Abstract
Coronavirus disease 2019 (COVID-19), an infectious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is responsible for the most threatening pandemic in modern history. The aim of this systematic review and meta-analysis was to investigate the associations between serum albumin concentrations and COVID-19 disease severity and adverse outcomes. A systematic literature search was conducted in PubMed, from inception to October 30, 2020. Sixty-seven studies in 19,760 COVID-19 patients (6141 with severe disease or poor outcome) were selected for analysis. Pooled results showed that serum albumin concentrations were significantly lower in patients with severe disease or poor outcome (standard mean difference, SMD: − 0.99 g/L; 95% CI, − 1.11 to − 0.88, p < 0.001). In multivariate meta-regression analysis, age (t = − 2.13, p = 0.043), publication geographic area (t = 2.16, p = 0.040), white blood cell count (t = − 2.77, p = 0.008) and C-reactive protein (t = − 2.43, p = 0.019) were significant contributors of between-study variance. Therefore, lower serum albumin concentrations are significantly associated with disease severity and adverse outcomes in COVID-19 patients. The assessment of serum albumin concentrations might assist with early risk stratification and selection of appropriate care pathways in this group.
Keywords: Albumin, Coronavirus 2, COVID-19, SARS
Introduction
Coronaviruses are a large family of enveloped positive-sense RNA viruses known to cause clinical symptoms ranging from the common cold to severe respiratory infections, such as the severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (MERS). The latter, caused by SARS-CoV and MERS-CoV, respectively, have caused recent epidemics with mortality rates ranging from 10 to 37% [1–4]. Coronavirus disease 2019 (COVID-19) is a recently identified infectious disease caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [5]. COVID-19 represents the most threatening pandemic outbreak in modern history, affecting some fifty million people and causing more than 1.2 million deaths globally as of November 12, 2020. Actions for the containment of the disease have included different degrees of lockdown strategies in many countries, generating unpredictable economic and social consequences. The spectrum of COVID-19 illness ranges from being asymptomatic or experiencing mild symptoms to important clinical manifestations such as severe pneumonia, which can further progress to acute respiratory distress syndrome (ARDS), multiple organ failure and, potentially, death [6, 7]. An increased risk of severe disease and adverse outcomes has been observed in older adults and patients of any age with comorbidities such as coronary heart disease, diabetes, respiratory disease and hypertension [6, 8]. There are ongoing efforts to better understand the pathophysiology, presentation and clinical outcomes of the disease, including the identification of biomarkers for diagnosis, risk stratification, disease monitoring and prognosis. Early studies in COVID-19 patients have reported alterations in routine laboratory tests, particularly white blood cell count, neutrophils, lymphocytes, platelets, alanine aminotransferase, aspartate aminotransferase, D-dimer, total bilirubin and creatinine [8–11]. Reductions in serum albumin concentrations have also been associated with disease severity [12, 13]. The aim of this present study was to appraise the available evidence regarding the associations between serum albumin concentrations, disease severity and adverse outcomes in COVID-19 patients.
Materials and methods
Search strategy, eligibility criteria and study selection
An electronic search was performed in Medline (PubMed interface) using the keywords “albumin” AND “coronavirus” OR “albumin” AND “COVID-19” from inception to October 30, 2020. The inclusion criteria were: (a) studies reporting continuous data on serum albumin concentrations in COVID-19 patients, (b) articles investigating COVID-19 patients with different disease severity or clinical outcomes, (c) articles in adult patients, (d) number of studied patients ≥ 10, (e) articles in English and (f) full-text article was available. Two investigators independently screened the abstracts to establish relevance. If relevant, the two investigators independently reviewed the full articles. Any disagreement between the reviewers was resolved by a third investigator. The reference list of the studies identified was also checked in order to identify additional studies. We used the Newcastle–Ottawa scale to assess the quality of each study [14]. The Newcastle–Ottawa scale evaluates the following components: cohort selection, cohort comparability on the basis of the design or analysis, how the exposure is determined and how the outcomes of interest are evaluated. Studies achieving a score of six or more were considered to be of high quality.
Endpoint
The study endpoint was the pooled SMD of serum albumin concentrations between patients with low versus high severity or good versus poor outcomes. Disease severity was based on symptoms, disease progression (from moderate to severe grade or critical grade), ICU admission, intubation and ARDS onset, whereas outcome was based on survival vs. death during the study period.
Statistical analysis
Standardized mean differences (SMD) were used to build forest plots of continuous data and to evaluate differences in serum albumin concentrations between COVID-19 patients with low versus high severity or good vs. poor outcomes. A p-value < 0.05 was considered statistically significant, and 95% confidence intervals (CIs) were reported. When required, the mean and standard deviation values were extrapolated from median and IQR as previously reported by Wan et al. [15] or median and range as reported by Hozo et al. [16]. Heterogeneity of SMD across studies was tested by using the Q statistic (significance level at p < 0.10). The I2 statistic, a quantitative measure of inconsistency across studies, was also calculated (I2 < 25%, no heterogeneity; I2 between 25 and 50%, moderate heterogeneity; I2 between 50 and 75%, large heterogeneity; and I2 > 75%, extreme heterogeneity). A random-effects model was used if heterogeneity was high. Sensitivity analysis was conducted to investigate the influence of individual studies on the overall risk estimate, by sequentially excluding one study at a time. To evaluate the presence of potential publication bias, the associations between study size and magnitude of effect were analysed by means of Begg’s adjusted rank correlation test and Egger’s regression asymmetry test at the p < 0.05 level of significance [17, 18]. Duval and Tweedie “trim and fill” procedure was performed to identify and correct for funnel plot asymmetry arising from publication bias [19]. Statistical analyses were performed using MedCalc for Windows, version 15.4 64 bit (MedCalc Software, Ostend, Belgium) and Stata 14 (STATA Corp., College Station, TX, USA).
Results
Electronic search results and characteristics of the included studies
A flow chart describing the screening process is presented in Fig. 1. We initially identified 421 studies. A total of 343 studies were excluded after the first screening because they were either duplicates or irrelevant. After full-text review of the remaining 78 articles, a further 11 studies were excluded because they did not meet the inclusion criteria. Thus, 67 studies were included in the meta-analysis [20–86]. The characteristics of these studies, all published in 2020, are presented in Table 1. A total of 19,760 COVID-19 patients were studied, 13,628 (49% males, mean age 53 years) with low severity or favourable outcome and 6141 (58% males, mean age 65 years) with high severity or poor outcome. Three studies were prospective [29, 41, 52], 51 retrospective [20–28, 30–34, 36–39, 42–46, 48–51, 53, 54, 57–60, 62, 63, 65, 66, 69–76, 78, 80, 81, 83–86], while 13 did not specifically declare the study design [23, 35, 40, 47, 55, 56, 61, 64, 67, 68, 77, 79, 82]. Fifty-two studies (77.6%) were performed in China [22, 24–26, 28–31, 33–35, 37–43, 46–57, 59, 62–64, 68–85] while the remaining 15 were conducted in the rest of the world [20, 21, 23, 27, 32, 36, 44, 45, 58, 60, 61, 65–67, 86]. Endpoints included disease severity based on current clinical guidelines (31 studies, 46%) [22, 24, 28, 32, 33, 35, 36, 38, 39, 44–47, 49–52, 54, 56, 58, 64, 66, 69, 72, 73, 75, 77, 79, 81–83], survival (19 studies, 28%) [20, 23, 26, 30, 31, 34, 42, 53, 59–62, 65, 67, 71, 76, 84–86], intensive care unit (ICU) admission (7 studies, 10%) [21, 27, 41, 48, 68, 70, 78] and other outcomes (10 studies, 15%) [25, 29, 37, 40, 43, 55, 57, 63, 74, 80]. Among the 67 retrieved studies, only Aloisio et al. [20] reported the lowest albumin concentrations throughout hospitalization, whereas all the remaining studies reported albumin concentrations measured within the first 24–48 h from admission.
