Abstract
Gut CD4 T cells are major targets of HIV-1 and are massively depleted early during infection. To better understand the mechanisms governing HIV-1-mediated CD4 T cell death, we developed the physiologically-relevant Lamina Propria Aggregate Culture (LPAC) model. The LPAC model is ideal for studying CD4 T cell death induced by clinically-relevant Transmitted/Founder (TF) HIV-1 strains and is also suitable for studying how enteric microbes and soluble factors (e.g., Type I Interferons) impact LP CD4 T cell death and function. Here, we detail the protocol to establish LP CD4 T cell infection using a process of spinoculation, the subsequent evaluation of infection levels using multicolor flow cytometry and the determination of overall LP CD4 T cell death using absolute LP CD4 T cell counts. We also describe the preparation of virus stocks of Transmitted/Founder (TF) HIV-1 infectious molecular clones that were successfully used in the LPAC model.
Keywords: Lamina Propria Aggregate Culture, Lamina Propria Mononuclear cells, HIV-1, Gut, T cells, Infection, Death
Background
CD4 T cell depletion is the hallmark pathogenic outcome of HIV-1 infection. However, the magnitude and kinetics of CD4 T cell depletion depends on the tissue compartment evaluated. The gastrointestinal tract is a major site of early HIV-1 replication and CD4 T cell death. At these early stages, the circulating HIV-1 strains primarily utilize CCR5 instead of CXCR4 as HIV co-receptors along with CD4. In fact, nearly all transmitted/founder (TF) HIV-1 strains utilize CCR5 ( Keele et al., 2008 ; Salazar-Gonzalez et al., 2009). Unfortunately, CD4 T cells from peripheral blood mononuclear cells (PBMCs) and tonsil histocultures (the Human Lymphoid Aggregate Culture or HLAC model) are generally resistant to cell death following infection by CCR5-tropic HIV-1, likely due to the lack of CCR5 expression. Moreover, blood and tonsil CD4 T cells are mostly of the naïve and central memory subsets, which are less prevalent in the gastrointestinal tract. By contrast, gut CD4+ T cells are mainly of the effector memory subset and express high levels of CCR5. Gut CD4+ T cells also have higher representation of Th17 and Th22 subsets, which protect the integrity of the intestinal barrier, than that of blood or tonsil CD4+ T cells. Thus, peripheral blood and the HLAC model have limited utility in understanding TF HIV-1-mediated CD4+ T cell depletion in the gut. This prompted our group to adapt principles from the HLAC model ( Doitsh et al., 2010 ; Doitsh et al., 2014 ) towards lamina propria mononuclear cells (LPMCs) ( Steele et al., 2014 ).
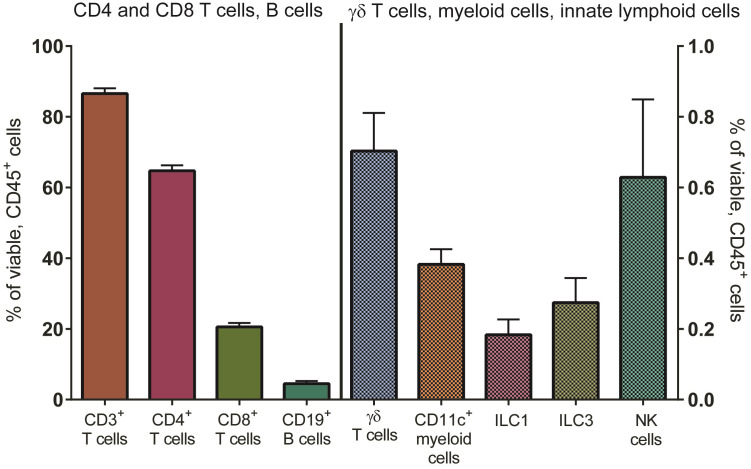
The protocol for the isolation of LP mononuclear cells (LPMC) from human small intestinal tissue in the LPAC model is detailed in previous publications, and described in more detail in Procedure A ( Howe et al., 2009 ; Dillon et al., 2010 , 2012 and 2016; Steele et al., 2014 ; Harper et al., 2015 ; Yoder et al., 2017 ). In brief, LPMC are isolated from surgically resected small intestinal samples by collagenase-digestion after the removal of muscle, fat, mucus and epithelial layers. LPMC are cryopreserved at 5 x 106 to 10 x 106 LPMC per cryovial and have excellent overall viability on thawing (mean ± SEM: 87 ± 0.8% of total leucocytes (CD45+); N = 69, see Procedure B). Jejunum LPMCs consist mostly of CD3 T cells. On average, CD4 T cells constitute 65 ± 1.5% of viable LP leucocytes with smaller percentages of CD8 T cells, B cells and innate immune cells present (Figure 1). On average, 74 ± 3% (N = 30) of LP CD4 T cells express CCR5 (data not shown).
Figure 1. Distribution of immune cell populations in LPMC isolated from human jejunum tissue.
Multi-color flow cytometry was used to determine frequencies of total CD3+ cells (n = 77), CD4+ cells (n = 75), CD8+ cells (n = 77), CD19+ B cells (n = 34) (left axis) and various innate immune cells including γδT cells (n = 45), CD11c+ myeloid cells (n = 50) and subsets of innate lymphoid cells (ILCs, n = 11) (right axis) within viable, leucocytes (CD45+).
