Abstract
Ectopic expression of transcription factor combinations has been recently demonstrated to reprogram differentiated somatic cells towards the dendritic cell (DC) lineage without reversion to a multipotent state. DCs have the ability to induce potent and long-lasting adaptive immune responses. In particular, conventional type 1 DCs (cDC1s) excel on antigen cross-presentation, a critical step for inducing CD8+ T cell cytotoxic responses. The rarity of naturally occurring cDC1s and lack of in vitro methodologies for the generation of pure cDC1 populations strongly hinders the study of cDC1 lineage specification and function. Here, we describe a protocol for the generation of induced DCs (iDCs) by lentiviral-mediated expression of the transcription factors PU.1, IRF8 and BATF3 in mouse embryonic fibroblasts. iDCs acquire DC morphology, cDC1 phenotype and transcriptional signatures within 9 days. iDCs generated with this protocol acquire functional ability to respond to inflammatory stimuli, engulf dead cells, process and cross-present antigens to CD8+ T cells. DC reprogramming provides a simple and tractable system to generate high numbers of cDC1-like cells for high content screening, opening new avenues to better understand cDC1 specification and function. In the future, faithful induction of cDC1 fate in fibroblasts may lead to the generation of patient-specific DCs for vaccination.
Keywords: Cell reprogramming, Transcription factor, Cell fate conversion, Dendritic cell, Antigen presenting cell, Immunity, Mouse embryonic fibroblast, Immunotherapy
Background
Dendritic cells (DCs) are professional antigen presenting cells specialized in the recognition, processing and presentation of antigens to T cells, playing a pivotal role in the induction of adaptive immune responses and immunological memory ( Merad et al., 2013 ). DCs can be divided into 4 main subsets: plasmacytoid DCs (pDC), producers of large amounts of type 1 interferons, monocyte-derived DCs, derived from circulating monocytes, and conventional DCs (cDC), a functionally heterogeneous subset further subdivided in conventional DCs type 1 (cDC1) and type 2 (cDC2). While cDC1s excel on priming cytotoxic CD8+ T cell responses by cross-presenting exogenous antigens on major histocompatibility complex (MHC) class I, cDC2s are specialized in the presentation of extracellular antigens to CD4+ T cells via MHC class II. This functional specialization results from stage-specific interplay between different transcription factors (TFs) that specify DC subsets. For instance, it has been shown that conditional deletion of E26 transformation-specific (ETS) family TF PU.1 in hematopoietic progenitors impacts the specification of the whole DC lineage ( Carotta et al., 2010 ). Interferon regulatory factor 8 (IRF8) knock-out mice lack pDC and cDC1 subsets, and basic leucine zipper ATF-Like transcription factor 3 (BATF3) deficient mice have impaired cDC1 specification ( Schiavoni et al., 2002 ; Hildner et al., 2008 ; Carotta et al., 2010 ).
Growing evidence supports the crucial role of cDC1s in initiating immune responses in the context of cancer (Bottcher and Reis, 2018), and their in vivo rarity has propelled attempts to generate cDC1 in vitro (Perez and De Palma, 2019; Wculek et al., 2019 ). Several strategies have been explored to generate bona fide DCs from blood monocytes or bone marrow (BM) precursors in the presence of different hematopoietic cytokines ( Inaba et al., 1992 ; Lu et al., 1995 ; Lutz et al., 1999 ; Son et al., 2002 ; Naik et al., 2005 ; Poulin et al., 2010 ; Proietto et al., 2012 ; Balan et al., 2014 ; Mayer et al., 2014 ; Lee et al., 2015 ; Balan et al., 2018 ; Kirkling et al., 2018 ). However, limiting numbers of BM progenitors, together with the generation of mixed populations of DC subsets with conflicting functions, strongly limits the use of these approaches to elucidate DC development and harness cDC1s for therapy (Yona and Mildner, 2018).
Cell reprogramming offers an exciting opportunity to overcome these challenges. Through enforced expression of TFs, it is possible to reprogram somatic cells into induced pluripotent stem cells (iPSCs) (Takahashi and Yamanaka, 2006; Takahashi et al., 2007 ). Protocols to differentiate DCs from iPSCs have been described ( Choi et al., 2009 ; Senju et al., 2009 and 2011; Horton et al., 2019 ). However, these rely on complex and long culture conditions that give rise to mixed populations of distinct DC subsets. Alternatively, somatic cells can be reprogrammed directly into other specialized cell types without transiting through pluripotency or multipotency. Mouse and human fibroblasts were directly reprogrammed into several cell types, such as neurons, cardiomyocytes and hematopoietic stem and progenitor cells, using TFs specifying target-cell identity ( Pereira et al., 2012 and 2013, Xu et al., 2015 ).
Here, we describe a direct cell reprogramming approach to generate cDC1-like cells (induced DCs, or iDCs) by enforced expression of the TFs PU.1, IRF8 and BATF3 in mouse embryonic fibroblasts (MEFs) ( Rosa et al., 2018 ). MEF-derived iDCs acquire DC morphology, cDC1 phenotype and transcriptional program characteristic of natural cDC1s. Functionally, iDCs acquire the ability to respond to toll-like receptor 3 (TLR3) and TLR4 stimuli, engulf protein and dead cells and, importantly, cross-present antigens to CD8+ T cells. Thus, the protocol described here enables the fast generation (within 9 days) of cDC1-like cells in vitro using a controlled and adaptable direct reprogramming approach suitable for screening studies aiming to better understand cDC1 developmental specification and functional maturation. This approach, coupled with pharmacological inhibition, gene knock-out or knock-down, will enable the discovery of regulators of DC specification acting individually or in combination ( Pires et al., 2019 ). Employing high-content screening methodologies based on recent CRISPR-Cas9 technologies in MEFs paves the way for defining novel critical regulators of cDC1 fate and how cross-presentation ability is established de novo in multiple cell types. In the future, DC reprogramming will serve as a platform to develop immunotherapies for cancer and infectious disease.
