Abstract
Ex vivo culture assays of biopsy specimens are advantageous for the experimental evaluation of human circadian characteristics. We developed a simple and non-invasive experimental evaluation method for monitoring the expression of circadian clock genes in an ex vivo culture assay using human hair follicles. This method imposes little burden on subjects. This assay is useful for validating correlations between circadian characteristics in hair follicles and intrinsic characteristics observed in physiological and behavioral studies. While they should be further validated, this ex vivo method constitutes a useful tool for estimating in vivo circadian characteristics.
Keywords: Circadian rhythm, Clock gene, Human, Hair follicle, Ex vivo, Non-invasive
Background
Living organisms exhibit circadian rhythms in physiology and behavior that are driven by the circadian clock (Young and Kay, 2001). The circadian clockwork consists of cell-autonomous and clock gene-driven negative feedback loops of transcription (Dunlap, 1999). In mammals, the transcription factors BMAL1 and CLOCK activate the transcription of clock and clock-related genes such as Period (Per) and Cryptochrome (Cry) via E-box elements. PER together with CRY, a potent transcriptional inhibitor, subsequently functions to negatively regulate this complex (Reppert and Weaver, 2002). The in vivo evaluation of individual intrinsic circadian characteristics in humans, either with a constant routine or forced desynchrony protocol, is expensive and labor-intensive. Therefore, evaluation using ex vivo culture assays to estimate in vivo circadian characteristics could present important advantages. For example, several studies have concluded that circadian characteristics in peripheral cells reflect individual circadian preferences, known as chronotype ( Brown et al., 2005 ; Hida et al., 2013 ). For a simple and non-invasive evaluation of cell-autonomous circadian performance in humans, including intrinsic period length, we developed a method to monitor clock gene expression in real time using an ex vivo culture of hair follicles ( Yamaguchi et al., 2017 ).
Materials and Reagents
(Optional) Keep-warm bag
Sterile sampling tubes (the volume size should be within 0.2 from 1.5 ml) (e.g., 0.2 ml tube, Bio-Bik, catalog number: 133003; 1.5 ml tube, VIOLAMO, catalog number: 1-1600-1)
35 mm dishes (e.g., IWAKI, catalog number: 1000-035)
DMEM without Phenol Red (Sigma-Aldrich, catalog number: D2902)
-
DMEM (Nacalai, catalog number: 08456-94)
Although DMEM from other suppliers is usable as a substitute for these DMEMs, the possibility cannot be excluded that the difference in concentrations of minor ingredients affects success rates and results slightly.
Sodium bicarbonate (Sigma-Aldrich, catalog number: S8761)
HEPES (Nacalai, catalog number: 17557-94)
D-Glucose (Sigma-Aldrich, catalog number: G8769)
Penicillin/streptomycin (Thermo Fisher Scientific, catalog number: 15070-063)
L-Glutamin (Nacalai, catalog number: 16948-04)
Sodium pyruvate (Sigma-Aldrich, catalog number: S8636)
Luciferin (WAKO, catalog number: 126-05116)
-
Silicone (Shin-Etsu, catalog number: KS-64)
Note: Reagents #6-13: Similar items from other suppliers serve as a substitute.
-
Adenovirus vectors carrying the luciferase (luc) gene driven by circadian promoter/enhancer elements (e.g., Bmal1-luc, Per2-luc and Per3-luc). Some entry vectors we constructed and deposited are available from the RIKEN bank as follows:
RDB15083 mouse Period2 (-2.8 ~ +0.1 kb) - Luciferase/pENTR-1A
RDB15084 human Period3 (-3.1 ~ +0.2 kb) - Luciferase/pENTR-1A
RDB15085 human Bmal1 (-1.7 ~ +0.1 kb) - Luciferase/pENTR-1A
The ViraPower Adenoviral Gateway Expression Kit (K4940-00, Thermo Fisher Scientific) is required to generate adenoviruses using these entry vectors. Procedures using adenoviruses must be performed inside P2 areas. Autoclave and discard any infectious wastes
Infection medium (see Recipes)
Pre-incubation medium (see Recipes)
Rinse medium (see Recipes)
Luciferin-containing medium (see Recipes)
Equipment
Non-slip, cosmetic-use tweezers (e.g., Shiseido, eyebrow nippers 211)
Tweezers (e.g., FST, catalog number: 18132-12)
Block incubator set to 36.5 °C (e.g., ASTEC, model: BI-516C)
-
Laminar flow cabinet (e.g., SANYO, model: MCV-B131F)
Note: Equipment #1-4: Similar items from other suppliers serve as a substitute.