Materials and Reagents
1.5 ml Eppendorf tube graduated (Fisher, catalog number: 05-408-137)
15 ml conical tubes (Thermo Fisher, catalog number: 339651)
50 ml conical tubes (Thermo Fisher Scientific, catalog number: 339652)
60 ml disposable syringe
Vinyl Specimen Molds (Tissue Tek Cryomold, catalog number: 4566)
Round-bottom Polystyrene tubes (“FACS tubes”) (Falcon, catalog number: 352052)
Syringe-driven filter (0.45 μm) (Millipore, catalog number: SLHP033RS)
Petri dish 100 mm x 20 mm (Fisher, catalog number: FB0875711Z)
Polycarbonate thick wall ultracentrifugation tube (Beckman Coulter, catalog number: 355631)
Cell strainer (70 μm) (Fisher, catalog number: 22363548)
Mega Cassette System Tissue Tek (Sakura, catalog number: 4173)
96 V-bottom culture plates (Corning, catalog number: 3894)
96-well white tissue culture-treated sterile microplate (PerkinElmer, catalog number: 6005680)
T175 culture flasks (Celltreat, catalog number: 229351)
TZM-bl cells (AIDS Reagent Program, catalog number: 8129)
-
TF infectious molecular clones:
CH040 (AIDS Reagent Program, catalog number: 11740)
CH058 (AIDS Reagent Program, catalog number: 11856)
CH077 (AIDS Reagent Program, catalog number: 11742)
CH470 (AIDS Reagent Program, catalog number: 12422)
APC anti-human CD8 (Clone RPA-T8) (Tonbo Biosciences, catalog number: 20-0088)
Bovine Serum Albumin (BSA) (Fisher Scientific, catalog number: BP1600-100)
293T cells (ATCC, catalog number: CRL-3216)
RD-1 HIV-1 core antigen (Clone KC57) (Beckman Coulter, catalog number: 6604667)
0.25% Trypsin (Corning, catalog number: MT25053Cl)
Collagenase D (Roche Diagnostics, catalog number 11088882001)
DNase I (Sigma-Aldrich, catalog number: DN25-1G)
Fetal Bovine Serum (FBS) (Life Technologies, catalog number: 26140079)
HIV p24 antigen ELISA (Perkin Elmer, catalog number: NEK05001KT)
Human AB Serum (Gemini Bioproducts, catalog number: 100-512)
One ShotTM Stbl3 competent cells (ThermoFisher, catalog number: C737303)
PE/Dazzle 594 anti-human CD3 (Clone UCHT1) (Biolegend, catalog number: 300450)
2 M CaCl2 (Sigma-Aldrich, catalog number: C1016)
10% Buffered Formalin Phosphate (Fisher Scientific, catalog number: SF100-4)
20% Sucrose solution in PBS
70% EtOH composed of Absolute EtOH200 proof (Fisher Scientific, catalog number: BP2818-4) diluted in Molecular Biology Grade Water (Hyclone, catalog number: SH30538.03)
Ampicillin Sodium Salt (Fisher Scientific, catalog number: BP1760-5)
BriteliteTM plus reagent (PerkinElmer, catalog number: 6066761)
Cryotube Vial (Thermo Scientific, catalog number: 363401)
Dextran Sulfate (Fisher Scientific, catalog number: BP1585-100)
Dimethyl sulfoxide (DMSO) (Sigma, catalog number: D2650-5X10mL)
DL-Dithiothreitol (DTT) (Sigma, catalog number: D9779-1G)
Dulbecco’s Modified Eagle Medium (DMEM) (Corning, catalog number: MT10013CV.
Dulbecco’s Phosphate Buffered Saline (DPBS) (GIBCO, catalog number: 14190-144)
EDTA (Sigma-Aldrich, catalog number: E6511-100G)
Fungizone (Amphotericin B) (Corning, catalog number: 30-003-CF)
Hanks’ Balanced Salt Solution 1x (HBSS) (Corning, catalog number: 21-021-CM)
HEPES (Sigma-Aldrich, catalog number: H0887-100mL)
Kanamycin Sulfate (Fisher Scientific, catalog number: BP906-5)
LB broth (Fisher scientific, catalog number: BP9723)
Medium A (Fixation Medium) (Beckman Coulter, catalog number: GAS001S100)
Medium B (Permeabilization Buffer) (Beckman Coulter, catalog number: GAS002S100)
Paraformaldehyde (Fisher Scientific, catalog number: T353-500)
Penicillin and streptomycin and L-glutamine (Gibco, catalog number: 10378-016)
Penicillin-Streptomycin (Corning, catalog number: 30-002-Cl)
Qiagen HiSpeed Plasmid Maxi Kit (Qiagen, catalog number: 12662)
OTC Compound (Tissue Tek, catalog number: 4583)
RNA Later Solution (Invitrogen, catalog number: AM7021)
RPMI 1640 (RPMI) (Gibco, catalog number: 11875-093)
SOC medium (Corning, catalog number: 46-003-CR)
Trypan Blue (Corning, catalog number: 25-900-CI)
Zombie Aqua Fixable Viability Dye (Biolegend, catalog number: 423102)
Zosyn (piperacillin/tazobactium)(Novaplus, catalog number: 44567-801-10)
KCl
NaCl
Na2PO4·7H2O
Dextrose
1% Paraformaldehyde (see Recipes)
Thaw Media (see Recipes)
2x HBS buffer (see recipes)
Complete DMEM Media (see Recipes)
Culture RPMI Media (see Recipes)
Flow-staining buffer (see Recipes)
Overnight holding media for jejunum tissue (see Recipes)
Mucous removal media (see Recipes)
Epithelium removal media (see Recipes)
Collagenase Media for LPMC isolation (see Recipes)
Preservative for isolated LPMCs (see Recipes)
Complete RPMI medium for TF virus concentration (see Recipes)
Modified epithelium removal media lacking BSA (see Recipes)
Equipment
37 °C water bath
37 °C incubator
Centrifuge (Beckman Coulter, model: Allegra6R)
Cool Cell LX Freezing Container (Biocision, catalog number: BCS-405)
Germinator 500 (Surgical Equipment sterilizer) (Cell Point Scientific Inc., catalog number: 11931)
Hemocytometer (Hausser Scientific, catalog number: 1492)
Luminometer plate reader (PerkinElmer 2030 Multlabel Reader, model: VictorTM X5)
Multi-channel pipette (Ranin 20-200 μl LTS, model: LM12-200)
Multi-color Flow Cytometer (BD Biosciences, LSRII)
Nutating Mixer Fixed Speed 120V (rotator) (Fisher Scientific, catalog number: 88-861-041)
Orbital shaker (Forma Scientific, model: 4518)
TC20 Automated Cell Counter (Bio-Rad, catalog number: 1450102)
Stratacooler (Stratagene Agilent, catalog number: 401349)
Surgical forceps (VWR, catalog number: 82027-388)
Surgical scissors (VWR, catalog number: 82027-590)
Software
FlowJo Software (Tree Star Inc.)
Procedure
-
Isolation of LPMCs from human jejunum
Jejunum tissue was obtained from the University of Colorado Hospital from patients undergoing elective abdominal surgery. The types of surgeries that provide jejunum tissue include Whipple procedures and bowel obstructions. Jejunum tissue received represents the surgical margins (tissue that would otherwise be discarded) and are macroscopically normal (verified by pathological assessment via histology). The time period between removal of jejunum tissue by surgeons and collection of tissue by laboratory personnel varies between 30 min and 3 h. The following protocol details how LPMCs are isolated from whole jejunum tissue, but in brief, tissue samples are obtained from the hospital, small pieces are removed for histology or RNA preservation, the remaining tissue is stored overnight in holding media, the following day the tissue is dissected to isolate the mucosa layer, followed by mechanical and enzymatic digestion to isolate LPMC for cryopreservation.
Acquire tissue from hospital. Generally, it is provided in a specimen container (depending on the tissue size).
Place whole tissue in a Petri dish and weigh; record the weight.
-
Using surgical forceps and scissors, cut off 3 small pieces of tissue (0.5 cm2 each), making sure to include the mucosa layer and any underlying tissue.
Note: Generally, the tissue is provided as a long tube with surgical sutures closing both ends. You may need to remove surgical sutures on one end in order to access the mucosa inside.
Place 1 tissue piece into a 1.5 ml Eppendorf tube containing 1 ml RNA later. Store the tube at -20 °C until use for gene analysis of whole tissue (e.g., expression of ccr5 or mucosa-associated 16S for bacteria phyla).
