Abstract
Heme oxygenase-1 (HO-1) is a stress responsive enzyme that metabolizes heme and releases free iron, carbon monoxide (CO), and biliverdin (BV), which rapidly undergoes conversion to bilirubin (BL). Estimation of bilirubin is the basis of HO-1 assay. HO-1 activity is widely employed to determine antioxidant response of cells under different physiological stress environment. Intra-macrophage infection often acts as such a stress inducer and measurement of HO-1 activity in infected cells indicates the ability of pathogens towards modulating oxidative response of host. The present protocol describes analysis of HO-1 activity in infected macrophages by spectrophotometric method, which is much less complex and therefore advantageous over other methods like high-performance liquid chromatography, radiochemical methods and detection of CO by gas chromatography. The main steps include: (1) Preparation of macrophage microsomal fraction containing HO-1 (2) Isolation of rat liver cytosolic fraction containing biliverdin reductase and (3) Assessment of heme oxygenase-1 activity by spectrophotometric detection of bilirubin. This method provides a simple and sensitive approach to measure cellular antioxidant response under infected condition.
Keywords: Heme oxygenase-1, Macrophage, Leishmania, Spectrophotometry, Bilirubin, Hemin
Background
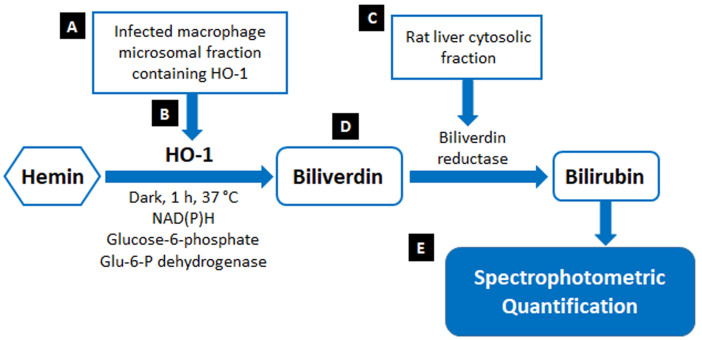
Reactive oxygen species (ROS) is one of the major host defense arsenals against invading pathogens used by macrophages (Missall et al., 2004). On the other hand, intra-macrophage pathogens neutralize early oxidative burst for their successful persistence within macrophages (Paiva and Bozza, 2014). In response to such oxidative stress, organisms can deploy antioxidant enzymes of host cells such as superoxide dismutase (SOD), catalase (CAT) glutathione peroxidase (GPX), and heme oxygenase-1 (HO-1) to scavenge ROS (Kathirvel et al., 2010). Intracellular parasite Leishmania donovani could effectively exploit host antioxidant enzyme HO-1 for ROS neutralization (Saha et al., 2019). HO-1 is a potent anti-oxidant enzyme catalysing the oxidative cleavage of heme to generate carbon monoxide (CO), ferrous iron (Fe2+), and biliverdin (BV). The biliverdin is further acted upon by another enzyme biliverdin reductase (BVR) to produce bilirubin (BL) (Tenhunen et al., 1969).There are several techniques to quantify the activity of HO-1 based on detection of one of its ultimate reaction product bilirubin via high-performance liquid chromatography (Lincoln et al., 1988; Ryter et al., 1999), visible spectrophotometry (Schacter, 1978; Tenhunen et al., 1969) and radiochemical methods (Sierra and Nutter, 1992). Detection of CO by gas chromatography (Vreman and Stevenson, 1988) has also been used to assay HO-1 activity, but because of its complexity and subsequent product analysis steps the protocol is not variedly applicable. Now, biliverdin is the primary metabolite of the heme degradation by HO-1. However, it has poor spectral properties with an extinction coefficient (ε) of ~8 to 10 mM-1 cm-1 (Kutty and Maines, 1981). Thus, most common HO-1 activity assays rely on the reduction of biliverdin to bilirubin. Original spectrophotometric quantification of bilirubin for detection of HO-1 activity was outlined by Tenhunen et al. (1969). In the method, bilirubin formation was monitored spectrophotometrically by the increase in absorbance at 468 nm (ε468 = 43.5 mM-1 cm-1), which is approximately 5-fold higher than that of biliverdin. Modifications of this main spectrophotometric assay for assessment of HO-1 activity was carried out and HO-1 activity was determined by monitoring bilirubin formation using the difference in absorbance at 464 to 530 nm (ε464-530 = 40 mM-1 cm-1) (Maines, 1996; Maines and Kappas, 1974).The current protocol (Figure 1) utilises the same principle but are performed with certain minor modifications to make it much more convenient.
