Abstract
The ability of the human fungal pathogen Candida albicans to disseminate into tissues is promoted by a switch from budding to invasive hyphal growth. This morphological transition is stimulated by multiple environmental factors that can vary at different sites of infection. To identify genes that promote invasive growth, C. albicans mutants can be screened for defects in growing invasively into solid agar medium as a substitute for studying tissue invasion. This in vitro approach has advantages in that it permits the media conditions to be varied to mimic different host environments. In addition, the concentration of agar can be varied to determine the effects of altering the rigidity of the matrix into which the cells invade, as this provides a better indicator of invasive growth than the ability to form hyphae in a liquid culture. Testing under multiple conditions can be used to identify mutant cells with the strongest defects. Therefore, protocols and media for analyzing invasive growth of C. albicans under different conditions will be described that are appropriate for testing a single strain or high-throughput analysis of a collection of mutant C. albicans strains.
Keywords: Hyphae, Pseudohyphae, Morphological switch, Invasive growth, Candida albicans
Background
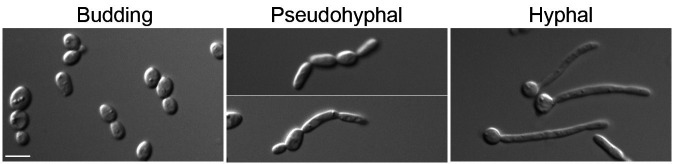
Candida albicans is a multimorphic fungal pathogen that can grow by forming buds (small spherical cells), pseudohyphal cells (chains of elongated cells), or hyphal cells (long filamentous chains of cells with parallel walls) (Figure 1) ( Noble et al., 2017 ). A wide range of environmental stimuli encountered in the host have been shown to promote the switch to invasive growth, including human body temperature (37 °C), alkaline pH, CO2, factors in serum, and the sugar GlcNAc (N-acetyl glucosamine) (Kornitzer, 2019). Budding cells are thought to facilitate bloodstream dissemination and colonization of the gastrointestinal tract (Pierce and Kumamoto, 2012; Witchley et al., 2019 ). In contrast, the filamentous pseudohyphal and hyphal forms of C. albicans are better able to promote invasive growth into tissues ( Noble et al., 2017 ). For example, our recent studies have shown a key role for invasive growth in establishing oral C. albicans infections ( Naseem et al., 2019 ).
Figure 1. Morphologies of C. albicans cells.
Scale bar, 5 μm.
Genetic screens have identified a large number of mutants that regulate the ability of C. albicans to switch from budding to hyphal growth ( Noble et al., 2010 ; O'Meara et al., 2015 ; Glazier et al., 2017 ; Kornitzer, 2019). However, a weak hyphal stimulus was often used in these studies as a way to sensitively detect a partial defect. Many of the previously reported mutants can still be induced by stronger stimuli, such as those encountered in the host ( Riggle et al., 1999 ; Noble et al., 2010 ; Naseem et al., 2011 ; Douglas et al., 2013 ). Other mutants are only defective in forming hyphae in liquid medium and can still invade into a solid substrate. This phenotype is likely caused by contact with a solid substrate, which triggers an independent hyphal pathway (Kumamoto, 2005 and 2008; Douglas et al., 2013 ). Therefore, to identify genes that are broadly important for invasive hyphal growth, we screened for C. albicans mutants that were defective in invading into solid agar media under a diverse set of strongly inducing conditions ( Naseem et al., 2019 ). Analysis of the strongest mutants in a mouse model of oral candidiasis revealed an important role for the RVS161 and RVS167 genes, which are known to promote the scission phase of endocytosis ( Naseem et al., 2019 ). The RVS genes are also important for systemic infections caused by C. albicans, further implicating endocytosis in virulence ( Douglas et al., 2009 ).
