Abstract
Expression of the Human Endogenous Retrovirus Type K (HERV-K), the youngest and most active HERV, has been associated with various cancers and neurodegenerative diseases. As in all retroviruses, a fraction of HERV-K transcripts is exported from the nucleus in unspliced or incompletely spliced forms to serve as templates for translation of viral proteins. In a fraction of HERV-K loci (Type 2 proviruses), nuclear export of the unspliced HERV-K mRNA appears to be mediated by a cis-acting signal on the mRNA, the RcRE, and the protein Rec—these are analogous to the RRE-Rev system in HIV-1. Interestingly, the HIV-1 Rev protein is able to mediate the nuclear export of the HERV-K RcRE, contributing to elevated HERV-K expression in HIV-infected patients. We aimed to understand the structural basis for HIV Rev-HERV-K RcRE recognition. We examined the conformation of the RcRE RNA in solution using small-angle X-ray scattering (SAXS) and atomic force microscopy (AFM). We found that the 433-nt long RcRE can assume folded or extended conformations as observed by AFM. SAXS analysis of a truncated RcRE variant revealed an “A”-shaped topological structure similar to the one previously reported for the HIV-1 RRE. The effect of the overall topology was examined using several deletion variants. SAXS and biochemical analyses demonstrated that the “A” shape is necessary for efficient Rev-RcRE complex formation in vitro and nuclear export activity in cell culture. The findings provide insight into the mechanism of HERV-K expression and a structural explanation for HIV-1 Rev-mediated expression of HERV-K in HIV-infected patients.
Importance
Expression of the human endogenous retrovirus type K (HERV-K) has been associated with various cancers and autoimmune diseases. Nuclear export of both HIV-1 and HERV-K mRNAs is dependent on the interaction between a small viral protein (Rev in HIV-1 and Rec in HERV-K) and a region on the mRNA (RRE in HIV-1 and RcRE in HERV-K). HIV-1 Rev is able to mediate the nuclear export of RcRE-containing HERV-K mRNAs, which contributes to elevated production of HERV-K proteins in HIV-infected patients. We report the solution conformation of the RcRE RNA—the first three-dimensional topological structure for a HERV molecule—and find that the RcRE resembles the HIV-1 nuclear export signal, RRE. The finding reveals the structural basis for the increased HERV-K expression observed in HIV-infected patients. Elevated HERV expression, mediated by HIV infection or other stressors, can have various HERV-related biological consequences. The findings provide structural insight for regulation of HERV-K expression.
Keywords: HERV-K, RcRE, small-angle X-ray scattering, atomic force microscopy, HIV-1 Rev
Graphical Abstract
Introduction
Retroviruses constitute an ancient and ubiquitous family of RNA viruses that replicate via a DNA intermediate, which integrates into the host’s genome. When retroviruses infect germ cells, the resulting integrated sequence is inherited by the offspring in a Mendelian manner and becomes endogenized—HERVs are a result of multiple ancient germ-line infections1 and constitute ~8% of the human genome2. A growing number of studies attribute HERVs with important physiological roles in placental development3, embryonic immunity4, and muscle formation5,6. However, HERVs have also been associated with a number of diseases, most notably germline cancers and autoimmune diseases. We direct the reader to several reviews on the role of HERVs in human health and disease7–12.
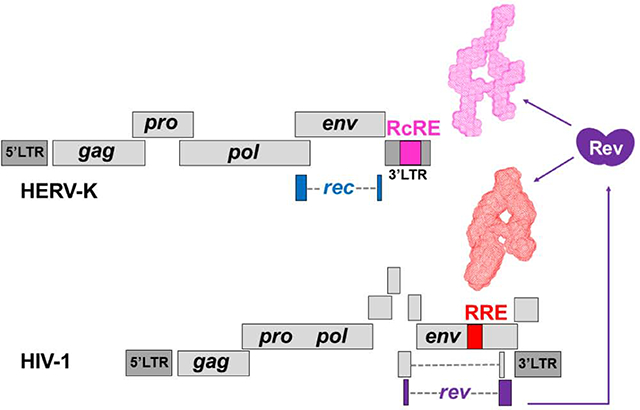
HERV-K entered the human lineage relatively recently (less than 1 million years ago)13. The full-length HERV-K genome encodes four genes (gag, pro, pol, and env) flanked by long terminal repeat (LTR) sequences that contain regulatory elements (Figure 1). Many HERV-K loci have deletions or inactivating mutations. Deletion events, most of which occur during homologous recombination, have resulted in about 1000 solo LTRs distributed throughout the genome14. Nevertheless, a number of the HERV-K loci encode functional proteins and LTRs can exert regulatory activity on neighboring genes9,15. The function of the HERV-K protein products is the subject of ongoing research. The HERV-K Gag protein can package HERV-K sequences into viral particles16,17. HERV-K Gag is homologous to the retrotransposon capsid-forming protein Arc, which encapsidates and transmits cellular mRNAs between neurons18. The Gag-Pro-Pol polyprotein is cleaved into the catalytic enzymes—protease, reverse transcriptase, and integrase. Env is a glycoprotein that has been proposed to facilitate oncogenesis via its ability to promote cell fusion and suppress immune responses (reviewed in 8).
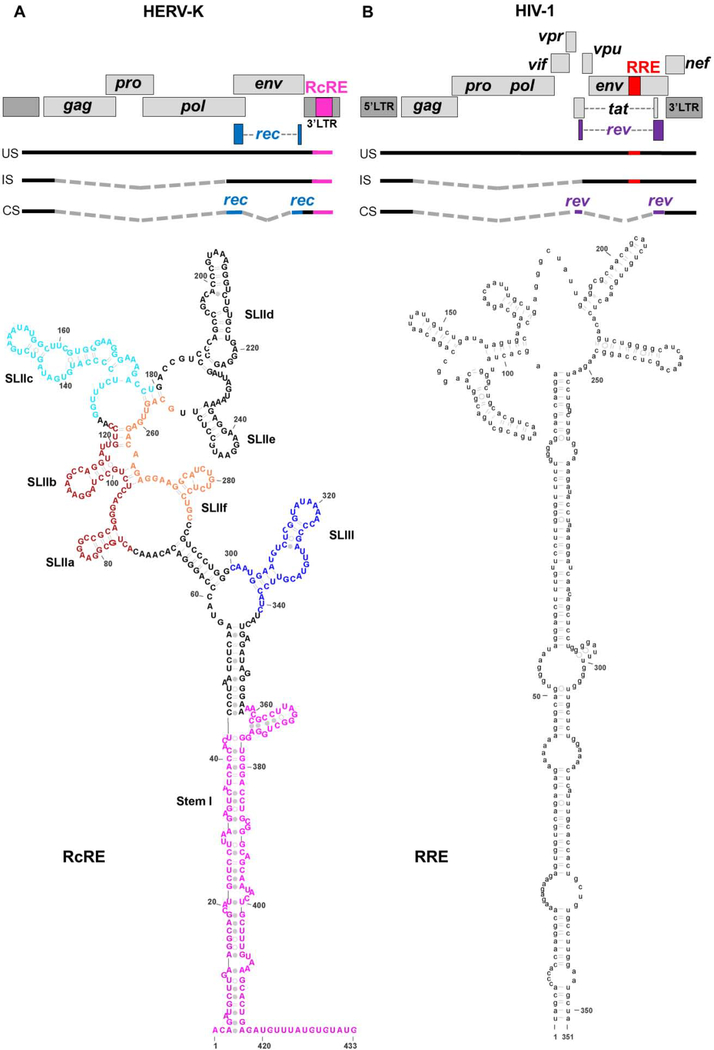
Figure 1.
Organization of the HERV-K and HIV-1 genomes and secondary structures of the HERV-K RcRE (A) and HIV-1 RRE (B). Above: Genetic organization of type 2 HERV-K and HIV-131,79. The 433-nt long RcRE is located in the ~1-kb 3’LTR and is part of the 3’ tail of the unspliced (US) and incompletely spliced (IS) mRNAs. A completely spliced (CS) mRNA codes for the Rec protein (blue coding regions). Gray, dashed lines represent regions excised during splicing. Though multiple splicing sites exist, only one schematic is shown to represent each mRNA group. Below: Secondary structure of the 433-mer HERV-K 108 RcRE (SHAPE-based structure from Langner et al.38) and of the 351-nt HIV-1 RRE37. The sequences were chosen based on previous mapping studies30,35. Stem regions that were deleted to prepare RcRE variants are shown in various colors. Secondary structure images prepared with VARNA80 and Adobe Illustrator.
In addition to the core elements, complex retroviruses also code for smaller, accessory proteins, which are products of alternative splicing (Figure 1). Synthesis of Gag, Gag-Pro-Pol, and Env polyproteins requires that unspliced or incompletely spliced HERV-K mRNAs are exported from the nucleus to the cytoplasm. Improperly processed cellular mRNAs are typically (though not always19) targeted for degradation, and viruses tend to evolve in order to evade these cellular quality control pathways20,21. Complex retroviruses code for a small protein—the product of a fully spliced message—which enters the nucleus upon translation and binds to a cis-acting region on the viral genome21,22. In HIV, the small protein is called Rev, and it binds to the Rev-response element (RRE) present on unspliced or incompletely spliced mRNAs. Rev-RRE binding initiates Rev oligomerization onto the RRE and this complex recruits the Crm-1/Ran-GTP cellular factors for nuclear transport (reviewed in 21). Cargo molecules are transported through nuclear core complexes that can mediate the egress and ingress of molecules that range widely in size, including large ribosomal units23. Other complex retroviruses, including HIV-224, HERV-K (see below), Human T-cell Lymphotropic Virus Type I (HTLV-I)25, Jaagsiekte Sheep Retrovirus (JSRV)26, Equine Infectious Anemia Virus (EIAV)27, and Mouse Mammary Tumor Virus (MMTV)28,29, use analogous protein-RNA pairs to mediate nuclear export of intron-containing mRNAs.