Figure 1. Schematic representation of HO-1 activity assay.
Materials and Reagents
Pipette tips (Tarsons, catalog numbers: 521020,521010,521000)
Tissue culture flasks, 50 ml (Falcon, catalog number: 353108)
Cell scraper (Falcon, catalog number: 353086)
Polypropelene conical tube, 50 ml (Falcon, catalog number: 352070)
Microcentrifuge tubes, 1.5ml (Falcon, catalog number: 500010)
0.2 µm syringe-driven filter unit (Millipore, catalog number: SLGP0033RS)
Quartz cuvette [10 mm, 1 ml volume] (Optiglass, catalog number: MCQ-254)
Tissue papers
26-gauge needle
10 ml syringe
Petri dish
RAW 264.7 cell (ATCC, catalog number: TIB-71)
Murine bone marrow derived macrophages (BMDM) (isolated from BALB/c mice) (for details, see Recipes)
Leishmania donovani (MHOM/IN/1983/AG83)
DMEM medium (Gibco, catalog number: 11885-084)
M199 medium (Gibco, catalog number: 12340-030)
FCS (Gibco, catalog number: 10082-147)
Antibiotic solution, 100x (Himedia, catalog number: A001A)
Protein assay dye reagent concentrate (Bio-Rad, catalog number: 5000006)
1 mM NAD(P)H (Santa Cruz, catalog number: sc-202725)
2 mM glucose-6-phosphate (Santa Cruz, catalog number: sc-210728)
1 U glucose-6-phosphate dehydrogenase (Sigma-Aldrich, catalog number: G-6378)
25 µM hemin (Sigma-Aldrich, catalog number: 51280)
Biliverdin hydrochloride (Sigma-Aldrich, catalog number: 30891)
NaCl (M.W.= 58.44 g/mol)
Na2HPO4 (M.W.= 141.96 g/mol)
KCl (M.W.= 74.55 g/mol)
KH2PO4 (M.W.= 136.08 g/mol)
K2HPO4 (M.W.= 174.18 g/mol)
MgCl2 (M.W.= 95.211 g/mol)
Sucrose (M.W. = 342.29 g/mol)
Sodium citrate (M.W. = 258.06 g/mol)
Glycerol
70% ethanol
Phosphate buffer saline/buffer (pH 7.4) (for details, see Recipes)
0.1 M potassium phosphate buffer (pH 7.4) containing 2 mM MgCl2 and complete protease inhibitor (see Recipes)
0.6 M sucrose solution (see Recipes)
0.1 M sodium citrate buffer (pH 5) containing 10% glycerol (see Recipes)
2 mg of rat liver cytosolic protein (see Recipes)
Equipment
250 ml bottle
-80 °C freezer
Sterile scissors and forceps
Variable volume pipettes (Tarsons, catalog numbers: 030050, 030040, 030020, 030000)
Incubator (Thermo Scientific)
Autoclave
Centrifuge (Thermo Scientific)
Ultracentrifuge (Thermo Scientific, model: WXUltra90)
Laminar air flow (NEO Equipments, model: LX80)
Vortex (Tarsons, catalog number: 3020)
pH meter (Sartorius, model: PB-11)
Magnetic stirrer (Tarsons, catalog number: 6030)
Spectrophotometer (Jasco, catalog number: V-630)
Procedure
-
Preparation of microsomal fraction containing HO-1 enzyme
Infect 80% confluent macrophage cells (RAW 264.7 or BMDM) in T25 flasks with Leishmania donovani promastigotes with a parasite to macrophage ratio (10:1) for the indicated time points (0, 0.25, 0.5, 1, 2 and 4 h).
After incubation,discard the media and carefully wash the cells with 1 ml PBS solution and the steps were carried out by placing all the flasks on ice throughout.
Add 1 ml potassium phosphate buffer to each of the flasks and gently scrape the cells using scrapers.
Transfer the scraped cells in a 50 ml Falcon pre-chilled by placing on ice.
Add 2 ml more potassium phosphate buffer.
Incubate the cells for 15 min on ice.
Briefly sonicate the cell suspensions at 80% amplitude for 15 s at 5 min interval for 3 times keeping on ice.
Add sucrose solution to the cell lysate to obtain the final concentration 0.25 M sucrose.
-
Centrifuge the solutions at 1,000 × g for 10 min at 4 °C.
Note: The pellet will contain the nuclei.
-
Centrifuge the supernatant at 12,000 × g for 15 min at 4 °C.
Note: The pellet will contain the mitochondria.
Ultracentrifuge the supernatant at 105,000 × g for 1 h at 4 °C.
