Abstract
Growing evidences suggest that peritubular capillaries pericytes are the main source of scar-forming myofibroblasts during chronic kidney disease (CKD), as well as early phases of acute kidney injury (AKI). In a swine model of sepsis and I/R (Ischemia Reperfusion) injury-induced AKI we demonstrated that renal pericytes are able to transdifferentiate toward α-SMA+ myofibroblasts leading to interstitial fibrosis. Even if precise pericytes identification requires transmission electron microscopy and the co-immunostaining of several markers (i.e., Gli, NG2 chondroitin sulphate proteoglycan, CD146, desmin or CD73) and emerging new markers (CD248 or TEM1, endosialin), previous studies suggested that PDGFR-β could be used as marker for renal pericytes characterization. Recently, double immunofluorescence staining of PDGFR-β and α-SMA was performed to identify the damage activated pericytes (PDGFR-β+/α-SMA+ cells) in the early phase of fibrosis development. Our data highlighted the crucial role of renal pericytes in the physiopathology of sepsis and I/R associated AKI. In this protocol, we describe the procedure for double immunofluorescence staining of PDGFR-β and α-SMA in swine Formalin-Fixed Paraffin-Embedded (FFPE) kidney biopsies and the method for image analysis and quantification.
Keywords: PDGFR-β+/α-SMA+ immunofluorescence , Pericyte-to-Myofibroblast Transition, Renal pericytes, Swine kidney biopsies, Renal I/R injury, Sepsis, AKI, Perivascular cells, AKI-to-CKD transition
Background
Renal fibrosis is considered the principal responsible for progression of renal disease and it is associated with a limited capacity of kidney to regenerate after injury. The principal source of interstitial fibrosis in progressive renal diseases ( Simone et al., 2014 ; Fiorentino et al., 2018 ) is represented by activated fibroblasts, named myofibroblasts, that are recognized by their ability to synthesize de novo α-smooth muscle actin (α-SMA) (Meran and Steadman, 2011; Hewitson, 2012). These cells derived from multiple sources, including not only resident fibroblasts, but also endothelial cells, tubular cells, circulating bone marrow–derived cells and pericytes (Di Carlo and Peduto, 2018). Several studies evaluated the critical role of capillary pericytes in chronic kidney disease (CKD) ( Lin et al., 2008 ; Grgic et al., 2012 ; Kramann et al., 2015 ) as well as early phases of acute kidney injury (AKI) (Leaf et al., 2016; Castellano et al., 2018 and 2019).
Recent studies revealed that dysfunctional pericytes are principally involved in sepsis-induced microvascular dysfunction and vascular leakage which are the key hallmarks of end-organ dysfunction and septic shock ( Goldenberg et al., 2011 ; Page and Liles, 2013; Castellano et al., 2014 ; Stasi et al., 2017 ; Fani et al., 2018 ). Consistent with these findings, we found that pigs challenged with LPS led to a dysfunctional response of pericytes in the microvasculature of renal parenchyma ( Castellano et al., 2019 ) (Figure 1) .
Figure 1. LPS induced PMT in a swine model of LPS-induced AKI.
A. Representative pictures of PDGRβ and α-SMA staining performed in swine renal sections of healthy and endotoxemic pigs.Pericytes were double-stained for PDGFRβ (green) and α-SMA (red). In healthy pigs (T9 CTR), α-SMA immunostaining signal was detected only within vascular smooth-muscle cells (dotted white arrow) and PDGFRβ+/αSMA+ perivascular cells were barely detectable. In endotoxemic pigs (T9 LPS) the number of PDGFRβ+/αSMA+ cells significantly increased (white arrows), as shown by the colocalization of the two markers in the interstitiumand at glomerular level. Magnification 630X. B. Quantitative data (n = 5 for each group, P = 0.001). Results are expressed as median ± IQR of five independent pigs for each group. The statistical analysis was performed using the Mann-Whitney test. Modified from Figure 1 in Castellano et al. (2019) . License: (http://creativecommons.org/licenses/by/4.0/).
