155151. doi: 10.1016/j.cyto.2020.155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verdecchia P., Cavallini C., Spanevello A., Angeli F. Eur. J. Intern. Med. 2020;76:14. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. Cell. 2020;181:271. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Proc. Natl. Acad. Sci. U.S.A. 2020;117:11727. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W.J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.S., Wang Q., Gao G.F., Yuan Z., Yan J. Nature. 2020;584:120. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 46.Choi J.R. Front. Chem. 2020;8:517. doi: 10.3389/fchem.2020.00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.GeurtsvanKessel C.H., Okba N.M.A., Igloi Z., Bogers S., Embregts C.W.E., Laksono B.M., Leijten L., Rokx C., Rijnders B., Rahamat-Langendoen J., van den Akker J.P.C., van Kampen J.J.A., van der Eijk A.A., van Binnendijk R.S., Haagmans B., Koopmans M. Nat. Commun. 2020;11:3436. doi: 10.1038/s41467-020-17317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaur M., Tiwari S., Jain R. Sens. Biosens. Res. 2020;29:100362. doi: 10.1016/j.sbsr.2020.100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kermali M., Khalsa R.K., Pillai K., Ismail Z., Harky A. Life Sci. 2020;254:117788. doi: 10.1016/j.lfs.2020.117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whetton A.D., Preston G.W., Abubeker S., Geifman N. J. Proteome Res. 2020;19:4219. doi: 10.1021/acs.jproteome.0c00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., Xiang X., Ren H., Xu L., Zhao L., Chen X., Long H., Wang Q., Wu Q. J. Infect. 2020;80:646. doi: 10.1016/j.jinf.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terpos E., Ntanasis-Stathopoulos I., Elalamy I., Kastritis E., Sergentanis T.N., Politou M., Psaltopoulou T., Gerotziafas G., Dimopoulos M.A. Am. J. Hematol. 2020;95:834. doi: 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahmoudi M. Mol. Pharm. 2020;1:1. [Google Scholar]
- 54.Xu Y., Xiao M., Liu X., Xu S., Du T., Xu J., Yang Q., Xu Y., Han Y., Li T., Zhu H., Wang M. Emerg. Microb. Infect. 2020;9:924. doi: 10.1080/22221751.2020.1752610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerard G.F., Fox D.K., Nathan M., D'Alessio J.M. Mol. Biotechnol. 1997;8:61. doi: 10.1007/BF02762340. [DOI] [PubMed] [Google Scholar]
- 56.Kubina R., Dziedzic A. Diagnostics. 2020;10:434. doi: 10.3390/diagnostics10060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshikawa R., Abe H., Igasaki Y., Negishi S., Goto H., Yasuda J. PLoS Neglected Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mautner L., Baillie C.-K., Herold H.M., Volkwein W., Guertler P., Eberle U., Ackermann N., Sing A., Pavlovic M., Goerlich O., Busch U., Wassill L., Huber I., Baiker A. Virol. J. 2020;17:160. doi: 10.1186/s12985-020-01435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagamine K., Hase T., Notomi T. Mol. Cell. Probes. 2002;16:223. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 61.Kashir J., Yaqinuddin A. Med. Hypotheses. 2020;141:109786. doi: 10.1016/j.mehy.2020.109786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacofsky D., Jacofsky E.M., Jacofsky M. J. Arthroplasty. 2020;35:S74. doi: 10.1016/j.arth.2020.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.di Mauro G., Scavone C., Rafaniello C., Rossi F., Capuano A. Int. Immunopharm. 2020;84:106519. doi: 10.1016/j.intimp.2020.106519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krajewski R., Gołębiowska J., Makuch S., Mazur G., Agrawal S. Clin. Chim. Acta. 2020;510:746. doi: 10.1016/j.cca.