-
Discard the supernatant.
Note:The pellet is the microsomal fraction containing HO-1 enzyme.
Resuspend the pellet in 500 µl of 0.1 M potassium phosphate buffer.
Store at -20 °C until use.
-
Estimation of protein concentration (Bradford, 1976)
Prepare 6 dilutions of bovine serum albumin (BSA) (10 mg/ml) as standard with a range of 0 to 50 µg protein in 995 µl diluted Bradford assay reagent (1:5) in duplicate and adding 5 µl from each of the test samples in 995 µl diluted Bradford assay reagent (1:5) in triplicate.
Mix all the samples by gently inverting 2 to 3 times.
Incubate all at room temperature for 5 min and read absorbance at 595 nm.
Note: Do not keep the prepared solutions for more than 1 h at room temperature.
-
Isolation of rat liver cytosolic fraction containing biliverdin reductase
Euthanize a Sprague Dawley rat.
Perfuse the liver with 0.9% NaCl solution in situ via hepatic portal vein until fully blanched.
Excise out the blanched liver and mince it completely with help of scissors.
Homogenize the liver sample in 5 ml of 0.1 M sodium citrate buffer using tissuelyzer (Bio-Rad)
Centrifuge the homogenate at 10,000 × g for 20 min at 4 °C.
Carefully pipette out the supernatant and ultracentrifuge the sup at 105,000 × g for 1 h at 4 °C.
The supernatant serves as the source of biliverdin reductase.
A portion of the obtained supernatant was tested for biliverdin reductase activity by assessing the amount of bilirubin produced from biliverdin via measuring the absorbance at 464 nm (bilirubin) and 670 nm (biliverdin). Five µg of protein from the obtained supernatant was incubated with 10 µM biliverdin (Sigma) and 400 µg/ml bovine serum albumin (BSA) in 100 mM potassium phosphate buffer (pH 7.4) at 37°C for 5 min in a total volume of 200 µl. The reaction was initiated by addition of 1 mM NADPH. The protein concentration of the supernatant was around 10 mg/ml.
-
Store at -80°C until use.
Note:Use the solution as early as possible as the enzymes lose their activity upon storage.
-
Assessment of heme oxygenase-1 activity
Incubate 600 µg of microsomal protein with a reaction mixture containing 1 mM NAD(P)H, 2 mM glucose-6-phosphate, 1 U glucose-6-phosphate dehydrogenase (Sigma-Aldrich), 25 µM hemin, 2 mg of rat liver cytosolic protein, and 100 mM potassium phosphate buffer (pH 7.4).
Adjust the final volume to 400 µl with potassium phosphate buffer (pH 7.4).
Place the tubes in the dark for 1 h at 37 °C.
After 1h, terminate the reaction by placing the tubes on ice for 2 min.
Determine HO-1 activity by measuring bilirubin concentration by spectrophotometer.
-
Spectrophotometric detection of bilirubin
Determine bilirubin concentration by the difference in absorption between 464 and 530 nm (extinction coefficient, 40 mM-1 cm-1 for BL).
Express HO-1 activity in picomoles of BL formed per milligram microsomal protein per hour (Figure 2).
Figure 2. HO-1 activity was measured in Leishmania donovani infected RAW 264.7 macrophages for the indicated time periods (0-4 h).
Data analysis
Calculate the difference between the two O.D.s.
A = A464 – A530
Incubation was performed for 1 h.
Therefore, ΔA = A h-1
Path length = 1 cm
Therefore, using Lambert Beers law [A= εcl];
[where, c = concentration, ε = extinction coefficient, 40 mM-1 cm-1 for bilirubin (BL), l = path length, 1 cm]
h-1/(mM-1cm-1 x cm)
h-1/mM-1
pmol µl-1 h-1[1 mM = 103 picomoles µl-1]
Total volume of reaction was 400 µl
Therefore, pmol h-1
pmol h-1
Amount of protein added was 600 µg.