Fig. 1.
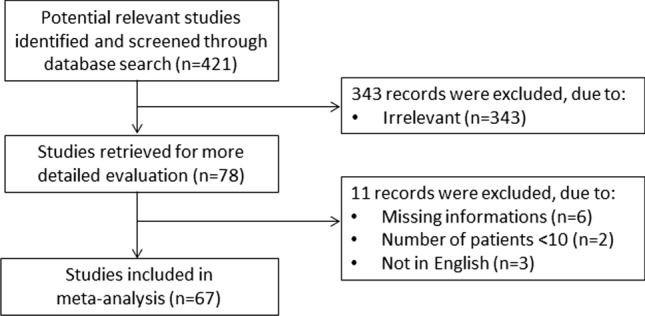
Flow chart illustrating the electronic search strategy and review
Table 1.
Summary of the studies in COVID-19 patients, according to disease severity or outcome, included in the meta-analysis
| Mild disease or good outcome | Severe disease or poor outcome | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| First Author, Country, Reference |
Study design |
Outcome | NOS (stars) |
n | Age (Years) |
Gender (M/F) |
Albumin (g/L, Mean ± SD) |
n | Age (Years) |
Gender (M/F) |
Albumin (g/L, Mean ± SD) |
|
Aloisio et al. Italy [20] |
R |
Survivor Non-Survivor |
7 | 338 | 58 | 223/115 | 28.3 ± 5.2 | 89 | 73 | 70/19 | 20.3 ± 5.4 |
|
Batsug et al. Turkey [21] |
R |
ICU Non-ICU |
8 | 145 | 43 | 81/64 | 47.5 ± 6.8 | 46 | 71 | 26/20 | 35.8 ± 7.8 |
|
Bi et al. China [22] |
R |
Severe Non-severe |
7 | 91 | 44 | 51/40 | 41.3 ± 4.0 | 22 | 54 | 13/9 | 38.8 ± 4.5 |
|
Bonetti et al. Italy [23] |
U |
Survivor Non-Survivor |
7 | 74 | 62 | 51/23 | 36.5 ± 4.2 | 70 | 78 | 45/25 | 34.1 ± 3.1 |
|
Cao et al. China [24] |
R |
Severe Non-severe |
8 | 53 | 44 | 22/31 | 36.8 ± 4.3 | 27 | 71 | 16/11 | 32.8 ± 6.5 |
|
Cheng et al. China [25] |
R |
No progression Poor prognoses* |
8 | 205 | 49 | 71/134 | 40.2 ± 5.9 | 251 | 60 | 140/111 | 37.9 ± 6.3 |
|
Cheng et al. China [26] |
R |
Survivor Non-Survivor |
8 | 53 | 54 | 29/24 | 39.2 ± 3.7 | 36 | 69 | 20/16 | 34.0 ± 5.2 |
|
de la Rica et al. Spain [27] |
R |
ICU Non-ICU |
7 | 27 | 66 | 18/9 | 39.2 ± 4.2 | 21 | 66 | 14/7 | 29.0 ± /5.2 |
|
Deng et al. China [28] |
R |
Severe Non-severe |
8 | 53 | 35 | 24/29 | 42.0 ± 3.2 | 12 | 33 | 12/0 | 40.9 ± 3.6 |
|
Feng et al. China [29] |
P |
Good outcome Poor outcome** |
8 | 94 | 63 | 58/36 | 35.6 ± 5.5 | 20 | 69 | 13/7 | 29.9 ± 4.4 |
|
Gan et al. China [30] |
R |
Survivor Non-Survivor |
7 | 56 | 62 | 30/26 | 34.9 ± 4.4 | 39 | 70 | 28/11 | 30.5 ± 4.5 |
|
Gao et al. China [31] |
R |
Survivor Non-Survivor |
8 | 175 | 70 | 79/96 | 36.2 ± 5.1 | 35 | 74 | 22/13 | 30.8 ± 6.1 |
|
Ghweil et al. Egypt [32] |
R |
Severe Non-severe |
7 | 36 | 56 | 28/8 | 29.0 ± 1.0 | 30 | 63 | 20/10 | 25.0 ± 2.0 |
|
Gong et al. China [33] |
R |
Severe Non-severe |
7 | 161 | 45 | 72/89 | 39.7 ± 4.3 | 28 | 64 | 16/12 | 34.2 ± 5.1 |
|
Guo et al. China [34] |
R |
Survivor Non-Survivor |
8 | 43 | 60 | 22/21 | 29.4 ± 1.4 | 31 | 68 | 21/10 | 26.5 ± 3.3 |
|
He et al. China [35] |
U |
Severe Non-severe |
7 | 32 | 42 | 15/17 | 41 ± 5.4 | 21 | 57 | 13/8 | 31.7 ± 4.0 |
|
Hirashima et al. Japan [36] |
R |
Severe Non-severe |
7 | 45 | 42 | 27/18 | 39.5 ± 4.5 | 16 | 63 | 13/3 | 31.0 ± 4.0 |
|
How et al. China [37] |
R |
Progression Improvement*** |
8 | 84 | 47 | 34/50 | 36.5 ± 5.2 | 17 | 72 | 10/7 | 32.3 ± 7.2 |
|
Hu et al. China [38] |
R |
Severe Non-severe |
8 | 19 | 43 | 11/8 | 41.6 ± 4.3 | 21 | 63 | 13/8 | 35.6 ± 5.1 |
|
Hu et al. China [39] |
R |
Severe Non-severe |
8 | 130 | 63 | 58/72 | 31.4 ± 6.2 | 52 | 64 | 42/10 | 27.2 ± 5.7 |
|
Hua et al. China [40] |
U |
Ventilation§ No ventilation |
7 | 204 | 67 | 108/96 | 32.7 ± 4.6 | 265 | 69 | 158/107 | 31.4 ± 6.1 |
|
Huang et al. China [41] |
P |
ICU Non-ICU |
8 | 28 | 49 | 19/9 | 33.8 ± 4.9 | 13 | 49 | 11/2 | 28.4 ± 3.8 |
|
Huang et al. China [42] |
R |
Survivor Non-Survivor |
7 | 283 | 53 | 149/134 | 37.6 ± 6.2 | 16 | 69 | 11/5 | 30.5 ± 4.0 |
|
Huang et al. China [43] |
R |
Critical or Dead Non-critical |
8 | 2,008 | 60 | 943/1,065 | 36.7 ± 5.3 | 615 | 66 | 380/235 | 32.0 ± 4.6 |
|
Hundt et al. USA [44] |
R |
Severe Non-severe |
8 | 1,175 | 63 | 574/601 | 34.0 ± 3.0 | 652 | 67 | 395/257 | 31.0 ± 2.7 |
|
Lee et al. Republic of Korea [45] |
R |
Severe Non-severe |
8 | 557 | 52 | 155/402 | 41.2 ± 3.9 | 137 | 71 | 57/80 | 35.3 ± 5.0 |
|
Lei et al. China [46] |
R |
Severe Non-severe |
8 | 50 | 62 | 22/28 | 35.5 ± 4.1 | 65 | 69 | 36/29 | 33.2 ± 4.9 |
|
Li et al. China [47] |
U |
Severe Non-severe |
7 | 41 | 53 | 20/21 | 40.0 ± 3.9 | 24 | 56 | 15/9 | 35.0 ± 5.9 |
|
Li et al. China [48] |
R |
ICU Non-ICU |
8 | 312 | 49 | 131/181 | 40.7 ± 3.7 | 211 | 62 | 119/92 | 36.3 ± 5.2 |
|
Li et al. China [49] |
R |
Severe Non-severe |
7 | 45 | 50 | 24/21 | 40.4 ± 3.5 | 89 | 64 | 51/38 | 36.0 ± 6.0 |
|
Lian et al. China [50] |
R |
Severe Non-severe |
8 | 140 | 66 | 62/78 | 38.4 ± 4.4 | 92 | 68 | 47/45 | 35.2 ± 5.6 |
|
Liang et al. China [51] |
R |
Severe Non-severe |
8 | 1,459 | 48 | 816/643 | 33.9 ± 8.9 | 131 | 66 | 88/43 | 32.6 ± 8.9 |
|
Liu et al. China [52] |