There are approximately 100 HERV-K loci in the human genome, some of which (known as type 2 HERV-K) code for a Rev/RRE-like system referred to as Rec and Rec-response element (RcRE) (the terms K-Rev, K-RRE, and cORF have also been used)30–33. The Rec protein interacts with the RcRE RNA in vitro32, and in cell culture this interaction is necessary for Crm1-dependent nuclear export of RcRE-containing RNAs32,33. Successful nuclear export of unspliced or incompletely spliced mRNAs enables translation of the viral Gag, Gag-Pro-Pol, and Env polyproteins33. Functional cross-talk studies between HIV-1 and HERV-K have demonstrated a non-reciprocal relationship between the two retroviral systems; namely, HIV-1 Rev can mediate nuclear export of RcRE-containing mRNAs30,32, but HERV-K Rec cannot induce transport HIV-1 RRE-containing mRNAs32. HIV-1 Rev-mediated trafficking of RcRE-containing mRNAs appears to vary with different RcRE loci34. Regardless, expression of the Rev protein in HIV-infected individuals (or in lentivirus-based therapeutic approaches) could alter the molecular composition of the cell by mediating the expression of both type 1 and type 2 HERV-K loci and neighboring genes.
Although the HIV-1 RRE and HERVK RcRE RNAs share little sequence similarity (Supplemental Figure 1), they have similar secondary structures characterized by a long stem and a cluster of several small stems centered around a central junction (Figure 1)30,35–38. We previously solved the three-dimensional topological structure of the HIV-1 RRE and found that it has a unique, flat and extended shape that resembles the letter “A”39. Based on the similarity of the secondary structures, we hypothesized that the two molecules, the RRE and the RcRE RNAs, have conserved three-dimensional structures allowing Rev to recognize both elements. To understand the structural basis of HIV Rev/HERV-K RcRE recognition, we examined the three-dimensional topological structure of the RcRE RNA in solution using small angle X-ray scattering (SAXS) and atomic force microscopy (AFM). We found that the RcRE exhibits striking similarities to the HIV-1 RRE attesting to the significance of the three-dimensional structure. The preservation of the three-dimensional topology was critical for Rev-binding in vitro and RcRE nuclear export activity in cell culture. The findings reveal the structural basis for the interaction between HIV-1 Rev and the HERV-K RcRE RNA and provide insight into the mechanism of Rev-mediated HERV-K expression.
Results
HIV-1 Rev binds to and mediates the nuclear export of RcRE-containing mRNAs.
Previous studies have demonstrated that the HIV-1 Rev protein is able to mediate the nuclear export of RcRE-containing mRNAs30,34,40. To compare the binding affinity of HIV-1 Rev to the RcRE versus the RRE, we performed electrophoretic mobility shift assays (EMSAs). The HERV-K 108 sequence (7p22.1) was chosen due to its use in previous RcRE (K-RRE) studies30,32, it is relatively well-studied, and its Env protein has been shown to inhibit HIV-1 virion production41. The RcRE sequence from this retrovirus has been mapped to a 433-nucleotide long region30, henceforth referred as the 433-mer RcRE.
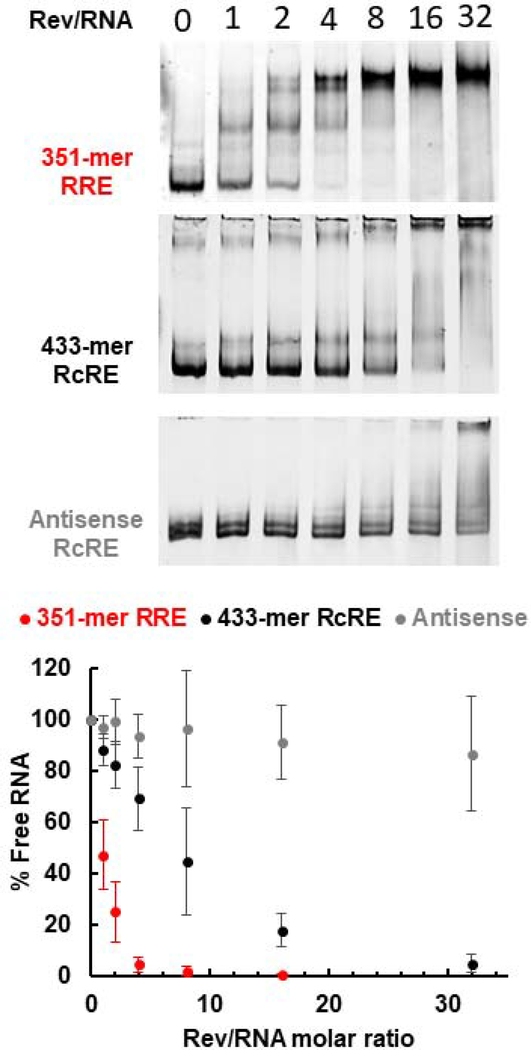
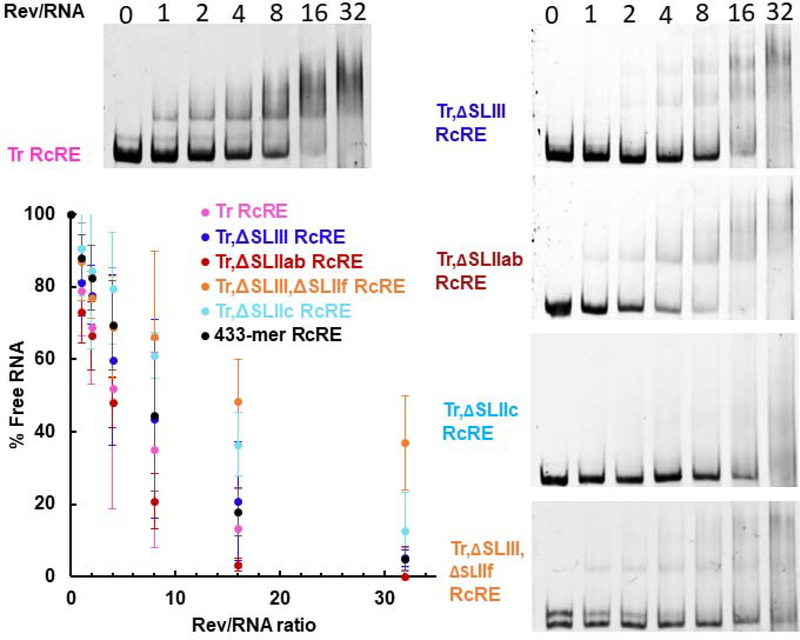
Figure 2 shows binding reactions of RRE and RcRE RNAs (in the presence of excess yeast tRNA) with increasing amounts of purified recombinant HIV-1 Rev protein. The fraction of unbound RNA was plotted as a function of Rev/RNA molar ratio. Rev binds to the RcRE but less efficiently than to the RRE: it took approximately eight times more Rev for 50% of unbound RNA to engage in complexes. In addition, while Rev-RRE complexes form distinct bands representing stable complexes, Rev-RcRE multimers appear to be heterogeneous and unstable resulting in a smear or weakly distinct bands. As a negative control, an RNA with the 433-mer RcRE antisense sequence did not form protein-RNA complexes in the same range as the RRE and the RcRE. Complexes with the antisense RNA were only formed in the presence of high Rev amounts, suggesting non-specific binding.
Figure 2.
Rev-RRE and Rev-RcRE electrophoretic mobility shift assays. The Rev/RNA molar ratio is indicated at the top of each lane. Representative gel images from at least three binding assays are shown. The percentage of unbound RNA is plotted as a function of Rev/RNA molar ratio. Error bars represent standard deviation of at least three independent experiments.
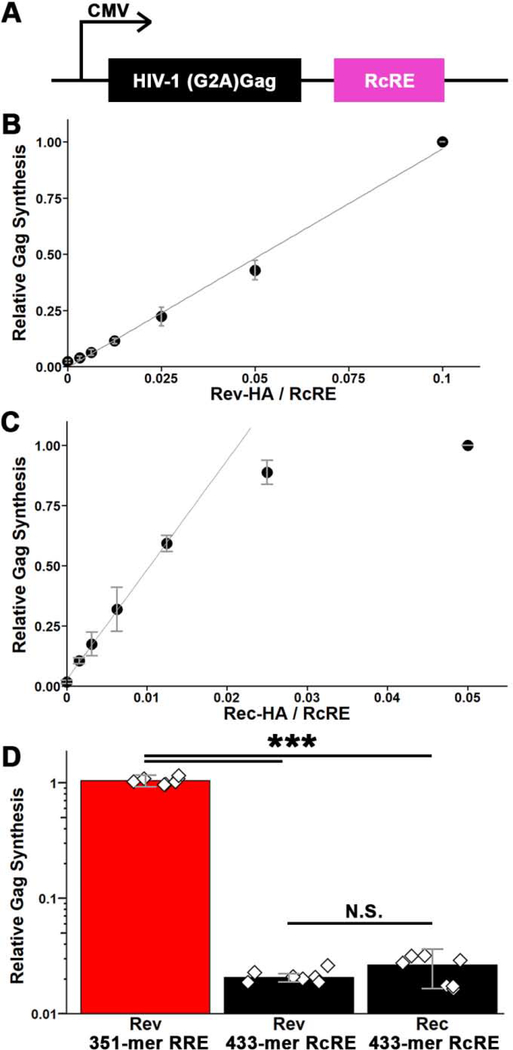
The binding assays were corroborated by nuclear export activity assays (Figure 3). To test Rev-RcRE nuclear export activity in cell culture, we modified a previously constructed Gag reporter vector39,42 in which the RRE sequence is fused to the HIV-1 Gag sequence. A G2A substitution in Gag prevents release of virus-like particles allowing for measurement of total Gag produced from transfected cells—obviating measurements in the cell culture media. The reporter plasmid was modified to replace the RRE with the RcRE (Figure 3A). An HA-tag was fused to the Rev-and Rec-expression vector to allow for normalization of reporter expression to protein expression levels. Comparative experiments using untagged and HA-tagged Rev indicated that the presence of the tag had no significant effect on activity (Supplemental Figure 2).
Figure 3.
Nuclear export activity of the RcRE in the presence of HIV-1 Rev protein. A) Schematic of the HIV-1 Gag reporter vector used to assess the nuclear export activity of the RcRE. B and C) 433-mer RcRE activity as a function of Rev (B) or Rec (C) concentration (mass ratio of protein- to RcRE-expression plasmid). Activity is reported relative to Gag detected in the presence of the highest Rec or Rev protein concentration and normalized to actin levels and transfection efficiency. Error bars represent standard deviation among 3 replicates. D) Comparison of the relative activity of RRE and RcRE with Rev or Rec proteins. The protein production was quantified (by anti-HA antibody) and used to normalize Gag signal. Error bars represent standard deviation among 7 replicates. Diamonds represent individual experiments. ***, p < 0.001; N.S., not statistically significant (p > 0.05).