Blocking endocytosis may have therapeutic value, as small molecules inhibitors of endocytosis and hyphal growth showed efficacy against oral candidiasis ( Pierce et al., 2015 ; Bar- Yosef et al., 2017 ; Romo et al., 2017 ; Vila et al., 2017 ). Furthermore, the rvsΔ and other C. albicans endocytosis mutants were much more susceptible to killing by copper (Douglas and Konopka, 2019), suggesting copper could enhance the therapeutic value of endocytosis inhibitors. Thus, C. albicans agar invasion assays can be useful for screening mutant strains and novel inhibitors for efficacy in blocking the invasive growth of C. albicans.
Materials and Reagents
Inoculating loop
Culture tubes (Sarstedt, catalog numbers: 62.554.205 for 15 ml size; 62.547.100 for 50 ml size)
2 L flasks (Pyrex, catalog number: 4980-2L)
Glass test tubes (Pyrex, catalog number: 9820)
Micropipette 20-200 μl tips (VWR, catalog number: 89079-478)
Micropipette 100-1,000 μl tips (Denville, catalog number: P2103-N)
Microcentrifuge tubes (Denville, catalog number: C2170)
Spectrophotometer cuvettes (Grainger, catalog number: 23M350)
10 cm-sized Petri dishes (Kord-Valmark, catalog number: 41141000)
96-well flat bottom polystyrene plate (Fisher Scientific, catalog number: 2420811)
Whatman filter paper (Fisher Scientific, catalog numbers: 1003-055 [55 mm], 1450-080 [80 mm], 1002-090 [90 mm], 1003-125 [125 mm])
Razor blade (Fisher Scientific, catalog number: 18100970)
C. albicans strains can be obtained from the Fungal Genetics Stock Center (http://www.fgsc.net)
Agar (Fisher Scientific, catalog number: BP9744-5)
BD Bacto Yeast extract (Fisher Scientific, catalog number: 212720)
Peptone (Fisher Scientific, catalog number: BP1420-2)
Dextrose (Fisher Scientific, catalog number: D16-10)
Bovine Calf Serum (Hyclone, catalog number: SH30073.03)
N-acetylglucosamine (GlcNAc) (Chem-Impex, catalog number: 01427)
Yeast Nitrogen Base: BD Bacto (Fisher Scientific, catalog number: 291920)
Adenine (Chem-Impex, catalog number: 00006)
Arginine (Fisher Scientific, catalog number: BP370-100)
Asparagine (Fisher Scientific catalog number: BP374-100)
Alanine (Fisher Scientific catalog number: BP369-100)
Citrulline (Argos, catalog number: 372-75-8)
Cysteine (Fisher Scientific, catalog number: BP376-100)
Glutamic acid (Fisher Scientific, catalog number: 194677)
Glycine (Fisher Scientific, catalog number: BP381-500)
Histidine (Fisher Scientific, catalog number: BP382-100)
Isoleucine (Fisher Scientific, catalog number: BP384-100)
Leucine (Fisher Scientific, catalog number: BP385-100)
Lysine (Fisher Scientific, catalog number: BP386-100)
Methionine (Fisher Scientific, catalog number: BP388-100)
Ornithine (Fisher Scientific, catalog number: BP389-100)
Phenylalanine (Fisher Scientific, catalog number: BP391-100)
Proline (Fisher Scientific, catalog number: BP392-100)
Serine (Chem-Impex, catalog number: 00279)
Tryptophan (Fisher Scientific, catalog number: BP395-100)
Threonine (Chem-Impex, catalog number: 00282)
Tyrosine (Fisher Scientific catalog number: BP396-100)
Uracil (Sigma, catalog number: U0750-100G)
Uridine (Chem-Impex, catalog number: 00339)
Valine (Fisher Scientific, catalog number: BP397-100)
p-Aminobenzoic acid (Sigma, catalog number: A-0254)
Sodium Phosphate (dibasic, Na2HPO4) (Sigma, catalog number: S9390)
HEPES (Millipore, catalog number: 391333-100GM)
Potassium phosphate (dibasic, K2HPO4) (Fisher Scientific, catalog number: P288-500)
Mannitol (Sigma, catalog number: M4125-500G)
GlcNAc Medium (see Recipes)
Serum Medium (see Recipes)
Spider Medium (see Recipes)
Alkaline pH Medium (150 mM HEPES pH 8) (see Recipes)
Synthetic Dextrose Medium (see Recipes)
Yeast Extract Peptone Dextrose (YEPD or YPD) Medium (see Recipes)
YP sucrose Medium (see Recipes)
10x supplemental solution for SD media (standard) (see Recipes)
10x supplemental solution for SD media (see Recipes)
Equipment
Roller
Centrifuge
Autoclave
Spectrophotometer (Thermo Fisher GENESYS 6, catalog number: 335908)
Pipet Aid (Drummond, catalog number: 4-000-100)
Micropipettor (Gilson, catalog numbers: FA10003M [2-20 μl], FA10005M [20-200 μl], FA10006M [100-1,000 μl])
Microplate Replicator, 48-Well Format (also called a pinner) (Boekel, catalog number: 140501)
-
A light microscope equipped with a 2x objective and also a 40x long working distance objective
Note: The model is not that important, but it will be essential to have a camera attached for recording images.