In recent years, the fate-tracing mapping and the ultrastructural analysis have shed more lights on the pericytes behavior in several mouse model of renal diseases ( Lin et al., 2008 ; Humphreys et al., 2010 ). In fairness, the identification of pericytes by criteria that requires elaborate techniques as fate-tracing analysis is not practical in large animal model. Since PDGFR-β is defined as the constitutive marker for isolation and characterization of renal pericytes ( Chen et al., 2011 ; Wang et al., 2017 ) and a-SMA is a marker associated with myofibroblasts (Hewitson, 2012), the double-labeling for PDGFR-β and α-SMA provide a “picture” of pericyte distribution and dysfunctional activation in renal parenchyma during AKI ( Guzzi et al., 2019 ). In accordance with other phenomena of cellular transdifferentation as EndMT, the loss of PDGFR-β and the increased level of α-SMA, Collagen I and MMP proteins is defined as Pericytes to Myofibroblast transition (PMT) ( Chang et al., 2012 ).
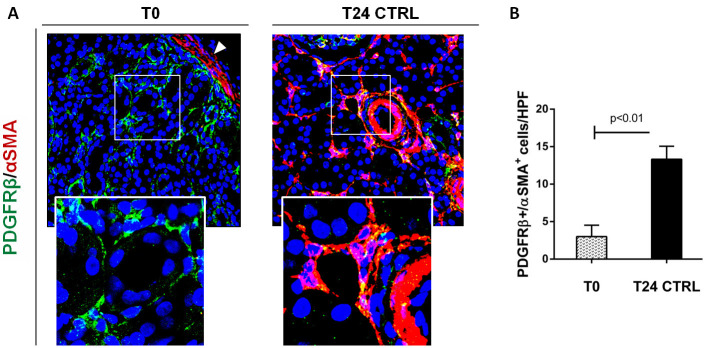
We, firstly, performed double immunofluorescent staining of PDGFR-β and α–SMA to study the PMT in swine and mice models of renal I/R injury (Figure 2) ( Castellano et al., 2018 ). Interestingly, in normal swine kidney biopsies we found the colocalization of the PDGFR-β and NG2 in peritubular capillaries, indicating the overall accuracy of PDGFR-β as capillary pericytes markers in pig models whereas glomerular mesangial cells were PDGFRβ+ but NG2−. After 24h of renal I/R we observed the downregulation of the constitute markers PDGFR-β and NG2 and the dramatic upregulation of α-SMA.
Figure 2. Complement activation induced PMT in a swine model of I/R injury.
A. Representative pictures of PDGRβ and α-SMA staining performed on paraffin sections at T0 (before ischemia) and 24 h after reperfusion (T24 CTRL). At T0, PDGFRβ+/αSMA+ cells were rarely detectable (zoomed image) and α-SMA was localized in the arterial wall (white arrow). After 24 h from I/R injury, the number of these cells dramatically increased (T24 CTRL). Magnification 630x. B. Results are expressed as median ± IQR of five independent pigs for each group. The statistical analysis was performed using the Mann-Whitney test. Modified from Figure 4 in Castellano et al. (2018) . License: (http://creativecommons.org/licenses/by/4.0/).
These data were also confirmed in our sepsis model of AKI and taken together we demonstrated that PDGFR-β+pericytes are able to synthesize pro-fibrotic markers acquiring the typical features of myofibroblasts, leading to extracellular matrix deposition and interstitial fibrosis ( Castellano et al., 2019 ). In accordance, we confirmed our data in vitro and we found that exposition of human placental derived pericytes to I/R injury (C5a) and sepsis stimuli (LPS) led to acquisition of α-SMA contractile stress fibers ( Castellano et al., 2018 and 2019).
Therefore, the protocol described here could be useful to characterize pericytes and their dysfunctional activation and will facilitate the research in the early acute kidney injury as well as other fields relating inflammation and fibrosis ( Castellano et al., 2018 and 2019).
