Abstract
Apolipoprotein E (APOE) e4 genotype, which increases the risk of Alzheimer’s disease (AD), predicted severe COVID-19 infection in one UK Biobank (UKB) cohort. Single nucleotide polymorphisms (SNPs) in the bridging integrator 1 (BIN1) gene indicate the second highest odds-ratios for sporadic AD, exceeded only by APOE variants. The objective of the present study was to evaluate the effects of BIN1 and the SNP rs744373 on COVID-19-relaetd survival using UKB-derived data. In addition, the interaction and alignment of BIN1 and SARS-Cov-2 were evaluated. For this purpose, the major (non-Alzheimer’s) BIN1 allele was designated as BIN and the SNP rs744373 minor (Alzheimer’s) allele as RS7. To evaluate the interaction and alignment of BIN1 and SARS-Cov-2, Protein Data Bank (pdb) entries were searched on the RCSB Protein Data Bank. The results revealed that the BIN RS7 heterozygote was associated with the lowest mortality rate (11.7%), followed by the BIN BIN homozygote (17.2%). The RS7 RS7 homozygote was associated with the highest mortality rate (28.1%). Logistic regression analysis was also performed using survival or mortality as the dependent variable, and sex, age, genotype, AD and coronary heart disease (CHD) as independent variables. The effects of sex, age and genotype were significant at the 95% level. The male sex and older-aged subjects were more likely to succumb to test-confirmed COVID-19 than females and younger subjects. The effects of AD and CHD were insignificant. Protein molecule alignment analyses suggested that the BIN allele may interfere with the replication of the SARs-Cov2 virus. The findings of the present study demonstrate that the risks for COVID-19 mortality are not simply related to an advanced chronological age or the comorbidities commonly observed in aged subjects, such as CHD and AD, but also with AD genes.
Keywords: dementia, BIN1, COVID-19
Introduction
In older-aged patients with COVID 19, pre-existing dementia is a major risk factor for the severity of COVID-19 infection (1,2). Apolipoprotein E (APOE) e4 genotype, which increases the risk of Alzheimer’s disease (AD), has predicted severe COVID-19 infection in one UK Biobank (UKB) cohort (3).
Single nucleotide polymorphisms (SNPs) in the bridging integrator 1 (BIN1) gene exhibit the second highest odds ratios for sporadic AD, exceeded only by APOE variants. The most frequently reported BIN1 AD risk variant is the SNP rs744373, which has a global allele frequency of 37% and is associated with an increase in AD risk with odds-ratio of 1.17–1.19 (4).
In the present study, UK Biobank (UKB)-derived data were used to evaluate the effects of BIN1 and the SNP rs744373 on the survival of patients with COVID-19 (5). In addition, the interaction and alignment of BIN1 and SARS-Cov-2 were evaluated.
Data and methods
UKB.
The UKB databases consist of data obtained from >500,000 community volunteers between 40–70 years of age at baseline (2006–2010), residing close to 22 assessment centers in the UK, Scotland and Wales. Baseline assessments on the database include demographics, the lifestyle of the subjects and the disease history of the subjects, with links available leading to electronic medical records. The UKB application for the present study was approved as UKB project 57245 (S.L. and P.H.R.).
Electronic links regarding age between the UKB records and the COVID-19 laboratory test results from the National Health Service in the UK are available from March 16 to April 26, 2020, including the peak of daily COVID-19 laboratory-confirmed cases due to the current pandemic. During this time period, the testing of subjects in older-aged groups was largely restricted to hospital inpatients exhibiting any clinical indications of infection; thus, test positivity is considered a good marker of severe COVID-19 infection (5).
The present study analyzed baseline (2006–2010) demographic characteristics and data for pre-existing diagnoses during follow-up available on the UKB database. No data from COVID-19 testing were available for the UKB assessment centers in Scotland and Wales. The present study included data from subjects who were alive and who tested positive for COVID-19, as well as data from and deceased subjects who had tested positive for COVID-19.
Data of medical diagnoses were available from baseline questionnaires (2006–2010) that recorded reports of the disease of participants diagnosed by doctors/medical professionals. Data for diagnoses of new disease since baseline were from linked electronic medical records to hospital inpatient routine data to March 2017, which were coded according to the International Classification of Diseases 10th revision (ICD-10; https://www.cdc.gov/nchs/icd/icd10.htm). The diagnoses which were considered were coronary heart disease (CHD) and AD. The effects of hypertension could not be evaluated, as only one hypertensive subject was available in the patient sample examined. Autopsy data were not available and parts of the respiratory system affected could not be evaluated. In addition, the expression of BIN1 rs744373 in the lung tissue of newly-infected patients with CHD could not be evaluated.