Materials and Reagents
100 mm tissue culture plates (Corning, catalog number: 430167)
150 mm tissue culture plates (Corning, catalog number: 430599)
1.5 ml microtubes (Sarstedt, catalog number: 72.690.001)
15 ml centrifuge tubes (Sarstedt, catalog number: 62.547.205)
50 ml centrifuge tubes (Sarstedt, catalog number: 62.547.205)
Polypropylene round-bottom FACS tubes (Falcon, catalog number: 352063)
150 ml bottle top vacuum filters, 0.45 μm low-protein binding (Corning, catalog number: 430625)
Amicon ultra-15 centrifugal filter units (Merck Millipore, catalog number: UFC903096)
Wild type mice (C57BL/6) or double homozygous Clec9aCre/Cre RosatdTomato/tdTomato (Clec9a-tdT) mice ( Rosa et al., 2018 )
Human embryonic kidney (HEK) 293T cell line (ATCC, catalog number: CRL-3216)
Sterile 40 µm cell strainers (Corning, catalog number: 431750)
Gelatin from porcine skin type A (Sigma-Aldrich, catalog number: G1890)
Polyethylenimine (PEI) transfection reagent (Sigma-Aldrich, catalog number: 408727)
Hexadimethrine bromide (Polybrene) (Sigma-Aldrich, catalog number: H9268)
Doxycycline hyclate (DOX) (Sigma-Aldrich, catalog number: D9891)
Trypsin-EDTA (0.25%), phenol red (Thermo Fisher Scientific, catalog number: 25200-056)
TrypLE Express (1x) no phenol red (Thermo Fisher Scientific, catalog number: 12604-021)
HyClone Phosphate Buffered Saline solution (PBS) (GE Healthcare, catalog number: SH30256.01)
HyClone high-glucose Dulbecco's Modified Eagles Medium (DMEM) (GE Healthcare, catalog number: SH30243.01)
Opti-MEM reduced serum medium (Thermo Fisher Scientific, catalog number: 31985070)
HyClone 10,000 U/ml Penicillin 10,000 μg/ml Streptomycin Solution (Pen/Strep) (GE Healthcare, catalog number: SV30010)
HyClone Fetal Bovine Serum (FBS) (GE Healthcare, catalog number: SV30160.03)
Rat serum (GeneTex, catalog number: GTX73226)
Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418)
Plasmid pMD2.G (Addgene, catalog number: 12259)
Plasmid psPAX2 (Addgene, catalog number: 12260)
Plasmid FUW-M2rtTA (Addgene, catalog number: 20342)
Plasmid pFUW-TetO-PIB containing a tri-cistronic cassette encoding PU.1, IRF8 and BATF3 (PIB) reprogramming factors ( Rosa et al., 2018 ). Cell Reprogramming in Hematopoiesis and Immunity Lab, Division of Molecular Medicine and Gene Therapy, Lund University, Sweden.
4’,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, catalog number: D9542)
Hoechst staining solution (Sigma-Aldrich, catalog number: H6024)
APC conjugated anti-mouse CD45 monoclonal antibody (Biolegend, catalog number: 103112)
PE-Cy7 conjugated anti-mouse MHC-II monoclonal antibody (Biolegend, catalog number: 107630)
DMEM Complete medium (see Recipes)
Dissociation solution (see Recipes)
0.1% Gelatin solution (see Recipes)
Wash solution (see Recipes)
Staining solution (see Recipes)
Freezing solution (see Recipes)
Equipment
Sterile dissecting scissors (Sigma-Aldrich, catalog number: S3146)
Sterile stainless steel forceps (Sigma-Aldrich, catalog number: Z168777)
Freezing container Nalgene Mr. Frosty (Sigma-Aldrich, catalog number: C1562)
Hemacytometer cell counting chamber (Marienfeld Superior, catalog number: 13444890)
4-16K Refrigerated centrifuge (Sigma)
FACSCanto II flow cytometry analyzer (8-color, blue/red/violet) (BD Biosciences)
BD FACSAria III cell sorter (BD Biosciences)
Heracell 150i CO2 incubator (Thermo Scientific)
ScanLaf Mars class 2 laminar flow hood (LaboGene)
IX70 inverted tissue culture microscope (Olympus)
Celldiscoverer 7 inverted microscope (Zeiss)
Software
GraphPad PRISM 6, https://www.graphpad.com/
FlowJo V10, https://www.flowjo.com/
ZEN 3.1 (blue edition), https://www.zeiss.com/
Adobe Photoshop CS6, https://www.adobe.com/se
Procedure
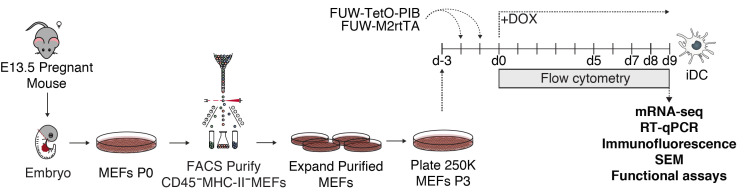
A schematic representation of the direct reprogramming protocol described herein can be found in Figure 1.
Figure 1. Schematic representation of the protocol for direct reprogramming of mouse embryonic fibroblasts to dendritic cells.
Mouse embryonic fibroblasts (MEFs) were isolated from C57BL/6 wild type 13.5 days-old embryos. Embryos were dissected to remove the head (grey), liver and heart (red) and dissociated to a single-cell suspension. MEFs were plated and allowed to grow to confluency (Passage 0, P0). Cells were dissociated and purified by fluorescence-activated cell sorting (FACS) to remove contaminant hematopoietic cells (CD45+ or MHC-II+) and expanded until passage 3 to 4. If a reporter was used such as the double homozygous Clec9aCre/Cre RosatdTomato/tdTomato, tdTomato+ cells were removed as well. 250,000 MEFs were plated per 100 mm gelatin-coated Petri dish and transduced twice with lentiviral particles encoding PU.1, IRF8 and BATF3 (PIB) reprogramming factors (FUW-TetO-PIB), and M2rtTA (FUW-M2rtTA). Doxycycline (DOX) was added to the culture media (Day 0, d0) to induce expression of PU.1, IRF8 and BATF3 and reprogramming was followed by flow cytometry at reprogramming Day 5 (d5), 7 (d7), 8 (d8) and 9 (d9). Emergent induced dendritic cells (iDCs) may be purified by FACS and characterized by mRNA-sequencing (mRNA-seq), reverse transcriptase quantitative real-time PCR (RT-qPCR), immunofluorescence, scanning electron microscopy (SEM) and functional assays at reprogramming day 9.