-
Digital camera for taking macroscopic images of colony morphology
Note: The type of camera is not that critical; a camera on a mobile phone will work fine.
Procedure
-
Invasion of cells spotted on top of agar
Notes:
If working with strains from a large collection of mutants, use PCR to confirm that the appropriate genes are deleted in the mutant strains.
-
To help distinguish between mutants that are defective in the initiation or maintenance of invasive growth, cells can be preinduced to form hyphae in liquid by incubation for 90 min at 37 °C in YPD plus 10% BCS or 50 mM GlcNAc prior to application onto the surface of an agar plate (Naseem et al., 2019). (Mutants that are defective in maintenance will not continue to form hyphae. Mutants with a defect in initiation, but not maintenance, can continue to form hyphae if they are first induced under conditions that will stimulate hyphal growth.)
Wild type and mutant C. albicans cells are streaked on to YPD plates from frozen stocks and incubated for 2 days at 30 °C.
A single colony from freshly streaked overnight culture grown on a YPD plate is transferred with an inoculating loop into 5 ml YPD liquid medium in a 15 ml glass tube. Grow cells overnight in a 30 °C incubator on a roller.
Harvest the cells from the overnight culture by centrifuging for 5 min at 1,000 × g in a 15 ml centrifuge tube. Resuspend the pellet in 10 ml PBS and centrifuge it again at 1,000 × g for 5 minutes. Repeat the same process twice and then resuspend the cells into 5 ml PBS.
Adjust the cells to 1 x 107 cells/ml. A convenient way to do this is to use a spectrophotometer at 600 nm wavelength. However, it is necessary to first set up a calibration curve to determine the relationship between cell number and optical density (OD). The relationship can vary between different spectrophotometers because a spectrophotometer measures light scattering, not absorbance. The final cell density can then be adjusted as follows: [Cell density desired] x [volume 1] = [Cell density initial] x [volume 2]. (Note that the volumes are indicating a ratiometric relationship. Volume 1 is the amount of cells that the investigator wants to use. This depends on how big of an experiment they are doing. Volume 2 is determined by volume 1.)
-
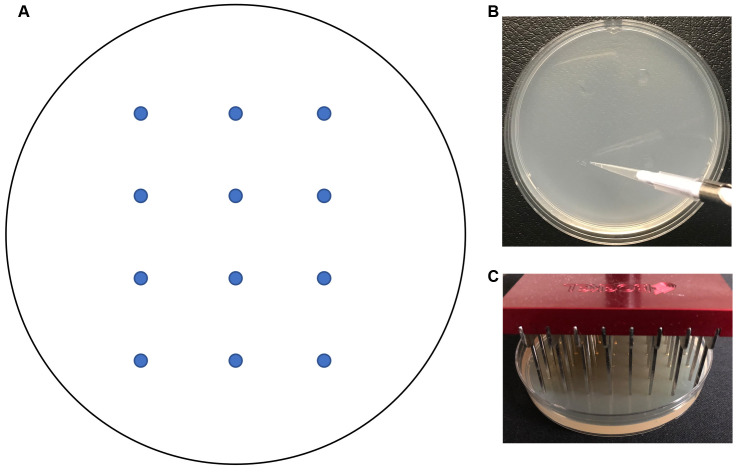
Use a micropipettor to spot a sample of each strain (3 μl of 1 x 107 cells/ml in PBS) onto the surface of a 10 cm agar plate (Figure 2B). Place the spots at least 2 cm apart to prevent inhibition of growth due to nutrient depletion. See Figure 2A for a template.