Data analysis.
Data processing was performed on Minerva, a Linux mainframe with Centos 7.6, at the Icahn School of Medicine at Mount Sinai. The present study used PLINK, a whole-genome association analysis toolset, to process the UKB chromosome 2 files (6). The present study also used The UK Biobank Data Parser (ukbb_parser), a python-based package that allows easy interfacing with the large UK Biobank dataset (7). The major (non-Alzheimer’s) BIN1 allele was designated as BIN and the SNP rs744373 minor (Alzheimer’s) allele as RS7.
Molecular structure analysis.
To evaluate the interaction and alignment of BIN1 and SARS-Cov-2, Protein Data Bank (pdb) entries were searched on the RCSB Protein Data Bank. In total, 4 entries were identified that allowed us to examine the association between BIN1 and SARS-Cov2 as follows: i) 6YUN: Crystal Structure of C-terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2 (deposited, April 27, 2020; released, May 20, 2020; method, X-ray diffraction; resolution, 1.44 Å) (8). ii) 7K7P: Structure of SARS-CoV-2 non-stuctural protein 1 (NSP1) (deposited September 23, 2020; released, September 30, 2020; method, X-ray diffraction; resolution, 1.77 Å). iii) 6WEY: High-resolution structure of the SARS-CoV-2 NSP3 Macro X domain (deposited, April 3, 2020; released, April 29, 2020; method, X-ray diffraction; resolution, 0.95 Å (9). iv) 2FIC: Crystal structure of the BAR domain from human Bin1/Amphiphysin II (deposited, December 29, 2005; released, November 14, 2006; method, X-ray diffraction; resolution, 1.99 Å (10). BAR domains are found in proteins that bind and remodel membranes and participate in cytoskeletal and nuclear processes. Amphiphysin II is a protein that in humans is encoded by the BIN1 gene. The protein structures were superimposed and aligned with Pymol v 2.3.4.
Statistical analysis.
Statistical analysis was performed using Fisher’s exact test and logistic regression analysis and using SPSS v22 software. Data are from UK Biobank. Values of P<0.05 were considered to indicate statistically significant differences.
Results
The present study analyzed data from 619 subjects from the UKB database (49% female, 51% male). The mean age was 54±9.2 years (means ± SD). In total, 115 (18.57%) patients succumbed to COVID-19 infection. A total of 10 subjects (1.6%) had AD, 33 (5.33%) had CHD (CHD) and 1 subject had hypertension (Table I).
Table I.
Demographics of the patient sample.
| Demographic | No./percentage of subjects |
|---|---|
| Total no. of subjects | 619 |
| Female | 49% |
| Male | 51% |
| Age, years (mean ± SD) | 54±9.2 |
| Deceased due to COVID-19 | 115 (18.5%) |
| Alzheimer’s disease | 10(1.6%) |
| Coronary heart disease | 33 (5.3%) |
| Hypertension | 1 |
Data pertaining to COVID-19-related survival vs. the BIN1 genotype are presented in Table II. The effect of genotype on survival was significant (P=0.004, two-tailed Fisher’s exact test). The BIN RS7 heterozygote was associated with the lowest mortality rate (11.7%), followed by the BIN BIN homozygote (17.2%). The RS7 RS7 homozygote was associated with the highest mortality rate (28.1%).
Table II.
COVID-19 survival versus BIN1 genotype.
| Genotype | Alive | Deceased | Total | % Deceased |
|---|---|---|---|---|
| BIN BIN | 308 | 64 | 372 | 17.2 |
| BIN RS7 | 250 | 33 | 283 | 11.7 |
| RS7 RS7 | 46 | 18 | 64 | 28.1 |
The effect of genotype on mortality was significant (P=0.004; two-tailed Fisher’s exact test).
A total of 2 COVID-19 outcomes were analyzed: i) Alive with test positivity; and ii) deceased with test-confirmed positivity for COVID-19. The COVID-19 outcome by multiple factors is presented in Table III. Logistic regression analysis was performed using survival (alive) or mortality (deceased) as dependent variables, and sex, age, genotype, AD and CHD as independent variables. The effects of sex, age and genotype were significant at the 95% level. Males and older-aged subjects with test-confirmed COVID-19 positivity were more likely to not survive than females and younger subjects. The effects of AD and CHD were insignificant.
Table III.