-
Isolation of mouse embryonic fibroblasts
Note: MEFs can be purchased from vendors such as Sigma and ScienCell. In this case, thaw and expand fibroblasts, purify (Step A26), and use them in reprogramming experiments until passage 3 (P3) or P4. Alternatively, MEFs can be isolated from pregnant mice as described in the protocol below.
-
Set up timed matings for wild-type (C57BL/6) or double homozygous Clec9aCre/Cre RosatdTomato/tdTomato (Clec9a-tdT) mice ( Rosa et al., 2018 ).
Note: If using cells isolated from Clec9a-tdT reporter mice, tracking of induction of DC fate during reprogramming will be facilitated by the activation of tdTomato expression.
-
The following morning, check for vaginal plugs. If these are detected, isolate plugged females in individual cages and consider this time-point as Embryonic day (E) 0.5.
Note: Plugs can be difficult to detect. As an alternative set up weekly matings keeping two females and one male overnight and removing the male the following day.
Check isolated plugged females at E13.5 to confirm pregnancy before MEF isolation.
Sacrifice pregnant females via CO2 inhalation followed by cervical dislocation.
Disinfect the abdomen of pregnant female with 70% ethanol. Dissect the abdominal cavity with sterile scissors and forceps and expose the abdominal wall by pulling the skin with both hands.
-
Using new scissors and forceps, cut the peritoneum and expose the uterine horns containing the embryo sacs. Isolate the uterus with scissors and transfer it to a 50 ml centrifuge tube with 30 ml of ice-cold PBS.
Note: Uterus can be kept on ice for several hours but PBS supplemented with 10% FBS should be used instead.
-
Inside a cell culture hood, wash uterus twice with sterile chilled PBS and transfer it to a 100 mm dish with 5 ml of sterile chilled PBS.
Note: Typically, 6-8 embryos are recovered per pregnant female.
-
With sterile scissors and forceps, make an incision along the uterine horns to isolate the sacs carrying the embryos.
Note: Don’t perform deep cuts in order to avoid piercing the embryos.
Using forceps, separate embryos from placentas and gently cut-open each individual sac.
Transfer the isolated embryos to a new 100 mm dish containing 5 ml of chilled PBS and leave on ice while completing the remaining dissections.
Using forceps, remove embryo heads. If needed, this tissue can be used for embryo genotyping.
Position the forceps underneath the red-colour visceral organs (fetal liver and heart) and remove them in order to avoid extensive contamination of MEF cultures with hematopoietic cells.
Transfer the dissected embryos to a new 100 mm dish without PBS. Using scissors cut the remaining embryonic tissue into small pieces. Add 1.5 ml of dissociation solution (see Recipes) per embryo, and transfer the tissue to a 50 ml conical tube. Rinse the plate with additional 1.5 ml of dissociation solution per embryo and incubate at 37 °C for 20 min.
Add additional 3 ml of dissociation solution per embryo, pipette up and down to help tissue dissociation and incubate at 37 °C for additional 20 min.
Add 6 ml of DMEM complete medium per embryo to inactivate trypsin and pipette up and down a few times to help with tissue dissociation.
Incubate for 5 min at room temperature (20-25 °C) without agitation to deposit cell debris. Filter the supernatant through a 40 μm sterile cell strainer into a new 50 ml conical tube.
Centrifuge at 350 × g for 5 min, discard supernatant and resuspend the cell pellet in 10 ml of fresh complete medium.
-
Count cell number and adjust cell concentration to 106 cells per ml of DMEM complete medium.
Note: On average, 107 cells are obtained per embryo. Alternatively, the cell counting step can be skipped and one embryo equivalent of cells plated per 100 mm dish.
Pre-coat a 100 mm tissue culture dish with 5 ml of 0.1% gelatin solution and incubate for 15 min at 37 °C.
Aspirate gelatin solution, transfer 10 ml of cell suspension (107 cells per dish, or one embryo equivalent) to the pre-coated 100 mm tissue culture dishes and incubate at 37 °C with 5% CO2 for 24 h.
-
Next day, change culture media to remove floating cells and check cell density under an inverted tissue culture microscope.
Note: It is expected to have some cell death due to trypsin digestion but the plates should be at least 60-70% confluent. If less cells are recovered, consider reducing the dissociation incubation times to 15 min. At this point, MEFs will have small size and 107 cells can be plated per dish. After passages, MEFs will increase the size and become flatter, and a reduced number of cells should be plated per dish.
-
When cells become confluent (consider this passage 0 or P0), remove medium, wash once with PBS, and dissociate with 3 ml of TrypLE Express per plate by incubating at 37 °C with 5% CO2 for 5 to 10 min. After cell detaching, add 7 ml of DMEM complete medium and resuspend cells by pipetting up and down a few times. Transfer cell suspension to a 15 ml canonical tube.
Note: MEFs P0 can be frozen at this time in freezing solution. It is recommended to resuspend 1 confluent plate of MEFs per ml of freezing solution. 1 ml of cell suspension should be transferred to a cryovial and frozen overnight at -80 °C using a Mr. Frosty freezing container. For long-term storage, cryovials should be transferred to -150 °C ultra low freezer or liquid nitrogen.
Count cells using a hemocytometer and centrifuge cells at 350 × g for 5 min.
Discard supernatant, resuspend the cell pellet in 100 μl of staining solution (see Recipes) per 106 cells and incubate for 30 min at 4 °C.
Wash cells in 10 ml of ice-cold wash buffer and centrifuge at 350 × g for 5 min.
Discard the supernatant and resuspend the cell pellet in 300 μl of wash buffer per 106 cells.
-
Purify CD45-MHC-II- MEFs or, in case of using a reporter such as Clec9a-tdT MEFs, exclude tdTomato+ cells as well, by Fluorescence-activated cell sorting (FACS) to remove rare contaminant hematopoietic cells.
Note: During FACS purification, we recommend using 100 micron nozzle and collecting purified MEFs in polypropylene round-bottom FACS tubes coated with 500 μl of FBS supplemented with Pen/Strep diluted 1:100. Typically, contaminant hematopoietic cells represent less than 1% of MEFs before purification (Rosa et al., 2018), but can be a confounding population during reprogramming experiments. After FACS sorting the MEF population should be > 99% CD45-MHC-II-.