P |
Severe Non-severe |
7 | 44 | 41 | 21/23 | 44.0 ± 4.6 | 17 | 56 | 10/7 | 41.8 ± 6.8 |
|
Liu et al. China [53] |
R |
Survivor Non-Survivor |
7 | 302 | 64 | 148/154 | 35.8 ± 5.2 | 34 | 74 | 21/13 | 27.6 ± 3.2 |
|
Liu et al. China [54] |
R |
Severe Non-severe |
8 | 561 | 43 | 288/273 | 41.9 ± 5.7 | 64 | 60 | 41/23 | 38.4 ± 5.5 |
|
Liu et al. China [55] |
U |
Progression Improvement*** |
7 | 67 | 37 | 32/35 | 41.3 ± 4.6 | 11 | 66 | 7/4 | 36.6 ± 6.6 |
|
Ma et al. China [56] |
U |
Severe Non-severe |
8 | 429 | 42 | 199/230 | 40.7 ± 5.2 | 94 | 50 | 35/59 | 38.9 ± 6.6 |
|
Mo et al. China [57] |
R |
Refractory General |
8 | 70 | 46 | 31/39 | 39.0 ± 4.5 | 85 | 61 | 55/30 | 36.0 ± 6.0 |
|
Mori et al. Japan [58] |
R |
Severe Non-severe |
8 | 23 | 69 | 13/10 | 34.7 ± 10.9 | 22 | 58 | 21/1 | 25.0 ± 8.7 |
|
Pan et al. China [59] |
R |
Survivor Non-Survivor |
8 | 35 | 65 | 18/17 | 29.1 ± 4.2 | 89 | 69 | 67/22 | 28.1 ± 3.7 |
| Pourabdollah Toutkaboni et al. Iran [60] | R |
Survivor Non-Survivor |
7 | 481 | 55 | 301/180 | 34.7 ± 4.1 | 89 | 64 | 68/21 | 30.0 ± 4.0 |
|
Recinella et al. Italy [61] |
U |
Survivor Non-Survivor |
8 | 66 | 79 | 32/34 | 31.9 ± 5.7 | 43 | 86 | 22/21 | 27.0 ± 5.2 |
|
Sheng et al. China [62] |
R |
Survivor Non-Survivor |
7 | 144 | 66 | NR | 37.5 ± 5.2 | 88 | 75 | NR | 31.1 ± 3.3 |
|
Shi et al. China [63] |
R |
ICUorDeath Non-ICU |
7 | 51 | 58 | 27/24 | 36.2 ± 5.1 | 36 | 66 | 22/14 | 29.3 ± 5.1 |
|
Sun et al. China [64] |
U |
Severe Non-severe |
7 | 44 | 42 | 25/19 | 40.2 ± 3.9 | 19 | 59 | 11/8 | 33.5 ± 5.4 |
|
Tsibouris et al. Greece [65] |
R |
Survivor Non-Survivor |
7 | 45 | NR | NR | 33.0 ± 6.1 | 16 | NR | NR | 26.0 ± 5.3 |
|
Varim et al. Turkey [66] |
R |
Severe Non-severe |
7 | 85 | 62 | 48/37 | 33.3 ± 5.4 | 59 | 69 | 31/28 | 29.8 ± 4.9 |
|
Violi et al. Italy [67] |
U |
Survivor Non-Survivor |
8 | 255 | 66 | 148/107 | 33.9 ± 5.3 | 64 | 77 | 45/19 | 30.3 ± 5.1 |
|
Wang et al. China [68] |
U |
ICU Non-ICU |
8 | 39 | 54 | 27/12 | 37.6 ± 4.0 | 46 | 65 | 18/28 | 33.2 ± 5.4 |
|
Wang et al. China [69] |
R |
Severe Non-severe |
7 | 72 | 44 | 29/43 | 38.6 ± 2.3 | 71 | 65 | 44/27 | 32.0 ± 3.0 |
|
Wang et al. China [70] |
R |
ICU Non-ICU |
8 | 14 | 66 | 11/3 | 35.0 ± 5.9 | 14 | 71 | 10/4 | 30.5 ± 4.3 |
|
Wang et al. China [71] |
R |
Survivor Non-Survivor |
7 | 175 | 64 | 86/89 | 34.9 ± 4.7 | 24 | 70 | 16/8 | 31.6 ± 4.6 |
|
Wang et al. China [72] |
R |
Severe Non-severe |
7 | 79 | 41 | 38/41 | 42.1 ± 5.1 | 26 | 59 | 18/8 | 37.7 ± 5.9 |
|
Wang et al. China [73] |
R |
Severe Non-severe |
7 | 230 | 45 | 102/128 | 41.0 ± 4.4 | 45 | 61 | 26/19 | 34.3 ± 3.8 |
|
Wu et al. China [74] |
R |
ARDS Non-ARDS |
7 | 117 | 48 | 68/49 | 33.7 ± 4.0 | 84 | 59 | 60/24 | 30.3 ± 4.7 |
|
Xue et al. China [75] |
R |
Severe Non-severe |
7 | 56 | 61 | 30/26 | 34.8 ± 3.8 | 58 | 64 | 34/24 | 28.0 ± 7.3 |
|
Yao et al. China [76] |
R |
Survivor Non-Survivor |
8 | 96 | 48 | 36/60 | 39.3 ± 3.6 | 12 | 63 | 7/5 | 31.1 ± 4.7 |
|
Yu et al. China [77] |
U |
Severe Non-severe |
7 | 799 | 61 | 384/415 | 35.7 ± 4.7 | 864 | 66 | 454/410 | 34.5 ± 5.1 |
|
Zeng et al. China [78] |
R |
ICU Non-ICU |
7 | 406 | 43 | 206/200 | 40.3 ± 5.4 | 55 | 60 | 33/22 | 35.6 ± 5.4 |
|
Zhang et al. China [79] |
U |
Severe Non-severe |
7 | 56 | 43 | 24/32 | 37.5 ± 3.9 | 24 | 65 | 9/15 | 32.8 ± 5.4 |
|
Zhang et al. China [80] |
R |
Normal Abnormal§§ |
7 | 72 | 35 | 33/39 | 42.5 ± 4.7 | 573 | 47 | 295/278 | 41.0 ± 4.5 |
|
Zhang et al. China [81] |
R |
Severe Non-severe |
7 | 84 | 44 | 29/55 | 40.4 ± 3.2 | 31 | 65 | 20/11 | 34.4 ± 4.1 |
|
Zheng et al. China [82] |
U |
Severe Non-severe |
7 | 55 | 44 | 26/29 | 33.9 ± 9.4 | 13 | 61 | 10/3 | 32.5 ± 3.2 |
|
Zhou et al. China [83] |
R |
Severe Non-severe |
8 | 95 | 35 | 38/57 | 42.4 ± 4.5 | 28 | 40 | 17/11 | 40.4 ± 4.8 |
|
Zhou et al. China [84] |
R |
Survivor Non-Survivor |
8 | 137 | 52 | 56/81 | 33.5 ± 4.4 | 54 | 69 | 16/38 | 29.0 ± 3.6 |
|
Zhou et al. China [85] |
R |
Survivor Non-Survivor |
7 | 51 | 73 | 31/20 | 33.6 ± 4.9 | 67 | 71 | 22/45 | 33.1 ± 5.0 |
|
Zinellu et al. Italy [86] |
R |
Survivor Non-Survivor |
8 | 77 | 68 | 27/50 | 34.7 ± 5.3 | 28 | 80 | 8/20 | 32.3 ± 3.9 |
ARDS Acute respiratory distress syndrome; ICU intensive care unit; Non-severe Patients with mild or moderate disease; NOS Newcastle–Ottawa quality assessment scale for case–control studies; NR Not Reported; P prospective; R retrospective; Severe Patients with severe or critical disease; U undeclared.