Note: On a typical 10 cm Petri plate we can spot at least 12 strains.
Repeat this step to spot the samples onto different types of media to test invasive growth under different conditions, including agar with Bovine Calf Serum (BCS), GlcNAc, alkaline pH, and spider medium. Recipes are described below. The concentration of agar can be varied when preparing the medium to test the effects of differing the rigidity of the surrounding matrix ( Douglas et al., 2013 ) (see Recipe 2 below). Additional types of media that stimulate invasive hyphal growth have also been described ( Liu et al., 1994 ; Biswas and Morschhauser, 2005; Vinces et al., 2006 ).
Invert the plates and incubate plates at 37 °C. It may be necessary to add a tray of water to the incubator to maintain humidity to prevent the plates from drying out if keeping plates for over 5 days.
Monitor the edge of the spots for invasive growth daily by eye (Figure 3A) or use a microscope (Figure 3B). It may be possible to detect initiation of invasive growth with the aid of a microscope after 1 day. Visible signs of invasive growth may take 2 days to one week.
-
Invasion of cells applied to surface of agar with a 48 prong pinner
To facilitate the analysis of a large set of strains, a 48 prong pinner can be used to transfer cells to the surface of an agar plate (e.g., a Boekel Microplate Replicator). A 48 prong pinner is used because it can transfer cells onto agar in a standard 10 cm-sized Petri plate.
Inoculate strains into wells of a 96-well flat bottom polystyrene plate containing 300 µl appropriate liquid medium. Incubate overnight at 30 °C until growth is saturated.
-
Sterilize 48 pin replicator.
Assemble four containers together in a row (empty Petri dishes or empty micropipette tip box lids may be used).
Fill containers with 25 ml sterile deionized H2O, 10% bleach, or 97% ethanol in the following order: #1 = H2O, #2 = 10% bleach, #3 = H2O, #4 = 97% ethanol.
First, dip replicator into H2O, agitate to clean pins, then blot replicator on Whatman filter paper.
Place replicator into 10% bleach for 20-40 s. Blot replicator on filter paper.
Transfer to second container of H2O, agitate to rinse pins, blot replicator on filter paper one last time.
Dip replicator into 97% ethanol (do not blot).
Shake off excess ethanol, and pass through the flame of a Bunsen burner.
Once flames have extinguished, turn replicator ninety degrees, dip into 97% ethanol and flame.
After cooling, turn replicator ninety degrees once again, dip into 97% ethanol, flame, and cool.
Insert cooled sterile replicator into the 48 wells on the left half of the 96-well strain plate.
Mix wells of plate with replicator pins, and then press replicator lightly onto Petri plate containing the desired medium (e.g., 2% agar containing 4% BCS). Repeat this procedure to pin cells onto plates containing additional types of media.
Repeat sterilization of replicator (Steps B2c to B2i) when switching to new strains.
Incubate pinned plates at 37 °C for 48 h and examine for invasive growth as described above (Step A8).
-
Embedding method for analysis of invasive hyphal growth
Embedding cells in agar is an excellent way to determine invasive growth since the cells are surrounded by agar in three dimensions. However, it can be more difficult to compare a set of mutant strains since they each have to be analyzed on a different Petri plate. Thus, a good approach is to screen mutants by spotting on the agar surface and then retesting those with mutant phenotypes by embedding into agar.
Grow overnight cultures of the desired strains to be analyzed. Transfer a single colony from a fresh culture grown on a YPD plate into into 5 ml YPD liquid medium in a 15 ml glass tube. Incubate on a tube roller or in a shaking incubator at 30 °C.