Place 1 tissue piece into a 1.5 ml Eppendorf tube containing 1 ml of 10% buffered formalin. Store the tube at RT for 24 h. After 24 h, remove the tissue and transfer to a new Eppendorf tube containing 1 ml 70% EtOH. Store at RT until paraffin embedding for use in histological analysis of whole tissue.
Place 1 tissue piece into a vinyl specimen mold containing OTC. Cover the surface of the tissue with more OTC and place the specimen mold into a mega cassette. Put the entire cassette into liquid nitrogen for 5 min to flash freeze the tissue. Transfer the cassette to -80 °C for storage until use in histological analysis of whole tissue.
Rinse the remaining tissue with 5 ml RT HBSS. Repeat rinsing 2-3x until tissue is clean (i.e., blood is no longer visible and any feces that were made visible by removing sutures to open one end of the tissue have been washed away).
Return the tissue to the specimen container and add enough overnight media (Recipe 1) to cover the entire tissue (approximately 10-30 ml).
Store at 4 °C overnight.
-
The following morning, transfer the whole tissue to a Petri dish for dissection.
Using surgical forceps and scissors, cut off all surgical sutures in order open both ends of the tissue “tube”.
Make an incision along the long length of the tissue “tube”.
-
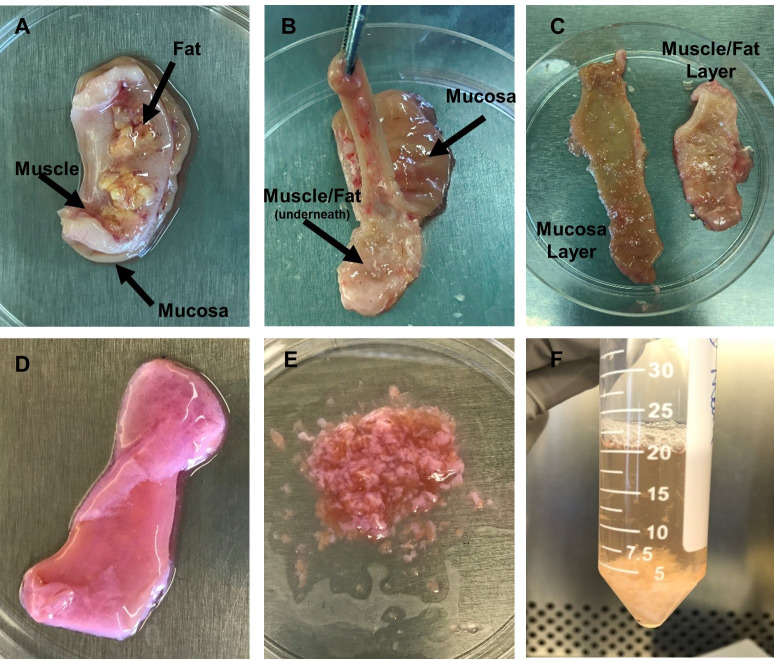
Lay the fully open tissue piece on the Petri dish with the mucosa side down and the muscle/fat side upwards (Figure 2A).
Note: Fat is usually yellowish/white in color and attached to the muscle. The muscle layer is hard and pink/red in color. Large bands of muscle can often be seen. The mucosa layer is soft and light brown in color.
Starting from one end, use the forceps to lift the muscle layer away from the mucosa and cut downwards to remove muscle and fat from the mucosa separating the layers as you proceed. The tissue can be inverted for easier access to the mucosa (Figure 2B). Removal of all muscle and fat reveals a smooth and distinct mucosa layer and a spate muscle/fat layer (Figure 2C).
Transfer the mucosa layer to a Petri dish and weigh; record the “mucosa only” weight.
Cut the mucosa into pieces weighing approximately 1 g. Place each mucosa piece into its own 50 ml conical tube (e.g., for 5 g mucosa therefore require five 50 ml conical tubes).
Add 30 ml mucous removal media (Recipe 2) to each tube.
Place tubes onto rotator positioned inside a 37 °C incubator and rotate for 45 min with caps tightly closed.
Remove liquid from each tube using a graduated pipette and discard directly into liquid waste.
Add 20 ml RT HBSS to each tube and vortex for 10 s to wash away any remaining mucous on the tissue. Remove wash liquid from each tube using a graduated pipette and discard directly into liquid waste.
Repeat wash 2-3 times until liquid is clear and viscous mucous has been removed.
Transfer tissue piece to a new 50 ml conical tube.
Add 30 ml epithelium removal media (Recipe 3) to each tube.
Place tubes onto rotator positioned inside a 37 °C incubator and rotate for 45 min with caps tightly closed.
Remove liquid from each tube using a graduated pipette and discard directly into liquid waste. Tissue will now appear light pink in color (Figure 2D).
Add 20 ml RT HBSS to each tube and firmly vortex for 10 s to further dislodge any loose epithelial cells. Remove liquid from each tube using a graduated pipette and discard directly into liquid waste. Repeat 2-3x until liquid is clear and no more epithelial cells are detached.
Add 30 ml epithelium removal media (Recipe 3) to the tubes containing the 1 g piece of mucosa.
Place tubes onto rotator positioned inside a 37 °C incubator and rotate for a secondary 45 min incubation with caps tightly closed.
-
Repeat Steps 18-19 to remove epithelial cell layer.
Note: If downstream examination of LPMCs suggests that epithelial cells were not sufficiently removed (e.g., presence of epithelial cells detected via microscope or majority of cells are CD45 negative after staining via flow cytometry), then an additional step is suggested.
Additional Step: Continuously vortex tube containing 1 g piece of mucosa for 5 min in the presence of 30 ml of modified epithelium removal media lacking BSA (Recipe 13). Remove liquid as described above and repeat vortexing as necessary 1-3x with fresh media.
Remove mucosa tissue from 1 tube and transfer to Petri dish. Using surgical forceps and scissors, mince tissue into small pieces (Figure 2E). Transfer minced tissue to a new 50 ml conical tube. Repeat for all tubes with mucosa tissue to prep tissue for enzymatic digestion.
Add 20 ml collagenase media for LPMC isolation (Recipe 4) to each tube.
Place tubes onto rotator positioned inside a 37 °C incubator and rotate for 60 min with caps tightly closed. Tissue fragments with now be white in color (Figure 2F).
Remove liquid from each tube using a graduated pipette and filter liquid through a cell strainer (70 μm) placed inside a new 50 ml conical collection tube (this liquid will contain LPMCs).
-
Add 20 ml RT HBSS to each tube and gently vortex for 5 s to further dislodge any loose LPMCs.
Remove liquid from each tube using a graduated pipette and filter through a cell strainer (70 μm) placed inside 50 ml conical collection tubes.
Repeat 2-3x until liquid is clear and no more LPMCs are detached.
For the 50 ml conical tubes that have been used to collect LPMCs, pellet cells by centrifugation (585 × g, 8 min, at 25 °C; break set to “high”).
