Abstract
Differential exposure of tumor cells to microenvironmental cues greatly impacts cell phenotypes, raising a need for position based sorting of tumor cells amenable to multiple OMICs and functional analyses. One such key determinant of tumor heterogeneity in solid tumors is its vasculature. Proximity to blood vessels (BVs) profoundly affects tumor cell phenotypes due to differential availability of oxygen, gradient exposure to blood-borne substances and inputs by angiocrine factors. To unravel the whole spectrum of genes, pathways and phenotypes impacted by BVs and to determine spatial domains of vascular influences, we developed a methodology for sorting tumor cells according to their relative distance from BVs. The procedure exemplified here using glioblastoma (GBM) model is based on differential uptake of intra-venously injected, freely-diffusing fluorescent dye that allows separation of stroma-free tumor cells residing in different, successive microenvironments amenable for subsequent OMICs and functional analyses. This reliable, easy to use, cost effective strategy can be extended to all solid tumors to study the impact of vasculature or the lack of it.
Keywords: Blood vessels, Tumor angiogenesis, Spatial transcriptomics, Tumor heterogeneity, Hypoxia
Background
Tumor dependence on adequate blood supply has provided a rationale for anti-angiogenic cancer therapy (Carmeliet and Jain, 2000; Vasudev et al., 2013 ). Traditionally attributed to provision of oxygen and other blood-borne substances, the roles of the tumor vasculature in tumor biology were recently extended to include perfusion-independent, paracrine inputs exerted by secreted endothelial-produced angiocrine factors ( Butler et al., 2010 ; Beck et al., 2011 ; Lu et al., 2013 ), as well as via direct tumor-BV contacts mediated by reciprocal Notch signaling ( Cao et al., 2014 ; Wieland et al., 2017 ). Because tumor neovascularization often fails to fully match increasing perfusion demands imposed by continual tumor growth, the relative distance of a tumor cell from the nearest perfused vessel is likely to greatly impact the nature and magnitude of vascular inputs operating through all indicated modes of tumor-BV communication. Correspondingly, differential exposures to vascular signals might facilitate emergence of certain tumor phenotypes and generate intra-tumor cell heterogeneity. To determine how vascular cells may impact tumor cells, tumor-endothelial cell (EC) co-cultures in both 2D and 3D configurations have been used ( Khodarev et al., 2003 ; Timmins et al., 2004 ). However, because ex-vivo systems do not reveal perfusion-dependent effects and are also unable to recapitulate the complex interactions prevailing in the tumor microenvironment, in vivo approaches are clearly needed. The latter has thus far been limited to using EC-specific manipulations of selected candidate genes ( Beck et al., 2011 ; Cao et al., 2014 and 2017). The need still existed for an integrative, high-throughput approach for unraveling the whole spectrum of genes, pathways and phenotypes affected by the vasculature and, moreover, a general approach to determine spatial ranges of vascular influences per each of these phenotypes.
The methodology described herein allows retrospective sorting of successive GBM tumor cells subpopulations residing at progressively increasing distances from the nearest BV and in a form suitable for subsequent transcriptomic and functional analyses. This methodology was used to understand how vasculature impacts cancer cell transcriptome and metabolome ( Kumar et al., 2019 ). This methodology is not limited to the above two omics, but can be further extended to any other omics such as to study epigenome or methylome. If integrated with single cell RNA sequencing, this assay can help in dissecting the characteristics of tumor cells as a function of blood vessels to a very fine resolution. Of particular relevance is the existence of diverse metabolic niches within a tumor due to differential availability of oxygen and other blood-borne substances. Using this methodology we showed that the classical 'Warburg Effect', i.e., the use of aerobic glycolysis for energy production instead of the more efficient mitochondrial respiration, in fact, reflects overall summation of different metabolic niches, and that perivascular cells still actively rely on mitochondrial respiration. While there is a growing awareness of intratumor metabolic compartmentalization, and even of an interplay between different compartments, it has thus far been dealt with in a dichotomous manner of distinguishing 'normoxic' vs. 'hypoxic' microenvironments ( Allen et al., 2016 ). Considering, however, existence of intermediate states shaped by the perfusion gradient and by differential exposure to angiocrine factors, our study showed that there is a continuum of diverse microenvironments. Ability to isolate such tumor subpopulations should aid in addressing open issues pertinent to the highly complex BV-tumor interplay. We believe this methodology can also be extended to other solid tumors too, as there exist an unveven distribution of vascular supply in most of the different types of solid tumors.