COVID-19-related survival.
| Demographic | OR | 95% LB | 95% UB | P-value |
|---|---|---|---|---|
| Sex | 1.681 | 1.046739 | 2.702886 | 0.032 |
| Age | 1.185 | 1.144216 | 1.227869 | <0.001 |
| Genotype | 1.628 | 1.132469 | 2.338711 | 0.008 |
| AD | 1.178 | 0.292433 | 4.76606 | 0.816 |
| CHD | 0.989 | 0.419169 | 2.333758 | 0.981 |
Data were analyzed by logistic regression. Survival, i.e. alive or deceased were used as dependent variables; sex, age, genotype, AD and CHD were used as independent variables. Odds ratios (OR) and 95% confidence intervals were calculated. LB, lower bound; UB, upper bound. The effects of sex, age, and genotype were significant at the 95% level. The effects of AD and CHD were insignificant. OR indicates that males were 1.68-fold as likely to die as females; the subjects’ risk of death increases by 1.185-fold for every year of age. As the genotype varies from BIN RS7 to BIN BIN to RS7 RS7, OR indicates that the risk of mortality rises by 1.628 with each genotype change.
For BIN1 and Crystal Structure of C-terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2, Pymol was used to perform 6 cycles of calculations on 119 atoms aligned, final root mean square deviation of atomic positions (RMSD)=1.850 Å for 101 atoms. Amino acid residues serine 81-tyrosine 100 of BIN1 aligned closely with residues asparagine 99-proline 118 of SARS-Cov2 (Fig. 1A and B).
Figure 1.
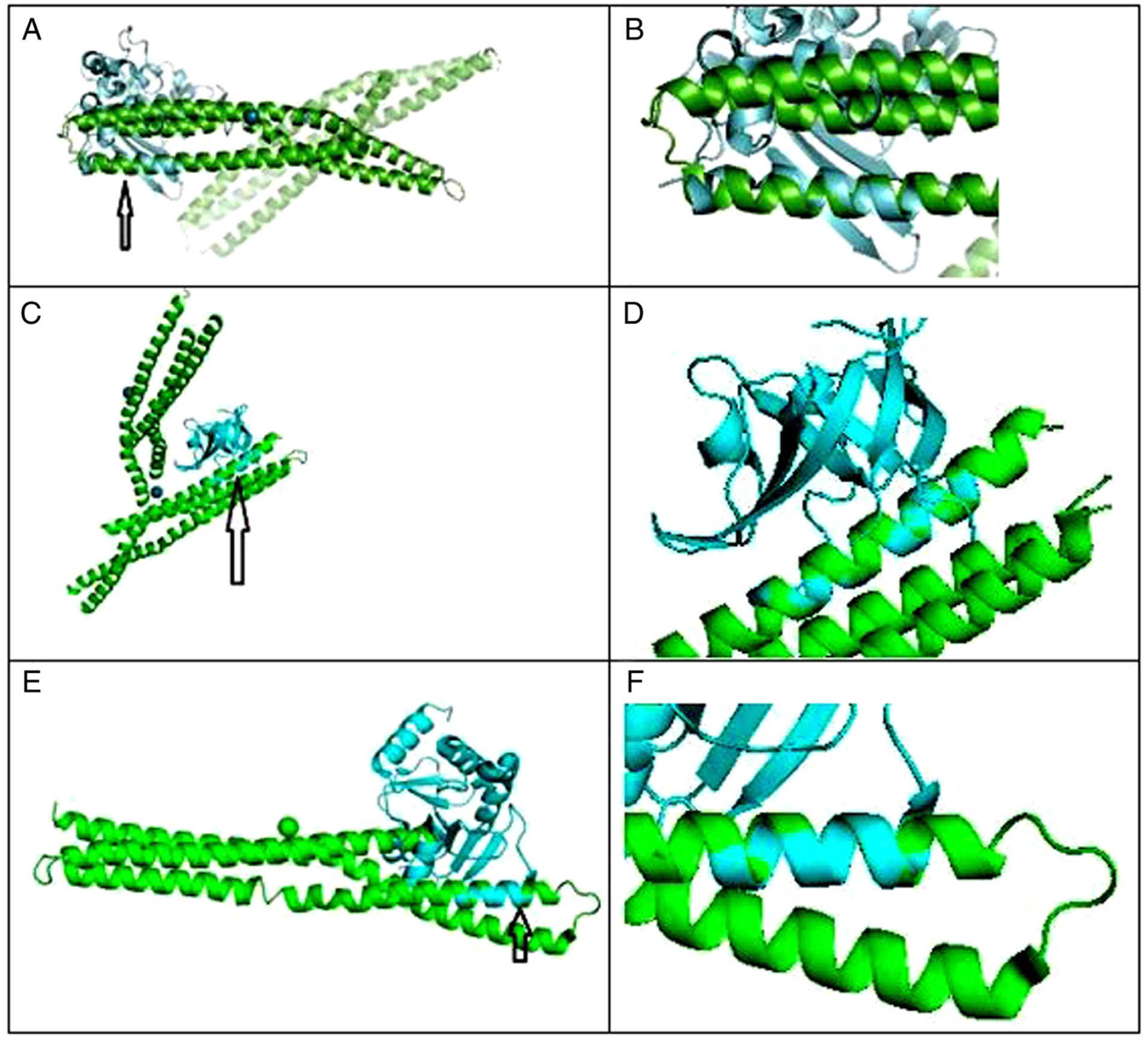
(A) C-terminal dimerization domain of nucleocapsid phosphoprotein from SARS-CoV2 aligned with BAR domain from human BIN1/amphiphysin II. Arrow points to closely aligned (RMSD=1.850Å) alpha helices of SARS-Cov2 (blue) and BIN1 (green). (B) Closeup of aligned α helices. Amino acid residues serine 81-tyrosine 100 of BIN1 aligned closely with residues asparagine 99-proline 118 of SARS-Cov2. (C) BAR domain from human BIN1/amphiphysin II (green) and SARS-CoV-2 NSP1 (blue); arrow points to closely aligned (RMSD=0.298 Å) α helices of SARS-Cov2 NSP1 and BIN1. Amino acid residues glutamic acid 229-asparagine 243 of BIN1 aligned closely with residues valine 26-glycine 40 of SARS-Cov2 NSP1. (D) Closeup view of closely aligned α helices in part C. (E) BAR domain from human BIN1/Amphiphysin II (green) and SARS-CoV-2 NSP3 (blue); arrow points to closely aligned (RMSD = 0.795 Å) α helices of SARS-Cov2 NSP3 and BIN1. Amino acid residues glutamic acid 168-valine 158 of BIN1 aligned closely with residues valine 267-arginine 277 of SARS-Cov2 NSP3. (F) Closeup view of closely aligned α helices in part E.