We first transfected HEK-293T/17 cells with this reporter vector together with increasing amounts of Rev or Rec expression plasmid to establish the range in which a linear dose-dependent response is observed (Figure 3B,C). Based on this, subsequent experiments were performed with 50 ng of Rev or 12.5 ng of Rec plasmid and 1.0 μg of reporter plasmid. Because the reporters measured the same protein product (HIV-1 Gag) and the activating proteins shared the HA epitope tag, it was possible to directly compare the activities of Rec and Rev on the RRE and RcRE response elements. The results showed that, per amount of protein, Rec and Rev had similar activities on the RcRE. Remarkably, however, Rev displayed almost 2 orders of magnitude higher activity on the RRE (Figure 3D). Thus, the HIV-1 Rev can mediate the nuclear export of RcRE-mRNAs just as well as Rec, but in vitro binding is ~8-fold less efficient.
The RcRE RNA is a flexible and extended molecule.
To understand the basis of Rev-RcRE recognition, we examined the RcRE RNA using three biophysical techniques. The 433-nt long RcRE RNA was prepared by in vitro transcription and isolated from a non-denaturing gel to preserve the original fold.
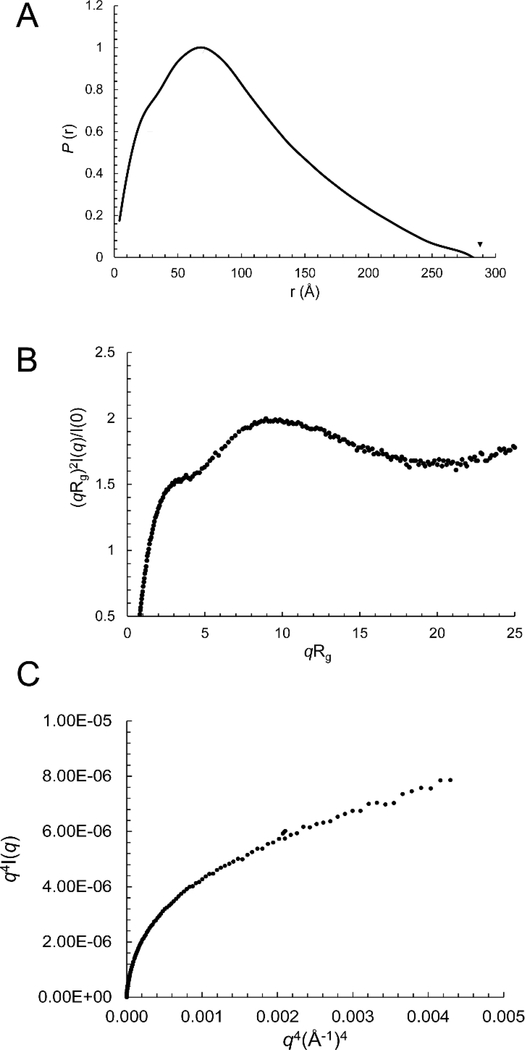
The purified 433-mer RcRE RNA was analyzed by dynamic light scattering (DLS) and SAXS complemented with wide-angle X-ray scattering (WAXS). DLS provides information about the degree of dispersity of a population of molecules in solution and an estimate of its hydrodynamic radius. The RcRE RNA was polydisperse with a hydrodynamic radius of 4.7 nm. SAXS allows for the examination of a molecule in solution and provides information about the molecule’s overall shape, size, and degree of flexibility43–46. Measurements of the intensity of the scattered X-rays (I) and the momentum transfer, q, can be used to calculate various structural parameters. To eliminate the concentration effect, we measured samples at three concentrations and extrapolated data to infinite dilution. Unless otherwise indicated, all structural parameters and three-dimensional envelope models were derived from data extrapolated to infinite dilution. The radius of gyration, Rg, can be derived from the Guinier plot and GNOM analysis.
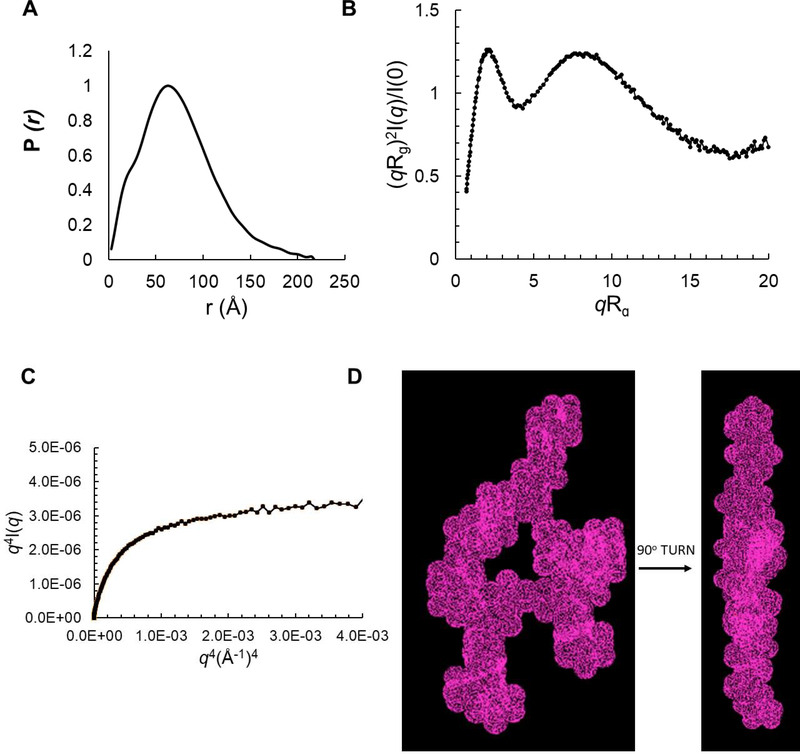
The Rg of the 433-mer RcRE was measured at 75±4 Å (Guinier plot) and 80±1 Å (GNOM analysis) (Table 1). The molecular weight estimated based on the SAXS data revealed that this sample (and all the RcRE variants described below) forms a monomer in solution (Table 1). A paired-distance distribution function (PDDF), P(r), can be calculated to show the histogram of distances between pairs of points within the particle and the maximum dimension of the particle, Dmax47. In such a plot, a spherical molecule results in a bell-shaped curve with the maximum peak in the middle, while an extended molecule exhibits an asymmetric curve with the peak shifted to the lower r region48. For the 433-mer RcRE RNA, the P(r) function is characteristic of an extended molecule (Figure 4A). The distances corresponding to the P(r) maxima represent the dimensions of the molecule, and for the 433-mer RcRE they are 25 Å (the typical width of an A-form RNA double helix), 70 Å, and 282 Å (Table 1 and Figure 4A). These dimensions suggest that the molecule is flat, extended and about 282 Å long.
Table 1.
SAXS-derived three-dimensional parameters for the six RcRE RNA variants described in this study.
| RcRE variant | Length (nucleotides) | Theoretical MW (kDa) | MW by SAXS (kDa) | Rg, Guinier (Å) | Rg, Real Space (Å) | Depth* (Å) | Width* (Å) | Dmax (Å) |
|---|---|---|---|---|---|---|---|---|
| 433-mer RcRE | 433 | 140.4 | 131.7 | 75±4 | 80±1 | ~28 | ~70 | 282 |
| Truncated (Tr) | 313 | 102.0 | 107.0 | 61±1 | 60 ± 1 | 23 | 62 | 218 |
| TrΔSLIII | 271 | 87.9 | 89.8 | 53±1 | 53.0 ± 0.8 | 25 | 60 | 192 |
| TrΔSLIIab | 265 | 86.2 | 87.7 | 50.2 ± 0.8 | 53.1 ± 0.7 | 25 | 62 | 185 |
| TrΔSLIII,IIf | 239 | 77.2 | 83.5 | 52±1 | 53.4±0.9 | 24 | 56 | 183 |
| TrΔSLIIc | 259 | 84.2 | 79.2 | 49±1 | 51.9±0.6 | 23 | 57 | 181 |
Rg Guinier and Rg in Real Space were produced from Guinier plots and GNOM, respectively. The radius of gyration (Rg, the radius about the axis of rotation of the molecule) was generated from Guinier plot of the data extrapolated to infinite dilution in the range of qRg < 1.3
Dmax, the molecule’s maximum dimension was calculated in GNOM.
Values estimated based on the P(r) function.
The molecular weight (MW) was obtained by sequence (theoretical) and by SAXS based on the correlation volume, Vc. Qmax in the range of 0.2–0.3 Å−1 was used for the estimation.
Figure 4.
The 433-nt long RcRE RNA is flexible and extended. A) Paired-distance distribution function, B) Rg-based dimensionless Kratky plot. C) Porod-Debye plot.
The Kratky plot analysis of the SAXS data can assess the flexibility and/or degree of unfolding in samples49. The dimensionless Kratky plot presented as (qRg)2I(q)/I(0) vs. qRg shows a curved shape with a high, open end that indicates that the 433-mer RcRE is extended and flexible (Figure 4B). The flexibility of the 433-mer RcRE is also confirmed by the Porod-Debye plot, which has a parabolic shape without the presence of the Porod plateau (Figure 4C); this also indicates that the molecule is flexible50. Thus, the 433-mer RcRE is quite dynamic; consequently, the reconstruction of a three-dimensional molecular model is not suitable for such a flexible system.
Stem I assumes folded or extended conformations.
The flexibility analysis of the 433-mer suggested that the RcRE may assume different conformations in solution. Interestingly, data from Doudna’s group suggested that the long stem of the HIV-1 RRE interconverts between folded and extended conformations depending on the presence of the Rev protein51. We hypothesized that the unique long stem I of the RcRE (Figure 1, red-colored nucleotides) may also assume folded and extended conformations, conferring the flexibility observed for the 433-mer. To visualize a population of RNA molecules at the single molecule level (in the same solution used for the SAXS experiments), we employed atomic force microscopy. AFM is a technique in which a probe (cantilever) scans the mica slide to which the molecule is attached and a topographical image is obtained based on the resistance imposed on the probe by the molecular forces. Individual molecules can be visualized this way, allowing for assessment of heterogeneity in a population of molecules.
