Abstract
Roseburia and Eubacterium species of the human gut microbiota play an important role in the maintaince of human health, partly by producing butyrate, the main energy source of our colonic epithelial cells. However, our knowledge of the biochemistry and physiology of these bacteria has been limited by a lack of genetic manipulation techniques. Conjugative transposons previously introduced into Roseburia species could not be easily modified, greatly limiting their applicability as genetic modification platforms. Modular plasmid shuttle vectors have previously been developed for Clostridium species, which share a taxonomic order with Roseburia and Eubacterium, raising the possibility that these vectors could be used in these organisms. Here, we describe an optimized conjugation protocol enabling the transfer of autonomously replicating plasmids from an E. coli donor strain into Roseburia inulinivorans and Eubacterium rectale. The modular nature of the plasmids and their ability to be maintained in the recipient bacterium by autonomous replication makes them ideal for investigating heterologous gene expression, and as a platform for other genetic tools including antisense RNA silencing or mobile group II interon gene disruption strategies.
Keywords: Conjugation, Gut microbiota, Lachnospiraceae, Gene transfer, Shuttle vector
Background
Roseburia and Eubacterium species are among the most abundant bacteria in the human gut microbiota ( Zhernakova et al., 2016 ), impacting human health by utilising dietary and host derived polysaccharides ( Scott et al., 2006 and 2011; Cockburn et al., 2015 ; Sheridan et al., 2016 ) and producing the health promoting metabolite butyrate as a fermentation end product ( Duncan et al., 2002 and 2006). Additionally, these species are capable of modulating host immunity via flagella ( Neville et al., 2013 ). The lack of genetic modification techniques for these organisms has prevented a more complete understanding of the complex interactions between these bacteria and their human host.
Previously, conjugative transposons were successfully transferred into Roseburia inulinivorans from Eubacterium cellulosolvens and Clostridium cf. saccharolyticum ( Scott et al., 2008 ). These large, novel mobile genetic elements could not be easily modified and thus were a suboptimal platform for detailed genetic modification. This work did however illustrate that conjugative mating was possible between Lachnospiraceae bacteria including Roseburia species. The development of easily modified conjugative plasmids for clostridial species ( Purdy et al., 2002 ; Heap et al., 2009 ) raised the possibility that these techniques could be adapted for Roseburia and Eubacterium species.
The detailed protocol presented here is based on procedures established in Sheridan et al. (2019) . In this work, the different conjugative plasmids developed for use in Clostridium species ( Heap et al., 2009 ) were tested for transferability into the Roseburia and Eubacterium rectale species. Plasmid pMTL83151 was successfully transferred into two strains of E. rectale, while pMTL83151 and pMTL82151 were transferred into Roseburia inulinivorans A2-194. Transfer frequencies of10-6-10-8 per potential recipient were obtained. These frequencies are similar to those observed when suicide vectors were introduced into other Gram-positive bacteria ( Williams et al., 1990 ; Aquino de Muro and Priest, 2000 ). The ability to add exogenous DNA to bacterial species opens up opportunities for genetically manipulation, including knockout mutagenesis. Alternatively, these plasmids could be modified as expression vectors for mobile group II interon gene disruption strategies, as has been demonstrated in several clostridial species ( Heap et al., 2007 ). Additionally, Plasmid pMTL83151 was shown to be a suitable vector for heterologous gene expression ( Sheridan et al., 2019 ), producing an enzymatically active Streptococcus glycoside hydrolase in both species and thus proving the utility of this technique in studying researcher-selected functional gains in these important bacteria. The protocol below is a stepwise guide to introducing foreign DNA to these bacteria.
Materials and Reagents
Pipette tips
Cuvettes (Bio-Rad, catalog number: 1652083)
Petri dishes (Greinerbio-one, catalog number: 633180)
Glass Pasteur pipettes (Fisher Scientific, catalog number: FB50261)
Nylon membranes (Roche, catalog number: 11417240001)
X-ray film (Fujifilm) (Fisher Scientific, catalog number: 12735325)
50 ml conical centrifuge tubes (Corning, catalog number: 10038980)
Strains (Table 1)
-
Plasmids (Table 1)
Note: Modular plasmids can be obtained from CHAINbiotools (http://clostron.com/pMTL80000.php).