After FACS purification, transfer purified MEFs to a 50 ml canonical tube with 10 ml of DMEM complete medium and centrifuge at 350 × g for 5 min.
Discard the supernatant and resuspend the cell pellet in fresh complete medium. Count cell number and adjust cell concentration to 106 cells per ml of DMEM complete medium.
Transfer cell suspension to gelatin pre-coated 100 mm tissue culture dishes with 9 ml of DMEM complete medium (106 cells per dish, final volume 10 ml) and incubate at 37 °C with 5% CO2 for 24 h. Consider this as passage 1.
-
Next day, replace culture media to remove floating dead cells and check cell density.
Note: Typically, 50-60% cell confluency is achieved at this step.
When confluent, split cells to new gelatin pre-coated 100 mm tissue culture dishes at 1:4 dilution. Consider this Passage 2. For reprogramming experiments use MEFs from passages 3-4 to avoid cellular senescence.
If required, purified MEFs can be frozen from confluent plates. Prepare 1 vial per confluent plate in 1 ml of freezing solution (as described in Step A22).
-
-
Lentiviral Production
Note: Perform lentiviral production in a biosafety level 2 laboratory in a dedicated laminar flow hood and place all viral contaminated waste (tubes, tips, plates) in an appropriate residue container for biohazardous materials.
-
Expand HEK 293T cells in 150 mm plates with 20 ml of DMEM complete medium. When confluent, remove the medium, wash once with PBS, and dissociate with 6 ml of TrypLE Express by incubating at 37 °C with 5% CO2 for 5 to 10 min.
Note: HEK 293T cells are semi-adherent so removal of medium and washing with PBS needs to be performed slowly to avoid cell detachment.
Add 6 ml of DMEM complete medium and resuspend cells by pipetting up and down a few times. Transfer cells into 50 ml canonical tubes and centrifuge at 350 × g for 5 min. Remove supernatant and split cells to new 150 mm tissue culture dishes at 1:6 dilution 48 h prior transfection.
-
In 15 ml tubes, add 10 μg of pFUW-TetO-PIB (inducible lentiviral construct with the tri-cistronic cassette encoding PU.1, IRF8 and BATF3 reprogramming factors) or FUW-M2rtTA, for each 150 mm dish to be transfected. To both tubes add 7.5 μg of packaging psPAX2 and 2.5 μg of envelope pMD2.G plasmids. Adjust the final volume to 2 ml by adding Opti-MEM. Add 60 μl of Polyethylenimine (PEI) transfection reagent (1 mg/ml), vortex thoroughly and incubate transfection mix for 30 min at room temperature to allow the formation of DNA-lipid complexes.
Note: The lentiviral transfer plasmid pFUW-TetO drives expression of PU.1, IRF8 and BATF3 under the control of a doxycycline-responsive promoter. The FUW-M2rtTA lentiviral plasmid drives expression of the reverse tetracycline transactivator M2rtTA under the control of a constitutively active human ubiquitin C promoter. In this system, when both vectors are co-expressed in the presence of doxycycline, M2rtTA protein bridges doxycycline to the doxycycline-inducible promoter, inducing the expression of PU.1, IRF8 and BATF3. The inducible system is recommended for DC reprogramming experiments because it allows timed induction of transgene expression.
Change HEK 293T cell culture media for 10 ml DMEM without FBS and Pen/Strep. Add transfection mix dropwise to the cells and incubate at 37 °C with 5% CO2 for 6 h.
Replace culture media of transfected HEK 293T cells with 20 ml of DMEM complete medium and incubate cells at 37 °C with 5% CO2 for 24 h.
On the following day, replace HEK 293T cell culture media with 12 ml of fresh DMEM complete medium and incubate cells at 37 °C with 5% CO2.
-
Harvest viral supernatants 36 h, 48 h and 60 h after transfection into 50 ml polypropylene tubes, store viral supernatants at 4 °C and replace culture media with 12 ml of fresh DMEM complete medium between harvests.
Note: In total, 36 ml of viral supernatant are harvested per 150 mm plate. Viral particles are not stable at 37 °C but last 1-2 weeks at 4 °C. By harvesting at three time points viral titers are maximized.
-
Filter harvested supernatants containing viral particles through a 150 ml bottle top 0.45 μm low-protein binding PES filter system.
Note: Low-protein binding filters should be used to minimize the loss of viral particles by filter adsorption.
Pipette 12 ml of filtered viral supernatant into an Amicon ultra-15 centrifugal filter unit, centrifuge at 2,000 × g at 4 °C for 30 min and discard flow-through liquid.
Add additional 12 ml of filtered viral supernatant to the concentrated viral volume in the filter unit, mix the tube content and centrifuge at 2,000 × g at 4 °C for an additional 30 min. Discard the flow-through.
-
Add the remaining filtered viral supernatant to the concentrated viral solution left in the filter, mix the contents by inverting the tube and centrifuge once again at 2,000 × g at 4 °C for 20 to 40 min. The final centrifugation time should be adjusted to obtain 600-800 μl of the total volume of concentrated viruses in the filter.
Note: If the final volume is larger than 800 μl, mix the concentrate and centrifuge again at 2,000 × g at 4 °C for an additional 10 to 20 min.
-
Prepare aliquots in microtubes on ice (50-200 μl depending on the size of future reprogramming experiments) of each type of concentrated lentiviruses and store at -80 °C for long-term storage (1-2 years) or at 4 °C for short-term storage (1-2 weeks). Concentrated or non-concentrated lentiviruses can also be used fresh.
Note: Freezing concentrated viruses at -80 °C once leads to a reduction of approximately 10-20% of viral titer. Do not freeze and thaw repeatedly as this results in reduced titer.
-
-
Dendritic Cell Reprogramming
Pre-coat 100 mm tissue culture dishes with 5 ml of 0.1% gelatin solution per plate and incubate for at least 15 min at 37 °C with 5% CO2.
Thaw MEF vials at 37 °C. As soon as the cell solution is thawed, add cells dropwise into 10 ml of DMEM complete medium pre-warmed at 37 °C and mix carefully.
Centrifuge at 350 × g for 5 min. Aspirate supernatant and resuspend the cell pellet in 10 ml of DMEM complete medium per vial.
-
Aspirate gelatin solution of pre-incubated 100 mm plates. Dispense 10 ml of MEF suspension per gelatin pre-coated 100 mm tissue culture dish and incubate overnight at 37 °C with 5% CO2. If required, expand fibroblasts to increase starting cell numbers until passage 3-4 is reached.