Materials and Reagents
Aluminium Foil (Carl Roth GmbH, Rotilabo®, catalog number: 0954.1)
StripetteTM Serological Pipets 5 ml, 10 ml (Corning, catalog numbers: 4051, 4488)
Dish 60 mm (Corning, catalog number: 3261)
Falcon® centrifuge tube 50 ml,15 ml (Corning, catalog numbers: 352070, 352096)
Safety safeshield scalpels (Biosigma srl, catalog number: 530050)
Metal steel base mold (Leica Biosystem, catalog number: 3803081)
Coverslips for optical microscopy 24 x 40 x 0.16 mm (Bio Optica SpA, catalog number: 09-2040)
Paper towel
Tissue Embedding Rings (Bio Optica SpA, catalog number: 07-7650)
Nail varnish
10% neutral buffered formalin (NBF) (Diapath SpA, catalog number: F0047)
Paraffin Lab O-Wax 56-58 °C (Histo-Line Laboratories srl, catalog number: R0040-20)
Sterile DPBS (Euroclone SpA, catalog number: ECM4053XL)
Xylene (Bio Optica SpA, catalog number: 06-1304Q)
Ethanol absolute (100%) (Bio Optica SpA, catalog number: 06-10099)
Deionized water
Microscopy slides Super Frost® Plus (Bio Optica SpA, catalog number: 09-OPLUS)
Normal Goat Serum (Sigma-Aldrich, catalog number: G9023, stored in aliquots at -20 °C)
Phosphate Buffered Saline Tablets (Sigma-Aldrich, catalog number: P4417)
α-SMA mouse monoclonal (Santa Cruz Biotechnology, catalog number: sc-32251)
PDGFR-β monoclonal antibody rabbit anti human (Abcam, catalog number: ab32570)
AlexaFluor goat anti-rabbit FITC-conjugated (488 nm) antibody (Molecular Probes, catalog number: A11070)
AlexaFluor goat anti-mouse TRITC conjugated (555 nm) antibody (Molecular Probes, catalog number: A21127)
TO-PRO3 IODIDE (Invitrogen, catalog number: T3605)
FluoroMount (Bio-Optica SpA, catalog number: K024)
Phosphate buffered saline tablet (Sigma, catalog number: P4417)
Acid citric (Sigma, catalog number: C759)
Sodium citrate (Sigma, catalog number S4641)
Normal Goat Serum (Sigma-Aldrich, catalog number G9023, stored in aliquots at -20 °C)
Unmasking Buffer (10 mM Sodium Citrate Buffer) (see Recipes)
Blocking solution (10% Normal Goat Serum) (see Recipes)
Buffer for diluting primary antibody (5% Normal Goat Serum) (see Recipes)
PBS 1x for the IF washing (see Recipes)
Equipment
Autoclave (AHSI S.p.A., model: FVA/A1)
Tweezers (Thermo Fisher Scientific, catalog number: 10303611)
Scissors for microscopy (Bio Optica SpA, catalog number: 32-703)
Gloves and Eye protection
Hot plate with magnetic stirrer
PMP Hellendhal Staining Jar (Kartell S.p.A., LABWARE Division, KartellTM, catalog number: 0035500)
Water bath
Fume cupboard (Euroclone SpA)
Pipettes (0.5-10, 10-100 and 100-1,000 μl) (Eppendorf, catalog numbers: 3111000.122, 3111000.149, 3111000.165)
Bard monopty biopsy instrument 16 G x 20 cm length (C.R. Bard.Inc., catalog number: 121620)
58 °C paraffin bath Cold plate (Bio Optica SpA, catalog number: PF100)
Microtome (Leica Biosystem, catalog number: RM2125 RTS)
Histology Bath (Bio Optica SpA, catalog number: WB100)
Microwave
Pap pen for immunostaining (Bio Optica SpA, catalog number: 11-100)
Orbital shaker (Thermo Scientific, catalog number: SHKA2000)
Humidified Chamber
pH meter (Thermo Scientific, catalog number: 3115101)
Software
GraphPad Prism® (version 5.0) (GraphPad Software)
Image analysis software (Leica QWin) (Leica)
Procedure
-
Collection of renal samples
-
Preparation of work tools for sample processing
Autoclave clean tweezers and scissors after wrapping them in aluminium foil.
-
Renal biopsies procedure
A Needle core biopsy was performed at the start of experimental procedure (T0) and at several intermediate time points in healthy and treated pigs. Renal biopsies was performed under direct ultrasound guidance with automated biopsy needles. After the sacrifice, kidneys were explanted and cuneiform biopsies of cortical and medullary regions were obtained with safety safeshield scalpels.
Each renal biopsy was delivered to the histological laboratory in a 50 ml Falcon containing 20ml of cold (~4 °C) sterile DPBS and was identified by a unique code for each animal.