Use a spectrophotometer to measure cell density at 600 nm. Adjust the cells to 5 x 105 cells/ml in 15 ml YPD broth (see Step A4).
Incubate in an incubator-shaker at 30 °C, 200-250 RPM, for 4 h to obtain log phase cells.
During incubation, prepare medium and cool in a water bath adjusted to 50 °C (approximately an hour may be required for cooling).
-
Types of media that may be used include 4% BCS, GlcNAc, alkaline pH, Spider medium, and YP sucrose as these activate a diverse set of different cell signaling pathways (Kornitzer, 2019). As described above, the concentration of agar can be varied to test the effects of differing the rigidity of the surrounding matrix.
Warning: Do not add serum to media before autoclaving, as it will denature.
After the 4 h incubation, measure density of log phase cells at 600 nm and calculate the volume of culture equivalent to 200 cells. Sample calculation: 2,000 cells/ml x 0.1 ml = 200 cells. See Step A4 for additional information on adjusting cell density.
Pour 25 ml of agar medium cooled to 50 °C into a 50 ml centrifuge tube.
For serum agar samples, add 1 ml of BCS, previously warmed to 50 °C.
Add volume of C. albicans culture equivalent to 200 cells.
Mix well (but avoid trapping bubbles) and then pour into a sterile 10 cm-sized Petri dish. Allow to solidify for 1 h.
Incubate plates at 37°C except for YP + Sucrose plates, which are incubated at room temperature or in a 25°C incubator.
Examine daily for evidence of invasive growth. Spots on agar were photographed with a digital camera (Figure 3A), or with a microscope using a 40x objective to view invasion extending from the edge of a spot (Figure 3B). The embedded colonies were photographed with a microscope through a 2x lens (Figure 4).
Figure 2. Spotting cells onto agar.
A. Template for spotting cells on a Petri plate. B. Spotting cells onto the surface of a Petri plate with a micropipettor. C. Transferring cells onto the surface of an agar plate with a pinner.
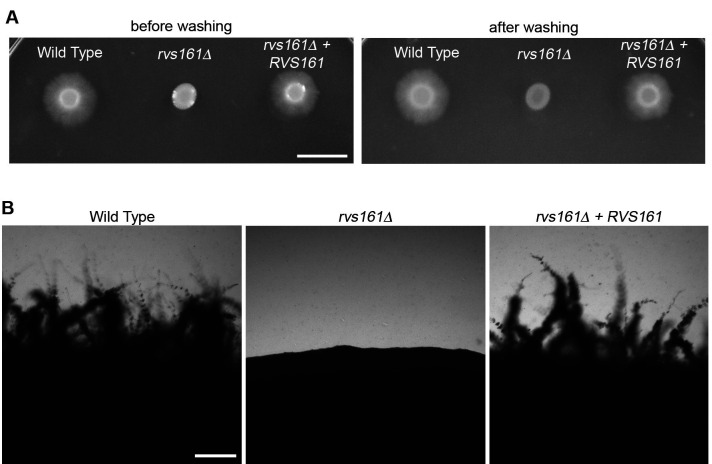
Figure 3. Invasive growth of C. albicans cells spotted on top of agar.
A. The indicated cell types were spotted onto the surface of an agar plate with 4% BCS, and then incubated at 37 °C for 7 days. The plate was then photographed before (left side) or after (right side) washing off the non-adherent cells. B. Higher magnification of the edges of spots showing invasive hyphal growth defect for the rvs161Δ mutant. Panel A: Scale bar = 1 cm; Panel B: Scale bar = 200 μm.
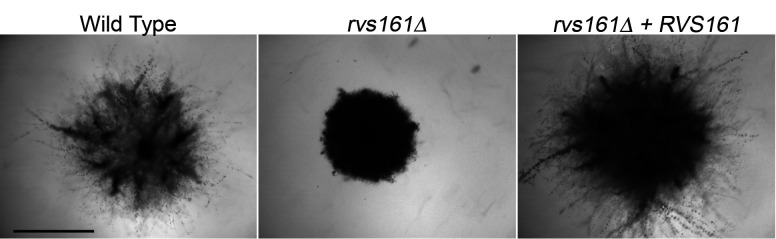
Figure 4. Invasive growth of cells embedded into agar.