Primers (Table 2)
PBS tablets (Sigma, catalog number: P4417)
Chloramphenicol (Sigma-Aldrich, catalog number: C0378) stock solution 10 µg/ml, stored at -20 °C
HindIII restriction endonuclease (NEB, catalog number: R0104S)
Wizard genomic DNA Purification kit (Promega, catalog number: A1120)
DIG High Prime DNA Labelling and Detection Starter Kit II (Roche, catalog number: 11585614910)
PCR reagents (Taq Polymerase kit Bioline, catalog number: BIO-21040 and dNTP’s Promega, catalog number: U1240)
Dipotassium phosphate, K2HPO4 (Fisher Scientific, catalog number: P/5240/53)
Potassium dihydrogen phosphate, KH2PO4 (Fisher Scientific, catalog number: P/4800/60)
Ammonium sulfate, (NH4)2SO4 (Fisher Scientific, catalog number: A/6480/53)
Sodium chloride, NaCl (Fisher Scientific, catalog number: S/3160/53)
Magnesium sulfate, MgSO4 (Sigma-Aldrich, catalog number: M7506)
Calcium chloride, CaCl2 (Sigma-Aldrich, catalog number: C1016)
Acetic acid (Fisher Scientific, catalog number: A/10400/PB17)
Propionic acid (Sigma-Aldrich, catalog number: P1386)
n-Valeric acid (Sigma-Aldrich, catalog number: V9759)
Iso-Valeric acid (Sigma-Aldrich, catalog number: I7128)
Iso-Butyric acid (Sigma-Aldrich, catalog number: I1754)
Biotin (Sigma-Aldrich, catalog number: B4501)
Cobalamin (Sigma-Aldrich, catalog number: V2876)
p-Aminobenzoic acid (Sigma-Aldrich, catalog number: A9878)
Folic acid (Sigma-Aldrich, catalog number: F7876)
Pyridoxamine (Sigma-Aldrich, catalog number: P9755)
Potassium hydroxide, KOH (Sigma-Aldrich, catalog number: P5958)
Ethanol 95% (Fisher Scientific, catalog number: E/0650DF/17)
Haemin (Sigma-Aldrich, catalog number: H5533)
Bacto tryptone (BD Diagnostics Systems, catalog number: 211705)
Yeast Extract (BD Diagnostics Systems, catalog number: 212750)
Potassium chloride, KCl (Sigma-Aldrich, catalog number: P3911)
Magnesium chloride hexahydrate, MgCl2·6H2O (Sigma-Aldrich, catalog number: M9272 )
Magnesium sulfate heptahydrate, MgSO4·7H2O (Sigma-Aldrich, catalog number: 230391)
Bacto casitone (BD Diagnostics Systems, catalog number: 225930)
Sodium bicarbonate, NaHCO3 (Sigma-Aldrich, catalog number: S576)
Glucose (Fisher Scientific, catalog number: G/0500/53)
Soluble starch (Sigma-Aldrich, catalog number: S2004)
Cellobiose (Sigma-Aldrich, catalog number: C7252)
Resazurine (Sigma-Aldrich, catalog number: R2127)
L-cysteine (Sigma-Aldrich, catalog number: C1276)
Thiamin (Sigma-Aldrich, catalog number: T1270)
Riboflavin (Sigma-Aldrich, catalog number: R9504)
Gas mix 10% Carbon Dioxide, 10% Hydrogen balance Nitrogen (Anaerobic) Cylinder (BOC, catalog number: 290564-L)
Agar (Oxoid, catalog number: LP0011)
Resazurin solution (see Recipes)
Anaerobic phosphate buffered saline (PBS) (see Recipes)
Mineral solution 1 (see Recipes)
Mineral solution 2 (see Recipes)
Short chain fatty acid solution (see Recipes)
Vitamin solution 1 (see Recipes)
Vitamin solution 2 (see Recipes)
Haemin solution (see Recipes)
SOC (see Recipes)
LB (see Recipes)
LA (see Recipes)
YCFAGSC and AMM (see Recipes)
Table 1. Strains and plasmids.
| Strain/plasmid | Relative characteristics | Source/Reference |
| Strains | ||
| Eubacterium rectale A1-86 | Butyrate producing, strict anaerobes of the Lachnospiraceae family. | ( Barcenilla et al., 2000 ) |
| Eubacterium rectale T1-815 | ( Barcenilla et al., 2000 ) | |
| Roseburia inulinivorans A2-194 | ( Duncan et al., 2006 ) | |
| Eubacterium rectale EAM3 | E. rectale A1-86 harbouring pMTL83151 | ( Sheridan et al., 2019 ) |
| Eubacterium rectale ETM3 | E. rectale T1-815 harbouring pMTL83151 | ( Sheridan et al., 2019 ) |
| Eubacterium rectale ETBglu | E. rectale T1-815 harbouring pMTL3β-glu | ( Sheridan et al., 2019 ) |
| Roseburia inulinivorans RAM2 | R. inulinivorans A2-194 harbouring pMTL82151 | ( Sheridan et al., 2019 ) |
| Roseburia inulinivorans RAM3 | R. inulinivorans A2-194 harbouring pMTL83151 | ( Sheridan et al., 2019 ) |
| Escherichia coli CA434 | Conjugative donor. Genotype: HB101 (thi-1 hsdS20 (r-B, m-B) supE44 recAB ara-14 leuB5proA2 lacY1 galK rpsL20 (strR) xyl-5 mtl-1) carrying R701 (Tra+, Mob+ conjugative plasmid) | ( Williams et al., 1990 ) |
| Plasmids* | ||
| pMTL82151 | ColE1, catP, traJ, MCS and pBP1 | ( Heap et al., 2009 ) |
| pMTL83151 | ColE1, catP, traJ, MCS and pCB102 | ( Heap et al., 2009 ) |
Table 2. Primers.