*Poor prognosis refers to progression from moderate to severe grade, critical grade or death.
**Patients that were discharged, those with non-severe condition, and those not requiring mechanical ventilation were considered to have a good outcome. Patients requiring mechanical ventilation and those who died were considered to have a poor outcome.
***Progression group: clinically advanced types; patients admitted to ICU; death during hospitalization. Improvement group: clinical status remained unchanged or improved, and patients discharged from the hospital.
§Both invasive and non-invasive ventilation.
§§Normal or abnormal imaging findings.
Meta-analysis
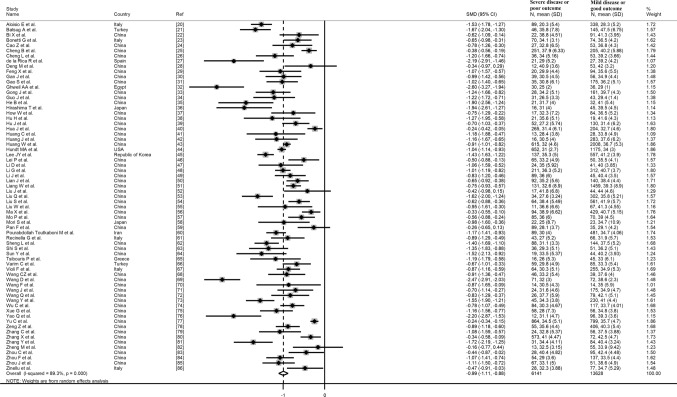
The overall standardized mean difference in serum albumin concentrations between COVID-19 patients with low versus high severity or good versus poor outcomes in the 67 studies is shown in Fig. 2. In all studies, patients with high disease severity or poor outcome had lower albumin concentrations compared to those with low severity or good outcome (mean difference range, − 0.16 to − 2.60) although the difference was not statistically significant in four studies [28, 52, 59, 82]. The pooled results confirmed that serum albumin concentrations were significantly lower in patients with high severity or poor outcome (SMD: − 0.99; 95% CI, − 1.11 to − 0.88, p < 0.001). Extreme heterogeneity between studies was observed (I2 = 89.3%; p < 0.001). Sensitivity analysis showed that the effect size was not modified when each study was in turn removed (effect size ranged between − 0.970 and − 1.007). Evidence of publication bias was provided by a funnel plot (Egger’s test, p = 0.004; Begg’s test, p = 0.081, Fig. 3). However, trim‐and‐fill analysis showed that no study was missing or should be added. To explore possible contributors to the between-study variance, we investigated the effects of age, gender, publication geographic area, outcome, the inflammation biomarkers white blood cell (WBC) count and C-reactive protein (CRP) and the liver function markers alanine aminotransferase (ALT) and aspartate aminotransferase (AST) on SMD by univariate meta-regression analysis. Both WBC (t = − 2.77, p = 0.008) and CRP (t = − 2.43, p = 0.019) were significantly related to the pooled SMD (Fig. 4). In addition, the pooled SMD value in Chinese studies (− 0.99, 95% CI − 1.05 to − 0.80, p < 0.001; I2 = 88.2%, p < 0.001) was lower than that observed in non-Chinese studies (− 1.22, 95% CI − 1.43 to − 1.01, p < 0.001; I2 = 84.5%, p < 0.001) and the difference was significant by meta-regression analysis (t = 2.09, p = 0.004). No statistically significant correlation was found between SMD and age (t = − 0.58, p = 0.56), gender (t = 0.46, p = 0.65), ALT (t = 0.34, p = 0.73) and AST (t = 0.40, p = 0.69) though a trend towards significance was observed with outcome (t = 1.72, p = 0.091). Multivariate meta-regression analysis, reported in Table 2, confirmed the significant association between effect size, WBC (t = − 2.10, p = 0.046) and CRP (t = − 2.28, p = 0.031) and also showed a significant relationship with age (t = − 2.13, p = 0.043) and publication geographic area (t = 2.16, p = 0.040).
Fig. 2.
Forest plot of SMD differences of serum albumin concentrations between COVID-19 patients with low/high severity and good/poor outcome
Fig. 3.
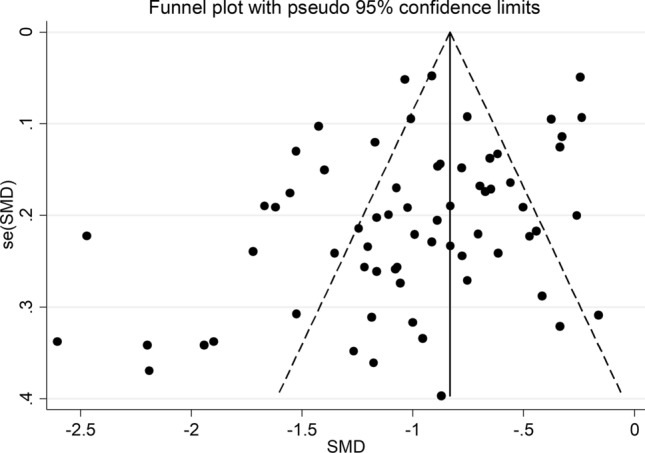
Funnel plot of studies examining albumin and severity and outcome in COVID-19
Fig. 4.