Materials and Reagents
T75 flasks
70 µm filter
Suturing thread and needle (Ethicon, catalog number: SXMD1B105)
Alcohol pads (Medi Plus, catalog number: KPN1528)
Insulin syringe, 31 G, 0.5 ml (Insumed, Pic solution)
GentleMACS C Tube (GentleMACSTM, catalog number: 130-093-237)
Mice (NOD/SCID, 8-10 weeks old, Male mice used in this particular study)
Human origin glioblastoma cell line–U87MG (ATCC, catalog number: HTB-14TM)
pLenti CMV GFP Puro (Addgene plasmid, catalog number: 17448)
Dulbecco’s Minimal Essential Medium (DMEM) High Glucose (Biological Industries, catalog number: 01-055-1)
Sodium Pyruvate (Biological Industries, catalog number: 03-042-B)
L-Glutamine (Biological Industries, catalog number: 03-020-1B)
Fetal Bovine Serum (Biological Industries, catalog number: 04-001-1A)
Penicillin-Streptomycin solution (Biological Industries, catalog number: 03-031-1B)
Trypsin EDTA Solution (Biological Industries, catalog number: 03-050-1A)
Phosphate buffered saline (PBS) (Biological Industries, catalog number: 02-020-1A)
2% Chlorhexidine solution (Ecolab, catalog number: 3059670)
Ketamine (Clorketam, Vetoquinol)
Rimadyl (Carprofen, catalog number: 141-053)
Eye-ointment (Pharmaderm, catalog number: 17033-211-38)
Xylazine (Sedaxylan, Eurovet animal health).
Ketamine-Xylaxine mixture, 140 mg/kg of Ketamine mixed with 10 mg/kg body wt of mice
Saline (Braun, catalog number: Ecolav PE Bottle 30 ml)
Hoechst 33342 (Sigma-Aldrich, catalog number: B2261)
Rat anti-mouse CD31 Antibody (BD PharMingen, catalog number: 555027)
Donkey anti-rat Cy5 (Jackson ImmunoResearch Laboratories Inc., catalog number: 712-605-153)
Trypan blue (Sigma-Aldrich, catalog number: T8154-100ML)
Isoflurane (TerrellTM, Piramal)
Tumor dissociation kit (Miltenyi Biotec, catalog number: 130-096-730)
RNeasy Mini Kit (Qiagen, catalog number: 74104)
FACS buffer (see Recipes)
Vascular perfusion dye (see Recipes)
Enzyme mix (see Recipes)
Equipment
Sterotactic machine (Stoelting, catalog number: 51500D)
High speed drill with drill bits (Stoelting, catalog number: 51555)
Surgical instrument set (World Precision Instruments, catalog number: 503407)
Sterotactic syringe (Hamilton, catalog number: CAL80301)
Tissue Dissociator (GentleMACSTM, catalog number: 130-093-235)
Fluorescent microscope (Nikon, model: SMZ25)
Procedure
-
Tumor cell preparation
Transduce U87MG cells with lentiviral particles encoding Enhanced Green Fluorescent Protein (eGFP) (Titer: 107 viral particles/ml) ( Tiscornia et al., 2006 ; Huang, 2011). Post 72 h of successful transduction, pure population can be obtained by FACS sorting.
Grow and expand U87MG-GFP cells in T75 flasks in DMEM supplemented with 10% fetal bovine serum (FBS) and 100 units/ml of penicillin/streptomycin (PEN-STREP) in a humidified chamber for 18-20 h at 37 °C and 5% CO2.
Trypsinize U87MG-GFP cells and centrifuge at 300 × g for 5 min, aspirate the media and resuspend the cells in 1x PBS. Check the viability and proceed only if the viability is > 95%. Prepare a sample containing 105 cells per µl.
-
Tumor implantation
Note: All procedures described below should be reviewed and approved by the appropriate Institutional Animal Use and Care Committee.
-
The surgical area must be disinfected using 2% chlorhexidine solution. The surgical room should contain the following items:
Sterotactic Machine with a heating pad.
High speed drill with drill bits.
Sterile tools containing scissors, scalpel, forceps, suturing needle with thread.
Anesthesia, Pain-reliever, Eye-ointment and Saline.
70% Ethanol swabs, Cotton buds and Gauze.