-
Incubator at 35-36.5 °C with 5% CO2 (e.g., ASTEC, model: SCA-165DRS)
Use a CO2 incubator that utilizes an infrared sensor that is not affected by humidity inside the chamber (see details in Procedure Step 18).
Photomultiplier tube (Hamamatsu, model: LM2400)
Luminescence microscope (Olympus, model: LV200)
Software
Cosinor software (freely downloadable from a website: https://www.circadian.org/softwar.html)
Procedure
Ex vivo culture of human hair follicles to investigate circadian gene expression:
Human studies must be conducted in accordance with the Declaration of Helsinki and approved by institutional review boards.
Recruit subjects and obtain informed consent from each subject or the subject's family.
(Optional) Although dependent on research aim, volunteers may be asked to follow a set schedule for one or several weeks. For example, wake-up time and meal times may be set based on the lifestyle habits of each volunteer. They may also be asked to refrain from consuming excess alcohol and snacks and taking long naps. Individual chronotypes can be assessed using the morningness-eveningness questionnaire (MEQ) or the Munich chronotype questionnaire (MCTQ) ( Zavada et al., 2005 ).
-
Prepare infection medium (see Recipes) by adding an adenovirus carrying the luciferase gene driven by circadian promoter/enhancer elements to fresh DMEM at a 1:20 dilution. The titer of our adenovirus stocks is within a range of 1.24 x 109-1.47 x 109 infectious units/ml. The use of adenovirus vectors carrying Per3-luc may be recommended (see the following note for this reason). For vector construction, we used the ViraPower Adenoviral Gateway Expression Kit ( Yamaguchi et al., 2017 ). Briefly, together with the luciferase gene, the transcription-regulatory region of a clock gene was subcloned into the pENTR-1A entry vector and subsequently introduced into the pAd/PL-DEST vector using LR recombination.
Note: We previously compared period length, robustness and standard deviation (SD) among three adenovirus vectors carrying Bmal1-luc, Per2-luc and Per3-luc (Yamaguchi et al., 2017). Although the average period length was almost identical among the three vectors, the SD of the period length of Per3 was consistently smaller than that of the others. Additionally, the period length of Per3 was the most robust at the smallest SD values. We therefore preferably used the adenovirus vector carrying Per3-luc in our experimental conditions.
Before sampling hair follicles, prepare sterile sampling tubes containing 80 μl of pre-incubation medium, rinse medium or infection medium (see Recipes for the composition of each medium). One tube containing the respective medium is required per hair follicle. Pre-warm and maintain medium at 36.5 °C using a block incubator.
(Optional) On cold winter days, warming the carotid arteries by attaching pocket warmers to both sides of the neck may improve the success rate of hair follicle sampling.
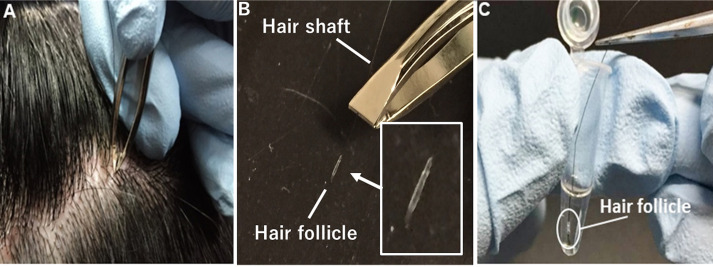
As shown in Figure 1, collect several strands of scalp (or facial) hair by firmly holding and pulling individual strands of hair by the root using non-slip tweezers (for cosmetic use). To avoid hair follicle cell death by drying, quickly immerse the hair follicle cells into pre-warmed pre-incubation medium without removing the hair shaft. Keep samples at 36.5 °C until the next step (infection: Step 11). Although hair collection does not necessarily require a sterile environment, procedures after Step 11 are recommended to be performed under sterile conditions.
Cut the hair shaft, leaving about 10 to 20 mm.