Note: Typically, 2 to 5 × 106 MEFs can be recovered from a confluent 100 mm plate, depending on the passage number.
-
To start DC reprogramming experiments, plate 250,000 MEFs per gelatin pre-coated 100 mm plate in 10 ml of DMEM complete medium and incubate overnight at 37 °C with 5% CO2.
Note: Depending on the required number of iDCs or purpose of the experiment, the reprogramming protocol can be scaled up by starting with a large number of 100 mm plates (i.e., 50 to 100 plates to purify iDCs for functional assays) or scaled down to 6-well plates (i.e., to test the influence of different culture conditions in DC reprogramming efficiency), adjusting cell number and viral volume according to the surface area.
-
On the afternoon of the following day, perform the first lentiviral transduction by replacing medium with 10 ml of DMEM complete medium supplemented with polybrene (8 μg/ml) and 30 to 60 μl of a 1:1 mix of FUW-TetO-PIB and FUW-M2rtTA concentrated lentiviruses. Incubate overnight at 37 °C with 5% CO2.
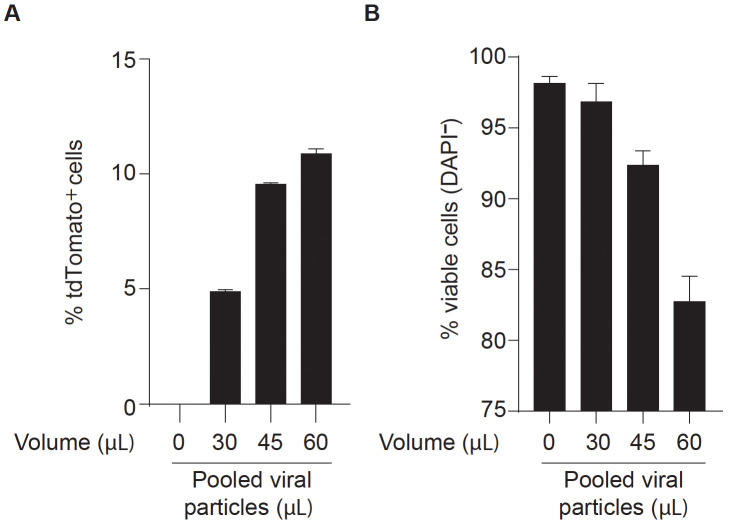
Note: For DC reprograming experiments it is recommended to define the optimal volume of lentiviral mix for efficient reprogramming without compromising cell viability (Figure 2). Depending on the aim of the experiment, this may be important to maximize the number of reprogrammed cells obtained in the end of the protocol. MEFs with higher passage number may require higher volumes of viruses.
After 16 h of incubation, replace media with fresh DMEM complete medium. Incubate for 6-8 h at 37 °C with 5% CO2 to allow cells to recover.
-
Perform second lentiviral transduction as described in Step C6. Incubate overnight at 37 °C with 5% CO2.
Note: The transduction mix (DMEM complete with FUW-TetO-PIB, FUW-M2rtTA and polybrene) can be prepared for both transductions and kept at 4 °C.
-
After 16 h of incubation, replace media with fresh DMEM complete medium supplemented with DOX (1 μg/ml).
Note: FUW-TetO-PIB and FUW-M2rtTA are a DOX inducible lentiviral system. Thus, adding DMEM complete medium supplemented with DOX will initiate expression of PIB reprogramming factors. Consider this as reprogramming Day 0.
-
Replace culture media with DMEM complete medium supplemented with DOX every 2-3 days for the duration of the DC reprogramming cultures (9 days).
Note: It is recommended to follow cell cultures during reprogramming under an inverted microscope. Reprogramming-associated cell death is expected in the first days after DOX supplementation, which depends on MEF passage and viral volume used. Cells will become progressively more confluent at later stages of reprogramming due to the proliferation of non-transduced and non-reprogrammed MEFs. When using Clec9a-tdT MEFs, tdTomato+ cells start being detected approximately 30 h after DOX supplementation (Rosa et al., 2018). iDCs remain adherent throughout the reprogramming process.
The emergence of iDCs during DC reprogramming can be analyzed at different time points. For this purpose, remove the medium, wash once with PBS, and dissociate with 3 ml of TrypLE Express per plate by incubating at 37 °C with 5% CO2 for 5 to 10 min.
After cell detaching, add 7 ml of DMEM complete medium and resuspend cells by pipetting up and down a few times. Transfer cell suspension to a 15 ml canonical tube and centrifuge at 350 × g for 5 min.
Discard the supernatant and resuspend 1 plate equivalent cell pellet in 200 μl of wash buffer for downstream analysis by flow cytometry or FACS sorting for microscopy, mRNA-sequencing or RT-qPCR analysis.
Figure 2. Optimization of lentiviral mix volume for efficient dendritic cell direct reprogramming.
Clec9a-tdT mouse embryonic fibroblasts (MEFs) were transduced with increasing volumes (30 to 60 μl) of concentrated pooled lentiviral particles encoding PU.1, IRF8 and BATF3 (PIB) transcription factors (FUW-TetO-PIB), together with M2rtTA (FUW-M2rtTA), at 1:1 ratio. Reprogrammed cells were analyzed at day 9 by flow cytometry to define an optimal volume of transduction for dendritic cell reprogramming. The percentage of tdTomato+ cells (A) indicates reprogramming efficiency and the percentage of DAPI-negative cells (B) indicates cell viability. Non-transduced Clec9a-tdT MEFs were used as negative control. In the example, 45 μl of pooled viral particles induces the maximum reprogramming efficiency without compromising cell viability.