Anaesthetize the mice (8-10 weeks old) using ketamine/xylazine mixture (140 mg/kg and 10 mg/kg body wt respectively). Place the mice post anesthetization (confirm by toe pinch) in the stereotactic machine on top of the heating pad and hook the mouse’s incisor teeth in the bite bar. Sterilize the dorsal area above parieto-occipital bone till the frontal bone of the mouse. Make a vertical long incision in the mid scalp using a scalpel and confirm the visualization of bregma. Ensure ophthalmic ointment is applied to avoid dryness of eyes during the procedure.
Drill a hole in the cerebral hemispheres at the following coordinates from Bregma: 2 mm anterior, 1.5 mm lateral and 3 mm deep to reach striatum of the brain (Figure 1).
Load around 8 µl of U87MG-GFP cells into a 10 µl syringe and using an injector pump, inject 2 µl at a rate of 0.5 µl/min on either lobes.
Post inoculation, leave the needle undisturbed for 5 min and then withdraw slowly over a period of 5 min to allow the cells to settle at the point of injection.
Close the burr hole in the skull with sterile bone wax and suture the skin atop. Inject analgesic Rimadyl (5 mg/kg) intradermally daily for three consecutive days.
Monitor the mice post-operatively until they retain normal activity.
-
-
Dye labeling and tumor harvesting
3 weeks post inoculation, tumor implanted mice should be injected with the vascular perfusion dye (Hoechst 33342, 5 mg/kg body wt) through the tail vein (intra venous).
To perform intra venous injection, cage of tumor bearing mice should be warmed using infra-red lamp for 5 min. Take one mouse and weigh it.
Prepare the Hoechst 33342 dye (5 mg/kg body wt of mice) in 1x PBS and load 100 µl into a 0.5 ml 31 G needle attached insulin syringe. Ensure that there are no air bubbles in the syringe containing the solution. A volume of 100-200 µl can be injected per mice.
Place the mice in the restrainer and swab the mice tail with an alcohol gauze. Warm the mice tail by holding it close to infra-red lamp for 10 s. Observe that the tail vein of the mice have enlarged (vasodilation).
Locate one of the two lateral veins and inject into the tail vein with the bevel of the needle facing upward and the needle almost parallel to the vein to ensure success (Figure 2).
If the needle is in the vein, there will be no resistance while injecting and the vein will blanch once injected. Inject the dye, retract the needle and ensure the bleeding has stopped by applying gentle pressure with sterile cotton gauze at the site of injection.
After a short incubation of 5-10 min depending on the extent of labeling required, anesthetize the mice with isoflurane and euthanize the mice by cervical dislocation. Quickly decapitate the head and isolate the brain by carefully opening the skull and place it in ice cold 1x PBS.
At this stage, the vascular perfusion dye would have diffused and stained the cells that are proximal to the tumor blood vessels. See Figure 3 for the illustration of diffusion of the perfusion dye in a U87MG-GFP xenograft labeling cells proximal to functional blood vessels post 5 min of dye labeling.
Under a fluorescent dissection microscope, separate the tumor region (fluorescently labelled) and proceed for tumor dissociation.
-
Tumor dissociation
Tumor dissociation must be performed using Tumor Dissociation Kit from MACS Miltenyi Biotec. Place the tumor tissue in ice cold DMEM and cut it into small pieces of 2-4 mm. Transfer the tumor tissue pieces into the gentleMACSC Tube containing the enzyme mix (Recipe 3) and mount it into a gentleMACSTM Dissociator.
Start the dissociation by selecting gentleMACS program h_tumor_01. After termination incubate the tumor sample for 30 min at 37 °C under continuous rotation using MACSmixTM tube rotator.
Once finished, run the gentleMACS program h_tumor_01 again. Transfer the contents through a 70 µm filter into a 50 ml tube and make up the volume to 30 ml with ice cold DMEM solution to stop the enzymatic reaction.
Centrifuge cell suspension at 300 × g for 7 min. Aspirate supernatant and resuspend the pellet in 1 ml of FACS Buffer. Proceed quickly for FACS sorting.
-
FACS sorting
Set up a sorting strategy to first exclude dead cells, debris and doublets from the mixture (see Figure 4 for gating strategy details). Gating must be done to exclude all the stromal elements and blood cells (by gating for GFP+ cells as the cancer cells are fluorescently labelled). Cancer cells may further be divided into different sub-fractions based on their Hoechst dye uptake which is proportional to their proximity to the BVs.