Therefore, pmoles of Billirubin (BL) (mg protein)-1 h-1
pmoles of Billirubin (BL) (mg protein)-1 h-1
Recipes
-
Phosphate buffer saline/buffer (pH 7.4)
Add 8 g of NaCl (M.W. = 58.44 g/mol) in a sterilized 1 L bottle
Add 1.44 g of Na2HPO4 (M.W. = 141.96 g/mol)
Add 0.2 g of KCl (M.W. = 74.55 g/mol)
Add 0.24 g of KH2PO4 (M.W. = 136.08 g/mol)
Dissolve the reagents completely in about 800 ml sterilized distilled water (dH2O)
Adjust the pH to 7.4 using 1 M HCl or 1 M NaOH (as required)
Make up the final volume to 1 L with sterilized distilled water
Sterilize the solution by autoclaving at 15 lbs pressure (121 °C) for 15 min
Store the solution at 4 °C
-
0.1 M potassium phosphate buffer containing 2 mM MgCl2 and complete protease inhibitor (pH 7.4)
Add 2.61 g of KH2PO4 (M.W. = 136.08 g/mol) in a sterilized 250 ml bottle
Add 2.04 g of K2HPO4 (M.W. = 174.18 g/mol)
Add 28.56 mg of MgCl2 (M.W. = 95.211 g/mol)
Dissolve the reagents completely in about 120 ml sterilized distilled water (dH2O)
Adjust the pH to 7.4 using 1 M HCl or 1 M NaOH (as required)
Make up the final volume to 150 ml with sterilized distilled water
Sterilize the solution by autoclaving as mentioned above
Store the solution at 4 °C
On the day of the experiment aliquot of 25 ml of the solution and add complete protease inhibitor just prior to use.
-
0.6 M sucrose solution
Add 20.53 g of sucrose (M.W. = 342.29 g/mol) to 80 ml sterilized dH2O in a sterilized 250 ml bottle
Dissolve the reagent completely
Make up the final volume to 100 ml and sterilize by autoclaving as mentioned above
Store at 4 °C
-
0.1 M sodium citrate buffer
Add 2.58 g of sodium citrate (M.W. = 258.06 g/mol) to 80 ml sterilized dH2O in a sterilized 250 ml bottle
Dissolve the reagent completely
Adjust the pH to 5.0 using 1 M HCl or 1 M NaOH (as required)
Add 10 ml of glycerol solution
Make up the volume to 100 ml and sterilize by autoclaving as mentioned above
Store at 4 °C
-
Rat liver cytosol
Euthanize a Sprague Dawley rat
Perfuse the liver with 0.9% NaCl solution in situ via hepatic portal vein until fully blanched (Wen et al., 2012)
Excise out the blanched liver and mince it completely with help of scissors
Homogenize in 5 ml of 0.1 M sodium citrate buffer pH 5 containing 10% glycerol
Centrifuge the obtained homogenate at 10,000 × g for 20 min
Collect the resulting supernatant carefully and ultracentrifuge at 105,000 × g for 1 h
Collect the obtained supernatant carefully and store in aliquots at -80 °C for use
Note:Use the prepared as early as possible as enzymes gradually loose their activity upon storage.
-
Bone marrow derived macrophages (BMDM)
Euthanize the mouse
Lay the mouse in a supine position and affix it by pinning the four legs through the mouse paw pads below the ankle joint
Thoroughly spray the mouse abdomen and hind legs with 70% ethanol
Make an incision at the top of each hind leg and pull the skin outwards exposing the muscle using sterile scissors and forceps
Use scissors to remove maximum muscles and connective tissues from the bones. Carefully isolate the entire femurs and tibia without cutting the bone ends or breaking the bones
Clean the bones from any attached tissues with help of tissue papers
Transfer the bones with help of forceps in a Petri dish containing 70% ethanol (EtOH) for one minute followed by air dry for about 5 min inside cell culture hood
-
Cut both ends of the isolated bones carefully so that the bones do not get shattered
Notes:
Hold the femur or tibia with help of forceps. Place the scissors just above the joint to prevent the bone from shattering. Turn the bone and repeat the procedure.
Do not let the cut ends of the bones touch anything as this leads to contamination of the marrow.
Flush out the bone marrow into a 50 ml Falcon tube by inserting a 26-gauge needle attached to a 10 ml syringe filled with 1x PBS (containing Antibiotic solution) at the knee side of both types of bone. Pass the PBS through the bone until the colour of the bone turns white from reddish, indicating that most of the marrow has been expelled
-
Discard the bone into an empty Petri dish
Notes:
Perform these steps as early as possible for each bone IMMEDIATELY following the cutting of the bone.
DO NOT put a bone down between cutting and flushing.