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu H., Zhang H., Ni S., Korabecna M., Yobas L., Neuzil P. Trends Anal. Chem. 2020;130:115984. doi: 10.1016/j.trac.2020.115984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu M., Wang D., Wang H., Zhang X., Liang T., Dai J., Li M., Zhang J., Zhang K., Xu D., Yu X. Clin. Transl. Med. 2020;10:e158. doi: 10.1002/ctm2.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Nat. Rev. Microbiol. 2020;1:1. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Afzal A. J. Adv. Res. 2020;26:149. doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lisboa Bastos M., Tavaziva G., Abidi S.K., Campbell J.R., Haraoui L.-P., Johnston J.C., Lan Z., Law S., MacLean E., Trajman A., Menzies D., Benedetti A., Ahmad Khan F. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Radiology. 2020;296:E41. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Radiology. 2020;296:E115. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Whiting P., Singatullina N., Rosser J.H. BJA Edu. 2015;15:299. [Google Scholar]
- 73.Lee E.Y.P., Ng M.-Y., Khong P.-L. Lancet Infect. Dis. 2020;20:384. doi: 10.1016/S1473-3099(20)30134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ye Z., Zhang Y., Wang Y., Huang Z., Song B. Eur. Radiol. 2020;30:4381. doi: 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Lancet Infect. Dis. 2020;20:425. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li X., Zeng X., Liu B., Yu Y. Radiology: Cardiothorac. Imaging. 2020;2 doi: 10.1148/ryct.2020200026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gong X., Song L., Li H., Li L., Jin W., Yu K., Zhang X., Li H., Ke H., Lu Z. PloS One. 2020;15 doi: 10.1371/journal.pone.0235134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muheim M., Weber F.J., Muggensturm P., Seiler E. BMJ Case Rep. 2020;13 doi: 10.1136/bcr-2020-237967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu J., Wu X., Zeng W., Guo D., Fang Z., Chen L., Huang H., Li C. Invest. Radiol. 2020;55 doi: 10.1097/RLI.0000000000000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mossa-Basha M., Medverd J., Linnau K.F., Lynch J.B., Wener M.H., Kicska G., Staiger T., Sahani D.V. Radiology. 2020;296:E26. doi: 10.1148/radiol.2019201326. [DOI] [PubMed] [Google Scholar]
- 81.Ding J., Fu H., Liu Y., Gao J., Li Z., Zhao X., Zheng J., Sun W., Ni H., Ma X., Feng J., Wu A., Liu J., Wang Y., Geng P., Chen Y. Eur. Radiol. 2020;30:3603. doi: 10.1007/s00330-020-06850-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu D., Wang J., Wu L., Huang Y., Zhang Y., Zhu M., Wang Y., Zhu Z., Yang C. Trac. Trends Anal. Chem. 2020;122:115701. [Google Scholar]
- 83.Cui F., Zhou H.S. Biosens. Bioelectron. 2020;165:112349. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mayeux R. NeuroRx. 2004;1:182. doi: 10.1602/neurorx.1.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.St John A., Price C.P. Clin. Biochem. Rev. 2014;35:155. [PMC free article] [PubMed] [Google Scholar]
- 86.CW Aref A., Kyoung M.Y., Hamed D., Ray T.C. arXiv. 2020;2008 [Google Scholar]
- 87.Ji T., Liu Z., Wang G., Guo X., Akbar Khan S., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H., Chen Y., Zhou Q. Biosens. Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mahapatra S., Chandra P. Biosens. Bioelectron. 2020;165:112361. doi: 10.1016/j.bios.2020.112361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Palestino G., Garcia-Silva I., Gonzalez-Ortega O., Rosales-Mendoza S. Expert Rev. Anti Infect. Ther. 2020:1. doi: 10.1080/14787210.2020.1776115. [DOI] [PubMed] [Google Scholar]
- 90.Chauhan G., Madou M.J., Kalra S., Chopra V., Ghosh D., Martinez-Chapa S.O. ACS Nano. 2020;14:7760. doi: 10.1021/acsnano.0c04006. [DOI] [PubMed] [Google Scholar]