Data analysis
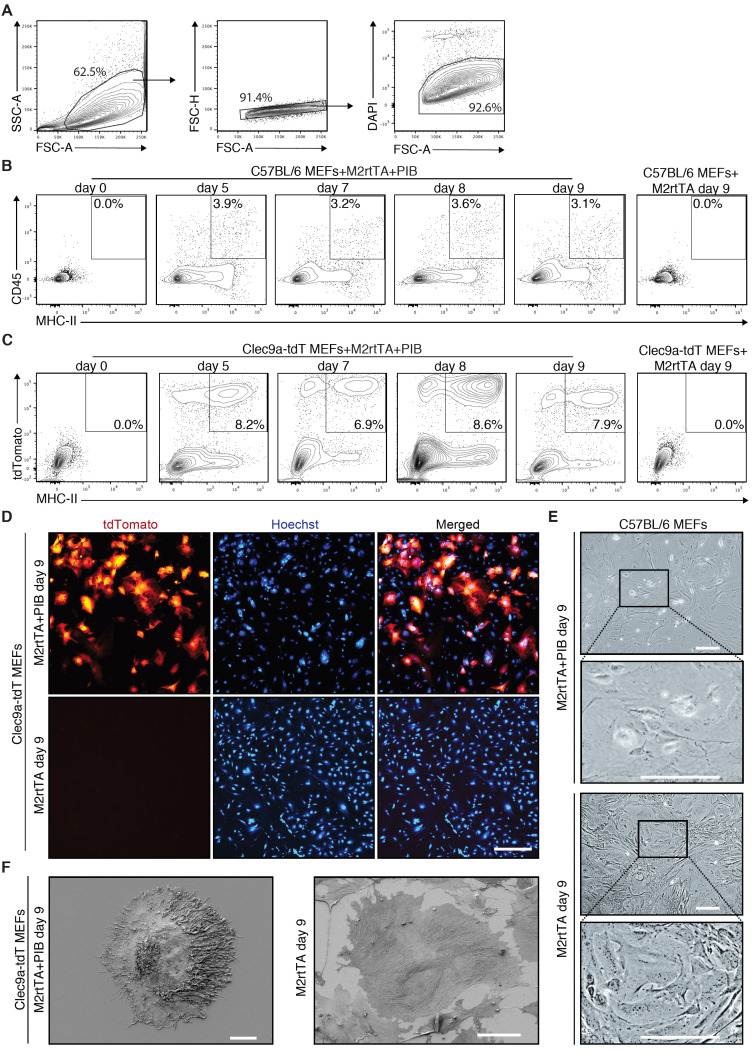
DC reprogramming can be evaluated at different time points between Day 0 and Day 9 (Figure 1). The emergence of CD45+MHC-II+ indicates iDC reprogramming efficiency and can be detected by flow cytometry at reprogramming Day 0, 5, 7, 8 and 9 (Figures 3A-3B). If using a reporter such as the Clec9a-tdT, the emergence of tdTomato+MHC-II+ iDCs can be followed by flow cytometry at the same time-points (Figure 3C). CD45, MHC-II or tdTomato are not detected in transduced MEFs at Day 0 or in M2rtTA-transduced MEFs at Day 9 (Figures 3B-3C). At Day 9, iDCs can be analyzed by microscopy (Figures 3D-3E). For more detailed morphology analysis, tdTomato+ cells can be purified by FACS and analyzed by scanning electron microscopy (SEM) (Figure 3F).
Figure 3. Flow cytometry and microscopy analysis of induced dendritic cells generated by direct reprogramming of mouse embryonic fibroblasts.
A. Gating strategy to analyze the emergence of induced dendritic cells (iDCs) by flow cytometry during reprogramming. Population was selected with FSC and SSC, followed by exclusion of doublets and dead cells (DAPI+). B. The emergence of CD45+MHC-II+ iDCs was quantified by flow cytometry 5 days (d5), 7 days (d7), 8 days (d8) and 9 days (d9) after induction of PU.1, IRF8 and BATF3 (PIB) in wild type (C57BL/6) mouse embryonic fibroblasts (MEFs). Transduced MEFs at Day 0 and M2rtTA-transduced MEFs at Day 9 were included as controls. CD45+MHC-II+ gating strategy was performed based on single-cell stainings for CD45 and MHC-II 9 days after induction of PIB in wild type MEFs. C. Flow cytometry analysis of tdTomato+MHC-II+ cells at d5, d7, d8 and d9 after induction of M2rtTA and PIB in Clec9a-tdT MEFs. Transduced MEFs at Day 0 and M2rtTA-transduced MEFs at Day 9 were included as controls. D. Immunofluorescence for tdTomato and Hoechst (blue) highlighting the emergence and frequency of tdTomato+ cells. M2rtTA-transduced MEFs at Day 9 were included as control. Scale bar = 500 μm. E. Bright-field micrographs of C57BL/6 MEFs 9 days after induction of M2rtTA or M2rtTA and PIB. Scale bars = 100 μm. F. Scanning electron microscopy analysis of a tdTomato+ cell 9 days after induction of M2rtTA and PIB in Clec9a-tdT MEFs. M2rtTA-transduced MEFs at Day 9 were included as control. Scale bars = 10 μm.
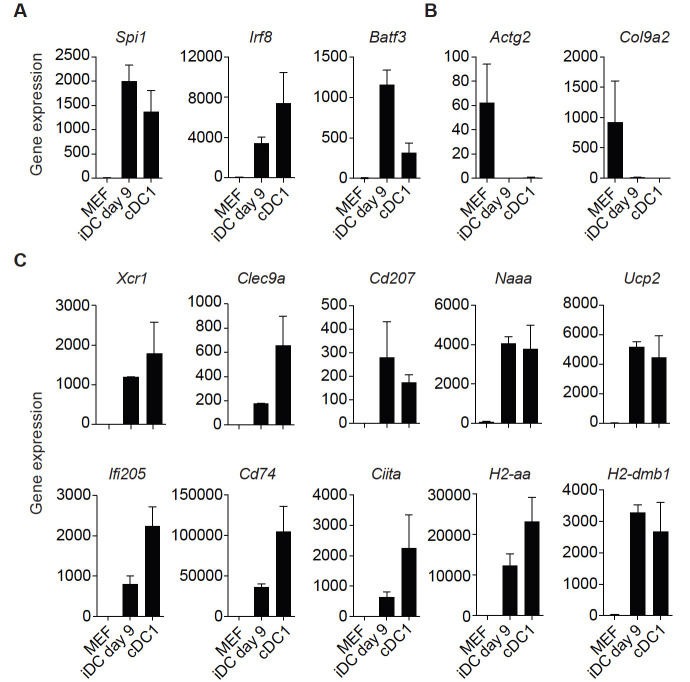
Additionally, mRNA-sequencing of purified tdTomato+CD45+MHC-II+ iDCs at reprogramming Day 9 (iDCs Day 9) and splenic MHC-II+CD11c+CD8a+ cDC1s can be performed to further validate successful reprogramming ( Rosa et al., 2018 ). High expression levels of genes encoding the reprogramming factors (Spi1 [encoding PU.1], Irf8 and Batf3) are detected in iDCs Day 9 and cDC1s (Figure 4A). The fibroblast-specific genes Actg2 and Col9a2 are down regulated in iDCs Day 9 and not expressed in cDC1s (Figure 4B). Expression of the DC genes Xcr1, Clec9a, Cd207, Naaa, Ucp2, Ifi205, Cd74, Ciita, H2-aa and H2-dmb1 is not detected in MEFs, is activated in iDCs Day 9 and expressed in splenic cDC1s (Figure 4C). This set of markers can be used to assess successful reprogramming either by mRNA-sequencing or RT-qPCR. Further functional evaluation of iDCs can be performed after FACS purification including: secretion of inflammatory cytokines, dead cell engulfment, antigen export to cytosol and antigen cross-presentation to OT-I CD8+ T cells ( Rosa et al., 2018 ).