Centrifuge the collected cell suspension at 1,500 rpm for 10 min at 4 °C
Wash the cell pellet again with 1x PBS
Resuspend the cells in RPMI 1640, supplemented with 100 U/ml penicillin and 100 µg/ml streptomycin, 10% FCS, and 25 ng/ml GM-CSF and were incubated at 5% CO2, 95% humidity, at 37 °C
On the 4th day, the media will be discarded and add new media containing 100 U/ml penicillin and 100 µg/ml streptomycin, 10% FCS, and 25 ng/ml GM-CSF
On Day 6, BMDM cells will be obtained for usage
Acknowledgments
This work was supported by Department of Biotechnology (BT/HRD/NBA/38/03/2018), Indo Israel Grant from the University Grants Commission [F. 6-10/2016(IC)], Department of Science and Technology (SB/SO/BB-0055/2013), Department of Biotechnology, West Bengal [221/BT(Estt)/RD-40/2014], and the University with Potential for Excellence II (Grant UGC/148/UPE/ST1). M.B. and S.S. received their fellowships from the University Grants Commission (New Delhi).The research was supported by Council of Scientific and Industrial Research, New Delhi, India.
Competing interests
The authors have no competing interests.
Ethics
Animal maintenance and the experiments were performed in accordance with the guidelines provided by the Committee for the Purpose of Control and Supervision of Experiments on Animals (New Delhi, India). The protocol was approved by the Institutional Animal Ethics Committee (IAEC) of Bose Institute (Kolkata, India) (IAEC approval no. IAEC/BI/82/2017).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Bradford M. M.(1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254. [DOI] [PubMed] [Google Scholar]
- 2.Kathirvel E., Chen P., Morgan K., French S. W. and Morgan T. R.(2010). Oxidative stress and regulation of anti-oxidant enzymes in cytochrome P4502E1 transgenic mouse model of non-alcoholic fatty liver. J Gastroenterol Hepatol 25(6): 1136-1143. [DOI] [PubMed] [Google Scholar]
- 3.Kutty R. K. and Maines M. D.(1981). Purification and characterization of biliverdin reductase from rat liver. J Biol Chem 256(8): 3956-3962. [PubMed] [Google Scholar]
- 4.Lincoln B. C., Mayer A. and Bonkovsky H. L.(1988). Microassay of heme oxygenase by high-performance liquid chromatography: application to assay of needle biopsies of human liver. Anal Biochem 170(2): 485-490. [DOI] [PubMed] [Google Scholar]
- 5.Maines M.(1996). Carbon monoxide and nitric oxide homology: differential modulation of heme oxygenases in brain and detection of protein and activity. Methods Enzymol 268: 473-488. [DOI] [PubMed] [Google Scholar]
- 6.Maines M. D. and Kappas A.(1974). Cobalt induction of hepatic heme oxygenase; with evidence that cytochrome P-450 is not essential for this enzyme activity. Proc Natl Acad Sci U S A 71(11): 4293-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Missall T. A., Lodge J. K. and McEwen J. E.(2004). Mechanisms of resistance to oxidative and nitrosative stress: implications for fungal survival in mammalian hosts. Eukaryot Cell 3(4): 835-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paiva C. N. and Bozza M. T.(2014). Are reactive oxygen species always detrimental to pathogens? Antioxid Redox Signal 20(6): 1000-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryter S., Kvam E. and Tyrrell R. M.(1999). Heme oxygenase activity determination by high-performance liquid chromatography. Methods Enzymol 300: 322-336. [DOI] [PubMed] [Google Scholar]
- 10.Saha S., Basu M., Guin S., Gupta P., Mitterstiller A. M., Weiss G., Jana K. and Ukil A.(2019). Leishmania donovani Exploits Macrophage Heme Oxygenase-1 To Neutralize Oxidative Burst and TLR Signaling-Dependent Host Defense. J Immunol 202(3): 827-840. [DOI] [PubMed] [Google Scholar]
- 11.Schacter B. A.(1978). Assay of microsomal heme oxygenase in liver and spleen. Methods Enzymol 52: 367-372. [DOI] [PubMed] [Google Scholar]
- 12.Sierra E. E. and Nutter L. M.(1992). A microassay for heme oxygenase activity using thin-layer chromatography. Anal Biochem 200(1): 27-30. [DOI] [PubMed] [Google Scholar]
- 13.Tenhunen R., Marver H. S. and Schmid R.(1969). Microsomal heme oxygenase. Characterization of the enzyme. J Biol Chem 244(23): 6388-6394. [PubMed] [Google Scholar]
- 14.Vreman H. J. and Stevenson D. K.(1988). Heme oxygenase activity as measured by carbon monoxide production. Anal Biochem 168(1): 31-38. [DOI] [PubMed] [Google Scholar]
- 15.Wen J. W., Olsen A. L., Perepelyuk M. and Wells R. G.(2012). Isolation of rat portal fibroblasts by in situ liver perfusion. J Vis Exp(64). pii: 3669. doi: 10.3791/3669. [DOI] [PMC free article] [PubMed] [Google Scholar]