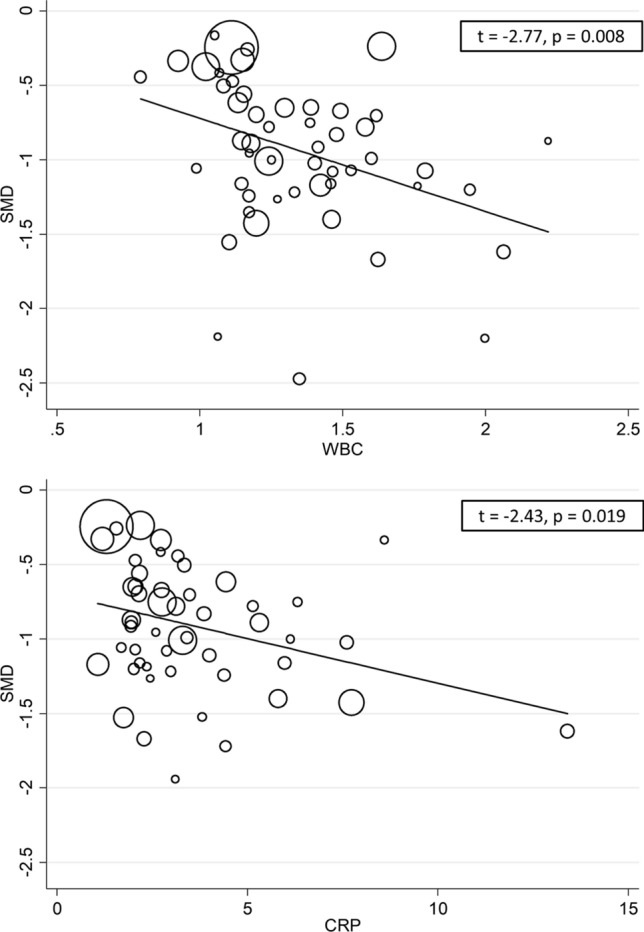
Univariate meta-regression analysis between WBC, CRP and effect size
Table 2.
Multivariate meta-regression analysis between effect size and possible contributors to heterogeneity
| t | p-value | |
|---|---|---|
| Age | − 2.13 | 0.043 |
| Gender | − 0.44 | 0.662 |
| Severity/outcome | 0.92 | 0.365 |
| Geographic area | 2.16 | 0.040 |
| ALT | 0.47 | 0.639 |
| AST | − 0.59 | 0.558 |
| WBC | − 2.10 | 0.046 |
| CRP | − 2.28 | 0.031 |
Discussion
The recently reported COVID-19 disease represents one of the worst pandemics in modern times. The disease started in China in December 2019 and spread rapidly through the globe [87]. This, the consequent lack of information regarding the pathophysiology and clinical progress, prevented the establishment and implementation of adequate public health responses. Several studies have described alterations in routine laboratory tests in patients affected by COVID-19, including a decrease in serum albumin concentrations [8–11]. However, no pooled analyses of the available evidence have been performed to accurately estimate the effect size of such reduction, to investigate its relationship with disease severity and outcomes and to evaluate which parameters may affect the effect size.
Our analysis demonstrated the presence of significantly lower serum albumin concentrations in COVID-19 patients with high disease severity or poor outcome when compared to those with low severity or good outcome. Albumin, a protein that exerts important homeostatic effects such as maintenance of the osmotic colloid pressure, intravascular transport of molecules, lipid metabolism, thrombosis and inflammation, is classically considered as a biomarker of malnutrition and poor health status [88, 89]. Hypoalbuminemia has been described as a negative prognostic factor in several diseases, [89–92]. Furthermore, low serum albumin concentrations have been shown to be associated with the severity of chronic inflammatory diseases, inflammatory bowel disease and diabetes mellitus [93], cirrhosis [94], as well as with the severity of surgical trauma [95], acute diseases [96] and sepsis [97]. In addition, in past SARS epidemics, hypoalbuminemia has been shown to be related with disease severity and increased hospital mortality [98, 99].
Extreme heterogeneity and a trend towards publication bias was observed. In multivariate meta-regression analysis, age, geographic area, WBC count and CRP were significantly associated with effect size. However, other factors not specifically investigated, such as nutritional status or assay preparation, might have also contributed to the observed heterogeneity. In particular, methodological issues with albumin determination might have influenced the heterogeneity observed between studies. It is well known that colorimetric methods for albumin determination, which are widely used in clinical institutions, are nonspecific and usually overestimate albumin concentrations when compared to the more specific and accurate immunoturbidimetric assays [100]. Unfortunately, as only few articles provided information regarding the assay used for albumin determination, we could not determine the impact of this factor on between-study variance by meta-regression analysis.
The mechanisms responsible for hypoalbuminemia in COVID‐19 have not been fully elucidated. Albumin is exclusively synthesized by the liver with a serum half‐life of approximately 21 days [101]. Notably, our analyses did not show any association between effect size and the liver function biomarkers ALT and AST, confirming previous observations that hypoalbuminemia in COVID-19 patients is not related to liver dysfunction [42]. Conversely, we found a relation between effect size and inflammation in accordance with previous studies by Huang et al. [42], which found that albumin concentrations were inversely correlated with WBC, neutrophil-to-lymphocyte ratio (NLR) and CRP, and by Huang et al. [43], that describe an inverse relationship between serum albumin and IL-6. As suggested by Huang et al. [42] hypoalbuminemia might be due to the presence of a systemic inflammatory state in COVID‐19. It is well known that inflammation may be responsible for the extravasation of serum albumin into the interstitial space due to an augmented capillary permeability, with an increased volume distribution of albumin [102]. However, it is also important to emphasize that serum albumin concentrations tend to decrease with advancing age in both sexes [103]. Therefore, the between-group differences in albumin concentrations may be in part explained by the higher disease severity and worse outcomes typically observed in older patients.
Although further research is required to investigate the relationship between albumin and COVID-19 disease outcomes, the identification of serum albumin as a marker of COVID-19 severity is biologically and clinically relevant. The determination of serum albumin concentrations, a relatively stable parameter that is strongly associated with key functional and health measures, using simple and relatively inexpensive analytical procedures, might provide rapid and useful information in regard to the overall homeostatic capacity of an individual. Consequently, the identification of relatively low serum albumin concentrations in hospitalized COVID-19 patients might assist with appropriate risk stratification and selection of suitable care pathways, even taking into consideration that age can be an important confounding factor.
In conclusion, our systematic review and meta-analysis showed that serum albumin concentrations in COVID-19 patients with high disease severity or poor outcomes are significantly lower when compared to those with milder disease. Age, geographical area and inflammation status are relevant contributors to the between-study variance. Further studies are required to investigate if albumin assessment may effectively help clinicians to early identify patients at high risk of poor outcome and if this parameter may be helpful also to successfully evaluate, at early stage, the response to treatment.
Funding
Open Access funding provided by Università degli Studi di Sassari. This research was supported by grants from the Sardinian Fondo di Sviluppo e Coesione (FSC) 2014–2020, Patto per lo Sviluppo della Regione Sardegna, L.R.7–2017 (RASSR82005) and by grants from Fondo di Ateneo per la Ricerca 2020.