To obtain a strong bioluminescence signal, use plucked scalp (or facial) hairs whose root surface is mostly or fully covered with hair follicle cells. A representative image showing successful infection of an adenovirus carrying Per2-luc to a hair root mostly covered with hair follicle cells is shown in our original paper (Figure 1B, Yamaguchi et al., 2017 ). We previously investigated the effect of differences in hair stage on circadian period length and found no significant differences in period length among stages ( Yamaguchi et al., 2017 ).
(Optional) In the case of out-of-lab sampling, maintain samples in a keep-warm bag without CO2 during transport to the laboratory. We confirmed that shipping duration up to approximately 10 h had no obvious effect on the circadian period length of clock gene expression.
Inside a laminar flow cabinet, use sterilized tweezers to pick up the hair shaft, immerse the hair follicles into pre-warmed rinse medium, and transfer and completely immerse the follicles into pre-warmed infection medium on a block incubator (36.5 °C). Procedures using adenoviruses must be performed inside P2 areas. Autoclave and discard any infectious wastes.
Close the lid of tubes and incubate them for 24 h at 36.5 °C with 5% CO2.
The next day, prepare sterile sampling tubes containing pre-warmed rinse medium (at 36.5 °C) for the removal of viruses. Two tubes are required for washing twice.
Syringe and push out silicone into a mound on the bottom of 35 mm culture dishes (Figure 2 and Video 1). Place the silicone somewhat off-center to facilitate locating the hair follicle at the center of the culture dish because reducing the distance between a PMT surface and infected cells enhances bioluminescence counts. Use one dish per hair follicle. Sterilization of silicone is not essential, but we recommend using silicone from an unopened packet designated for culture use. In addition, prepare and pre-warm luciferin-containing medium (see Recipes for the composition of each medium) at a concentration of 0.1 mM.
Use sterile tweezers to pick up the hair by the shaft and transfer the infected hair follicles into 80 μl of pre-warmed rinse medium. After one-minute incubation, again transfer infected hair follicles into the same volume of fresh rinse medium and incubate them for the same duration to remove adenoviruses as completely as possible.
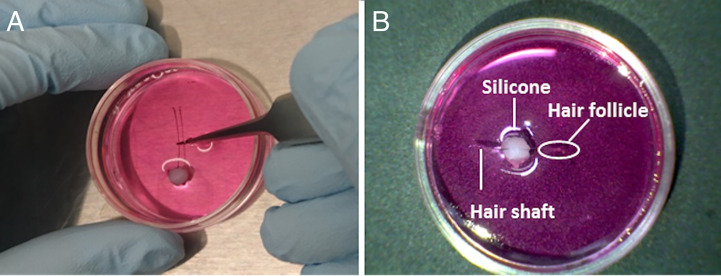
Transfer the hair follicle to the silicone mound in a 35 mm culture dish for bioluminescence monitoring (Video 2). To avoid floating during measurement, push the hair shaft into the silicone so that the shaft is stuck to the silicone and is fixed on the bottom of the dish (Figure 2). Position the hair follicle near to the center of the dish for efficient bioluminescence detection.
Completely cover the immobilized hair follicle with 2-3 ml pre-warmed luciferin-containing medium.
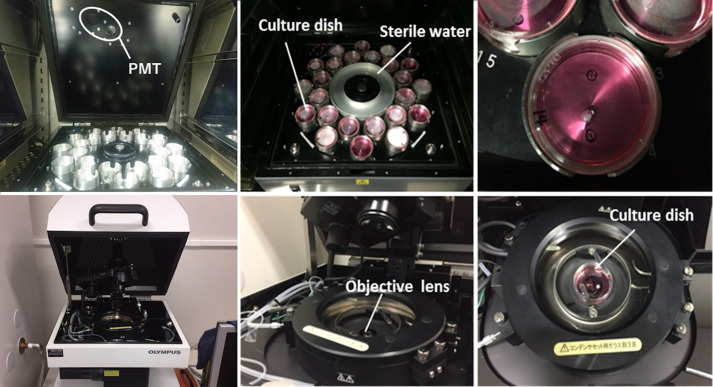
As shown in Figure 3, measure bioluminescence in real-time using a photomultiplier tube inside a dark box specifically designed to reduce background noise to detect ultra-weak photon emissions (LM2400, Hamamatsu, Japan) or a luminescence microscope optimized for single-cell imaging (Olympus, Japan) at 35 °C with 5% CO2. To avoid rust formation, the LM2400 is located inside a CO2 culture incubator under low humidity conditions. To prevent the culture medium from drying out, instead of using the water tray from the CO2 incubator, use the one inside the LM2400. Use a CO2 incubator that utilizes an infrared sensor that is not affected by humidity inside the chamber. For measurement, open the top of the LM2400 and simply place culture dishes on the metal tray. Culture dishes without hair follicles should provide reads of 5,000-10,000 counts per minute (background control). In successful cases, bioluminescence from the hair follicles should be greater than the background counts and show circadian oscillation over more than four to five days.