For BIN1 and Structure of SARS-CoV-2 NSP1, Pymol was used to perform 6 cycles of calculations on 76 atoms aligned, final root mean square deviation of atomic positions (RMSD)=0.208 Å for 64 atoms. Amino acid residues glutamic acid 229-asparagine 243 of BIN1 aligned closely with residues valine 26-glycine 40 of SARS-Cov2 NSP1 (Fig. 1C and D).
For BIN1 and Structure of SARS-CoV-2 NSP3, Pymol was used to perform 5 cycles of calculations on 119 atoms aligned, final root mean square deviation of atomic positions (RMSD)= 0.795 Å for 79 atoms. Amino acid residues glutamic acid 168-valine 158 of BIN1 aligned closely with residues valine 267-arginine 277 of SARS-Cov2 NSP3 (Fig. 1E and F).
Discussion
BIN1 was first identified as a tumor suppressor and a mediator of muscle development (4). BIN1 has been shown to exert protective effects against small cell lung cancer (11), and is moderately expressed in pneumocytes. The interaction between BIN1 and SARs-Cov2 could be related in part to the alignment of α helices shown in Fig. 1 and may explain the increased survival of the BIN RS7 heterozygotes.
The decreased mortality of the heterozygotes (11.7%) compared to the BIN BIN homozygotes (17.2%) (Table II) may be due to heterozygote advantage (12). The heterozygous genotype has a higher relative fitness than either the homozygous dominant or homozygous recessive genotypes. In humans, heterozygotes for sickle cell trait are resistant to malaria. Heterozygotes for cystic fibrosis may be more resistant to tuberculosis and diarrheal diseases. A significantly lower proportion of HLA-DRB1 heterozygosity exists among Hepatitis C Virus infected cases than uninfected cases (13).
In reference to BIN1 and SARs-Cov2, the BIN allele may interfere with the replication of the SARs-Cov2 virus in 2 ways. By binding to the SARs-Cov2 nucleocapsid phosphoprotein, BIN1 could impede viral infection via the following: i) By conjugating to SARs-Cov2 NSP1, BIN1 could interfere with viral infection (14); and ii) the SARs-Cov2 NSP3 folds into a tunnel, which then plugs itself into cell membranes, allowing newly produced viral RNA to exit (15). By conjugating to NSP3, BIN1 could interfere with this process.