Compliance with ethical standards
Conflicts of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 2.Kuiken T, Fouchier RA, Schutten M, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Groot RJ, Baker SC, Baric RS. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 5.Adhikari SP, Meng S, Wu YJ, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty. 2020;9:29. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rauf A, Abu-Izneid T, Olatunde A, et al. COVID-19 pandemic: epidemiology, etiology, conventional and non-conventional Therapies. Int J Environ Res Public Health. 2020;17:E8155. doi: 10.3390/ijerph17218155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Zheng Y, Gou X, et al. Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. Int J Infect Dis. 2020;94:91–95. doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lippi G, Plebani M. Laboratory abnormalities in patients with COVID-2019 infection. Clin Chem Lab Med. 2020;58:1131–1134. doi: 10.1515/cclm-2020-0198. [DOI] [PubMed] [Google Scholar]
- 9.Paliogiannis P, Zinellu A, Scano V, et al. Laboratory test alterations in patients with COVID-19 and non COVID-19 interstitial pneumonia: a preliminary report. J Infect Dev Ctries. 2020;14:685–690. doi: 10.3855/jidc.12879. [DOI] [PubMed] [Google Scholar]
- 10.Paliogiannis P, Mangoni AA, Dettori P, Nasrallah GK, Pintus G, Zinellu A. D-dimer concentrations and COVID-19 severity: a systematic review and meta-analysis. Front Public Health. 2020;8:432. doi: 10.3389/fpubh.2020.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paliogiannis P, Zinellu A. Bilirubin levels in patients with mild and severe Covid-19: a pooled analysis. Liver Int. 2020;40:1787–1788. doi: 10.1111/liv.14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Shi L, Wang Y, Duan G, Yang H. Albumin and total bilirubin for severity and mortality in coronavirus disease 2019 patients. J Clin Lab Anal. 2020;34:e23412. doi: 10.1002/jcla.23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parohan M, Yaghoubi S, Seraji A. Liver injury is associated with severe coronavirus disease 2019 (COVID-19) infection: a systematic review and meta-analysis of retrospective studies. Hepatol Res. 2020;50:924–935. doi: 10.1111/hepr.13510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wells G, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of non-randomised studies in meta-analyses. 2013. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
- 15.Wan X, Wang W, Liu J, Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol. 2014;14:135. doi: 10.1186/1471-2288-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hozo SP, Djulbegovic B, Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res Methodol. 2005;5:13. doi: 10.1186/1471-2288-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 18.Sterne JA, Egger M. Funnel plots for detecting bias in meta-analysis: guidelines on choice of axis. J Clin Epidemiol. 2001;54:1046–1055. doi: 10.1016/s0895-4356(01)00377-8. [DOI] [PubMed] [Google Scholar]
- 19.Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 20.Aloisio E, Chibireva M, Serafini L, et al. A comprehensive appraisal of laboratory biochemistry tests as major predictors of COVID-19 severity. Arch Pathol Lab Med. 2020 doi: 10.5858/arpa.2020-0389-SA. [DOI] [PubMed] [Google Scholar]
- 21.Bastug A, Bodur H, Erdogan S, et al. Clinical and laboratory features of COVID-19: Predictors of severe prognosis. Int Immunopharmacol. 2020;88:106950. doi: 10.1016/j.intimp.2020.106950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bi X, Su Z, Yan H, et al. Prediction of severe illness due to COVID-19 based on an analysis of initial fibrinogen to albumin ratio and platelet count. Platelets. 2020;31:674–679. doi: 10.1080/09537104.2020.1760230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonetti G, Manelli F, Patroni A, et al. Laboratory predictors of death from coronavirus disease 2019 (COVID-19) in the area of Valcamonica, Italy. Clin Chem Lab Med. 2020;58:1100–1105. doi: 10.1515/cclm-2020-0459. [DOI] [PubMed] [Google Scholar]
- 24.Cao Z, Li T, Liang L, et al. Clinical characteristics of coronavirus disease 2019 patients in Beijing, China. PLoS One. 2020; 15:e0234764. doi: 10.1371/journal.pone.0234764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng B, Hu J, Zuo X, et al. Predictors of progression from moderate to severe coronavirus disease 2019: a retrospective cohort. Clin Microbiol Infect. 2020;26:1400–1405. doi: 10.1016/j.cmi.2020.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng L, Yang JZ, Bai WH, et al. Prognostic value of serum amyloid A in patients with COVID-19. Infection. 2020;48:715–722. doi: 10.1007/s15010-020-01468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de la Rica R, Borges M, Aranda M, et al. Low albumin levels are associated with poorer outcomes in a case series of COVID-19 patients in spain: a retrospective cohort study. Microorganisms. 2020;8:1106. doi: 10.3390/microorganisms8081106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng M, Qi Y, Deng L, et al. Obesity as a potential predictor of disease severity in young COVID-19 patients: a retrospective study. Obesity. 2020;28:1815–1825. doi: 10.1002/oby.22943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng X, Li P, Ma L, et al. Clinical characteristics and short-term outcomes of severe patients with COVID-19 in Wuhan, China. Front Med. 2020;7:491. doi: 10.3389/fmed.2020.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gan J, Li J, Li S, Yang C. Leucocyte subsets effectively predict the clinical outcome of patients with COVID-19 pneumonia: a retrospective case-control study. Front Public Health. 2020;8:299. doi: 10.3389/fpubh.2020.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao S, Jiang F, Jin W, et al. Risk factors influencing the prognosis of elderly patients infected with COVID-19: a clinical retrospective study in Wuhan, China. Aging. 2020; 12:12504–12516. doi: 10.18632/aging.103631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghweil AA, Hassan MH, Khodeary A, et al. Characteristics, outcomes and indicators of severity for COVID-19 among sample of ESNA quarantine hospital's patients, egypt: a retrospective study. Infect Drug Resist. 2020;13:2375–2383. doi: 10.2147/IDR.S263489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong J, Ou J, Qiu X, et al. A tool for early prediction of severe coronavirus disease 2019 (COVID-19): a multicenter study using the risk nomogram in wuhan and guangdong, China. Clin Infect Dis. 2020;71:833–840. doi: 10.1093/cid/ciaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo J, Zhou B, Zhu M, et al. CURB-65 may serve as a useful prognostic marker in COVID-19 patients within Wuhan, China: a retrospective cohort study. Epidemiol Infect. 2020;148:e241. doi: 10.1017/S0950268820002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He B, Wang J, Wang Y, et al. The metabolic changes and immune profiles in patients With COVID-19. Front Immunol. 2020;11:2075. doi: 10.3389/fimmu.2020.02075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirashima T, Arai T, Kitajima H, et al. Factors significantly associated with COVID-19 severity in symptomatic patients: a retrospective single-center study. J Infect Chemother. 2020;27(1):76–82. doi: 10.1016/j.jiac.2020.