| Primer | Sequence (5'-3') | Target | Reference |
| PS#MTL-for | TATCTATGATACCGTGGTCAAC | pMTL80000 series plasmids | ( Sheridan et al., 2019 ) |
| PS#MTL-rev | CTGCTGAAGCCAGTTACC | pMTL80000 series plasmids | ( Sheridan et al., 2019 ) |
| FD1 | AGAGTTTGATCCTGGCTCAG | Full 16S rRNA gene | ( Wood et al., 1998 ) |
| RP2 | ACGGCTACCTTGTTACGACTT | Full 16S rRNA gene | ( Wood et al., 1998 ) |
| 519R | GWATTACCGCGGCKGCTG | 16S rRNA gene (universal) | ( Turner et al., 1999 ) |
| 926F | ACTCAAAGGAATTGACGG | 16S rRNA gene (universal) | ( Muyzer et al., 1995 ) |
Equipment
Pipettes Gilson P1000 (Gilson, catalog number: F123602)
Pipettes Gilson P200 (Gilson, catalog number: F123601)
Pipettes Gilson P20 (Gilson, catalog number: F123600)
Pipettes Gilson P2 (Gilson, catalog number: F144801)
Concept Plus Anaerobic Workstation, Ruskinn Technology
Hungate tubes (Sciquip, catalog number: 2047-00125)
Hungate lids butyl rubber septa (Sciquip, catalog numbers: 2047-11600 and 2047-16000)
Wheaton bottles (Merck, catalog number: 33110-U)
CO2 hooks (made in-house) and CO2 piped gas supply
Shaking Incubator for E. coli growth (Sanyo Orbital incubator)
Static Incubator for anaerobic bacteria (Sciquip incu-160S)
Gene pulser (Bio-Rad, model: 1652078)
Platform rocker (Stuart, model: STR6)
UV-linker, Bio-Rad GS Gene linker UV chamber (UVP CL-1000 Ultraviolet Crosslinker)
Hybridizer (UVP laboratory products HB 1000 hybridizer)
Blot transfer pump (Hybaid Vacu-aid blot processing pump, DA7C.VAC/T)
Centrifuge (Jouan MR1822)
Transilluminator (UVtec BXT-20.L)
Procedure
-
Electroporation of plasmids into E. coli CA434
Pre-chill 1 mm electroporation cuvette in refrigerator overnight.
Dilute 1 µl of plasmid solution (10 µg/ml) in 4 µl of dH2O (final concentration 2 µg/ml).
Thaw 55 µl electrocompetent E. coli CA434 (prepared in house following standard protocols, https://www.protocols.io/view/Making-your-own-electrocompetent-cells-imsv6m) on ice and add the plasmid solution, mixing gently.
Transfer the mixture to the cuvette and place in Gene Pulser.
Electroporate at 1.8 kV, 200 Ohms, 25 µF.
Transfer into 1 ml SOC medium, pre-warmed to 37 °C.
Incubate for 1 h at 37 °C, 200 RPM.
Dilute in sterile dH2O in ten-fold dilutions (101, 102 and 103).
Spread 50 µl of these dilutions onto LA plates (see Recipes) supplemented with chloramphenicol 10 µg/ml (Cm10) and incubate at 37 °C for 24 h or until colonies appear.
Pick individual colonies into LB Cm10 and incubate at 37 °C, 200 RPM for 24 h.
-
Anaerobic culturing of recipient bacterium
Prepare anaerobic media YCFAGSC and AMM in broth as 7.5 ml aliquots in Hungate tubes, sealed with butyl rubber septa and 2% agar solutions divided into 100 ml aliquots in Wheaton bottles, with all dispensing and inoculating carried out under CO2 using the Hungate technique as described by Bryant, 1972 (Figure 1).
Incubate anaerobic YCFAGSC liquid cultures at 37 °C, without agitation in Hungate tubes.