The indicated strains were embedded into agar containing 4% BCS and then incubated at 37 °C for one week. Scale bar = 1 mm.
Data analysis
Visually inspect the plates for signs of invasion into the agar surrounding the original spot of cells. Note the extensive invasive growth emanating from the original spot of cells seen for the wild type sample but is absent for the rvs161Δ mutant (Figure 3A).
To enhance detection of invasive growth, the agar plates can be gently washed with a stream of water to remove non-invasive cells (samples on the right in Figure 3A). Wear gloves and gently use a finger to dislodge non-adherent cells without damaging the agar.
At early times of incubation, or to check for even a very limited extent of invasive growth, the edges of the spots of cells can be viewed through a microscope with a 40x lens and photographed (Figure 3B). If appropriate, the zone of invasive growth can be measured with a ruler to quantify the extent of invasive growth.
Alternatively, the agar can be cut with a razor blade through the center of the spot and then turned on its side to visualize the depth of invasive growth ( Warenda et al., 2003 ). However, better results can be obtained by studying embedded samples.
Embedded samples can be analyzed similar to samples spotted on the surface. A sample of cells embedded into agar containing 4% BCS is shown in Figure 4. Note how the wild type and the complemented rvs161Δ + RVS161 strains showed extensive invasive hyphal growth into the agar that was absent for the rvs161Δ mutant.
The extent of invasive growth should be analyzed on a daily basis for at least a week. Some mutant strains may show a strong defect at early time points, but will start to show significant invasive growth after ~7 days ( Naseem et al., 2019 ).
Recipes
Note: In many recipes the agar and other components are autoclaved separately and then combined later.
-
GlcNAc Medium (per liter)
6.7 g Yeast Nitrogen Base in 400 ml water in 1 L flask
18 g Agar in 400 ml water in 2 L flask
Autoclave these two separately at 121°C for 40 min, when cooled to 55 °C, add 100 ml sterile 10x Amino Acid Supplement (if required), and 100 ml of filter sterilized 500 mM GlcNAc to the 2 L flask containing agar solution
Mix well and pour into Petri dishes
Note: GlcNAc is heat sensitive, add when agar is cooled to 55 °C (final concentration 50 mM), then pour into Petri plates.
-
Serum Medium (per liter)
-
Agar mass to desired % w/v required in 2 L flask
Note: Higher concentrations of agar (4% and above) are difficult to work with as they become, making it difficult to pour and easier to trap bubbles. Use a flask that is at least four times the volume of the medium inside.
Bovine Calf Serum volume filter sterilized separately, added to desired % v/v
1 L dH2O (adjust volume as needed)
Autoclave flask 121°C 40 min
Note: Bovine Calf Serum is heat sensitive. Add appropriate volume at 55 °C (to final concentration and volume). Stir to mix, then pour into Petri plates.
-
-
Spider Medium (per liter)
13.5 g Agar 2 L flask
2 g K2HPO4, 10 g Yeast Nitrogen Base and 10 g Mannitol filter sterilized separately, adjust to pH 7.2
1 L dH2O (adjust volume as needed)
Autoclave flask 121°C 40 min and pour contents of smaller container into larger flask. Stir to mix, cool to 55 °C, then pour into Petri plates
-
Alkaline pH Medium (150 mM HEPES pH 8) (per liter)
18 g Agar
6.7 g Yeast Nitrogen Base
20 g Dextrose
35.75 g HEPES
100 ml 10x Amino Acid Supplement (if required)
Place the 18 g agar in a 2 L flask with 500 ml deionized H2O
In a separate flask, add 6.7 g Yeast Nitrogen Base, 20 g Dextrose, 35.75 g HEPES and 400 ml deionized H2O
Note: A stock solution of HEPES can be made and sterile filtered ahead of time.