Add 1 ml culture media to resuspend each LPMC pellet (Recipe 7) and combine cell pellets into 1 tube. Store at 4 °C until time to freeze down cells.
Add 20 ml collagenase media for LPMC isolation (Recipe 4) to each tube.
Place tubes onto rotator positioned inside a 37 °C incubator and rotate for a secondary 60 min with caps tightly closed.
Repeat Steps A26-A27 to collect additional LPMCs.
Discard any remaining tissue using appropriate biowaste disposal. These tissue fragments will be white in color and fibrous, indicating that full digestion of jejunum tissue is complete.
In the tube containing collected LPMCs, bring the volume up to 25 ml using HBSS. Make sure the cells are fully resuspended and homogenous within the liquid, otherwise cell counts will be affected.
-
Remove 15 μl of cell suspension and add to 15 μl 0.4% Trypan blue.
Load 10 μl of cell mixture and count LPMCs using a hemocytometer.
Calculate the total number of LPMCs recovered from tissue processing.
LPMCs should be frozen at 5 to 10 million cells per cryovial, therefore label the appropriate number of cryovials needed to divide total LPMCs among cryovials.
Pre-cool labeled cryovials in a cold StrataCooler.
Pellet cells by centrifugation (585 × g, 8 min, at 25 °C; break set to “high”).
Remove liquid from tube using a graduated pipette and discard directly into liquid waste.
-
To the LPMC cell pellet, add 500 μl per cryovial of Culture RPMI media (Recipe 7). Make sure the cells are fully resuspended and homogenous within the liquid, otherwise cell distribution to cryovials will be uneven.
Note: If total LPMCs will be split into 4 cryovials, then add 500 μl x 4= 2,000 μl culture RPMI media to cell pellet.
-
Slowly add 500 μl per cryovial (total amount added equal in volume to previous step) of preservative (Recipe 5). Gently swirl to mix.
Note: If total LPMCs will be split into 4 cryovials, then add 500 μl x 4 = 2,000 μl preservative to cell pellet.
Transfer 1 ml LPMCs to each cryovial. (Example used in previous steps: tube now contains LPMCs resuspended in 4,000 μl composed of 2,000 μl Culture RPMI media + 2,000 μl preservative for a total of 4 ml to be divided amongst 4 cryovials.)
Transfer cryovials containing LPMCs to Cool Cell freezing container.
Place freezing container in -80 °C for 24 h.
Transfer cryopreserved LPMCs to liquid nitrogen for storage until use.
-
Thawing cryopreserved LPMC
Place Thaw Media (Recipe 6) in a 37 °C water bath for at least 30 min.
Add 100 μl Thaw Media to a 15 ml conical tube. 1 x 15 ml conical per 1 cryovial of LPMC (cryopreserved 5-10 x 106 cells/vial).
Place cryovial of frozen LPMC in the 37 °C water bath and thaw quickly.
Transfer thawed LPMC into a 15 ml conical containing 100 μl thaw media (Step A2).
Wait 1 min.
Add 200 μl Thaw Media to the 15 ml conical tube, wait 1 min.
Add 400 μl Thaw Media to the 15 ml conical tube, wait 1 min.
Add 800 μl Thaw Media to the 15 ml conical tube, wait 1 min.
Add 1.6 ml Thaw Media to the 15 ml conical tube, wait 1 min.
Add 3.2 ml Thaw Media to the 15 ml conical tube, wait 1 min.
Add additional Thaw Media for a final volume of 10 ml.
Pellet cells by centrifugation (585 × g, 8 min) at 25 °C (RT; break set to “high”).
Remove supernatant and flick tube to dislodge pellet.
Resuspend LPMC in 1 ml Thaw Media and gently pipette up and down, add 9 ml Thaw Media.
Pellet cells by centrifugation (585 × g, 8 min, at 25 °C; break set to “high”).
Remove supernatant and flick tube to dislodge pellet.
-
Resuspend LPMC in 1 ml RT DPBS and filter through Cell strainer (70 μm).
Tip for filtering LPMC:
Pre-wet filter with DPBS.
Add LPMC/DPBS.
Rinse 15 ml conical with DPBS and filter.
Add 1 ml DPBS directly to filter to rinse.
Count the LPMC in Trypan Blue using a hemocytometer.
Pellet cells by centrifugation (585 × g, 8 min) at 25 °C (RT; break set to “high”).
Remove supernatant and flick tube to dislodge pellet.
Resuspend LPMC in Culture Media (Recipe 7) at 2.5 x 106 LPMC/ml.
-
Infection protocol
This protocol has utilized both the laboratory-adapted R5-tropic HIV-1Ba-L ( Steele et al., 2014 ; Dillon et al., 2016 ) and the Transmitted/Founder (TF) HIV-1 infectious molecular clones (IMC) CH058, CH470 and CH040 ( Harper et al., 2015 ; Yoder et al., 2017 ) (see Procedures E and F).
-
Divide LPMC into 15 ml conical tube/s for “mock” and “HIV” conditions with the total number of tubes based on the following ‘infection by spinoculation’ criteria (HIV p24 per 2.5 x 106 LPMC in 1 ml final volume of culture media):
500 ng HIV p24 antigen (p24) of TF strains or 25 ng p24 HIV-1Ba-L per 2.5 x 106 LPMC per ml with no more than 2.5 x 106 LPMC (i.e., 1 ml) per 15 ml conical tube. HIV p24 concentrations of virus stocks need to be calculated for each virus preparation using an HIV p24 antigen ELISA following manufacturers protocol.
If multiple 15 ml conical tubes are needed, evenly distribute the LPMC over tubes: e.g., if 6 x 106 total LPMC are needed for infection, distribute 2 x 106 cells per 15 ml tube.
Mock-infect with matched LPMC/ml per 15 ml conical.
Note: If using GFP-expressing virus strains, add 1.5 x 106 LPMC into 15 ml conical to be used as GFP compensation control when evaluating GFP-expressing LP CD4 T cells using multi-color flow.
Pellet cells by centrifugation (585 × g, 8 min) at 25 °C (RT; break set to “high”).
Remove all supernatant and flick tube to dislodge pellet.
-
Resuspend LPMC in Culture Media and then add HIV or mock as calculated (Step B1a). An example calculation is detailed below (Table 1).
-
Table 1 illustrates the calculations used to infect 2.5 x 106 LPMC with 500 ng p24 in a total of 1 ml of culture media. The specific column headings (denoted A-E) are detailed to assist with the calculations.
For example:
The total amount of p24 needed (Column B) is calculated based on the number of cells per 15 ml tube (Column A) when LPMC are infected with 500 ng p24 per 2.5 x 106 LPMC. The total final volume for spinoculation is based on infecting no more than 2.5 x 106 LPMC/ml per 15 ml tube and is calculated in Column C. The total volume of virus stock added is based on a final amount of 500 ng HIV p24 per 2.5 x 106 LPMC and is therefore dependent on the p24 concentration of the stock HIV. This example details the calculation when the stock concentration is 20,000 ng p24/ml and is based on Column B value (i.e., the total p24 needed) and Column C (i.e., the total final volume for spinoculation). The volume of culture media to add is based on the difference between the total volume (Column C) and the volume of p24 added (Column D). The volume of mock is matched to the volume of p24 added to the HIV tube.