Collect the fractions into tubes containing DMEM, centrifuge at 300 × g for 5 min to collect the cells.
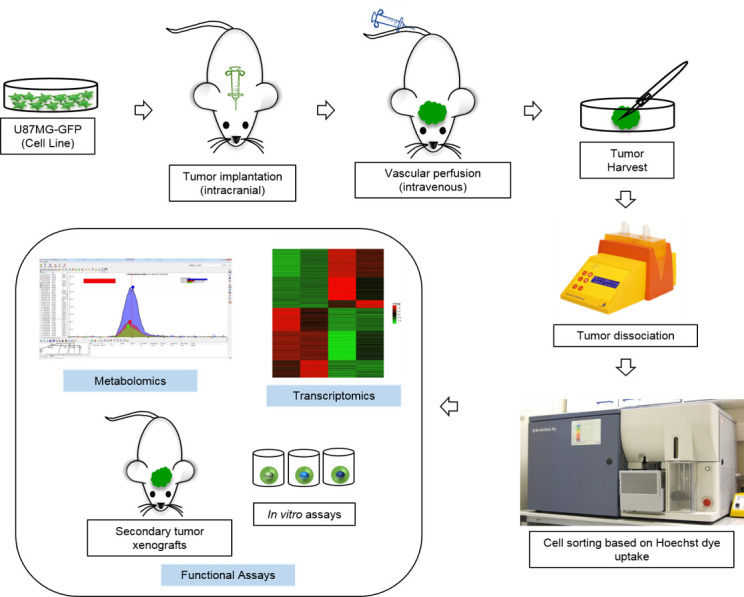
The cells are now ready for downstream assays. This includes preparation of RNA for transcriptomics, metabolites for metabolomics etc. In addition, the cells can also be used for in vitro or in vivo functional assays (Figure 5).
-
Sample preparation
For transcriptomics: At least 50,000 cells is a pre-requisite for good quality RNA. Total RNA extraction is done using RNeasy mini kit. Quality of the RNA can be analyzed by TapeStation before proceeding to generate RNA Sequencing library.
For metabolomics: Cells must be quickly quenched in a suitable solvent [e.g., ice cold solution of Methanol:Acetonitrile:Water (5:3:2)] centrifuged at 20,000 × g for 20 min and protein free supernatant extract was used for chromatographic separation.
For in vitro assays, the cells have to be cultured in DMEM + 10% FBS media. Please ensure that all the procedures prior to this are done in sterile conditions if the goal is to use the cells for in vitro assays.
For secondary xenograft, count the viable number of cells post sorting and inject 106 cells subcutaneously or 105 cells intracranially into NOD/SCID mice.
Figure 1. Intracranial injection schematic.
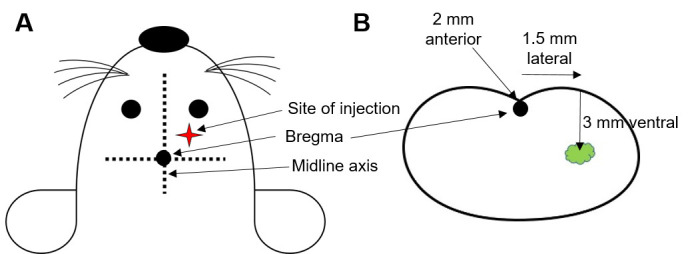
A. Site of injection is determined by using bregma as the point of reference, which lies on the middle axis between the eyes and ears at the intersection of coronal and sagittal sutures. B. Co-ordinates for the site of injection from bregma as shown through a coronal section of mouse brain.
Figure 2. Tail Vein Injection in mice.
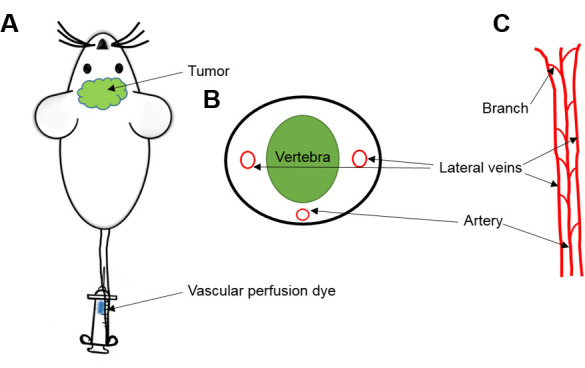
A. Injection of vascular perfusion dye (Hoechst 33342) through the tail vein of mouse. B. Cross section. C. Anatomy of mice tail circulation to help and identify one of the two lateral veins through which dye must be infused.