Adjust the pH to pH 8
If adding the Amino Acid Supplement Solution, use 100 ml less water
Autoclave flasks 121 °C 40 min and combine the contents into one flask. Stir to mix, cool to 55 °C, then pour into Petri plates
-
Synthetic Dextrose Medium (per liter)
6.7 g Yeast Nitrogen Base in 1 L flask
18 g Agar in 2 L flask
100 ml 10x Amino Acid Supplement (if required)
20 g Dextrose autoclaved separately
800 ml dH2O distributed among flasks (adjust volume as needed)
Autoclave flasks 121 °C 40 min and then pour contents of smaller flasks into the larger flask. Stir to mix, cool to 55 °C, then pour into Petri plates
-
Yeast Extract Peptone Dextrose (YEPD or YPD) Medium (per liter)
20 g Dextrose in 1 L flask
18 g Agar, 10 g Yeast Extract, 20 g Peptone, 120 mg Adenine in 2 L flask
1 L dH2O distributed among flasks (adjust volume as needed)
Autoclave flasks 121°C 40 min and then pour contents of smaller flasks into the larger flask. Stir to mix, cool to 55 °C, then pour into Petri plates
-
YP sucrose Medium (per liter)
20 g sucrose in 1 L flask
18 g Agar, 10 g Yeast Extract, 20 g Peptone, 120 mg Adenine in 2 L flask
1 L dH2O distributed among flasks (adjust volume as needed)
Autoclave flasks 121°C for 40 min and pour contents of smaller flasks into the larger flask. Stir to mix, cool to 55 °C, then pour into Petri plates
-
10x supplemental solution for SD media (standard) (1 L)
Mix the following into 1 L dH2O in beaker:
Adenine 0.4 g
Uracil 0.4 g
Tryptophan 0.2 g
Histidine 0.2 g
Arginine 0.2 g
Methionine 0.2 g
Tyrosine 0.3 g
Leucine 0.3 g
Isoleucine 0.3 g
Lysine 0.3 g
Phenylalanine 0.5 g
Glycine 1 g
Asparagine 1 g
Valine 1.5 g
Threonine 2 g
Serine 4 g
Add 100 ml amounts into bottles and autoclave 121 °C 40 min
-
10x supplemental solution for SD media (1 L)
Mix the following into 1 L dH2O in beaker:
Alanine 0.5 g
Asparagine 0.5 g
Cysteine 0.5 g
Glutamic acid 0.5 g
Glycine 0.5 g
Proline 0.5 g
p-Aminobenzoic acid 0.05 g
Sodium Phosphate (Dibasic) 4 g
Add 100 ml amounts into each of 10 bottles and autoclave 121 °C 40 min
Acknowledgments
This work was supported by Public Health Service grants from the National Institutes of Health awarded to J.B.K. from the (R01GM116048, and R01AI047837).
The methods for studying invasive growth were briefly described in Naseem et al. (2019).