-
Centrifuge at 1,200 × g, 2 h, room temp (RT; break set to “off”).
Discard supernatant containing free virus and resuspend in 1 ml of Culture Media. Mock tubes are similarly treated with removal of supernatant and addition of Culture Media.
Centrifuge at 585 × g, 8 min at 25 °C (RT; break set to “high”).
Resuspend in Culture Media for a final concentration of 1 x 106 cells/ml. If additional stimulation conditions are required, divide cells into 15 ml conical tubes and add stimuli as appropriate.
Plate LPMC into 96 V-bottom culture plate (200 μl per well).
Add 200 μl DPBS to surrounding wells to minimize evaporation of Culture Media.
Incubate for 4 days at 37 °C, 5% CO2 and 95% humidity.
-
-
Multi-color flow cytometry protocol to determine percentage of HIV-infected LP CD4 T cells
Collect cultured LPMC and pellet cells by centrifugation (585 × g, 8 min, RT; break set to “high”).
Resuspend LPMC in DPBS and count in trypan blue using an automated cell counter. Record the number of viable, LPMC for all conditions.
-
Prepare an antibody cocktail containing a viability dye (1 μl per 1 x 106 LPMC) and fluorochrome-labeled antibodies directed against CD3 and CD8 in Flow-staining buffer (Recipe 8) (4 °C). Concentrations of fluorochrome-labeled antibodies should be titrated for optimal performance by available flow cytometer.
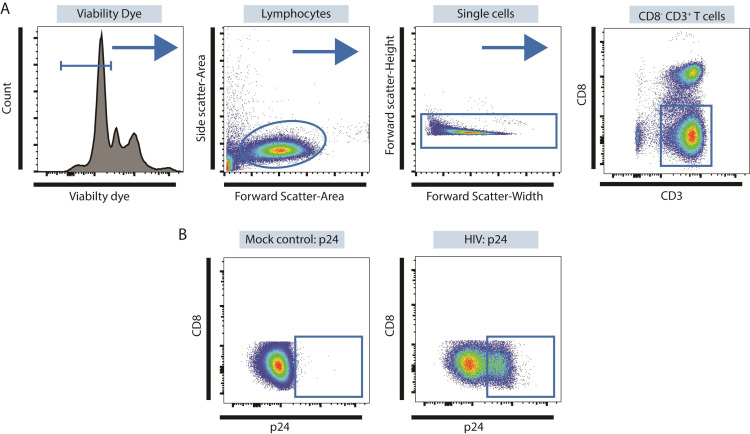
HIV down-regulates CD4 expression therefore LP CD4 T cells are identified as CD3+ CD8- T cells following in vitro infection with HIV-1 (Figure 2).
Specific viability dyes and fluorochrome-labelled CD3 and CD8 antibodies appropriate for multi-color flow cytometry can be user-specific and optimized for available flow cytometers. The example shown (Figure 3) uses Zombie Aqua Fixable Viability Dye (1 μl per 1 x 106 LPMC), PE/Dazzle 594 anti-human CD3 (0.125 μg per 1 x 106 LPMC) and APC anti-human CD8 (0.125 μg per 1 x 106 LPMC) and were acquired on an LSRII Flow Cytometer (BD Biosciences).
Transfer LPMC to FACS tubes and pellet cells by centrifugation (585 x g, 5 min, 4 °C; break set to “high”).
Remove supernatant and flick tube to dislodge pellet.
Resuspend LPMC in antibody cocktail.
Incubate in the dark, 30 min, 4 °C.
Add 4 ml DPBS (4 °C) and pellet cells by centrifugation (585 × g, 5 min, 4 °C; break set to “high”).
Remove supernatant and flick tube to dislodge pellet.
Resuspend LPMC in 100 μl Medium A (fixation medium).
Incubate in the dark, 20 min, 4 °C.
Add 4 ml Flow cytometry buffer (4 °C) and pellet cells by centrifugation (585 x g, 5 min, 4 °C; break set to “high”).
Remove supernatant and flick tube to dislodge pellet.
Resuspend LPMC in 100 μl Medium B (permeabilization buffer) containing 5 μl RD-1 HIV-1 core antigen.
Incubate in the dark, 30 min, 4 °C.
Add 4 ml Flow cytometry buffer (4 °C) and pellet cells by centrifugation (585 × g, 5 min, 4 °C; break set to “high”).
Remove supernatant and flick tube to dislodge pellet.
Resuspend LPMC in 400 μl 1% paraformaldehyde (Recipe 9).
Acquire on flow cytometer within 4 h.
-
Preparation of Transmitted/Founder (TF) HIV-1 infectious molecular clone stocks
The protocol below is used to specifically prepare TF HIV-1 virus stocks from infectious molecular clones (IMCs) CH040, CH058, CH077, and CH470.
-
Preparation of TF HIV-1 plasmids
-
Transformation of TF plasmids
Set the water bath to 42 °C.
Thaw a vial of OneShotTM Stbl3 competent cells on ice for 5 min.
Add 100-200 ng of stock TF plasmid to the thawed Stbl3 cells, mix by gently flicking the tube a few times.
Incubate the cells on ice for 30 min. Put the Stbl3 cell tube on a floater and heat shock the cells at 42 °C for 45 s.
Chill the cells on ice for 5 min immediately after the heat shock.
Add 800 μl of SOC medium to the cells.
Shake the cells at 30 °C in an orbital shaker for 1 h at 200 rpm.
Add 300-400 μl of cell mixture onto a LB agar plate with compatible antibiotics (Table 2) and spread evenly using standard method.
Incubate the LB plate at 30 °C overnight (16-18 h).
-
Maxi-prep
Pick a single colony and inoculate in 6 ml of LB broth containing compatible antibiotics (Table 2).
Shake the LB broth at 30 °C for 16-18 h at 200 rpm.
Transfer 1 ml of culture and inoculate into 500 ml of the LB broth with compatible antibiotics (Table 2).
Again, shake the 500 ml of LB broth at 30 °C for 16-18 h at 200 rpm.
Perform Maxi-prep according to maxi-prep kit’s instruction.
-
Sequence verification of newly generated TF HIV-1 plasmids
Because the TF HIV-1 molecular clones contain identical repeats on both ends, it is not rare that the plasmids would lose most of the virus genome insert during the transformation and plasmid amplification process. Therefore, it is critical to fully sequence the maxi-prep products before generating the live virus stocks. Included here is a set of 16 sequencing primers to ensure coverage of the entire range of the virus genome insert in the plasmid (Table 3).
-
-
Transfection of TF HIV-1 infectious molecular clones
Start the 293T cell culture a week before transfection so that the cells can be passaged at least twice. This will help the cells acclimate to the growth conditions and improve transfection efficiency.