Figure 3. Confocal image of tumor section from U87MG-GFP tagged tumor xenograft retrieved post 5 min of perfusion dye (Hoechst 33342) injection and stained for BVs (CD31).
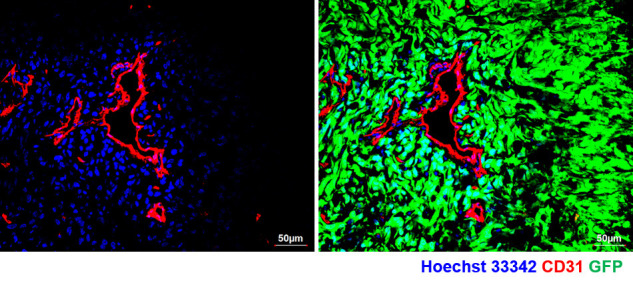
Note the spread of perfusion dye adjacent to BVs (left panel) and absent in non-perfused region of the tumor tissue. Right panel shows the presence of tumor in the whole field (GFP+ cells).
Figure 4. Gating strategy for FACS sorting of dissociated tumor tissue.
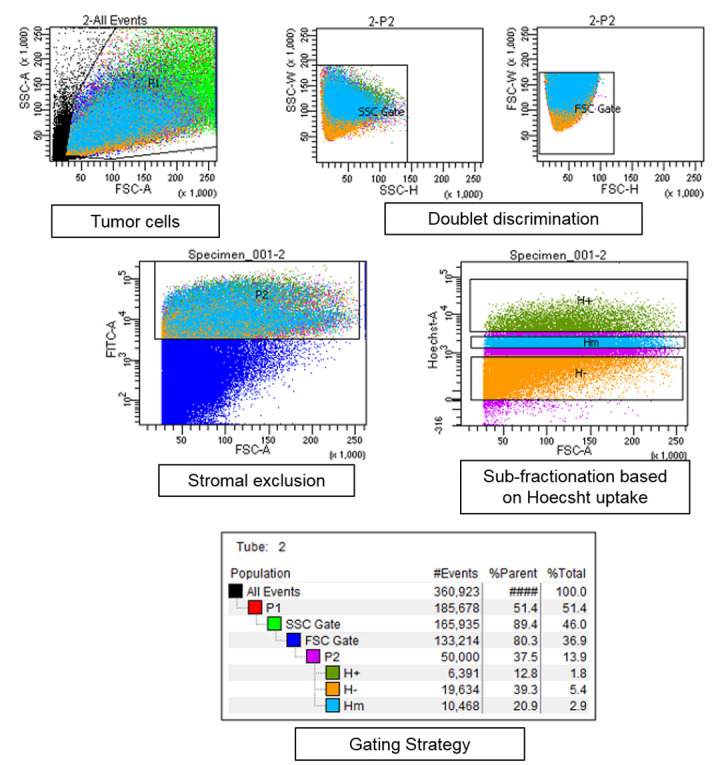
Tumor cells were gated to exclude dead cells and debris (marked black in first panel). Doublet exclusion is ensured by gating strategy–plotting the height and width of side scatter followed by forward scatter for tumor cell population. Fractionation is done on pure cancer cell population according to the Hoechst dye uptake. As a representative example, 3 cancer subpopulations are shown labelled as H+ the subpopulation with highest Hoechst staining and is closest to the BVs, followed by Hm–Hoechst medium and H–Hoechst negative population.
Figure 5. Schematic outline of the protocol and its possible applications.
Recipes
-
FACS buffer
2% FBS in 1x PBS and 1 mM EDTA
-
Vascular perfusion dye
Hoechst 33342 5 mg/ml in 1x PBS
-
Enzyme mix
100 µl of Enzyme H, 50 µl of Enzyme R and 12.5 µl of Enzyme A in 2.2 ml of DMEM
Incubate the mix in water bath at 37 °C 10 min prior to use
Acknowledgments
We thank members of the Eli Keshet’s laboratory for helpful suggestions during the standardization of this protocol. This work was supported by a DKFZ-MOST research cooperation grant (CA-178), a European Research Council Advance ERC Grant (322692-VASNICHE) and the Israel Cancer Research Fund Professorship Award to Eli Keshet. This protocol was adapted from previously published work ( Kumar et al., 2019 ).