All renal biopsies have to be immersed in ice for a half an hour at the most.
-
Preparation of renal biopsies before fixation
Each renal biopsy has to be transferred in a 60 mm dish containing 3 ml cold (~5 °C) DPBS and cleaned from any residual connective or adipose tissue by sterile tweezers.
The specimen size can be reduced to 0.5 x 0.5 cm with a sterile mono-use scalpel.
The renal specimen has to be transferred in another 60 mm dish containing 3 ml of cold (~4 °C) DPBS.
-
-
Paraffin slides processing
-
Renal specimen fixation
The renal specimen has to be transferred into 15 ml conical tube with 10% neutral buffered formalin (NBF) at room temperature for 8 h but no longer than 24 h. Make sure you have enough fixative to cover tissues. Fixative volume should be 5-10 times of tissue volume.
Note: The renal specimen is not hold up in paraffin cassettes. Tissue processing was carried out manually to mimic the individual cumulative steps of automated processing.
-
Renal specimen dehydration
After fixation, formalin has to be discarded.
-
The renal specimen has to be dehydrated moving to alcohol grades steps:
10 ml 50% ethanol for 40 min (max 5 days) at room temperature.
10 ml 70% ethanol for 40 min at room temperature.
10 ml of 95% ethanol for 25 min at room temperature.
10 ml of 95% ethanol for 25 min at room temperature.
10 ml of 100% ethanol for 25 min at room temperature.
10 ml of 100% ethanol for 25 min at room temperature.
Note: These steps should be completed fully (with sufficient time set aside for these steps) because insufficient dehydration can lead to tissue degradation.
-
Renal specimen clarification
The renal specimen has to be cleared with xylene:
10 ml of xilene for 15 min.
10 ml of xilene for 15 min.
-
Renal specimen infiltration by paraffin
The renal specimen has to be transferred into a becker containing liquid Paraffin in 58 °C paraffin bath: 10 ml of liquid Paraffin for 2 h in an oven at 56 °C-58 °C (the melting temperature of paraffin).
-
Embedding renal tissues in paraffin blocks
Small amount of molten paraffin has to be put in mold, dispensing from paraffin reservoir.
Using warm forceps, the renal specimen has to be transferred into the well of the metal steel base mold in health block, placing cut side down.
The embedding ring has to be put into the steel base mold (at 56 °C). The paraffin ribbon is floated onto the surface of a warm water bath, where it spreads out and flattens perfectly.
The mold has to be transferred quickly to cold plate (-20 °C), and gently press tissue flat. Paraffin will solidify in a thin layer that holds the tissue in position.
Hot paraffin has to be added to the mold from the paraffin dispenser. Be sure there is enough paraffin to cover the tissue. The paraffin block should solidify in 30 min at -20 °C.
When the wax is completely cooled and hardened the paraffin block has to be easily popped out of the mold; the wax blocks should not stick. If the wax cracks or the tissues are not aligned well, simply melt them again and start over.
-
Preparation of paraffin sections: Sectioning protocol:
-
The paraffin block has to be sectioned at the desired thickness (usually 4-5 µm) on a microtome and float on a 40 °C water bath containing distilled water.
Notes:
Keep record of the orientation and sequence of the sections.
The histology bath has to reach a temperature up to 56 °C to allow the paraffin sections to flatten.
The paraffined sections have to be transferred onto Super Frost Plus Microscope slide.
The slides have to dry overnight and store slides at room temperature until ready for use.
-
-
-
Immunofluorescence staining
-
The paraffin sections have to deparaffinated and rehydrated (The entire process must be set up in the fume cupboard)
-
The slides have to be deparaffinized in pure xylene for 15 min.
Note: Xylene removes the excess of wax around the tissue and deparaffinates the paraffined sections.
-
The slides have to rehydrated using graded alcohol series for few minutes and rinse in deionized water:
Ethanol 100% for 6 min.
Ethanol 95% for 1 min.
Ethanol 70% for 1 min.
Ethanol 50% for 1 min.
Deionized water for 5 min (for three times).
-
-
Heat-induced Antigen unmasking
-
The slides have to be transferred into a plastic vertical staining jar (suitable for microwave), containing 100 ml of sodium citrate buffer 10 mM at pH = 6 (filled to the brim) and put in a water bath.