- 91.Nikaeen G., Abbaszadeh S., Yousefinejad S. Nanomedicine. 2020;15:1501. doi: 10.2217/nnm-2020-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Narasimhan A.K., Chakaravarthi G., Rao M.S.R., Arunachalam K. Electromagn. Biol. Med. 2020:1. doi: 10.1080/15368378.2020.1762637. [DOI] [PubMed] [Google Scholar]
- 93.Narasimhan A.K., Lakshmi B S., Santra T.S., Rao M.S.R., Krishnamurthi G. RSC Adv. 2017;7:53822. [Google Scholar]
- 94.Sepúlveda B., Angelomé P.C., Lechuga L.M., Liz-Marzán L.M. Nano Today. 2009;4:244. [Google Scholar]
- 95.Zhao J., Zhang X., Yonzon C.R., Haes A.J., Van Duyne R.P. Nanomedicine. 2006;1:219. doi: 10.2217/17435889.1.2.219. [DOI] [PubMed] [Google Scholar]
- 96.Petryayeva E., Krull U.J. Anal. Chim. Acta. 2011;706:8. doi: 10.1016/j.aca.2011.08.020. [DOI] [PubMed] [Google Scholar]
- 97.Illath K., Narasimhan A.K., Shinde P., Wankhar S., Nagai M., Santra T.S. IEEE NEMS. 2020;1:448. [Google Scholar]
- 98.Hussein H.A., Hassan R.Y.A., Chino M., Febbraio F. Sensors. 2020;20 doi: 10.3390/s20154289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wen T., Huang C., Shi F.J., Zeng X.Y., Lu T., Ding S.N., Jiao Y.J. Analyst. 2020;145:5345. doi: 10.1039/d0an00629g. [DOI] [PubMed] [Google Scholar]
- 100.Pan Y., Li X., Yang G., Fan J., Tang Y., Zhao J., Long X., Guo S., Zhao Z., Liu Y., Hu H., Xue H., Li Y. J. Infect. 2020;81:e28. doi: 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. J. Med. Virol. 2020;9:1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.De Silva I.W., Nayek S., Singh V., Reddy J., Granger J.K., Verbeck G.F. Analyst. 2020;145:5725. doi: 10.1039/d0an01074j. [DOI] [PubMed] [Google Scholar]
- 103.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Science. 2020;367:1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nag P., Sadani K., Mukherji S. Trans. Indian Natl. Acad. Eng. 2020;5:233. doi: 10.1007/s41403-020-00128-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gowri A., Sai V.V.R. Sensor. Actuator. B Chem. 2016;230:536. [Google Scholar]
- 106.Chen C., Wang J. Analyst. 2020;145:1605. doi: 10.1039/c9an01998g. [DOI] [PubMed] [Google Scholar]
- 107.Jana J., Ganguly M., Pal T. RSC Adv. 2016;6:86174. [Google Scholar]
- 108.Murugan D., Bhatia H., Sai V.V.R., Satija J. Trans. Indian Natl. Acad. Eng. 2020;5:211. doi: 10.1007/s41403-020-00122-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. RNA. 2020;26:771. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. ACS Nano. 2020;14:5268. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 111.Shi L., Sun Q., He J., Xu H., Liu C., Zhao C., Xu Y., Wu C., Xiang J., Gu D., Long J., Lan H. Bio Med. Mater. Eng. 2015;26(Suppl 1):S2207. doi: 10.3233/BME-151526. [DOI] [PubMed] [Google Scholar]
- 112.Loczechin A., Seron K., Barras A., Giovanelli E., Belouzard S., Chen Y.T., Metzler-Nolte N., Boukherroub R., Dubuisson J., Szunerits S. ACS Appl. Mater. Interfaces. 2019;11:42964. doi: 10.1021/acsami.9b15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Manivannan S., Ponnuchamy K. Appl. Organomet. Chem. 2020:e5887. doi: 10.1002/aoc.5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huang J.C., Chang Y.F., Chen K.H., Su L.C., Lee C.W., Chen C.C., Chen Y.M., Chou C. Biosens. Bioelectron. 2009;25:320. doi: 10.1016/j.bios.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lin Q., Wen D., Wu J., Liu L., Wu W., Fang X., Kong J. Anal. Chem. 2020;92:9454. doi: 10.1021/acs.analchem.0c01635. [DOI] [PubMed] [Google Scholar]
- 116.Kai W R.S., Diqing S., Venkatramana D.K., Jinming L., Maxim C.J.C., Jian-Ping W. arXiv. 2020;1:1. [Google Scholar]
- 117.Xianyu Y., Wang Q., Chen Y. Trac. Trends Anal. Chem. 2018;106:213. [Google Scholar]