293T culture medium: complete Dulbecco’s Modified Eagle Medium (DMEM) with 10% FBS and 1% Penicillin-Streptomycin.
Transfer the 293T cells into T175 culture flasks during passages. The cells are ready for transfection at about 60-70% confluence.
-
Calcium Phosphate Transfection.
Reagents:
2 M CaCl2
2x HBS buffer (Recipe 11)
Sterilize both reagents by filtering through 0.22 µm filter, and store at 4 °C until use.
Transfection:
Composition of the transfection mixture (per T175 flask): TF plasmid (60 μg), H2O, 2x HBS buffer (2 ml), and 2 M CaCl2 (256 μl). The total volume is 4 ml.
Mix TF plasmid and H2O (total volume: 1,744 μl) in a 15 ml conical tube.
Add 2 ml 2x HBS buffer and mix well.
Gently add CaCl2 (dropwise) into the mix.
Incubate at room temperature for 30 min without mixing.
Add the transfection mix slowly to the medium in the 293T flask, mix by gently swirling the flask.
-
Transfer the flask into cell culture incubate (37 °C) in a BSL3 facility.
Note: Due to the high risk working with high concentrations of TF HIV-1 virus, it is recommended to carry out subsequent steps in BSL3 facility if possible.
After 16 h of incubation, replace half of medium (~20 ml) in the transfected flask with fresh complete DMEM medium (Recipe 10) to minimize the toxic effect of Calcium Phosphate to the cells.
Incubate for an additional 32 h before harvest.
-
Harvest the virus supernatant
Transfer the 293T flask into biosafety hood.
Decant the medium into a 50 ml conical, and centrifuge at 500 x g for 5 min to remove dead cells and cellular debris.
Attach a 0.45 μm syringe-driven filter to a 60 ml disposable syringe with the plunger removed.
Decant the centrifuged medium into the syringe, then reinsert the plunger and push the liquid into a new 50 ml conical tube.
The virus supernatant can be aliquoted and stored at -80 °C until use. However, to ensure higher virus titer, it is recommended to concentrate the supernatant via ultracentrifugation.
-
Concentrating the TF virus
Add 26 ml of virus supernatant into a polycarbonate thick wall ultracentrifugation tube.
-
Slowly add 4 ml of 20% sucrose solution (in PBS) under the virus supernatant at the bottom of the tube (underlaying).
Note: for details on the underlay technique, please check the Youtube video ( https://youtu.be/-SqlIMLUfcI ) at 0:43. Although the video shows Ficoll underlayed in whole blood, the same principle applies to underlaying 20% sucrose in the virus supernatant.
Centrifuge the tubes at 100,000 × g for 2 h (break set to high).
Gently decant the supernatant, and put the tubes upside down on layers of absorbent papers to drain the excess fluid for 5 min.
Add 300-400 μl of complete RPMI medium (Recipe 12) with 10% FBS and 1% Penicillin-Streptomycin.
Seal the tubes with Parafilm and store at 4 °C overnight to loosen the virus pellet.
Pipette up and down gently. Avoid generating too many bubbles to homogenize the mixture, then aliquot to small tubes and store at -80 °C until use.
-
-
Titration of the infectivity of the virus stocks (TZM-bl assay)
Start TZM-bl cell culture in complete DMEM medium a week before the assay.
Thaw virus quickly at 37 °C in a water bath, then keep cool on ice.
In a biosafety hood, make serial dilutions of the viruses in a round-bottom 96-well plate. For concentrated virus, a 1:100 dilution (5 µl virus into 495 µl media) to start is recommended, then a subsequent 1:1000 dilution (10 µl of 1:100 dilution into 990 µl media) then five 5-fold dilutions (100 µl of the 1:100 dilution into 400 µl media, then the same ratio subsequently). Keep the virus solutions on ice while preparing the TZM-bl cells. Make sure to include a pre-titrated HIV-1 stock as positive control.
Trypsin treat the cell culture flask to lift the TZM-bl cells.
Count the cell concentration in the supernatant using a cell counter.
Using 10,000/well density to calculate the volume required based on the number wells needed for the assay.
Add complete DMEM medium to adjust the total volume to equivalent to 160 μl/well.
Add 18.75 μg/ml of Dextran Sulfate into the cell mixture, mix well then transfer 160 μl to each well on a 96-well white tissue culture-treated sterile microplate using a multi-channel pipette.
Transfer 40 μl of the virus dilutions to corresponding wells and mix well by pipetting up and down. The final concentration for Dextran Sulfate should be 15 µg/ml after the addition of the virus dilution.
Fill each unused well with 200 μl of sterile PBS to reduce the volume lost due to evaporation during incubation.
Cover lid, and incubate for 48 h.
Prepare the BriteLite plus reagent by dissolving the lyophilized powder with 10 ml of BriteLite buffer and let warm up at room temperature.
In a biosafety hood, remove 95 μl of spent medium from each well using a multi-channel pipette.
Dispense 100 μl of BriteLite plus reagent to each well and mix well by pipetting up and down.
Let the plate incubate for 2-3 min then read with a luminometer plate reader (at default luminescence setting for 1 s).
A luminescence readout below 2 million is considered a valid value. The viral infectivity is reported as relative light unit (RLU), which is calculated by multiplying the luminescence readout with the corresponding dilution factor.
Figure 2. Images of tissue dissection and LPMC isolation.
A. Tissue is placed in a Petri dish with mucosa side down. The muscle layer and fat layer are facing upwards. B. Tissue orientation is inverted with muscle/fat layer facing downwards and mucosa layer facing upwards. Forceps are holding the mucosa layer (light brown in color) and pulling it away from the muscle/fat layer as scissors are used to separate the layers. C. Depiction of the two separated layers after dissection is complete. D. Tissue after epithelium has been removed during incubation with EDTA. Mucosa is now light pink in color. E. Minced tissue to prep for treatment with collagenase to enzymatically remove LPMCs. F. Tissue fragments and cells after incubation in collagenase. (Images by Kaylee Mickens.)
Table 1. Example calculations using an HIV virus strain with a concentration of 20,000 ng p24 per ml.
| A | B | C | D | E | |
|---|---|---|---|---|---|
| LPMC (x 106) per 15 ml conical tube | Total p24 (Column A/2.5 x 500) | Total volume(Column A /2.5*1000 μl) | Volume HIV/mock(Column B/20,000 x Column C) | Volume of culture media(Column C-Column E) | |
| HIV | 2.5 | 500 ng | 1000 μl | 25 μl | 975 μl |
| Mock | 2.5 | “500 ng” | 1000 μl | 25 μl | 975 μl |
Figure 3. Identifying productively-infected LP CD4 T cells using multi-color flow cytometry.
A. Identification of CD3+ CD8- LP CD4 T cells. Viable cells are identified as Viability Dye- and then lymphocytes identified within this population based on Forward Side Scatter-Area versus Side Scatter-Area properties as shown. Doublets are excluded based on Forward Scatter-Width versus Forward-Scatter Height properties and CD3+ CD8- T cells identified within these single cells. B. Expression of p24 in CD3+ CD8- LP CD4 T cells using mock conditions as controls.