The findings of the present study demonstrate that risks for COVID-19 mortality are not simply related to an advanced chronological age or the comorbidities commonly observed in aging, such as CHD and AD, but to AD genetics. Further investigation of the possible SARs-Cov2 antiviral properties of BIN1 may lead to the development of novel treatments for COVID-19.
Acknowledgements
The present study was supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Funding
The research reported in the present study was supported by the Office of Research Infrastructure of the National Institutes of Health under award numbers S10OD018522 and S10OD026880. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Availability of data and materials
All data generated or analyzed during this study are included in this published article or are available from UK Biobank.
Ethics approval and consent to participate
The UK Biobank application for the present study was approved as UKB project 57245 (SL and PHR).
Competing interests
The authors declare that they have no competing interests.
References
- 1.Docherty AB, Harrison EM, Green CA, Hardwick HE, Pius R, Norman L, Holden KA, Read JM, Dondelinger F, Carson G, et al. : Features of 20 133 UK patients in hospital with COVID-19 using the ISARIC WHO Clinical Characterisation Protocol: Prospective observational cohort study. BMJ 369: m1985, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perry G: Alzheimer’s disease patients in the crosshairs of COVID-19. J Alzheimers Dis 76: 1, 2020. [DOI] [PubMed] [Google Scholar]
- 3.Kuo CL, Pilling LC, Atkins JL, Masoli JAH, Delgado J, Kuchel GA and Melzer D: APOE e4 genotype predicts severe COVID-19 in the UK Biobank community cohort. J Gerontol A Biol Sci Med Sci 75: 2231–2232, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franzmeier N, Rubinski A, Neitzel J, Ewers M and Alzheimer’s Disease Neuroimaging Initiative (ADNI): The BIN1 rs744373 SNP is associated with increased tau-PET levels and impaired memory. Nat Commun 10: 1766, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atkins JL, Masoli JA, Delgado J, Pilling LC, Kuo CL, Kuchel GA and Melzer D: Preexisting comorbidities predicting COVID-19 and mortality in the UK Biobank community cohort. J Gerontol A Biol Sci Med Sci 75: 2224–2230, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM and Lee JJ: Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4: 7, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu A, Salminen LE,Thompson PM and Jahanshad N: The UK Biobank Data Parser: A tool with built in and customizable filters for brain studies. Organization for Human Brain Mapping Rome, Italy, June 9–13, 2019, 2019. [Google Scholar]
- 8.Zinzula L, Basquin J, Bohn S, Beck F, Klumpe S, Pfeifer G, Nagy I, Bracher A, Hartl FU and Baumeister W: High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the COVID-19 severe acute respiratory syndrome coronavirus 2. Biochem Biophys Res Commun: Oct 3, 2020 (Epub ahead of print). doi: 10.1016/j.bbrc.2020.09.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frick DN, Virdi RS, Vuksanovic N, Dahal N and Silvaggi NR: Molecular basis for ADP-Ribose binding to the Mac1 domain of SARS-CoV-2 nsp3. Biochemistry 59: 2608–2615, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casal E, Federici L, Zhang W, Fernandez-Recio J, Priego EM, Miguel RN, DuHadaway JB, Prendergast GC, Luisi BF and Laue ED: The crystal structure of the BAR domain from human Bin1/amphiphysin II and its implications for molecular recognition. Biochemistry 45: 12917–12928, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Jia Y, Zhao S, Zhang X, Wang X, Han X, Wang Y, Ma M, Shi J and Liu L: BIN1 reverses PD-L1-mediated immune escape by inactivating the c-MYC and EGFR/MAPK signaling pathways in non-small cell lung cancer. Oncogene 36: 6235–6243, 2017. [DOI] [PubMed] [Google Scholar]
- 12.Abramovs N, Brass A and Tassabehji M: Hardy-Weinberg equilibrium in the large scale genomic sequencing era. Front Genet 11: 210, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hraber P, Kuiken C and Yusim K: Evidence for human leukocyte antigen heterozygote advantage against hepatitis C virus infection. Hepatology 46: 1713–1721, 2007. [DOI] [PubMed] [Google Scholar]
- 14.Lapointe CP, Grosely R, Johnson AG, Wang J, Fernández IS and Puglisi JD: Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. bioRxiv: 2020.2008.2020.259770, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolff G, Limpens R, Zevenhoven-Dobbe JC, Laugks U, Zheng S, de Jong AWM, Koning RI, Agard DA, Grünewald K, Koster AJ, Snijder EJ and Bárcena M: A molecular pore spans the double membrane of the coronavirus replication organelle. Science 369: 1395–1398, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]