- 118.Zhao Z., Cui H., Song W., Ru X., Zhou W., Yu X. BioRxiv. 2020;1:1. [Google Scholar]
- 119.Pietschmann J., Vöpel N., Spiegel H., Krause H.-J., Schröper F. BioRxiv. 2020;1:1. [Google Scholar]
- 120.Thévenot D.R., Toth K., Durst R.A., Wilson G.S. Biosens. Bioelectron. 2001;16:121. doi: 10.1016/s0956-5663(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 121.Wang Y., Xu H., Zhang J., Li G. Sensors. 2008;8 doi: 10.3390/s8042043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Grieshaber D., MacKenzie R., Vörös J., Reimhult E. Sensors. 2008;8:1400. doi: 10.3390/s80314000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wilson M.S. Anal. Chem. 2005;77:1496. doi: 10.1021/ac0485278. [DOI] [PubMed] [Google Scholar]
- 124.Tripathy S., Singh S.G. Trans. Indian Natl. Acad. Eng. 2020:1. doi: 10.1007/s41403-020-00103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mahari S., Roberts A., Shahdeo D., Gandhi S. bioRxiv. 2020;1:1. 2020.04.24.059204. [Google Scholar]
- 126.Singh K.K., Chaubey G., Chen J.Y., Suravajhala P. Am. J. Physiol. Cell Physiol. 2020;319:C258. doi: 10.1152/ajpcell.00224.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miripour Z.S., Sarrami-Forooshani R., Sanati H., Makarem J., Taheri M.S., Shojaeian F., Eskafi A.H., Abbasvandi F., Namdar N., Ghafari H., Aghaee P., Zandi A., Faramarzpour M., Hoseinyazdi M., Tayebi M., Abdolahad M. Biosens. Bioelectron. 2020;165:112435. doi: 10.1016/j.bios.2020.112435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Goran J.M., Phan E.N., Favela C.A., Stevenson K.J. Anal. Chem. 2015;87:5989. doi: 10.1021/acs.analchem.5b00059. [DOI] [PubMed] [Google Scholar]
- 129.Cheng M.L., Weng S.F., Kuo C.H., Ho H.Y. PloS One. 2014;9:e113234. doi: 10.1371/journal.pone.0113234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Goran J.M., Phan E.N.H., Favela C.A., Stevenson K.J. Anal. Chem. 2015;87:5989. doi: 10.1021/acs.analchem.5b00059. [DOI] [PubMed] [Google Scholar]
- 131.Miripour Z.S., Sarrami-Forooshani R., Sanati H., Makarem J., Taheri M.S., Shojaeian F., Eskafi A.H., Abbasvandi F., Namdar N., Ghafari H., Aghaee P., Zandi A., Faramarzpour M., Hoseinyazdi M., Tayebi M., Abdolahad M. Biosens. Bioelectron. 2020;165:112435. doi: 10.1016/j.bios.2020.112435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kintzios S., Pistola E., Konstas J., Bem F., Matakiadis T., Alexandropoulos N., Biselis I., Levin R. Biosens. Bioelectron. 2001;16:467. doi: 10.1016/s0956-5663(01)00161-0. [DOI] [PubMed] [Google Scholar]
- 133.Mavrikou S., Moschopoulou G., Tsekouras V., Kintzios S. Sensors. 2020;20 doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. ACS Nano. 2020;14:5135. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 135.Rabiee N., Bagherzadeh M., Ghasemi A., Zare H., Ahmadi S., Fatahi Y., Dinarvand R., Rabiee M., Ramakrishna S., Shokouhimehr M., Varma R.S. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21145126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tu J., Torrente-Rodríguez R.M., Wang M., Gao W. Adv. Funct. Mater. 2019;30:1906713. [Google Scholar]
- 137.Mujawar M.A., Gohel H., Bhardwaj S.K., Srinivasan S., Hickman N., Kaushik A. Mater. Today Chem. 2020;17:100306. doi: 10.1016/j.mtchem.2020.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang D., Liu Q. Biosens. Bioelectron. 2016;75:273. doi: 10.1016/j.bios.2015.08.037. [DOI] [PubMed] [Google Scholar]
- 139.Stojanović R., Škraba A., Lutovac B. IEEE MECO. 2020;1:1. [Google Scholar]
- 140.Tavakoli M., Carriere J., Torabi A. Adv. Intell. Syst. 2020;2:2000071. [Google Scholar]