Table 2. TE molecular clone vector antibiotic resistance.
| HIV-1 Molecular Clones | AIDs Reagent Program Cat # | Vector Antibiotic Resistance | Antibiotics Concentration (µg/ml) |
|---|---|---|---|
| CH040 | 11740 | Kanamycin | 50 |
| CH058 | 11856 | Kanamycin | 50 |
| CH077 | 11742 | Kanamycin | 50 |
| CH470 | 12422 | Ampicillin | 200 |
Table 3. TF HIV-1 sequencing primers.
| Primer | NT sequence | Orientation | Location CH040TF (JN944905) |
|---|---|---|---|
| P1 | CTCTAGCAGTGGCGCCCGAAC | F | 627-647 |
| P2 | TTCAGCAAGCCGAGTCCTGCG | R | 710-690 |
| P3 | AGACACYATCAATGAGGAAGCTGC | F | 1394-1417 |
| P4 | TTAAAGGTACYTGAGGTCTGACTGG | R | 8990-8966 |
| P5 | CTGARAGACAGGCTAATTTTTTAGG | F | 2068-2092 |
| P6 | GAAGTTCAAYTAGGAATACCACATC | F | 2810-2834 |
| P7 | GRYCAATGGACATATCAAATWTATC | F | 3548-3572 |
| P8 | ATAAATGTCAGCTAAAAGGAGAAGC | F | 4347-4371 |
| P9 | GCCACACAATCATCACCTGCCATC | R | 5070-5047 |
| P10 | TAGTGACATAAARGTAGTGCCAAGA | F | 4987-5011 |
| P11 | TGGGTGTCRWCATAGCAGAATAGGC | F | 5776-5800 |
| P12 | ACGCCTGTGTACCCACAGACC | F | 6431-6451 |
| P13 | CAGCACAGTRCAATGYACACATGGA | F | 6933-6957 |
| P14 | GGGAGCWGCAGGAAGCACTATGG | F | 7758-7780 |
| P15 | GCAGTAGCTGARRGRACAGATAG | F | 8650-8672 |
| P16 | AGTGGGBTTYCCAGTCAGACCTC | F | 8966-8990 |
Data analysis
-
Determination of percentages of infected LP CD4 T cells based on p24 expression
To determine percentages of productively infected LP CD4 T cells (Figure 2), viable cells are identified based on lack of expression of a viability dye and then total lymphocytes within this population identified based on forward and side scatter properties. Doublets are excluded based on forward-scatter-width versus forward-scatter-height properties. Total T cells are identified by expression of CD3. HIV down-regulates CD4 expression therefore CD4 T cells are identified as CD8- T cells with total CD3+ T cells. To establish specific expression of HIV-1 core antigen (p24), mock conditions are used as controls. Profiles evaluating co-expression of p24 and GFP are detailed in Yoder et al. (2017) .
-
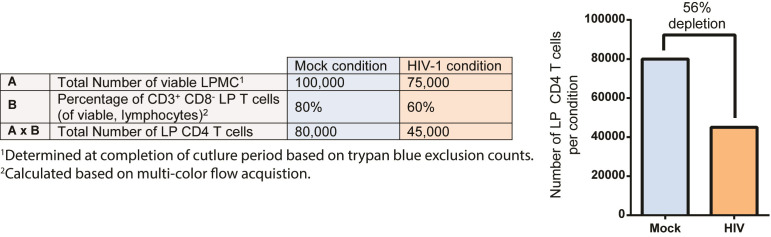
Quantification of LP CD4 T cell depletion
To quantify HIV-specific LP CD4 T cell depletion after 4 days in culture, the difference in the number of viable LP CD4 T cells in HIV-infected cultures versus matched mock-infected cultures is determined (Figure 4). To enumerate the number of viable LP CD4 T cells, the total viable LPMC collected at the completion of the culture period (determined by trypan blue exclusion) is multiplied by the percentage of CD3+ CD8- T cells in viable LPMC acquired using multi-color flow cytometry. The percent depletion relative to mock is then calculated.
-
Assay design, reproducibility and statistical analysis
A minimum of 6 x 105 cells per condition are plated in triplicate wells at the initiation of the culture. At the completion of the culture period, LPMC from triplicate wells are pooled together, counted and evaluated for infection and depletion. Assays are performed on at least 6 different donor samples. Parametric tests are performed.
Figure 4. Example calculations to determine LP CD4 T cell depletion using the LPAC model.
Line A reflects the total number of viable LPMC determined at the completion of the culture period based on trypan blue exclusion counts for both Mock condition and HIV-1 conditions. Line B reflects the percentage of CD3+CD8- LP T cells as a fraction of viable lymphocytes enumerated using multi-color flow cytometry. The total absolute number of LP CD4 T cells in each condition is then calculated by multiplying A with B. The graph reflects the number of LP CD4 T cells in mock and in HIV conditions with the percent depletion in HIV relative to mock shown, calculated as number in HIV-1 condition/number in mock condition (%).
Notes
This assay is highly reproducible within samples from the same donor; however, variability exists between samples from different donors. This can be minimized by restricting the use of samples to those with > 60% viability and > 50% LP CD4 T cells (as a fraction of total LPMC) at baseline (freshly thawed). Furthermore, we recommend assessment of CCR5 genotype/phenotype for each sample to limit low infection rates due to CCR5 expression.
Recipes
-
Overnight holding media for jejunum tissue
RPMI supplemented with:
10% Human AB Serum
1% Penicillin/streptomycin/L-glutamine
500 μg/ml Zosyn
1.25 μg/ml Fungizone
1 M HEPES
Mucous removal media (incubate in 37 °C water bath before use)
HBSS supplemented with:
1.67 mM DTT
500 μg/ml Zosyn
1.25 μg/mL Fungizone
-
Mucous removal media (incubate in 37 °C water bath before use)
HBSS supplemented with:
1.67 mM DTT
500 μg/ml Zosyn
1.25 μg/mL Fungizone
-
Epithelium removal media (incubate in 37 °C water bath before use)
HBSS supplemented with:
1 mM EDTA
500 μg/ml Zosyn
1.25 μg/ml Fungizone
0.1% BSA
-
Collagenase Media for LPMC isolation (incubate in 37 °C water bath before use)
RPMI supplemented with:
10% BSA
1% Penicillin/streptomycin/L-glutamine
500 μg/ml Zosyn
1.25 μg/ml Fungizone
10 μg/ml DNase I
0.5 mg/ml Collagenase D
Note: Adding more collagenase D could cause unwanted cleavage of cell surface proteins, which may affect identification of specific cells types during downstream techniques such as flow cytometry. Incubating mucosa tissue for longer than 2 periods of 60 min recommended by this protocol could also cause unwanted cleavage of cell surface proteins. New lots of collagenase D should be compared side-by-side for enzymatic activity and their effect on cell surface protein expression. Avoid multiple freeze/thaw cycles of Collagenase D as this will greatly impact enzymatic activity.