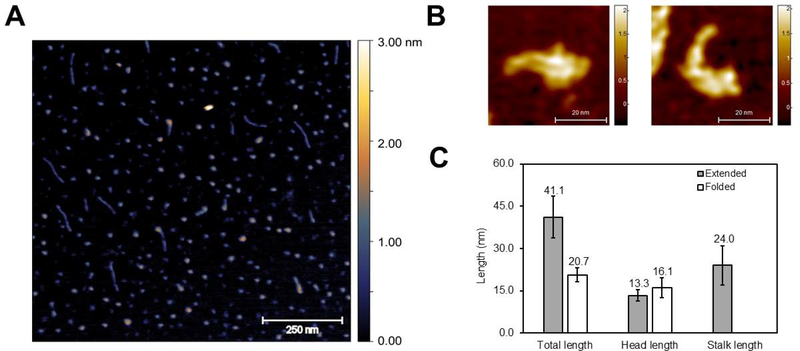
The 433-mer RcRE was attached to spermine-coated mica slides as was done previously by Kjems’s group for the HIV-1 RRE RNA52. Figure 5A is a representative image showing that most molecules are globular with an approximate Dmax of 21 nm (Figure 5C), while a small fraction consisted of longer species characterized by a long stalk and a wider region. This latter subpopulation had an average length of ~41 nm (Figure 5C) and exhibited a striking resemblance in shape and size to the 351-nt long HIV-1 RRE molecules previously analyzed by AFM52. These shapes were not observed in control experiments containing either buffer or denatured RNA (in the presence of 8M urea) (Supplemental Figure 3). Interestingly, high-resolution images of the 433-mer RcRE on 1-(3-aminopropyl) silatrane (APS)-coated mica showed the long stem I in extended or folded conformations (Figure 5B), consistent with the idea that the long stem I is dynamic.
Figure 5.
Atomic force microscopy on the 433-mer RcRE RNA. A) Representative AFM image. Imaging was performed twice using independently transcribed and purified RNAs. The scale on the right side of the image indicates height (distance on the axis perpendicular to the microscope slide). B) High-resolution images focusing on individual RcRE molecules. C) Measurements of the total length for extended (n=9) and globular (n=15) molecules. “Head” and “stalk” refer to the wide vs thin region of the molecule, respectively. The length of the “stalk” was measured only for extended molecules for which a long, thin feature was apparent.
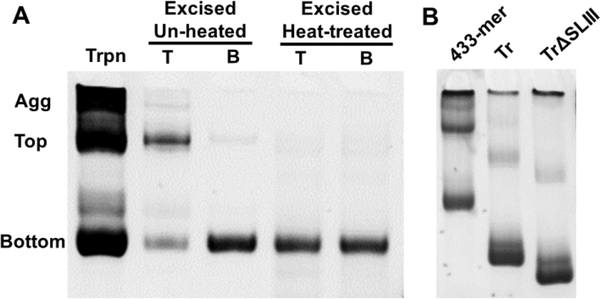
An assessment of the transcription reaction on a non-denaturing gel revealed two major bands (labelled bottom and top) and extensive aggregation for the 433-mer (Figure 6), despite the presence of a single band under denaturing conditions (Supplemental Figure 3). When the slow-migrating (top) species extracted from the gel is heat-treated, it migrates similarly to the bottom species suggesting that the top band may be a dimer of the bottom band. Truncation of ~40 base pairs from Stem I (Figure 1, magenta-colored nucleotides) results in much less aggregation during transcription, with the ratio of the top/bottom bands decreasing by ~7-fold (Figure 6B), suggesting that Stem I promotes intermolecular interactions in vitro.
Figure 6.
Non-denaturing PAGE of RcRE RNAs. A) The 433-mer RcRE is transcribed as a mixture of two bands (top and bottom) and aggregates (Agg). The top and bottom bands present in the transcription reaction (Trpn) were purified from a 6% native gel by excision. Migration of the gel-excised molecules was examined with and without heat treatment (85°C, 5 min followed by snap-cooling). B) Stem I is responsible for the formation of slow-migrating bands. Aliquots of in vitro transcription reactions prior to gel purification for the 433-mer, truncated (Tr), and truncated with SLIII deletion RcRE (TrΔSLIII). The bottom band is enriched in the variants in which the bottom half of Stem I is truncated.
The three-dimensional SAXS envelope of a truncated RcRE resembles the “A”-shaped HIV RRE.
We constructed a RcRE variant in which the characteristic long stem was truncated by ~40 base pairs (Figure 1: magenta-colored bases; Table 1, Supplemental Figure 4). The SAXS data for this variant were subjected to the same analyses for Rg, pair distance distribution function, and flexibility as described above for the 433-mer. The PDDF function shows that the truncated RcRE is extended with a maximum dimension of 218 Å (Figure 7A). The curved shape in the dimensionless Kratky plot (Figure 7B) and presence of the Porod plateau (Figure 7C) suggest that the truncated RcRE is much less flexible than the 433-mer (compare with Figure 4B and C). The PDDF also reveals a shoulder peak at ~20 Å and a second peak at ~62 Å. The first peak is related to the dimensions across an RNA duplex, the diameter of which is about 20 Å. The second peak suggests the presence of a structural feature in which two major structural segments are separated by ~57–67Å. Based on these dimensions and a Dmax value of 218 Å, a parallelepiped search volume of 240×150×60 Å3 was used to reconstruct a 3D molecular envelope model using the Dammin software in expert slow mode. The final model is shown in Figure 7D. The shape of the truncated, 313-mer RcRE is reminiscent of the unique HIV RRE “A” shape determined for HIV-1 and modeled for HIV-239,53,54 (Supplemental Figure 5A).
Figure 7.
SAXS analysis of a RcRE variant truncated by ~40 base pairs at SLI. A) Paired-distance distribution function with r maxima indicated by arrows. B) Rg-based dimensionless Kratky plot, C) Porod-Debye plot, D) Ab initio molecular envelope model (topological structure) generated for the truncated RcRE (313-mer) generated with the Dammin software81 using expert mode and parallelepiped search volume of 240×150×60 Å3.
We also performed EMSA analysis with Rev on the truncated RcRE. We found that the truncated RcRE (Tr RcRE) had similar binding efficiency as the 433-mer RcRE: Tr RcRE required ~0.5-fold less Rev for 50% of the unbound RNA to engage in protein-RNA complexes compared to the 433-mer RcRE (Figure 8, compare magenta and black-colored plots). On the other hand, the truncated RcRE exhibited a 2-fold decrease in nuclear export activity both in the presence of Rev and Rec (Figure 9, magenta-colored bars). Overall, truncation of Stem I had relatively subtle effects on in vitro binding and nuclear export activity in cultured cells.
Figure 8.
Electrophoretic mobility shift assays of in vitro transcribed RcRE deletion variants in the presence of purified recombinant HIV-1 Rev. The Rev/RNA molar ratio is indicated at the top each lane. Each well contains 0.25 pmol of RNA. The percentage of unbound RNA is plotted as a function of Rev/RNA molar ratio. Binding assays were performed at least three times: gels are representative images and error bars represent standard deviation.
Figure 9.
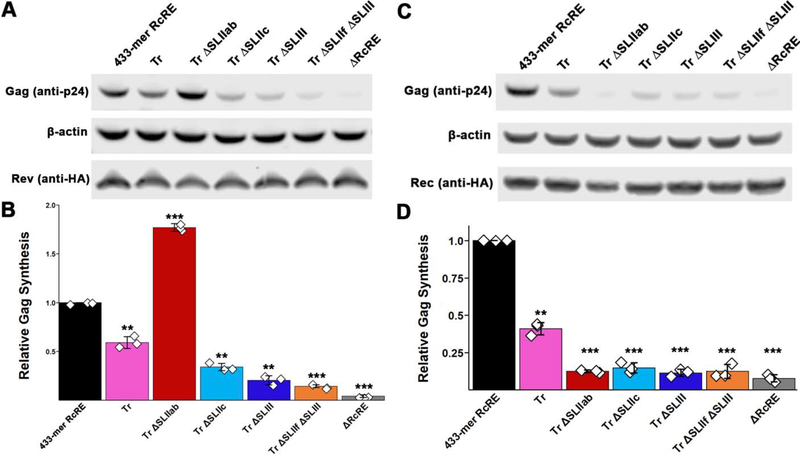
Nuclear export activity of RcRE deletion variants in the presence of HIV-1 Rev or HERV-K Rec. A,C) Representative images of western blots (A: Rev, C:Rec). B,D) Gag synthesis was normalized to actin and Rev or Rec levels (B and D, respectively) detected by anti-actin and anti-HA antibodies, as well as to Gaussia Luciferase levels to account for differences in transfection efficiency. Error bars represent standard deviation among 3 replicates. Diamonds represent individual experiments. **, p < 0.01; ***, p < 0.001.
The three-dimensional architecture of the RcRE is necessary for optimal Rev binding and nuclear export activity.
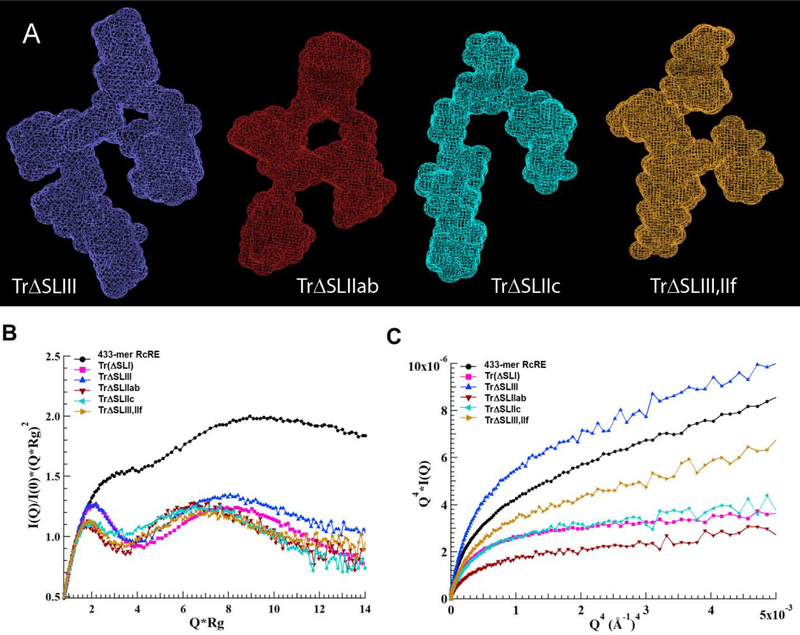
Previous studies demonstrated that SLIII and, to a lesser extent, SLIIf serve as putative binding sites on the RcRE for the HIV-1 Rev protein30,40. In contrast, SLIIa and SLIIb did not seem to contribute to Rev binding30,38. To gain insight into the structural basis of Rev-RcRE interactions, we constructed four RcRE variants in which SLIII (TrΔSLIII, indigo-colored bases in Figure 1), SLIIab (TrΔSLIIab, red), SLIIc (TrΔSLIIc, cyan), or SLIII in combination with SLIIf (orange) were deleted. Stem I was also truncated in all these variants (Figure 1, pink-colored bases were removed in all stem deletion constructs). Predicted secondary structures of these variants are shown in Supplemental Figure 4. The topological structure and flexibility of each deletion variant was analyzed by SAXS. Deletion of SLIII resulted in a distorted three-dimensional conformation (Figure 10A). Rev binding to this construct was only slightly less effective (~0.5-fold) than its parent as determined by the disappearance of free RNA in the EMSAs (Figure 8, compare indigo- and magenta-colored variants).