Competing interests
The authors declare no competing financial interests.
Ethics
All experimental procedures on animals were approved by the authority for biological and biomedical models of the Hebrew University of Jerusalem and comply with IACUC guidelines.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Allen E., Mieville P., Warren C. M., Saghafinia S., Li L., Peng M. W. and Hanahan D.(2016). Metabolic symbiosis enables adaptive resistance to anti-angiogenic therapy that is dependent on mtor signaling. Cell Rep 15(6): 1144-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beck B., Driessens G., Goossens S., Youssef K. K., Kuchnio A., Caauwe A., Sotiropoulou P. A., Loges S., Lapouge G., Candi A., Mascre G., Drogat B., Dekoninck S., Haigh J. J., Carmeliet P. and Blanpain C.(2011). A vascular niche and a VEGF-Nrp1 loop regulate the initiation and stemness of skin tumours. Nature 478(7369): 399-403. [DOI] [PubMed] [Google Scholar]
- 3. Butler J. M., Kobayashi H. and Rafii S.(2010). Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat Rev Cancer 10(2): 138-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cao Z., Ding B. S., Guo P., Lee S. B., Butler J. M., Casey S. C., Simons M., Tam W., Felsher D. W., Shido K., Rafii A., Scandura J. M. and Rafii S.(2014). Angiocrine factors deployed by tumor vascular niche induce B cell lymphoma invasiveness and chemoresistance. Cancer Cell 25(3): 350-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao Z., Scandura J. M., Inghirami G. G., Shido K., Ding B. S. and Rafii S.(2017). Molecular checkpoint decisions made by subverted vascular niche transform indolent tumor cells into chemoresistant cancer stem cells. Cancer Cell 31(1): 110-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carmeliet P. and Jain R. K.(2000). Angiogenesis in cancer and other diseases. Nature 407(6801): 249-257. [DOI] [PubMed] [Google Scholar]
- 7. Huang Y.(2011). Lentivirus and Retrovirus Transfection. Bio-101 e38. [Google Scholar]
- 8. Khodarev N. N., Yu J., Labay E., Darga T., Brown C. K., Mauceri H. J., Yassari R., Gupta N. and Weichselbaum R. R.(2003). Tumour-endothelium interactions in co-culture: coordinated changes of gene expression profiles and phenotypic properties of endothelial cells. J Cell Sci 6): 1013-1022. [DOI] [PubMed] [Google Scholar]
- 9. Kumar S., Sharife H., Kreisel T., Mogilevsky M., Bar-Lev L., Grunewald M., Aizenshtein E., Karni R., Paldor I., Shlomi T. and Keshet E.(2019). Intra-tumoral metabolic zonation and resultant phenotypic diversification are dictated by blood vessel proximity. Cell Metab 30(1): 201-211 e206. [DOI] [PubMed] [Google Scholar]
- 10. Lu J., Ye X., Fan F., Xia L., Bhattacharya R., Bellister S., Tozzi F., Sceusi E., Zhou Y., Tachibana I., Maru D. M., Hawke D. H., Rak J., Mani S. A., Zweidler-McKay P. and Ellis L. M.(2013). Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell 23(2): 171-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Timmins N. E., Dietmair S. and Nielsen L. K.(2004). Hanging-drop multicellular spheroids as a model of tumour angiogenesis. Angiogenesis 7(2): 97-103. [DOI] [PubMed] [Google Scholar]
- 12. Tiscornia G., Singer O. and Verma I. M.(2006). Production and purification of lentiviral vectors. Nat Protoc 1(1): 241-245. [DOI] [PubMed] [Google Scholar]
- 13. Vasudev N. S., Goh V., Juttla J. K., Thompson V. L., Larkin J. M., Gore M., Nathan P. D. and Reynolds A. R.(2013). Changes in tumour vessel density upon treatment with anti-angiogenic agents: relationship with response and resistance to therapy. Br J Cancer 109(5): 1230-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wieland E., Rodriguez-Vita J., Liebler S. S., Mogler C., Moll I., Herberich S. E., Espinet E., Herpel E., Menuchin A., Chang-Claude J., Hoffmeister M., Gebhardt C., Brenner H., Trumpp A., Siebel C. W., Hecker M., Utikal J., Sprinzak D. and Fischer A.(2017). Endothelial notch1 activity facilitates metastasis. Cancer Cell 31(3): 355-367. [DOI] [PubMed] [Google Scholar]