Figure 1. Collection of strands of scalp hair.
A. Firmly hold and pull individual strands of hair by the root using non-slip tweezers. B. Use plucked hairs whose root surface is mostly or fully covered with hair follicle cells. C. Quickly immerse hair follicle cells in pre-warmed pre-incubation medium without removing the hair shaft.
Figure 2. Fixing a hair follicle to the bottom of a culture dish.
A. Transfer a hair follicle to a silicone mound in a 35 mm culture dish. Push the hair shaft into the silicone so that the shaft is stuck to the silicone and is fixed to the bottom of the dish. B. Position the hair follicle near to the center of the dish.
Video 1. Placing silicone in a mound on the bottom of the culture dishes.

Video 2. Transferring a hair follicle to a silicone mound.
Figure 3. Monitoring bioluminescence in real time.
Bioluminescence can be measured in real time using a photomultiplier tube (top three panels, LM2400) or a luminescence microscope optimized for single-cell imaging (bottom three panels, LV200) at 35 to 36.5 °C with 5% CO2. Left panels, wide views of the instruments; middle panels, views of sample holders; right panels, views of culture dishes on the sample holders.
Data analysis
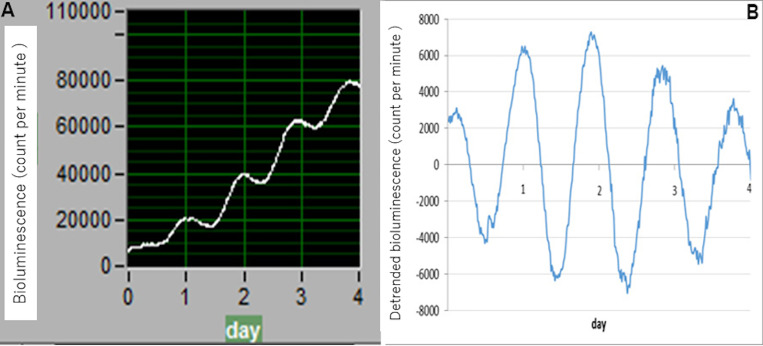
Data collection is performed using the associated software “Software for Photon Detection Unit C10749 (LM2400v21)-JP”. To analyze circadian parameters, baseline changes need to be removed. As shown in Figure 4, data sets are therefore detrended using Microsoft Excel by subtracting the 24 h running average from the raw data (A, raw data; B, detrended data). Circadian robustness, circadian phase (angle) and circadian period length are calculated using Cosinor software, provided as a gift by Dr. Refinetti. Oscillation data are considered reliable and useful not only when clear circadian oscillation persists for ≥ 4 days but also when circadian robustness is 70% or greater.
Figure 4. Example data.
Data sets are detrended by subtracting the 24 h running average from the raw data. A. Raw data (software, Photon Detection Unit C10749 (LM2400v21)-JP); B. detrended data (software, Microsoft Excel).
Notes
Although the absolute value of circadian period length in cultured hair follicles differs from those obtained from physiological and behavioral studies, we confirmed that the relative difference in period length among mouse genotypes is similar between clock gene expression in hair follicles and locomotor activity ( Yamaguchi et al., 2017 ). Our ex vivo method may therefore be a useful tool for estimating in vivo circadian characteristics.