Figure 10.
A) Topological structures of the RcRE variants in which various stems are deleted. All variants are a derivative of the Stem I-truncated variant shown in Figure 5 (magenta-colored bases are removed). From left to right: TrΔSLIII (blue-colored bases in Figure 1 are deleted), TrΔSLIIab (dark red-colored bases in Figure 1 are deleted), TrΔSLIIc (cyan-colored bases in Fig 1), and TrΔSLIII,ΔSLIIf (blue- and orange-colored bases in Figure 1 are deleted. Ab initio molecular envelope models were generated with the Dammin software using expert mode and parallelepiped search volume of 220×150×60 Å3 (TrΔSLIII) and 210×150×60 Å3 (TrΔSLIIab, TrΔSLIIc, and TrΔSLIII,ΔSLIIf). SAXS envelopes of the variants superimposed onto the topological structure of the Stem I-truncated RcRE are shown in Supplemental Figure 5. B) Kratky and C) Debye-Porod plots of the RcRE variants examined in this study.
The nuclear export activity of the TrΔSLIII construct was more dramatically reduced, however, with Gag synthesis being ~5-fold lower than in the presence of the 433-mer RcRE (Figure 9). The difference in the binding and nuclear export assays may be attributed to the marked flexibility of the TrΔSLIII variant, as determined by the Kratky and Porod-Debye plots (Figure 10B and 10C, indigo-colored plots). It is plausible that the inherent flexibility of this molecule allows for efficient Rev binding, giving rise to a mixture of complexes of varying nuclear export activity. Overall, the data suggest that in addition to providing a binding site, SLIII is also necessary for the maintenance of the “A”-shaped structure, which allows for effective Rev-RcRE multimerization and subsequent nuclear export.
The significance of the overall structure was further validated by two more deletion variants. Deletion of SLIIc (cyan) or concomitant deletion of SLIII and SLIIf (orange) resulted in distorted topological structures (Figure 10A). Rev-binding and nuclear export activity were significantly impaired for both RNAs, particularly for the TrΔSLIII,ΔSLIIf double mutant (Figures 8 and 9).
In stark contrast, deletion of SLIIab (dark red-colored bases in Figure 1) resulted in a compact structure that resembled the parent “A” shape (Figure 10A). This variant is shorter than the parent construct (185 vs. 218 Å, Table 1), possibly due to structural rearrangement, such as the remaining stem loops filling in the space previously occupied by SLIIab. As indicated by the Porod-Debye plot in Figure 10C, this construct is also the least flexible of all six structures examined in this study. Interestingly, deletion of SLIIab does not negatively affect HIV-1 Rev-binding or Rev-mediated nuclear export (Figure 8, 9), just as previously reported30. In fact, TrΔSLIIab exhibited even higher affinity for the Rev protein in vitro and ~2-fold higher nuclear export activity than the 433-mer RcRE. Perhaps this variant results in a more homogeneous population of molecules (see low flexibility in Figure 10C) that is enriched in RNAs with high-affinity for Rev. Overall, the SAXS, in vitro binding, and nuclear export activity assays strongly suggest that maintenance of the overall structure contributes significantly to successful HIV-1 Rev activity.
We also performed nuclear export activity assays for all six RcRE constructs in the presence of the Rec expression plasmid. Interestingly, all deletion variants, including TrΔSLIIab, exhibited reduced Gag synthesis (Figure 9C). The data suggest that the “A” shape is not sufficient for Rec-mediated export under the conditions tested and that the two proteins may employ different structural features or mechanisms of RNA-protein recognition, complex formation, and/or recruitment of cellular factors.
Discussion
The expanding field of study on human endogenous retroviruses is increasingly shedding light on the little understood roles of HERVs in human health and disease. In this work, we report the solution conformation of the HERV-K RcRE RNA (also known as K-RRE), the first three-dimensional topological structure of a HERV molecule. We found that: 1) the overall shape, assessed by SAXS and AFM, is flat and extended and resembles that of the HIV-1 RRE, 2) the long Stem I confers flexibility to the molecule and that the RcRE RNA may oscillate between folded and extended conformations, and 3) RcRE deletion variants that exhibit distorted three-dimensional topologies also exhibit reduced in vitro binding and nuclear export activity in cultured cells, but a variant that maintains the “A” shape has similar or better activity as the 433-mer RcRE.
The conservation of the RRE and RcRE morphologies provides a structural explanation for the observation that HIV-1 Rev is able to bind to and mediate the nuclear export of both elements (32,34,40 and Figures 2 and 3). HIV-1 Rev binding to the RcRE can result in increased nuclear export and, consequently, synthesis of HERV-K proteins in HIV-infected cells. Our results show that Rev can mediate the export of RcRE-containing mRNAs just as well as Rec (Figure 3D). Given that Rev is produced at high levels in an HIV-1-infected cell, it is conceivable that Rev contributes to HERV-K expression (or mRNAs associated with solo LTR’s) even more than Rec. On the other hand, Rev activates RRE-containing mRNAs at ~50-fold higher levels than RcRE-containing mRNAs (Figure 3). Although we cannot explain the mechanism behind this difference, we hypothesize that HIV-1, being a lytic virus, is expressed at much higher levels than non-lytic betaretroviruses, like HERV-K. Attenuation of RcRE binding regions during evolution may have resulted in weaker interactions and activity.
A number of studies have demonstrated cross-talk between HIV and HERV-K elements. HERV-K expression, in particular, can be activated by HIV-1 infection (34,57–61; reviewed in56). HIV-1 Tat and Vif have been shown to induce expression of HERV-K RNA62, but the level of HERV-K RNAs observed in the plasma of HIV-infected patients is much higher than could be explained by HIV-1 Tat and Vif activation alone63. Thus, it is plausible that HIV Rev binding to the RcRE—driven by structural similarity—could contribute to the increased HERV-K levels observed in HIV-1 patients. In fact, a recent high throughput sequencing study by Gray et al. found the expression of several HERV-K loci is increased significantly in the presence of HIV-1 Rev34. However, it should be noted that the group found that HIV-1 Rev does not exhibit similar activity with all HERV-K RcRE loci. Thus, we cannot exclude the possibility that there are structural differences between the various RcRE loci that result in the observed functional differences. Whether the topological structure of the RcRE is conserved among its loci remains to be determined.
HIV-induced HERV expression has various implications56. Monde et al. found that co-expression of HERV-K Gag with HIV-1 Gag reduces HIV infectivity by interfering with virus particle assembly41,64,65 and HERV-K Env has been shown to be incorporated into HIV-1 particles41,66 limiting their production41. Thus, HERV-K expression could augment the immune response against the virus. However, HERV-K Env has also been demonstrated to promote HIV infection by sequestering the cellular restriction factor tetherin67. Moreover, it has been suggested that HERV expression is implicated in HIV-associated cancer, amyotrophic lateral sclerosis, and dementia7,68. Finally, as Gray et al. intriguingly suggested, HIV Rev or HERV-K Rec can alter the cytoplasmic transcriptome and proteome by acting in trans on the RcRE of hundreds of solo LTR’s which reside nearby or within introns of cellular genes34.
AFM and SAXS analyses of the 433-mer RcRE RNA (Figures 4 and 5) suggest that the RcRE may assume a folded conformation that has the potential to extend. Stem I appears to impart flexibility to the molecule may be involved in inter- or intra-molecular interactions (Figure 6). Interestingly, Bai et al. found that, in the HIV-1 RRE, the long stem participates in intramolecular interactions with the core of the molecule and that these interactions are disrupted in the presence of the Rev protein51. Their results supported a mechanism in which initial Rev binding causes the long stem to unfold and reveal an additional Rev-binding site that promotes cooperative oligomerization. Like the RRE, the RcRE Stem I has several bulges (Figure 1); these could serve as hinge sites to allow dynamic, folding motions of the duplex. Truncation of Stem I does not appear to affect Rev binding (Figure 8), but has a 2-fold effect on the nuclear export activity of the RcRE (Figure 9). The biological significance, if any, of these observations remains to be determined.
The HERV-K RcRE is characterized by multiple stem loops (Figure 1). SLIII and, partially, SLIIf have been suggested to serve as Rev binding sites30,40. We found that three deletion variants that result in a distorted structure (TrΔSLIII, TrΔSLIII,ΔSLIIf, and TrΔSLIIc) have decreased in vitro binding and nuclear export activities in the presence of Rev (Figures 8 and 9). However, TrΔSLIIab, which retains the “A” shape and is the least flexible structure, has Rev-binding and nuclear export activity similar to or better than the 433-mer RcRE. Thus, the individual stems contribute not only as interaction sites for Rev molecules, but also as structural pillars for the RcRE RNA architecture, as was demonstrated previously for its HIV-1 counterpart54. The fact that the RcRE, unlike the RRE, does not lie in a coding region provides support for the hypothesis that the conservation of structural features in the RcRE is not necessarily linked to the env gene coding and that viral evolution is partly controlled by maintenance of the overall structure.
The structural conservation of the two cis-acting signals explains why Rev is able to mediate export of RcRE-containing mRNAs, but it is still unclear why the HERV-K Rec does not communicate with the HIV-1 RRE32,34. Our functional assays suggest that the “A” shape is not sufficient for successful Rec-mediated export—the “A”-shaped TrΔSLIIab exhibits decreased Rec-mediated Gag synthesis like the variants that have lost the original shape (Figure 9C and 9D). Thus, it appears that the differences are protein-specific.