Note: The water bat is obtained put slide jar into a glass of water (suitable for microwave) and put the entire thing into the microware. This reduces the boiling inside the jar .
The slides have to be subjected to three microwave (750 W) cycles of 5 min.
The volume of antigen retrieval buffer has to be top up with distilled water at each cycle in order to avoid evaporation during boiling.
-
The slides have to be let slowly cool.
Note: The number of slides must be the same of the positions available in the staining jar at each antigen unmasking process so that the efficiency of unmasking is always the same. If less slides are used, fill up the empty spaces with empty glass slides. This ensures a similar heat distribution to each slide and reduces the differences in antigen retrieval.
-
-
Immunofluorescence staining:
The cool slides have to be transferred into glass vertical staining jar with lid filled with deionized water. Wash the slides in deionized water for 5 min on aorbital shaker at room temperature.
The slides have to be rinsed in three changes of PBS 1x for 5 min each with agitations at room temperature.
The slides have to be placed with kidney sections facing up in the humidified chamber.
Each section on the slides has to be limited with a PAP-pen to prevent overflows.
100-200 μl of blocking buffer (10% Normal Goat Serum diluted in PBS 1x) has to be added to each section and incubated for 1 h, at room temperature, in the humidified box.
The staining mix has to be prepared combined and diluted appropriately the two different antibodies (α-SMA 1:100, PDGFR-β 1:100) in the blocking mix (5% of Goat Serum), respecting the dilution tested individually before.
The blocking buffer has to be removed gently (e.g., by tipping the slide sideways onto a paper towel), without rinsing before adding primary antibody.
100 μl of primary antibodies mix has to be added to each section and incubated in the humidified chamber overnight at +4 °C.
Negative control of the IF reaction has to be obtained incubating the isotype control antibody instead of the primary antibodies mix on a well delimited section in one of the slides.
The slides have to be rinsedin PBS 1x for three times, 5 min each.
A mix of the two secondary antibodies, respectively to bind the two primary antibodies, has to be prepared combined and diluted appropriately: Alexa Fluor goat anti-mouse 555 (1:200) and Alexa Fluor goat anti-rabbit 488 (1:200) in PBS 1x in the dark.
The secondary antibodies mix (the volume ranges from 50-100 µl depending of the biopsies area) has to be applied on the sections and incubate for 1 h in the humidified box in the dark.
The slides have to be submersed in PBS 1x for 5 min for 3 times.
To counterstain the slides, a solution of TOPRO3 (dilution 1:3,000) in PBS 1x has to be prepared and added on each section. The slides have to be incubated in the humidified box and in the dark for 10 min.
TOPRO3 solution has to be drained off from the slides, and without rising, a drop of fluoromount has to be added on the slides to mount a coverslip. Make sure that no air bubbles are formed.
The slides have to be sealed with a nail polish to preserve staining and stored for a long time in the dark.
The slides has to be acquired by confocal microscope Leica TCS SP2 (Leica).
-
Data analysis
Confocal microscopy was performed using the Leica TCS SP2 (Leica), equipped with argon-krypton (488 nm), green neon (543 nm), and helium-neon (633 nm) lasers. Confocal images were taken at 500-nm intervals through the z-axis of the section, encompassing a total of 7 µm in depth. Images from individual optical planes and multiple serial optical sections were analyzed, and the images were sequentially scanned in all three laser channels. The images were captured using a 63x objective lens and exported as TIFF files as showed in Figures 1 and 2.
Two independent observers blinded to the origin of the slides counted the number of α-SMA+ and PDGFR-cells in at least 10 consecutive high-power (630x) fields (0.0567 mm2 HPF/section) for each biopsy. The values were then averaged. For each field, positive cells were counted only in the cortical area (peritubular and glomerular capillaries). Subcapsular fibrotic areas, arterial adventitia and medullary areas were excluded from the region of interest. Cell count was determined according to nuclear staining. The final reported count was the mean of the two observer measures. In no case was the interobserver variability greater than 20%.
Results are expressed as median ± interquartile range (IQR) Statistically significant differences were assessed by the Mann-Whitney test. A P value < 0.05 was significant. Statistical analysis was performed using GraphPad Prism Software 5 as indicated in Figure 3.