-
Preservative for isolated LPMCs
20% DMSO
80% Human AB Serum
-
Thaw Media
RPMI supplemented with:
10% FBS
1% Penicillin/streptomycin/L-glutamine
10μg/ml DNase I
-
Culture RPMI Media
RPMI supplemented with:
10% Human AB Serum
1% Penicillin/streptomycin/L-glutamine
500μg/ml Zosyn
-
Flow-staining buffer
DPBS supplemented with:
1% Bovine Serum Albumin
2 mM EDTA
-
1% Paraformaldehyde
DPBS supplemented with:
1% paraformaldehyde
-
Complete DMEM Media
DMEM supplemented with:
10% FBS
1% Penicillin-Streptomycin
-
2x HBS buffer
50 mM HEPES, pH 7.05
10 mM KCl
12 mM dextrose
280 mM NaCl
1.5 mM Na2PO4
For 1 L:
Dissolve 11.92 g HEPES, 745.60 mg KCl, 2.16 g dextrose, 16.36 g NaCl, 40.2 g Na2PO4·7H2O in 950 ml H2O
Adjust pH to 7.05 with 1 N HCl, bring up to 1 L with H2O
Sterilize the solution by filtering through a 0.22 µm filter, and store at 4 °C until use
-
Complete RPMI medium for TF virus concentration
RPMI supplemented with:
10% FBS
1% Penicillin-Streptomycin
-
Modified epithelium removal media lacking BSA
HBSS supplemented with:
1 mM EDTA
500 μg/ml Zosyn
1.25 μg/ml Fungizone
Acknowledgments
This work was supported by National Institute of Health Grant R01 AI108404 (to C.C.W) and R01 AI134220 and AI145428 (to M.L.S. and C.C.W). We would like to thank Dr. Amanda Steele and Eric Lee for assistance with the development of the LPAC model, Kaylee Mickens for the pictures of gut processing in Figure 1, and Christine Purba for critical review of this protocol. We would also like to express our gratitude to the many laboratory members who processed the intestinal tissue samples used in the LPAC model and to the patients for agreeing to the use of their discarded surgical tissue for research purposes.
Competing interests
The authors declare no conflicts of interest.
Ethics
LPMC are obtained from discarded surgical tissue samples with patients providing informed consent to have these samples used for research purposes. All samples are de-identified to laboratory personnel and provided an ID code when received in the laboratory. This research was reviewed by the Colorado Multiple Institutional Review Board (IRB) (COMIRB) and met criteria, as defined by their policies in accordance with OHRP and FDA regulations, to be deemed “Not Human Research”.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Dillon S. M., Rogers L. M., Howe R., Hostetler L. A., Buhrman J., McCarter M. D. and Wilson C. C.(2010). Human intestinal lamina propria CD1c+ dendritic cells display an activated phenotype at steady state and produce IL-23 in response to TLR7/8 stimulation . J Immunol 184(12): 6612-6621. [DOI] [PubMed] [Google Scholar]
- 2. Dillon S. M., Manuzak J. A., Leone A. K., Lee E. J., Rogers L. M., McCarter M. D. and Wilson C. C.(2012). HIV-1 infection of human intestinal lamina propria CD4+ T cells in vitro is enhanced by exposure to commensal Escherichia coli . J Immunol 189(2): 885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dillon S. M., Lee E. J., Donovan A. M., Guo K., Harper M. S., Frank D. N., McCarter M. D., Santiago M. L. and Wilson C. C.(2016). Enhancement of HIV-1 infection and intestinal CD4+ T cell depletion ex vivo by gut microbes altered during chronic HIV-1 infection . Retrovirology 13: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Doitsh G., Cavrois M., Lassen K. G., Zepeda O., Yang Z., Santiago M. L., Hebbeler A. M. and Greene W. C.(2010). Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell 143(5): 789-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Doitsh G., Galloway N. L., Geng X., Yang Z., Monroe K. M., Zepeda O., Hunt P. W., Hatano H., Sowinski S., Muñoz-Arias I. and Greene W. C.(2014). Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505(7484): 509-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harper M. S., Guo K., Gibbert K., Lee E. J., Dillon S. M., Barrett B. S., McCarter M. D., Hasenkrug K. J., Dittmer U., Wilson C. C. and Santiago M. L.(2015). Interferon-alpha subtypes in an ex vivo model of acute HIV-1 infection: expression, potency and effector mechanisms . PLoS Pathog 11(11): e1005254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Howe R., Dillon S., Rogers L., McCarter M., Kelly C., Gonzalez R., Madinger N. and Wilson C. C.(2009). Evidence for dendritic cell-dependent CD4+ T helper-1 type responses to commensal bacteria in normal human intestinal lamina propria . Clin Immunol 131(2): 317-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keele B. F., Giorgi E. E., Salazar-Gonzalez J. F., Decker J. M., Pham K. T., Salazar M. G., Sun C., Grayson T., Wang S., Li H., Wei X., Jiang C., Kirchherr J. L., Gao F., Anderson J. A., Ping L. H., Swanstrom R., Tomaras G. D., Blattner W. A., Goepfert P. A., Kilby J. M., Saag M. S., Delwart E. L., Busch M. P., Cohen M. S., Montefiori D. C., Haynes B. F., Gaschen B., Athreya G. S., Lee H. Y., Wood N., Seoighe C., Perelson A. S., Bhattacharya T., Korber B. T., Hahn B. H. and Shaw G. M.(2008). Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105(21): 7552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salazar-Gonzalez J. F., Salazar M. G., Keele B. F., Learn G. H., Giorgi E. E., Li H., Decker J. M., Wang S., Baalwa J., Kraus M. H., Parrish N. F., Shaw K. S., Guffey M. B., Bar K. J., Davis K. L., Ochsenbauer-Jambor C., Kappes J. C., Saag M. S., Cohen M. S., Mulenga J., Derdeyn C. A., Allen S., Hunter E., Markowitz M., Hraber P., Perelson A. S., Bhattacharya T., Haynes B. F., Korber B. T., Hahn B. H. and Shaw G. M.(2009). Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med 206(6): 1273-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steele A. K., Lee E. J., Manuzak J. A., Dillon S. M., Beckham J. D., McCarter M. D., Santiago M. L. and Wilson C. C.(2014). Microbial exposure alters HIV-1-induced mucosal CD4+ T cell death pathways ex vivo . Retrovirology 11: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoder A. C., Guo K., Dillon S. M., Phang T., Lee E. J., Harper M. S., Helm K., Kappes J. C., Ochsenbauer C., McCarter M. D., Wilson C. C. and Santiago M. L.(2017). The transcriptome of HIV-1 infected intestinal CD4+ T cells exposed to enteric bacteria . PLoS Pathog 13(2): e1006226. [DOI] [PMC free article] [PubMed] [Google Scholar]