Unlike the HIV-1 Rev protein for which several structural studies have been reported69–72, the Rec protein is not nearly as well characterized. Both proteins are small (~15 kDa) and have an RNA-binding domain (that also serves as a nuclear localization signal) and a nuclear export domain. The RNA-binding domain is an arginine-rich motif and, in the Rev protein, it fits into a widened major groove on the HIV-1 RRE73. Rev has two oligomerization domains which assume alpha-helical conformations that enable Rev-Rev interactions to form a multimolecular complex capable of interacting with the Crm-1 factor. Rec appears to only have one oligomerization domain, located downstream of the RNA-binding sequence74. In addition, it has been reported that the Rec protein assumes mainly a tetrameric quaternary structure38, while Rev appears mainly in monomeric or dimeric states (although trimers and tetramers have also been reported75). A thorough and systematic in silico analysis of all known Rev-like proteins by Umunakwe et al. predicted that the HIV Rev protein is more hydrophobic than the Rec protein76, a property that may impart Rev with increased oligomerization efficiency and, consequently, increased nuclear export activity even with non-self targets such as the RcRE. Further characterization of the Rec protein and its interactions with the RcRE RNA are underway and will help to elucidate the mechanism of Rec-RcRE complex formation.
In conclusion, our analysis of the three-dimensional shape of the HERV-K RcRE using SAXS and AFM demonstrates that the RRE and RcRE are structurally conserved allowing the HIV-1 Rev protein to recognize both elements. This appears to affect the transcriptomic and proteomic landscape of HIV-1 infected cells. Moreover, the proposed structural model for the RcRE RNA can provide insight for targeting the regulation of HERV-K expression.
Materials and Methods
Plasmid construction and RNA preparation.
A plasmid containing the 433-nucleotide long HERV-K 108 RcRE (K-RRE) sequence (GenBank accession number AF179225 30) flanked by the T7 promoter and a XhoI site was synthesized by Integrated DNA Technologies, Inc. Deletions were introduced by PCR mutagenesis using primers flanking the regions to be deleted. The RcRE was truncated by first deleting nucleotides 1–43 using the primers 5’-/Phos/TCCCTAATCTCAAGTACCCAGGG-3’ and 5’-CCTATAGTGAGTCGTATTATGGACTAGC-3’, followed by deletion of nucleotides 357–433 using the primers 5’-/Phos/CTCGAGATCAGTTCTGGACCAG-3’ and 5’-TCCCTAATCTCAGTAGATGGAATG-3’. SLIII was deleted using the primers 5’-CCCAGGGACGGGCAGGAGA-3’ and 5’-/Phos/TACTGAGATAGGGGACTCGAG-3’. SLIIab was deleted using the primers 5’-GTTTGTGTCCCTGGGTACTTGAGATTAGGG-3’ and 5’-/Phos/AAGGTTTCTCCCCATGTGATAGTCTGAAAT-3’. SLIIc was deleted using the primers /5Phos/GACCGTCCCCCAGCCCGACACCCGTAAAGG and TTGGACAATACCTGGCTTTCCTAGGCAGAG. SLIIf was deleted using the primers /5Phos/CCGTCCCTGGGCAATGGAATGTCTCGGTAT and AAGAGGCATTCCTTCCTCTTTTACTAATCC. Template DNA was digested with DpnI and E. coli TOP10 cells (Invitrogen) were used to transform the PCR products.
Codon-optimized sequences of Rev and Rec proteins with C-terminal HA-tag (amino acid sequence: YPYDVPDYA) were ordered from Integrated DNA Technologies, Inc. and inserted using inversion PCR and In-Fusion cloning (Takara Bio) into pcDNA3.1+(Zeo) vector backbone.
RNA was prepared by in vitro transcription. Plasmids were digested with XhoI and digestion reactions were purified using a PCR purification kit (New England Biolabs). Antisense RcRE DNA template was prepared by PCR. In vitro transcription was performed using T7 RNA synthesis kit (New England Biolabs) according to manufacturer’s instructions and the reactions were incubated overnight at 37 °C. The template DNA was digested with DNase and the RNA was resolved overnight on a long (28-cm) non-denaturing gel to allow for effective separation of bands. Tris-borate buffer containing 0.5 mM MgCl2 was used for both the gel and the running buffer. RNA was visualized by UV absorption on a phosphorescent plate, isolated by excision, and eluted by shaking gently overnight in 10mM Tris-HCl at pH 8.0, 100mM KCl, and 0.5mM MgCl₂ at 4 °C. RNA was concentrated using Amicon Ultra centrifugal filter units and washed several times in elution buffer. RNase inhibitor was added to the sample, which was shipped for SAXS data collection. All samples were prepared approximately 1 week prior to data collection to maximize freshness and were never frozen.
Small-angle X-ray scattering (SAXS) data collection and analysis.
SAXS measurements were performed at the 12ID-B beamline of the Advanced Photon Source, Argonne National Laboratory. Photon energy was 13.3-keV and sample-to-detector distance was 1.9 m to achieve a useable q range of 0.005 < q < 0.88 Å−1, where q = (4π/λ)sinθ, and 2θ is the scattering angle. Concentration series measurements for the same sample were carried out to remove the scattering contribution due to interparticle interactions and to extrapolate the data to infinite dilution. The sample concentrations were from 0.25 to 1.25 mg/ml. Thirty or forty-five 2D-SAXS intensity maps were recorded with a Pilatus 2 M pixel detector for each matching buffer and sample solution using a flow cell, with the exposure time of 0.5–2 seconds to minimize radiation damage and to yield optimal signal-to-noise ratio. The 2D images were reduced to 1D scattering profiles, and outliers were eliminated before averaging using the MATLAB scripts developed by the beamline.
The buffer background subtraction and intensity extrapolation to infinite dilution were carried out using MATLAB script developed by the 12-ID-B beamline. The radius of gyration (Rg) was generated from Guinier plot of the data extrapolated to infinite dilution in the range of qRg < 1.3. For comparison, Rg was also calculated in real and reciprocal spaces using program GNOM in q range up to 0.30 Å−1 77. The pair-distance distribution function P(r) and maximum dimension (Dmax) were also calculated using GNOM. The molecular weights were estimated based on method of correlation volume, Vc, using formula for RNA45,50. Thirty-two ab-initio shape reconstructions (molecular envelopes) were generated independently using DAMMIN in expert mode with parallelepiped search volume then averaged and filtered. The SAXS scattering profiles, structure parameters and model fitting can be found at https://www.sasbdb.org/project/1037/7bmegp49uk/.
Ex Situ Atomic Force Microscopy (AFM) Imaging Conditions and Analysis.
To study the morphology of RcRE RNA under ex situ conditions, freshly cleaved muscovite mica (Grade V5) (Ted Pella, Redding, CA) was coated with 100 μg/μL spermine solution for 10 min52. To remove excess spermine, mica was rinsed with nuclease-free water and dried gently in open air as described by Pallesen et al.52. RcRE RNA at 1 ng/μl was spotted onto the spermine-coated mica, washed with nuclease-free water, and further dried in open air. AFM measurements were performed immediately after sample preparation to avoid any dehydration. Images were obtained using two independently prepared samples. Images were obtained with a Nanoscope V MultiMode 8 scanning probe microscope (Bruker, Santa Barbara, CA) equipped with a closed-loop vertical engage E-scanner and operated in the ScanAsyst-Air mode (a PeakForce tapping mode). Images were taken with a Scanasyst-Air (Bruker, Santa Barbara, CA) silicon nitride cantilever of triangular geometry with a nominal spring constant of 0.4 N/m, a nominal tip radius of 2 nm and a nominal resonant frequency of 70 kHz. Scan rates were set at 0.5–1 Hz. ScanAsyst mode continuously monitors image quality and makes automatic appropriate image parameter adjustments. Quantitative AFM image processing and measurements (length, stalk length, width) were completed using Gwyddion 2.50 free software78 (Czech Metrology Institute).
To obtain high-resolution AFM images, freshly cleaved muscovite mica (Grade V1) (Ted Pella, Redding, CA) was coated with 1-(3-aminopropyl)silatrane (APS) solution (50 mM APS stock was diluted 300-fold in water before use) for 30 min. To remove excess APS, mica was rinsed with pico-pure water and dried gently in filtered nitrogen flow. RcRE RNA at 5 nM was spotted onto the APS-coated mica for ~5 min and washed with 200 μl buffer (10 mM Tris, 100 mM KCl, 0.5 mM MgCl2, pH 8.0). Images were obtained with AC mode in solution at 15 °C using a Cypher VRS AFM instrument. High resolution images were recorded at scan sizes of 500×500 nm2, 1024×1024 pixels and scanning rate of 2.44 Hz. Biolever mini AFM probes with spring constant of 0.09 N/m, tip radius of 8 nm and resonant frequency of 110 kHz were used. Images were processed using SPIP (Scanning Probe Image Processor) software.
HIV-1 Rev purification and binding assays.
The rev gene sequence was amplified from a pCMV-Rev plasmid (from B. Felber, NCI-Frederick) using primers flanked by NdeI and SalI restriction sites. The product was subcloned cloned into the pCR2.1 TOPO vector using the TOPO TA cloning kit (ThermoFisher Scientific) and subsequently into the pET28a vector (NdeI and XhoI sites) and verified by sequencing. The hexahistidine-tagged protein was produced recombinantly in E. coli BL21(DE3) cells. A 200-mL culture was induced with 1 mM Isopropyl β-d-1-thiogalactopyranoside (IPTG) at OD600 ~0.5 and cultured at room temperature overnight. Cells were harvested by centrifugation and the pellet was resuspended in 2 mL buffer A (50 mM Tris-HCl at pH 8.0, 0.5 M NaCl, 5 mM imidazole) containing 1 mM phenylmethylsulfonyl fluoride (PMSF), lysed by sonication on ice (3 sets of 20 sec on/30 sec off) and clarified by centrifugation. The crude extract was filtered (0.22 μm pore) and loaded onto a 5-mL His-Trap column (GE) pre-equilibrated in buffer A. The column was subsequently washed with 5 volumes of buffer A and the protein was eluted using a 100-mL, 5–500 mM imidazole gradient, at 5 mL/min using a refrigerated GEAkta Purifier 10 FPLC system. Fractions containing the Rev protein were pooled and concentrated in Rev buffer: 10 mM HEPES-KOH at pH 7.5, 150 mM KCl, 1% Triton X-100, 10% glycerol, 1 mM MgCl2, and 0.5 mM EDTA.