Figure 3. Statistical analysis by GraphPad Prism Software.
A. Open GraphPad Prism and select the parameters indicated by the red arrows. B. Start to insert the values in each group indicated by the red arrow. C-D. Click analyze and choose the test for the analysis. Given the small sample (five independent pigs for each group) we choose non-parametric test, Mann-Whitney test and presented the data as Medians and IQRs.
Recipes
-
Unmasking Buffer (10 mMSodium Citrate Buffer) (500 ml)
Citric Acid Solution (10.5 g acid citric dissolved in 500 ml distilled water) (41 ml)
Sodium Citrate Solution (14.7g sodium citrate dissolved in 500 ml distilled water) (9 ml)
Distilled water (500 ml)
Mix to dissolve. Adjust pH to 6.0 with 1 N HCl and mix well. Store this solution at 4 °C for longer storage
-
Blocking solution (10% Normal Goat Serum)
5 ml PBS
500 μl Normal Goat Serum
Store at 4 °C for 1 week
-
Buffer for diluting primary antibody (5% Normal Goat Serum)
5 ml PBS 1x
250 μl Normal Goat Serum
Store at 4 °C for 1 week
-
PBS 1x for the IF washing
Dissolve one Phosphate buffered saline tablet in 200 ml of deionized water yields 0.01 M phosphate buffer, 0.0027 M potassium chloride and 0.137 M sodium chloride, pH 7.4 at 25 °C
Store at 25 °C for 1 week
Acknowledgments
These studies were supported by University of Bari “Aldo Moro”, the Italian Ministry of Health (Ricerca Finalizzata 2009 and Giovani Ricercatori 2011-2012 [GR-2011-02351027] granted to Giuseppe Castellano), a Regional Strategic Grant (Apulia Region (PSR 094) granted to Loreto Gesualdo) and an unrestricted research grant from Pharming Group.
Competing interests
Authors have no conflicts of interest or competing interests to disclose.
Ethics
These studies were approved by the ethical committee of the Ministry of Health, Italy.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Castellano G., Franzin R., Stasi A., Divella C., Sallustio F., Pontrelli P., Lucarelli G., Battaglia M., Staffieri F., Crovace A., Stallone G., Seelen M., Daha M. R., Grandaliano G. and Gesualdo L.(2018). Complement Activation During Ischemia/Reperfusion Injury Induces Pericyte-to-Myofibroblast Transdifferentiation Regulating Peritubular Capillary Lumen Reduction Through pERK Signaling. Front Immunol 9: 1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Castellano G., Stasi A., Franzin R., Sallustio F., Divella C., Spinelli A., Netti G. S., Fiaccadori E., Cantaluppi V., Crovace A., Staffieri F., Lacitignola L., Grandaliano G., Simone S., Pertosa G. B. and Gesualdo L.(2019). LPS-Binding Protein Modulates Acute Renal Fibrosis by Inducing Pericyte-to-Myofibroblast Trans-Differentiation through TLR-4 Signaling. Int J Mol Sci 20(15): 3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Castellano G., Stasi A., Intini A., Gigante M., Di Palma A. M., Divella C., Netti G. S., Prattichizzo C., Pontrelli P., Crovace A., Staffieri F., Fiaccadori E., Brienza N., Grandaliano G., Pertosa G. and Gesualdo L.(2014). Endothelial dysfunction and renal fibrosis in endotoxemia-induced oliguric kidney injury: possible role of LPS-binding protein. Crit Care 18(5): 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang F. C., Chou Y. H., Chen Y. T. and Lin S. L.(2012). Novel insights into pericyte-myofibroblast transition and therapeutic targets in renal fibrosis. J Formos Med Assoc 111(11): 589-598. [DOI] [PubMed] [Google Scholar]
- 5. Chen Y. T., Chang F. C., Wu C. F., Chou Y. H., Hsu H. L., Chiang W. C., Shen J., Chen Y. M., Wu K. D., Tsai T. J., Duffield J. S. and Lin S. L.(2011). Platelet-derived growth factor receptor signaling activates pericyte-myofibroblast transition in obstructive and post-ischemic kidney fibrosis. Kidney Int 80(11): 1170-1181. [DOI] [PubMed] [Google Scholar]