Culture medium composition reportedly affects period length ( Lee et al., 2011 ; Noguchi et al., 2012 ). For example, we have found that the absence of phenol red results in damping and relatively longer periods ( Yamaguchi et al., 2017 ). It is therefore important to use identical medium composition and product lot numbers throughout all sets of experiments for comparison of relative differences between subjects.
Dexamethasone (DEX) is a well-known synchronizer for peripheral clocks. Peripheral clock gene expression is often monitored in vitro and ex vivo after DEX treatment. However, adenoviral infection can also synchronize clock gene expression. To simplify the experimental protocol and reduce cellular stress, DEX treatment is therefore excluded when using adenovirus-infected hair follicles.
Recipes
-
Infection medium (80 μl or more per hair follicle)
DMEM (Nacalai, Japan)
1% penicillin/streptomycin
5% Adenovirus
-
Pre-incubation medium (80 μl or more per hair follicle)
DMEM without Phenol Red (Sigma-Aldrich)
0.035% sodium bicarbonate
10 mM HEPES
4.5 g/L D-Glucose
1% penicillin/streptomycin
1 mM L-Glutamin
1 mM sodium pyruvate
-
Rinse medium (80 μl or more for each washing)
DMEM (Nacalai, Japan)
1% penicillin/streptomycin
-
Luciferin-containing medium (2-3 ml per hair follicle)
DMEM (Nacalai, Japan)
1% penicillin/streptomycin
0.1 mM luciferin
Acknowledgments
We thank Ai Yamaguchi, Akihiko Okamoto, Ritsuko Matsumura, Miho Sato, Rie Okamitsu and Junko Sumino for their expert technical assistance. We also acknowledge the support of fellowships from the Yamaguchi Gerontology Research Institute, the Akaeda Medical Research Foundation, the SENSHIN Medical Research Foundation, and the Japan Society for the Promotion of Science. The authors declare no competing financial interests. This protocol was originally developed in Yamaguchi et al., 2017 .
Competing interests
The authors declare no conflicts of interest or competing interests.
Ethics
This study was conducted in accordance with the Declaration of Helsinki and was approved by the institutional review boards of Yamaguchi University (approval number: H25-81-4). Informed consent was obtained from all subjects.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Brown S. A., Fleury-Olela F., Nagoshi E., Hauser C., Juge C., Meier C. A., Chicheportiche R., Dayer J. M., Albrecht U. and Schibler U.(2005). The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol 3(10): e338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dunlap J. C.(1999). Molecular bases for circadian clocks. Cell 96(2): 271-290. [DOI] [PubMed] [Google Scholar]
- 3. Hida A., Kitamura S., Ohsawa Y., Enomoto M., Katayose Y., Motomura Y., Moriguchi Y., Nozaki K., Watanabe M., Aritake S., Higuchi S., Kato M., Kamei Y., Yamazaki S., Goto Y., Ikeda M. and Mishima K.(2013). In vitro circadian period is associated with circadian/sleep preference . Sci Rep 3: 2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee S. K., Achieng E., Maddox C., Chen S. C., Iuvone P. M. and Fukuhara C.(2011). Extracellular low pH affects circadian rhythm expression in human primary fibroblasts. Biochem Biophys Res Commun 416(3-4): 337-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Noguchi T., Wang C. W., Pan H. and Welsh D. K.(2012). Fibroblast circadian rhythms of PER2 expression depend on membrane potential and intracellular calcium. Chronobiol Int 29(6): 653-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reppert S. M. and Weaver D. R.(2002). Coordination of circadian timing in mammals. Nature 418(6901): 935-941. [DOI] [PubMed] [Google Scholar]
- 7. Yamaguchi A., Matsumura R., Matsuzaki T., Nakamura W., Node K. and Akashi M.(2017). A simple method using ex vivo culture of hair follicle tissue to investigate intrinsic circadian characteristics in humans . Sci Rep 7(1): 6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Young M. W. and Kay S. A.(2001). Time zones: a comparative genetics of circadian clocks. Nat Rev Genet 2(9): 702-715. [DOI] [PubMed] [Google Scholar]
- 9. Zavada A., Gordijn M. C., Beersma D. G., Daan S. and Roenneberg T.(2005). Comparison of the munich chronotype questionnaire with the horne-ostberg's morningness-eveningness score. Chronobiol Int 22(2): 267-278. [DOI] [PubMed] [Google Scholar]