Ten-μL binding reactions contained: 0.25 pmol HIV RRE or HERV-K RcRE RNA, 4 units RNAse inhibitor, 1 μg yeast tRNA, 4 μL 5X binding buffer (50 mM HEPES-KOH at pH 7.5, 500 mM KCl, 5 mM MgCl2, 2.5 mM EDTA, and 50% glycerol), and 10 μL of Rev serially diluted in the Rev buffer (containing 100 μg/mL BSA) described above. For EMSAs, RNA samples were freshly prepared as described above for SAXS. Samples did not undergo refolding to maintain the original conformation assumed during in vitro transcription. Also, since the RNAs were not labeled, they did not undergo a denaturation step so refolding was not necessary. Reaction mixtures were incubated at room temperature for 25 min and resolved on a continuously running 6% non-denaturing TBE gel at 150 V at room temperature. Bands stained with SYBR-Gold (Life Technologies) were quantified using the ImageStudio software.
Nuclear export activity assays.
Functional assays were performed as described previously54 with several modifications. Briefly, HEK293T/17 cells (ATCC® CRL-11268™; 5 × 105 cells in a 6-well plate) were transfected with 1 μg of the Gag reporter plasmid (433-mer RcRE or mutants), 1 ng of Gaussia Luciferase expression plasmid (a kind gift from Dr. Stanislaw Kaczmarczyk, Protein Expression Laboratory, SAIC-Frederick), and 0.050 μg of pcDNA3.1+(Zeo)-Rev-HA or 0.0125 μg of pcDNA3.1+(Zeo)-Rec-HA, where applicable. Forty-eight hours after transfection, culture media were collected and assayed for Gaussia Luciferase using Pierce™ Gaussia Luciferase Flash Assay Kit (Thermo Fisher Scientific), as a measure of transfection efficiency. Cells were collected in 1× NuPAGE™ LDS Sample Buffer (Thermo Fisher Scientific) and sonicated 2 × 10 s. The amount of Gag in the cell lysates was measured by near infra-red quantitative western blots using Odyssey CLx imager (LI-COR Biosciences). Membranes (Immobilon-FL PVDF Membrane, MilliporeSigma) were incubated overnight with a mixture of goat anti-p24 (a kind gift from Dr. David Ott, AIDS and Cancer Virus Program, SAIC-Frederick) and monoclonal rabbit anti-HA (Cell Signalling, #3724) antibodies at 1:20,000 dilutions. Subsequently, the membranes were incubated for 30 minutes with monoclonal mouse anti-β-actin (Abgent, AM1021A) antibody at 1:1,000 dilution. IRDye 800CW donkey anti-goat (1:20,000), IRDye 680LT donkey anti-mouse (1:20,000), and IRDye 680LT donkey anti-rabbit (1:20,000) were used as secondary antibodies. Gag detected by anti-p24 was normalized to cellular actin levels and to transfection efficiency as measured by the Gaussia Luciferase assay. P-values were obtained by two-tailed Student’s t-test.
Supplementary Material
Highlights for Structural mimicry drives HIV Rev-mediated HERV-K expression.
HIV-1 Rev binds to and mediates the nuclear export of the HERV-K RcRE.
The topological structure of the HERV-K RcRE resembles that of the HIV-1 RRE.
The unique long stem oscillates between folded and extended conformations.
RcRE variants defective of Rev-binding and nuclear export activity have distorted structures.
The first topological structure for a HERV molecule is reported.
Acknowledgements
We are grateful to Demetria Harvin, Dr. Xiaobing Zuo, Dr. Marzena Dyba, Leah Duke, LCDR Douglas Marks, MIDN 1/C Benjamin Phelps, and Janine Avante for technical assistance; Dr. Leighanne Basta for providing the pET28b vector for cloning and E.coli BL21(DE3) cells; Dr. Virginia Smith for thoughtful review of the manuscript and Dr. Danny Morse for insightful discussions. The work was supported by a Defense Threat Research Agency CB Technologies Service Academy Research Initiative grant to I.P.O and E.Y., Naval Academy Research Council grant to I.P.O., Dr. Ned Garrigues Scholarship to E.K.M, the John F. Kelley Biosciences Fund and Midshipman Research Support Grant to I.P.O. We acknowledge the use of the SAXS Core facility of Center for Cancer Research (CCR), National Cancer Institute (NCI). The SAXS core has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. This research used 12-ID-B beamline of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Abbreviations:
- HERV-K
human endogenous retrovirus type K
- RcRE
Rec-response element
- RRE
Rev-response element
- SAXS
small-angle X-ray scattering
- AFM
atomic force microscopy
Footnotes
Declarations of interest: none.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Belshaw R, Pereira V, Katzourakis A, Talbot G, Pačes J, Burt A, and Tristem M (2004). Longterm reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci U S A 101, 4894–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 3.Dupressoir A, Lavialle C, and Heidmann T (2012). From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta 33, 663–671. [DOI] [PubMed] [Google Scholar]
- 4.Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, et al. (2015). Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fei C, Atterby C, Edqvist P-H, Pontén F, Zhang WW, Larsson E, and Ryan FP (2014). Detection of the human endogenous retrovirus ERV3-encoded Env-protein in human tissues using antibody-based proteomics. J R Soc Med 107, 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Redelsperger F, Raddi N, Bacquin A, Vernochet C, Mariot V, Gache V, Blanchard-Gutton N, Charrin S, Tiret L, Dumonceaux J, et al. (2016). Genetic Evidence That Captured Retroviral Envelope syncytins Contribute to Myoblast Fusion and Muscle Sexual Dimorphism in Mice. PLOS Genetics 12, e1006289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Montojo M, Doucet-O’Hare T, Henderson L, and Nath A (2018). Human endogenous retrovirus-K (HML-2): a comprehensive review. Crit. Rev. Microbiol. 44, 715–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grandi N, and Tramontano E (2018). HERV Envelope Proteins: Physiological Role and Pathogenic Potential in Cancer and Autoimmunity. Front. Microbiol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grandi N, and Tramontano E (2018). Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses. Front Immunol 9, 2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Küry P, Nath A, Créange A, Dolei A, Marche P, Gold J, Giovannoni G, Hartung H-P, and Perron H (2018). Human Endogenous Retroviruses in Neurological Diseases. Trends in Molecular Medicine 24, 379–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Löwer R, Löwer J, and Kurth R (1996). The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci U S A 93, 5177–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryan FP (2004). Human endogenous retroviruses in health and disease: a symbiotic perspective. J R Soc Med 97, 560–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson WE (2019). Origins and evolutionary consequences of ancient endogenous retroviruses. Nat. Rev. Microbiol. 17, 355–370. [DOI] [PubMed] [Google Scholar]
- 14.Subramanian RP, Wildschutte JH, Russo C, and Coffin JM (2011). Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H-S (2012). Genomic impact, chromosomal distribution and transcriptional regulation of HERV elements. Mol Cells 33, 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Contreras-Galindo R, Kaplan MH, Dube D, Gonzalez-Hernandez MJ, Chan S, Meng F, Dai M, Omenn GS, Gitlin SD, and Markovitz DM (2015). Human Endogenous Retrovirus Type K (HERV-K) Particles Package and Transmit HERV-K–Related Sequences. Journal of Virology 89, 7187–7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruprecht K, Ferreira H, Flockerzi A, Wahl S, Sauter M, Mayer J, and Mueller-Lantzsch N (2008). Human Endogenous Retrovirus Family HERV-K(HML-2) RNA Transcripts Are Selectively Packaged into Retroviral Particles Produced by the Human Germ Cell Tumor Line Tera-1 and Originate Mainly from a Provirus on Chromosome 22q11.21. Journal of Virology 82, 10008–10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pastuzyn ED, Day CE, Kearns RB, Kyrke-Smith M, Taibi AV, McCormick J, Yoder N, Belnap DM, Erlendsson S, Morado DR, et al. (2018). The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein that Mediates Intercellular RNA Transfer. Cell 172, 275–288.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rekosh D, and Hammarskjold M-L (2018). Intron Retention in Viruses and Cellular Genes: Detention, Border Controls and Passports. Wiley Interdiscip Rev RNA 9, e1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogg JR (2016). Viral Evasion and Manipulation of Host RNA Quality Control Pathways. J. Virol. 90, 7010–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karn J, and Stoltzfus CM (2012). Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med 2, a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cullen BR (2000). Nuclear RNA Export Pathways. Molecular and Cellular Biology 20, 4181–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohr D, Frey S, Fischer T, Güttler T, and Görlich D (2009). Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J 28, 2541–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulous S, and Emerman M (1992). Functional mapping of the rev-responsive element of human immunodeficiency virus type 2 (HIV-2): influence of HIV-2 envelope- encoding sequences on HIV-1 gpl20 expression in the presence or absence of Rev. Journal of General Virology 73, 1773–1780. [DOI] [PubMed] [Google Scholar]
- 25.Bogerd HP, Huckaby GL, Ahmed YF, Hanly SM, and Greene WC (1991). The type I human T-cell leukemia virus (HTLV-I) Rex trans-activator binds directly to the HTLV-I Rex and the type 1 human immunodeficiency virus Rev RNA response elements. PNAS 88, 5704–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nitta T, Hofacre A, Hull S, and Fan H (2009). Identification and Mutational Analysis of a Rej Response Element in Jaagsiekte Sheep Retrovirus RNA. Journal of Virology 83, 12499–12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belshan M, Park GS, Bilodeau P, Stoltzfus CM, and Carpenter S (2000). Binding of equine infectious anemia virus rev to an exon splicing enhancer mediates alternative splicing and nuclear export of viral mRNAs. Mol. Cell. Biol. 20, 3550–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müllner M, Salmons B, Günzburg WH, and Indik S (2008). Identification of the Rem-responsive element of mouse mammary tumor virus. Nucleic Acids Res 36, 6284–6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mertz JA, Simper MS, Lozano MM, Payne SM, and Dudley JP (2005). Mouse Mammary Tumor Virus Encodes a Self-Regulatory RNA Export Protein and Is a Complex Retrovirus. Journal of Virology 79, 14737–14747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, Bogerd H, Le SY, and Cullen BR (2000). The human endogenous retrovirus K Rev response element coincides with a predicted RNA folding region. RNA 6, 1551–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Löwer R, Tönjes RR, Korbmacher C, Kurth R, and Löwer J (1995). Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J Virol 69, 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Bogerd HP, Peng S, Wiegand H, Truant R, and Cullen BR (1999). An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. PNAS 96, 13404–13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magin C, Hesse J, Löwer J, and Löwer R (2000). Corf, the Rev/Rex homologue of HTDV/HERV-K, encodes an arginine-rich nuclear localization signal that exerts a trans-dominant phenotype when mutated. Virology 274, 11–16. [DOI] [PubMed] [Google Scholar]