Figure 4. Gene signature to assess successful dendritic cell reprogramming.
A. Genes encoding the reprogramming factors PU.1, IRF8 and BATF3 (PIB) are expressed in induced dendritic cells (iDCs) at reprogramming Day 9 (tdTomato+CD45+MHC-II+) and in conventional dendritic cells type 1 (cDC1s, MHC-II+CD11c+CD8a+). B. Fibroblast-associated genes Actg2 and Col9a2 are down regulated in iDCs at reprogramming Day 9 and are not expressed in cDC1s. C. Dendritic cell genes Xcr1, Clec9a, Cd207, Naaa, Ucp2, Ifi205, CD74, Ciita, H2-aa and H2-dmb1 are expressed in iDCs at reprogramming Day 9 and in cDC1s. After induction of PIB in mouse embryonic fibroblasts (MEF), iDCs (tdTomato+CD45+MHC-II+) and splenic cDC1s (CD11c+MHC-II+CD8α+) were FACS sorted and expression of genes quantified by mRNA-sequencing ( Rosa et al., 2018 ; GSE103618). Gene expression is displayed in bar plots as mean of gene counts ± standard deviation. As an alternative, RT-qPCR can be used to quantify the expression of this gene set and assess successful reprogramming.
Recipes
-
DMEM Complete medium
DMEM supplemented with 10% (v/v) heat-inactivated FBS and 1:100 dilution of 10,000 U/ml Penicillin, 10,000 μg/ml Streptomycin stock solution
-
Dissociation solution
Dilute 0.25% trypsin-EDTA solution in the same volume of PBS (dilution 1:2) for a final concentration of 0.125% trypsin-EDTA
-
0.1% Gelatin solution
Dissolve 0.1 g of gelatin in 100 ml of MilliQ-grade water and autoclave
-
Wash solution
PBS supplemented with 2.5% FBS
-
Staining solution
Wash solution supplemented with rat serum (1/100), anti-mouse CD45 monoclonal antibody (0.25 µg per 100 μl) and anti-mouse MHC-II monoclonal antibody (0.25 µg per 100 μl)
-
Freezing solution
Heat-inactivated FBS supplemented with 10% DMSO
Acknowledgments
This project was co-funded by Cancerfonden (CAN 2017/745), the Swedish research council (2018-02042), Crafoord Foundation (20190561), NovoNordisk Fonden (0056527) and FCT (CENTRO-01-0145-FEDER-030013). The Knut and Alice Wallenberg foundation, the Medical Faculty at Lund University and Region Skåne are acknowledged for generous financial support. F.F.R and C.F.P. and F.F.R. are supported by FCT doctoral (SFRH/BD/130845/2017) and postdoctoral (SFRH/BPD/121445/2016) fellowships, respectively. This protocol was adapted from Rosa et al., 2018 .
Competing interests
Fábio F. Rosa, Cristiana F. Pires and Carlos-Filipe Pereira have filed a Patent Cooperation Treaty (PCT) protecting the intellectual property described here on 2018-04-05.
Ethics
This protocol was performed according to Lund University's research ethics committee guidelines and should be done in accordance with individual institutional guidelines. Procedures involving animal experimentation were approved by the Swedish Ethical Review board (5.8.18-19343/2017).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Balan S., Arnold-Schrauf C., Abbas A., Couespel N., Savoret J., Imperatore F., Villani A. C., Vu Manh T. P., Bhardwaj N. and Dalod M.(2018). Large-scale human dendritic cell differentiation revealing notch-dependent lineage bifurcation and heterogeneity. Cell Rep 24(7): 1902-1915 e1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balan S., Ollion V., Colletti N., Chelbi R., Montanana-Sanchis F., Liu H., Vu Manh T. P., Sanchez C., Savoret J., Perrot I., Doffin A. C., Fossum E., Bechlian D., Chabannon C., Bogen B., Asselin-Paturel C., Shaw M., Soos T., Caux C., Valladeau-Guilemond J. and Dalod M.(2014). Human XCR1+ dendritic cells derived in vitro from CD34+ progenitors closely resemble blood dendritic cells, including their adjuvant responsiveness, contrary to monocyte-derived dendritic cells . J Immunol 193(4): 1622-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bottcher J. P. and Reis E. S. C.(2018). The role of Type 1 conventional dendritic cells in cancer immunity. Trends Cancer 4(11): 784-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carotta S., Dakic A., D'Amico A., Pang S. H., Greig K. T., Nutt S. L. and Wu L.(2010). The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity 32(5): 628-641. [DOI] [PubMed] [Google Scholar]
- 5. Choi K. D., Vodyanik M. A. and Slukvin II(2009). Generation of mature human myelomonocytic cells through expansion and differentiation of pluripotent stem cell-derived lin-CD34+CD43+CD45+ progenitors . J Clin Invest 119(9): 2818-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hildner K., Edelson B. T., Purtha W. E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B. U., Unanue E. R., Diamond M. S., Schreiber R. D., Murphy T. L. and Murphy K. M.(2008). Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity . Science 322(5904): 1097-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Horton C., Davies T. J., Lahiri P., Sachamitr P. and Fairchild P. J.(2019). iPS cells reprogrammed from primary dendritic cells provide an abundant source of immunostimulatory dendritic cells for use in immunotherapy. Stem Cells 38(1):67-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Inaba K., Inaba M., Romani N., Aya H., Deguchi M., Ikehara S., Muramatsu S. and Steinman R. M.(1992). Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med 176(6): 1693-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kirkling M. E., Cytlak U., Lau C. M., Lewis K. L., Resteu A., Khodadadi-Jamayran A., Siebel C. W., Salmon H., Merad M., Tsirigos A., Collin M., Bigley V. and Reizis B.