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou W, Zhang W, Jin R, Liang L, Xu B, Hu Z. Risk factors for disease progression in hospitalized patients with COVID-19: a retrospective cohort study. Infect Dis. 2020;52:498–505. doi: 10.1080/23744235.2020.1759817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu H, Du H, Li J, et al. Early prediction and identification for severe patients during the pandemic of COVID-19: a severe COVID-19 risk model constructed by multivariate logistic regression analysis. J Glob Health. 2020;10:020510. doi: 10.7189/jogh.10.020510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu J, Zhou J, Dong F, et al. Combination of serum lactate dehydrogenase and sex is predictive of severe disease in patients with COVID-19. Medicine. 2020;99:e22774. doi: 10.1097/MD.0000000000022774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hua J, Qian C, Luo Z, Li Q, Wang F. Invasive mechanical ventilation in COVID-19 patient management: the experience with 469 patients in Wuhan. Crit Care. 2020;24:348. doi: 10.1186/s13054-020-03044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J, Cheng A, Kumar R, et al. Hypoalbuminemia predicts the outcome of COVID-19 independent of age and co-morbidity. J Med Virol. 2020 doi: 10.1002/jmv.26003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang W, Li C, Wang Z, et al. Decreased serum albumin level indicates poor prognosis of COVID-19 patients: hepatic injury analysis from 2,623 hospitalized cases. Sci China Life Sci. 2020;63:1678–1687. doi: 10.1007/s11427-020-1733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hundt MA, Deng Y, Ciarleglio MM, Nathanson MH, Lim JK. Abnormal liver tests in COVID-19: a retrospective observational cohort study of 1827 patients in a major U.S. hospital network. Hepatology. 2020 doi: 10.1002/hep.31487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JY, Hong SW, Hyun M, et al. Epidemiological and clinical characteristics of coronavirus disease 2019 in Daegu, South Korea. Int J Infect Dis. 2020;98:462–466. doi: 10.1016/j.ijid.2020.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lei P, Zhang L, Han P, et al. Liver injury in patients with COVID-19: clinical profiles, CT findings, the correlation of the severity with liver injury. Hepatol Int. 2020;14:733–742. doi: 10.1007/s12072-020-10087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li D, Liu C, Liu J, Hu J, Yang Y, Zhou Y. Analysis of risk factors for 24 patients With COVID-19 developing from moderate to severe condition. Front Cell Infect Microbiol. 2020;10:548582. doi: 10.3389/fcimb.2020.548582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li G, Zhou CL, Ba YM. Nutritional risk and therapy for severe and critical COVID-19 patients: a multicenter retrospective observational study. Clin Nutr. 2020;S0261–5614(20):30512–30514. doi: 10.1016/j.clnu.2020.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Li M, Zheng S, et al. Plasma albumin levels predict risk for nonsurvivors in critically ill patients with COVID-19. Biomark Med. 2020;14:827–837. doi: 10.2217/bmm-2020-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lian J, Jin C, Hao S, et al. High neutrophil-to-lymphocyte ratio associated with progression to critical illness in older patients with COVID-19: a multicenter retrospective study. Aging. 2020;12:13849–13859. doi: 10.18632/aging.103582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang W, Liang H, Ou L, et al. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med. 2020;180:1081–1089. doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J, Liu Y, Xiang P, et al. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J Trans Med. 2020;18:206. doi: 10.1186/s12967-020-02374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Q, Song NC, Zheng ZK, Li JS, Li SK. Laboratory findings and a combined multifactorial approach to predict death in critically ill patients with COVID-19: a retrospective study. Epidemiol Infect. 2020;148:e129. doi: 10.1017/S0950268820001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu S, Luo H, Wang Y, et al. Clinical characteristics and risk factors of patients with severe COVID-19 in Jiangsu province, China: a retrospective multicentre cohort study. BMC Infect Dis. 2020;20:584. doi: 10.1186/s12879-020-05314-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu W, Tao ZW, Wang L, et al. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin Med J. 2020;133:1032–1038. doi: 10.1097/CM9.0000000000000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma X, Li A, Jiao M, et al. Characteristic of 523 COVID-19 in henan province and a death prediction model. Front Public Health. 2020;8:475. doi: 10.3389/fpubh.2020.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mo P, Xing Y, Xiao Y, et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mori S, Ai T, Otomo Y. Characteristics, laboratories, and prognosis of severe COVID-19 in the Tokyo metropolitan area: a retrospective case series. PLoS One. 2020;15:e0239644. doi: 10.1371/journal.pone.0239644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan F, Yang L, Li Y, et al. Factors associated with death outcome in patients with severe coronavirus disease-19 (COVID-19): a case-control study. Int J Med Sci. 2020;17:1281–1292. doi: 10.7150/ijms.46614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pourabdollah Toutkaboni M, Askari E, et al. Demographics, laboratory parameters and outcomes of 1061 patients with coronavirus disease 2019: a report from Tehran, Iran. New Microbes New Infect. 2020;38:100777. doi: 10.1016/j.nmni.2020.100777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Recinella G, Marasco G, Serafini G, et al. Prognostic role of nutritional status in elderly patients hospitalized for COVID-19: a monocentric study. Aging Clin Exp Res. 2020;8:1–7. doi: 10.1007/s40520-020-01727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheng L, Wang X, Tang N, Meng F, Huang L, Li D. Clinical characteristics of moderate and severe cases with COVID-19 in Wuhan, China: a retrospective study. Clin Exp Med. 2020:1–5. [DOI] [PMC free article] [PubMed]
- 63.Shi S, Liu X, Xiao J, et al. Prediction of adverse clinical outcomes in patients with coronavirus disease 2019. J Clin Lab Anal. 2020:e23598. [DOI] [PMC free article] [PubMed]
- 64.Sun Y, Dong Y, Wang L, et al. Characteristics and prognostic factors of disease severity in patients with COVID-19: the Beijing experience. J Autoimmun. 2020;112:102473. doi: 10.1016/j.jaut.2020.102473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsibouris P, Ekmektzoglou K, Agorogianni A, et al. Gastrointestinal involvement in COVID-19 patients: a retrospective study from a Greek COVID-19 referral hospital. Ann Gastroenterol. 2020;33:465–472. doi: 10.20524/aog.2020.0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Varim C, Yaylaci S, Demirci T, et al. Neutrophil count to albumin ratio as a new predictor of mortality in patients with COVID-19 ınfection. Rev Assoc Med Bras. 2020;66:77–81. doi: 10.1590/1806-9282.66.S2.77. [DOI] [PubMed] [Google Scholar]
- 67.Violi F, Cangemi R, Romiti GF, et al. Is albumin predictor of mortality in COVID-19? Antioxid Redox Signal. 2020 doi: 10.1089/ars.2020.8142. [DOI] [PubMed] [Google Scholar]