- 34.Gray LR, Jackson RE, Jackson PEH, Bekiranov S, Rekosh D, and Hammarskjöld M-L (2019). HIV-1 Rev interacts with HERV-K RcREs present in the human genome and promotes export of unspliced HERV-K proviral RNA. Retrovirology 16, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mann DA, Mikaélian I, Zemmel RW, Green SM, Lowe AD, Kimura T, Singh M, Jonathan P, Butler G, Gait MJ, et al. (1994). A Molecular Rheostat: Co-operative Rev Binding to Stem I of the Rev-response Element Modulates Human Immunodeficiency Virus Type-1 Late Gene Expression. Journal of Molecular Biology 241, 193–207. [DOI] [PubMed] [Google Scholar]
- 36.Sherpa C, Rausch JW, Le Grice SFJ, Hammarskjold M-L, and Rekosh D (2015). The HIV-1 Rev response element (RRE) adopts alternative conformations that promote different rates of virus replication. Nucleic Acids Research 43, 4676–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, and Weeks KM (2009). Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460, 711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langner JS, Fuchs NV, Hoffmann J, Wittmann A, Brutschy B, Löwer R, and Suess B (2012). Biochemical Analysis of the Complex between the Tetrameric Export Adapter Protein Rec of HERV-K/HML-2 and the Responsive RNA Element RcRE pck30. J Virol 86, 9079–9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang X, Wang J, O’Carroll IP, Mitchell M, Zuo X, Wang Y, Yu P, Liu Y, Rausch JW, Dyba MA, et al. (2013). An unusual topological structure of the HIV-1 Rev response element. Cell 155, 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magin-Lachmann C, Hahn S, Strobel H, Held U, Löwer J, and Löwer R (2001). Rec (Formerly Corf) Function Requires Interaction with a Complex, Folded RNA Structure within Its Responsive Element rather than Binding to a Discrete Specific Binding Site. J Virol 75, 10359–10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terry SN, Manganaro L, Cuesta-Dominguez A, Brinzevich D, Simon V, and Mulder LCF (2017). Expression of HERV-K108 envelope interferes with HIV-1 production. Virology 509, 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis N, Williams J, Rekosh D, and Hammarskjöld ML (1990). Identification of a cis-acting element in human immunodeficiency virus type 2 (HIV-2) that is responsive to the HIV-1 rev and human T-cell leukemia virus types I and II rex proteins. J. Virol. 64, 1690–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernadó P, and Svergun DI (2012). Structural analysis of intrinsically disordered proteins by small-angle X-ray scattering. Mol Biosyst 8, 151–167. [DOI] [PubMed] [Google Scholar]
- 44.Fang X, Stagno JR, Bhandari YR, Zuo X, and Wang Y-X (2015). Small-angle X-ray scattering: a bridge between RNA secondary structures and three-dimensional topological structures. Curr. Opin. Struct. Biol. 30, 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rambo RP, and Tainer JA (2013). Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Zuo X, Yu P, Xu H, Starich MR, Tiede DM, Shapiro BA, Schwieters CD, and Wang Y-X (2009). A method for helical RNA global structure determination in solution using small-angle x-ray scattering and NMR measurements. J. Mol. Biol. 393, 717–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glatter O (1977). A new method for the evaluation of small-angle scattering data. J Appl Crystallogr 10, 415–421. [Google Scholar]
- 48.Svergun DI, and Koch MHJ (2003). Small-angle scattering studies of biological macromolecules in solution. Rep. Prog. Phys. 66, 1735–1782. [Google Scholar]
- 49.Doniach S (2001). Changes in Biomolecular Conformation Seen by Small Angle X-ray Scattering. Chem. Rev. 101, 1763–1778. [DOI] [PubMed] [Google Scholar]
- 50.Rambo RP, and Tainer JA (2011). Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 95, 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bai Y, Tambe A, Zhou K, and Doudna JA (2014). RNA-guided assembly of Rev-RRE nuclear export complexes. Elife 3, e03656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pallesen J, Dong M, Besenbacher F, and Kjems J (2009). Structure of the HIV-1 Rev response element alone and in complex with regulator of virion (Rev) studied by atomic force microscopy. FEBS J. 276, 4223–4232. [DOI] [PubMed] [Google Scholar]
- 53.Lusvarghi S, Sztuba-Solinska J, Purzycka KJ, Pauly GT, Rausch JW, and Grice SFJL (2013). The HIV-2 Rev-response element: determining secondary structure and defining folding intermediates. Nucleic Acids Res 41, 6637–6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O’Carroll IP, Thappeta Y, Fan L, Ramirez-Valdez EA, Smith S, Wang Y-X, and Rein A (2017). Contributions of individual domains to function of the HIV-1 Rev response element. Journal of Virology, JVI.00746–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magin C, Löwer R, and Löwer J (1999). cORF and RcRE, the Rev/Rex and RRE/RxRE Homologues of the Human Endogenous Retrovirus Family HTDV/HERV-K. Journal of Virology 73, 9496–9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Kuyl AC (2012). HIV infection and HERV expression: a review. Retrovirology 9, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bhardwaj N, Maldarelli F, Mellors J, and Coffin JM (2014). HIV-1 infection leads to increased transcription of human endogenous retrovirus HERV-K (HML-2) proviruses in vivo but not to increased virion production. J. Virol. 88, 11108–11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Contreras-Galindo R, Kaplan MH, Markovitz DM, Lorenzo E, and Yamamura Y (2006). Detection of HERV-K(HML-2) viral RNA in plasma of HIV type 1-infected individuals. AIDS Res. Hum. Retroviruses 22, 979–984. [DOI] [PubMed] [Google Scholar]
- 59.Contreras-Galindo R, López P, Vélez R, and Yamamura Y (2007). HIV-1 infection increases the expression of human endogenous retroviruses type K (HERV-K) in vitro. AIDS Res. Hum. Retroviruses 23, 116–122. [DOI] [PubMed] [Google Scholar]
- 60.Jones RB, Garrison KE, Mujib S, Mihajlovic V, Aidarus N, Hunter DV, Martin E, John VM, Zhan W, Faruk NF, et al. (2012). HERV-K-specific T cells eliminate diverse HIV-1/2 and SIV primary isolates. J. Clin. Invest. 122, 4473–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young GR, Terry SN, Manganaro L, Cuesta-Dominguez A, Deikus G, Bernal-Rubio D, Campisi L, Fernandez-Sesma A, Sebra R, Simon V, et al. (2018). HIV-1 Infection of Primary CD4+ T Cells Regulates the Expression of Specific Human Endogenous Retrovirus HERV-K (HML-2) Elements. Journal of Virology 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gonzalez-Hernandez MJ, Swanson MD, Contreras-Galindo R, Cookinham S, King SR, Noel RJ, Kaplan MH, and Markovitz DM (2012). Expression of human endogenous retrovirus type K (HML-2) is activated by the Tat protein of HIV-1. J. Virol. 86, 7790–7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Contreras-Galindo R, Kaplan MH, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Ferlenghi I, Giusti F, Lorenzo E, Gitlin SD, Dosik MH, Yamamura Y, et al. (2012). Characterization of Human Endogenous Retroviral Elements in the Blood of HIV-1-Infected Individuals. Journal of Virology 86, 262–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Monde K, Contreras-Galindo R, Kaplan MH, Markovitz DM, and Ono A (2012). Human Endogenous Retrovirus K Gag Coassembles with HIV-1 Gag and Reduces the Release Efficiency and Infectivity of HIV-1. Journal of Virology 86, 11194–11208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Monde K, Terasawa H, Nakano Y, Soheilian F, Nagashima K, Maeda Y, and Ono A (2017). Molecular mechanisms by which HERV-K Gag interferes with HIV-1 Gag assembly and particle infectivity. Retrovirology 14, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brinzevich D, Young GR, Sebra R, Ayllon J, Maio SM, Deikus G, Chen BK, Fernandez-Sesma A, Simon V, and Mulder LCF (2014). HIV-1 Interacts with Human Endogenous Retrovirus K (HML-2) Envelopes Derived from Human Primary Lymphocytes. J Virol 88, 6213–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lemaître C, Harper F, Pierron G, Heidmann T, and Dewannieux M (2014). The HERV-K human endogenous retrovirus envelope protein antagonizes Tetherin antiviral activity. J. Virol. 88, 13626–13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Christensen T (2015). Human endogenous retroviruses in neurologic disease. - PubMed - NCBI. APMIS 124, 116–126. [DOI] [PubMed] [Google Scholar]
- 69.Daugherty MD, Liu B, and Frankel AD (2010). Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat Struct Mol Biol 17, 1337–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, and Grimes JM (2010). Implications of the HIV-1 Rev dimer structure at 3.2 Å resolution for multimeric binding to the Rev response element. Proc Natl Acad Sci U S A 107, 5810–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DiMattia MA, Watts NR, Cheng N, Huang R, Heymann JB, Grimes JM, Wingfield PT, Stuart DI, and Steven AC (2016). The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly. Structure 24, 1068–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watts NR, Eren E, Zhuang X, Wang Y-X, Steven AC, and Wingfield PT (2018). A new HIV-1 Rev structure optimizes interaction with target RNA (RRE) for nuclear export. J. Struct. Biol. 203, 102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Battiste JL, Mao H, Rao NS, Tan R, Muhandiram DR, Kay LE, Frankel AD, and Williamson JR (1996). Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science 273, 1547–1551. [DOI] [PubMed] [Google Scholar]
- 74.Bogerd HP, Wiegand HL, Yang J, and Cullen BR (2000). Mutational Definition of Functional Domains within the Rev Homolog Encoded by Human Endogenous Retrovirus K. Journal of Virology 74, 9353–9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zapp ML, Hope TJ, Parslow TG, and Green MR (1991). Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: a dual function for an arginine-rich binding motif. Proc Natl Acad Sci U S A 88, 7734–7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Umunnakwe CN, Dorman KS, Dobbs D, and Carpenter S (2017). Identification of a homogenous structural basis for oligomerization by retroviral Rev-like proteins. Retrovirology 14, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Svergun DI (1992). Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr 25, 495–503. [Google Scholar]
- 78.Nečas D, and Klapetek P (2012). Gwyddion: an open-source software for SPM data analysis. centr.eur.j.phys. 10, 181–188. [Google Scholar]
- 79.Lee YN, and Bieniasz PD (2007). Reconstitution of an Infectious Human Endogenous Retrovirus. PLoS Pathog 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Darty K, Denise A, and Ponty Y (2009). VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Svergun DI (1999). Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.