(2018). Notch signaling facilitates in vitro generation of cross-presenting classical dendritic cells . Cell Rep 23(12): 3658-3672 e3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee J., Breton G., Oliveira T. Y., Zhou Y. J., Aljoufi A., Puhr S., Cameron M. J., Sekaly R. P., Nussenzweig M. C. and Liu K.(2015). Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J Exp Med 212(3): 385-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu L., McCaslin D., Starzl T. E. and Thomson A. W.(1995). Bone marrow-derived dendritic cell progenitors(NLDC 145+, MHC class II+, B7-1dim, B7-2-) induce alloantigen-specific hyporesponsiveness in murine T lymphocytes . Transplantation 60(12): 1539-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lutz M. B., Kukutsch N., Ogilvie A. L., Rossner S., Koch F., Romani N. and Schuler G.(1999). An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods 223(1): 77-92. [DOI] [PubMed] [Google Scholar]
- 13. Mayer C. T., Ghorbani P., Nandan A., Dudek M., Arnold-Schrauf C., Hesse C., Berod L., Stuve P., Puttur F., Merad M. and Sparwasser T.(2014). Selective and efficient generation of functional Batf3-dependent CD103+ dendritic cells from mouse bone marrow . Blood 124(20): 3081-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Merad M., Sathe P., Helft J., Miller J. and Mortha A.(2013). The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol 31: 563-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Naik S. H., Proietto A. I., Wilson N. S., Dakic A., Schnorrer P., Fuchsberger M., Lahoud M. H., O'Keeffe M., Shao Q. X., Chen W. F., Villadangos J. A., Shortman K. and Wu L.(2005). Cutting edge: generation of splenic CD8+ and CD8- dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures . J Immunol 174(11): 6592-6597. [DOI] [PubMed] [Google Scholar]
- 16. Pereira C. F., Chang B., Qiu J., Niu X., Papatsenko D., Hendry C. E., Clark N. R., Nomura-Kitabayashi A., Kovacic J. C., A. Ma'ayan, Schaniel C., Lemischka I. R. and Moore K.(2013). Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell 13(2): 205-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pereira C. F., Lemischka I. R. and Moore K.(2012). Reprogramming cell fates: insights from combinatorial approaches. Ann N Y Acad Sci 1266: 7-17. [DOI] [PubMed] [Google Scholar]
- 18. Perez C. R. and De Palma M.(2019). Engineering dendritic cell vaccines to improve cancer immunotherapy. Nat Commun 10(1): 5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pires C. F., Rosa F. F., Kurochkin I. and Pereira C.-F.(2019). Understanding and modulating immunity with cell reprogramming. Frontiers in Immunology 10(2809). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poulin L. F., Salio M., Griessinger E., Anjos-Afonso F., Craciun L., Chen J. L., Keller A. M., Joffre O., Zelenay S., Nye E., Le Moine A., Faure F., Donckier V., Sancho D., Cerundolo V., Bonnet D. and Reis e Sousa C.(2010). Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8α+ dendritic cells . J Exp Med 207(6): 1261-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Proietto A. I., Mittag D., Roberts A. W., Sprigg N. and Wu L.(2012). The equivalents of human blood and spleen dendritic cell subtypes can be generated in vitro from human CD34+ stem cells in the presence of fms-like tyrosine kinase 3 ligand and thrombopoietin . Cell Mol Immunol 9(6): 446-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rosa F. F., Pires C. F., Kurochkin I., Ferreira A. G., Gomes A. M., Palma L. G., Shaiv K., Solanas L., Azenha C., Papatsenko D., Schulz O., C. Reis e Sousa and Pereira C. F.(2018). Direct reprogramming of fibroblasts into antigen-presenting dendritic cells. Sci Immunol 3(30). [DOI] [PubMed] [Google Scholar]
- 23. Schiavoni G., Mattei F., Sestili P., Borghi P., Venditti M., Morse H. C. 3rd Belardelli F. and Gabriele L.(2002). ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8α+ dendritic cells . J Exp Med 196(11): 1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Senju S., Haruta M., Matsumura K., Matsunaga Y., Fukushima S., Ikeda T., Takamatsu K., Irie A. and Nishimura Y.(2011). Generation of dendritic cells and macrophages from human induced pluripotent stem cells aiming at cell therapy. Gene Ther 18(9): 874-883. [DOI] [PubMed] [Google Scholar]
- 25. Senju S., Haruta M., Matsunaga Y., Fukushima S., Ikeda T., Takahashi K., Okita K., Yamanaka S. and Nishimura Y.(2009). Characterization of dendritic cells and macrophages generated by directed differentiation from mouse induced pluripotent stem cells. Stem Cells 27(5): 1021-1031. [DOI] [PubMed] [Google Scholar]
- 26. Son Y. I., Egawa S., Tatsumi T., Redlinger R. E. Jr. Kalinski P. and Kanto T.(2002). A novel bulk-culture method for generating mature dendritic cells from mouse bone marrow cells. J Immunol Methods 262(1-2): 145-157. [DOI] [PubMed] [Google Scholar]
- 27. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. and Yamanaka S.(2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131(5): 861-872. [DOI] [PubMed] [Google Scholar]
- 28. Takahashi K. and Yamanaka S.(2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4): 663-676. [DOI] [PubMed] [Google Scholar]
- 29. Wculek S. K., Cueto F. J., Mujal A. M., Melero I., Krummel M. F. and Sancho D.(2019). Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol. [DOI] [PubMed] [Google Scholar]
- 30. Xu J., Du Y. and Deng H.(2015). Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell 16(2): 119-134. [DOI] [PubMed] [Google Scholar]
- 31. Yona S. and Mildner A.(2018). Good things come in threes. Sci Immunol 3(30). [DOI] [PubMed] [Google Scholar]