- 141.Seshadri D.R., Davies E.V., Harlow E.R., Hsu J.J., Knighton S.C., Walker T.A., Voos J.E., Drummond C.K. Front. Digit. Health. 2020;2 doi: 10.3389/fdgth.2020.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dias D., Paulo Silva Cunha J. Sensors. 2018;18 doi: 10.3390/s18082414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jeong H., Rogers J.A., Xu S. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abd4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Atangana E., Atangana A. Results Phys. 2020;19:103425. doi: 10.1016/j.rinp.2020.103425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schünemann H.J., Akl E.A., Chou R., Chu D.K., Loeb M., Lotfi T., Mustafa R.A., Neumann I., Saxinger L., Sultan S., Mertz D. Lancet Respir. Med. 2020;8:954. doi: 10.1016/S2213-2600(20)30352-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liu X., Zhang S. Influenza Other Respir. Viruses. 2020;14:472. doi: 10.1111/irv.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang Y., Hu Y., He Q., Yan J., Xiong H., Wen N., Cai S., Peng D., Liu Y., Liu Z. Biosens. Bioelectron. 2020;169:112604. doi: 10.1016/j.bios.2020.112604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Seo S.E., Tabei F., Park S.J., Askarian B., Kim K.H., Moallem G., Chong J.W., Kwon O.S. J. Ind. Eng. Chem. 2019;77:1. [Google Scholar]
- 149.Huang X., Xu D., Chen J., Liu J., Li Y., Song J., Ma X., Guo J. Analyst. 2018;143:5339. doi: 10.1039/c8an01269e. [DOI] [PubMed] [Google Scholar]
- 150.Chandra P. Sensors Int. 2020;1:100019. doi: 10.1016/j.sintl.2020.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu J., Geng Z., Fan Z., Liu J., Chen H. Biosens. Bioelectron. 2019;132:17. doi: 10.1016/j.bios.2019.01.068. [DOI] [PubMed] [Google Scholar]
- 152.Roda A., Michelini E., Zangheri M., Di Fusco M., Calabria D., Simoni P. Trac. Trends Anal. Chem. 2016;79:317. [Google Scholar]
- 153.Russell S.M., Alba-Patino A., Baron E., Borges M., Gonzalez-Freire M., de la Rica R. ACS Sens. 2020;5:1506. doi: 10.1021/acssensors.0c00979. [DOI] [PubMed] [Google Scholar]
- 154.Alba-Patiño A., Russell S.M., Borges M., Pazos-Pérez N., Álvarez-Puebla R.A., de la Rica R. Nanoscale Adv. 2020;2:1253. doi: 10.1039/d0na00026d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhao H., Liu F., Xie W., Zhou T.C., OuYang J., Jin L., Li H., Zhao C.Y., Zhang L., Wei J., Zhang Y.P., Li C.P. Sensor. Actuator. B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kaushik A., Yndart A., Kumar S., Jayant R.D., Vashist A., Brown A.N., Li C.Z., Nair M. Sci. Rep. 2018;8:9700. doi: 10.1038/s41598-018-28035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.de Campos da Costa J.P., Bastos W.B., da Costa P.I., Zaghete M.A., Longo E., Carmo J.P. IEEE Sensor. J. 2019;19:10701. [Google Scholar]
- 158.Hsu Y.P., Li N.S., Chen Y.T., Pang H.H., Wei K.C., Yang H.W. Biosens. Bioelectron. 2020;151:111960. doi: 10.1016/j.bios.2019.111960. [DOI] [PubMed] [Google Scholar]
- 159.Xia Y., Chen Y., Tang Y., Cheng G., Yu X., He H., Cao G., Lu H., Liu Z., Zheng S.Y. ACS Sens. 2019;4:3298. doi: 10.1021/acssensors.9b01927. [DOI] [PubMed] [Google Scholar]
- 160.Wu D., Zhang J., Xu F., Wen X., Li P., Zhang X., Qiao S., Ge S., Xia N., Qian S., Qiu X. Microfluid. Nanofluidics. 2017;21 [Google Scholar]
- 161.Rong Z., Wang Q., Sun N., Jia X., Wang K., Xiao R., Wang S. Anal. Chim. Acta. 2019;1055:140. doi: 10.1016/j.aca.2018.12.043. [DOI] [PubMed] [Google Scholar]
- 162.Minagawa Y., Ueno H., Tabata K.V., Noji H. Lab Chip. 2019;19:2678. doi: 10.1039/c9lc00370c. [DOI] [PubMed] [Google Scholar]
- 163.Sun F., Ganguli A., Nguyen J., Brisbin R., Shanmugam K., Hirschberg D.L., Wheeler M.B., Bashir R., Nash D.M., Cunningham B.T. Lab Chip. 2020;20:1621. doi: 10.1039/d0lc00304b. [DOI] [PubMed] [Google Scholar]