Competing interests
The authors declare that they do not have any competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Bar-Yosef H., Vivanco Gonzalez N., Ben-Aroya S., Kron S. J. and Kornitzer D.(2017). Chemical inhibitors of Candida albicans hyphal morphogenesis target endocytosis . Sci Rep 7(1): 5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Biswas K. and Morschhauser J.(2005). The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans . Mol Microbiol 56(3): 649-669. [DOI] [PubMed] [Google Scholar]
- 3. Douglas L. M. and Konopka J. B.(2019). Plasma membrane architecture protects Candida albicans from killing by copper . PLoS Genet 15(1): e1007911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Douglas L. M., Martin S. W. and Konopka J. B.(2009). BAR domain proteins Rvs161 and Rvs167 contribute to Candida albicans endocytosis, morphogenesis, and virulence . Infect Immun 77(9): 4150-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Douglas L. M., Wang H. X. and Konopka J. B.(2013). The MARVEL domain protein Nce102 regulates actin organization and invasive growth of Candida albicans . mBio 4(6): e00723-00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glazier V. E., Murante T., Murante D., Koselny K., Liu Y., Kim D., Koo H. and Krysan D. J.(2017). Genetic analysis of the Candida albicans biofilm transcription factor network using simple and complex haploinsufficiency . PLoS Genet 13(8): e1006948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kornitzer D.(2019). Regulation of Candida albicans hyphal morphogenesis by endogenous signals . J Fungi(Basel) 51: 10.3390/jof5010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumamoto C. A.(2005). A contact-activated kinase signals Candida albicans invasive growth and biofilm development . Proc Natl Acad Sci U S A 102(15): 5576-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumamoto C. A.(2008). Molecular mechanisms of mechanosensing and their roles in fungal contact sensing. Nat Rev Microbiol 6(9): 667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu H., Kohler J. and Fink G. R.(1994). Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog . Science 266(5191): 1723-1726. [DOI] [PubMed] [Google Scholar]
- 11. Naseem S., Douglas L. M. and Konopka J. B.(2019). Candida albicans rvs161 and rvs167 endocytosis mutants are defective in invasion into the oral cavity . mBio 106: 10.1128/mBio.02503-02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Naseem S., Gunasekera A., Araya E. and Konopka J. B.(2011). N-acetylglucosamine(GlcNAc) induction of hyphal morphogenesis and transcriptional responses in Candida albicans are not dependent on its metabolism . J Biol Chem 286(33): 28671-28680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Noble S. M., French S., Kohn L. A., Chen V. and Johnson A. D.(2010). Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity . Nat Genet 42(7): 590-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Noble S. M., Gianetti B. A. and Witchley J. N.(2017). Candida albicans cell-type switching and functional plasticity in the mammalian host . Nat Rev Microbiol 15(2): 96-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O'Meara T. R., Veri A. O., Ketela T., Jiang B., Roemer T. and Cowen L. E.(2015). Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat Commun 6: 6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pierce C. G., Chaturvedi A. K., Lazzell A. L., Powell A. T., Saville S. P., McHardy S. F. and Lopez-Ribot J. L.(2015). A novel small molecule inhibitor of Candida albicans biofilm formation, filamentation and virulence with low potential for the development of resistance . NPJ Biofilms Microbiomes 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pierce J. V. and Kumamoto C. A.(2012). Variation in Candida albicans EFG1 expression enables host-dependent changes in colonizing fungal populations . mBio 3(4): e00117-00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riggle P. J., Andrutis K. A., Chen X., Tzipori S. R. and Kumamoto C. A.(1999). Invasive lesions containing filamentous forms produced by a Candida albicans mutant that is defective in filamentous growth in culture . Infect Immun 67(7): 3649-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Romo J. A., Pierce C. G., Chaturvedi A. K., Lazzell A. L., McHardy S. F., Saville S. P. and Lopez-Ribot J. L.(2017). Development of anti-virulence approaches for Candidiasis via a novel series of small-molecule inhibitors of Candida albicans filamentation . mBio 8(6): e01991-01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vila T., Romo J. A., Pierce C. G., McHardy S. F., Saville S. P. and Lopez-Ribot J. L.(2017). Targeting Candida albicans filamentation for antifungal drug development . Virulence 8(2): 150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vinces M. D., Haas C. and Kumamoto C. A.(2006). Expression of the Candida albicans morphogenesis regulator gene CZF1 and its regulation by Efg1p and Czf1p . Eukaryot Cell 5(5): 825-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Warenda A. J., Kauffman S., Sherrill T. P., Becker J. M. and Konopka J. B.(2003). Candida albicans septin mutants are defective for invasive growth and virulence . Infect Immun 71(7): 4045-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Witchley J. N., Penumetcha P., Abon N. V., Woolford C. A., Mitchell A. P. and Noble S. M.(2019). Candida albicans morphogenesis programs control the balance between gut commensalism and invasive infection . Cell Host Microbe 25(3): 432-443 e436. [DOI] [PMC free article] [PubMed] [Google Scholar]