- 68.Wang CZ, Hu SL, Wang L, et al. Early risk factors of the exacerbation of coronavirus disease 2019 pneumonia. J Med Virol. 2020;92:2593–2599. doi: 10.1002/jmv.26071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang D, Li R, Wang J, et al. Correlation analysis between disease severity and clinical and biochemical characteristics of 143 cases of COVID-19 in Wuhan, China: a descriptive study. BMC Infect Dis. 2020;20:519. doi: 10.1186/s12879-020-05242-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang F, Yang Y, Dong K, et al. Clinical characteristics of 28 patients with diabetes and covid-19 in wuhan, China. Endocr Pract. 2020;26:668–674. doi: 10.4158/EP-2020-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang J, Zhang H, Qiao R, et al. Thrombo-inflammatory features predicting mortality in patients with COVID-19: The FAD-85 score. J Int Med Res. 2020;48:300060520955037. doi: 10.1177/0300060520955037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Q, Zhao H, Liu LG, et al. Pattern of liver injury in adult patients with COVID-19: a retrospective analysis of 105 patients. Mil Med Res. 2020;7:28. doi: 10.1186/s40779-020-00256-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, Liao B, Guo Y, et al. Clinical characteristics of patients infected with the novel 2019 Coronavirus (SARS-Cov-2) in Guangzhou, China. Open Forum Infect Dis. 2020;7:ofaa187. doi: 10.1093/ofid/ofaa187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu C, Chen X, Cai Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xue G, Gan X, Wu Z, et al. Novel serological biomarkers for inflammation in predicting disease severity in patients with COVID-19. Int Immunopharmacol. 2020;89:107065. doi: 10.1016/j.intimp.2020.107065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yao Q, Wang P, Wang X, et al. A retrospective study of risk factors for severe acute respiratory syndrome coronavirus 2 infections in hospitalized adult patients. Pol Arch Intern Med. 2020;130(5):390–399. doi: 10.20452/pamw. [DOI] [PubMed] [Google Scholar]
- 77.Yu C, Lei Q, Li W, Wang X, Li W, Liu W. Epidemiological and clinical characteristics of 1663 hospitalized patients infected with COVID-19 in Wuhan, China: a single-center experience. J Infect Public Health. 2020;13:1202–1209. doi: 10.1016/j.jiph.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zeng Z, Ma Y, Zeng H. Simple nomogram based on initial laboratory data for predicting the probability of ICU transfer of COVID-19 patients: multicenter retrospective study. J Med Virol. 2020 doi: 10.1002/jmv.26244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang C, Qin L, Li K, et al. A novel scoring system for prediction of disease severity in COVID-19. Front Cell Infect Microbiol. 2020;10:318. doi: 10.3389/fcimb.2020.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang X, Cai H, Hu J, et al. Epidemiological, clinical characteristics of cases of SARS-CoV-2 infection with abnormal imaging findings. Int J Infect Dis. 2020;94:81–87. doi: 10.1016/j.ijid.2020.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, Zheng L, Liu L, Zhao M, Xiao J, Zhao Q. Liver impairment in COVID-19 patients: a retrospective analysis of 115 cases from a single centre in Wuhan city, China. Liver Int. 2020;40:2095–2103. doi: 10.1111/liv.14455. [DOI] [PubMed] [Google Scholar]
- 82.Zheng M, Gao Y, Wang G, et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou C, Huang Z, Tan W, et al. Predictive factors of severe coronavirus disease 2019 in previously healthy young adults: a single-center, retrospective study. Respir Res. 2020;21:157. doi: 10.1186/s12931-020-01412-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou J, Huang L, Chen J, et al. Clinical features predicting mortality risk in older patients with COVID-19. Curr Med Res Opin. 2020;36:1753–1759. doi: 10.1080/03007995.2020.1825365. [DOI] [PubMed] [Google Scholar]
- 86.Zinellu A, Arru F, De Vito A, et al. The De Ritis ratio as prognostic biomarker of in-hospital mortality in COVID-19 patients. Eur J Clin Invest. 2020 doi: 10.1111/eci.13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anderson CF, Wochos DN. The utility of serum albumin values in the nutritional assessment of hospitalized patients. Mayo Clin Proc. 1982;57:181–184. [PubMed] [Google Scholar]
- 89.Nazha B, Moussaly E, Zaarour M, Weerasinghe C, Azab B. Hypoalbuminemia in colorectal cancer prognosis: nutritional marker or inflammatory surrogate? World J Gastrointest Surg. 2015;7:370–377. doi: 10.4240/wjgs.v7.i12.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arques S. Human serum albumin in cardiovascular diseases. Eur J Int Med. 2018;52:8–12. doi: 10.1016/j.ejim.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 91.Ye J, Liao B, Jiang X, et al. Prognosis value of platelet counts, albumin and neutrophil-lymphocyte ratio of locoregional recurrence in patients with operable head and neck squamous cell carcinoma. Cancer Manage Res. 2020;12:731–741. doi: 10.2147/CMAR.S234618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li N, Qiao H, Guo JF, et al. Preoperative hypoalbuminemia was associated with acute kidney injury in high-risk patients following non-cardiac surgery: a retrospective cohort study. BMC Anesthesiol. 2019;19:171. doi: 10.1186/s12871-019-0842-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Din H, Anderson AJ, Ramos Rivers C, et al. Disease characteristics and severity in patients with inflammatory bowel disease with coexistent diabetes mellitus. Inflamm Bowel Dis. 2020;26:1436–1442. doi: 10.1093/ibd/izz305. [DOI] [PubMed] [Google Scholar]
- 94.Spinella R, Sawhney R, Jalan R. Albumin in chronic liver disease: structure, functions and therapeutic implications. Hepatol Int. 2016;10:124–132. doi: 10.1007/s12072-015-9665-6. [DOI] [PubMed] [Google Scholar]
- 95.Hübner M, Mantziari S, Demartines N, Pralong F, Coti-Bertrand P, Schäfer M. Postoperative albumin drop is a marker for surgical stress and a predictor for clinical outcome: a pilot study. Gastroenterol Res Pract. 2016;2016:8743187. doi: 10.1155/2016/8743187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sahin A, Turkoglu S, Tunc N, et al. Is ischemia-modified albumin a reliable tool for the assessment of acute pancreatitis? Ther Clin Risk Manage. 2018;14:627–635. doi: 10.2147/TCRM.S162690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Godinez-Vidal AR, Correa-Montoya A, Enríquez-Santos D, Pérez-Escobedo SU, López-Romero SC, Gracida-Mancilla N. Is albumin a predictor of severity and mortality in patients with abdominal sepsis? Cir Cir. 2019;87:485–489. doi: 10.24875/CIRU.180003903. [DOI] [PubMed] [Google Scholar]
- 98.Chan JC, Tsui EL, Wong VC. Hospital Authority SARS Collaborative Group. Prognostication in severe acute respiratory syndrome: a retrospective time-course analysis of 1312 laboratory-confirmed patients in Hong Kong. Respirology. 2007;12:531–42. doi: 10.1111/j.1440-1843.2007.01102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leong HN, Earnest A, Lim HH, et al. SARS in Singapore–predictors of disease severity. Ann Acad Med Singap. 2006;35:326–331. [PubMed] [Google Scholar]
- 100.Infusino I, Panteghini M. Serum albumin: accuracy and clinical use. Clin Chim Acta. 2013;419:15–18. doi: 10.1016/j.cca.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 101.Rothschild MA, Oratz M, Schreiber SSJH. Serum albumin. Hepatology. 1988;8:385–401. doi: 10.1002/hep.1840080234. [DOI] [PubMed] [Google Scholar]
- 102.Soeters PB, Wolfe RR, Shenkin A. Hypoalbuminemia: pathogenesis and clinical significance. JPEN J Parenter Enter Nutr. 2019;43:181–193. doi: 10.1002/jpen.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Veering BT, Burm AG, Souverijn JH, Serree JM, Spierdijk J. The effect of age on serum concentrations of albumin and alpha 1-acid glycoprotein. Br J Clin Pharmacol. 1990;29:201–206. doi: 10.1111/j.1365-2125.1990.tb03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]