- 6. Di Carlo S. E. and Peduto L.(2018). The perivascular origin of pathological fibroblasts. J Clin Invest 128(1): 54-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fani F., Regolisti G., Delsante M., Cantaluppi V., Castellano G., Gesualdo L., Villa G. and Fiaccadori E.(2018). Recent advances in the pathogenetic mechanisms of sepsis-associated acute kidney injury. J Nephrol 31(3): 351-359. [DOI] [PubMed] [Google Scholar]
- 8. Fiorentino M., Grandaliano G., Gesualdo L. and Castellano G.(2018). Acute Kidney Injury to Chronic Kidney Disease Transition. Contrib Nephrol 193: 45-54. [DOI] [PubMed] [Google Scholar]
- 9. Goldenberg N. M., Steinberg B. E., Slutsky A. S. and Lee W. L.(2011). Broken barriers: a new take on sepsis pathogenesis. Sci Transl Med 3(88): 88ps25. [DOI] [PubMed] [Google Scholar]
- 10. Grgic I., Duffield J. S. and Humphreys B. D.(2012). The origin of interstitial myofibroblasts in chronic kidney disease. Pediatr Nephrol 27(2): 183-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guzzi F., Cirillo L., Roperto R. M., Romagnani P. and Lazzeri E.(2019). Molecular Mechanisms of the Acute Kidney Injury to Chronic Kidney Disease Transition: An Updated View. Int J Mol Sci 20(19): 4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hewitson T. D.(2012). Fibrosis in the kidney: Is a problem shared a problem halved? Fibrogenesis Tissue Repair 1): S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Humphreys B. D., Lin S. L., Kobayashi A., Hudson T. E., Nowlin B. T., Bonventre J. V., Valerius M. T., McMahon A. P. and Duffield J. S.(2010). Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol 176(1): 85-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kramann R., Schneider R. K., DiRocco D. P., Machado F., Fleig S., Bondzie P. A., Henderson J. M., Ebert B. L. and Humphreys B. D.(2015). Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell 16(1): 51-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leaf I. A., Nakagawa S., Johnson B. G., Cha J. J., Mittelsteadt K., Guckian K. M., Gomez I. G., Altemeier W. A. and Duffield J. S.(2017). Pericyte MyD88 and IRAK4 control inflammatory and fibrotic responses to tissue injury. J Clin Invest 127(1): 321-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin S. L., Kisseleva T., Brenner D. A. and Duffield J. S.(2008). Pericytes and perivascular fibroblasts are the primary source of collagen-producing cells in obstructive fibrosis of the kidney. Am J Pathol 173(6): 1617-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meran S. and Steadman R.(2011). Fibroblasts and myofibroblasts in renal fibrosis. Int J Exp Pathol 92(3): 158-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Page A. V. and Liles W. C.(2013). Biomarkers of endothelial activation/dysfunction in infectious diseases. Virulence 4(6): 507-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simone S., Rascio F., Castellano G., Divella C., Chieti A., Ditonno P., Battaglia M., Crovace A., Staffieri F., Oortwijn B., Stallone G., Gesualdo L., Pertosa G. and Grandaliano G.(2014). Complement-dependent NADPH oxidase enzyme activation in renal ischemia/reperfusion injury. Free Radic Biol Med 74: 263-273. [DOI] [PubMed] [Google Scholar]
- 20. Stasi A., Intini A., Divella C., Franzin R., Montemurno E., Grandaliano G., Ronco C., Fiaccadori E., Pertosa G. B., Gesualdo L. and Castellano G.(2017). Emerging role of Lipopolysaccharide binding protein in sepsis-induced acute kidney injury. Nephrol Dial Transplant 32(1): 24-31. [DOI] [PubMed] [Google Scholar]
- 21. Wang N., Deng Y., Liu A., Shen N., Wang W., Du X., Tang Q., Li S., Odeh Z., Wu T. and Lin H.(2017). Novel Mechanism of the Pericyte-Myofibroblast Transition in Renal Interstitial Fibrosis: Core Fucosylation Regulation. Sci Rep 7(1): 16914. [DOI] [PMC free article] [PubMed] [Google Scholar]