Pour anaerobic agar media into Petri plate (20 ml per plate) within anaerobic workstation (gas mix CO2:N2:H2, ratio 7:11:2) approximately 24 h before use, enabling agar to adjust to anaerobic atmosphere.
Incubate anaerobic agar cultures at 37 °C in an anaerobic workstation.
-
Mating of donor E. coli CA434 and recipient bacterium
-
Prepare 7.5 ml overnight culture of recipient. Donor, recipients and plasmids are described in Table 1.
*pBP1, pCB102, pCD6 and pIM13 (replicons of these plasmids). ColE1 (Gram-negative replicon), catP (chloramphenicol resistance gene), traJ (origin of transfer) and MCS (multiple cloning site).
Inoculate donor E. coli CA434 in 40 ml of LB Cm10 in 50 ml conical centrifuge tubes and incubate at 37 °C, 200 RPM overnight.
Centrifuge donor overnight culture at 1,200 × g for 10 min.
Decant supernatant and resuspend pellet in 20 ml of anaerobic PBS.
Centrifuge donor again at 1,200 × g for 10min.
Decant supernatant in anaerobic cabinet.
Transfer overnight culture of recipient bacterium (grown as described in Steps B1 and B2) to anaerobic cabinet.
Resuspend donor pellet in 1 ml of recipient overnight culture in anaerobic cabinet.
Spot 100 µl of this solution on the centre of AMM agar plate and incubate in the anaerobic cabinet at 37 °C for 48 h.
Scrape mating mix off centre of AMM plate with sterile loop and suspend in 500 µl of anaerobic PBS.
Spread 50 µl of this solution on to the YCFAGSC plates supplemented with either 5 or 7.5 µg/ml chloramphenicol and incubate anaerobically at 37 °C until colonies appear (usually 48 to 96 h).
Restreak single colonies on fresh YCFAGSC plates supplemented with either 5 or 7.5 µg/ml chloramphenicol and incubate anaerobically at 37 °C until single colonies appear. This may take 3 days incubation.
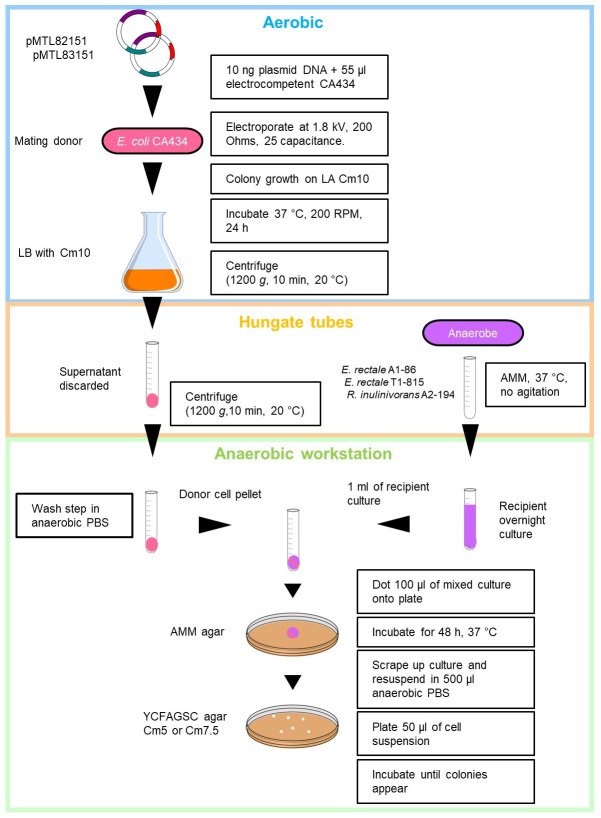
Diagrammatic representation of the optimised conjugation protocol shown in Figure 2.
-
-
Verification of putative transconjugants
It is good practice to perform various tests to confirm the validity of transconjugants. Some simple tests can readily eliminate bacteria that are not transconjugants, and can save time and expense.
Incubate putative transconjugants aerobically on AMM agar at 37 °C. The recipient bacterium is incapable of aerobic growth while the donor E. coli bacterium will grow.
Gram-stain using standard procedures. Coccus species are common contaminants in gut microbiology and are easily differentiated from the rod-like Roseburia and Eubacterium by Gram-staining.
-
PCR amplify the 16S rRNA gene directly from colonies or liquid culture using the universal bacterial 16S rRNA gene primers FD1 and RP2 (Table 2) (annealing temp. 52 °C), generating an amplicon of 1,495 bp. PCR amplification conditions are provided in Table 3.
Nucleotide code: Guanine (G), adenine (A), thymine (T), cytosine (C), adenine or thymine (W) and guanine or thymine (K)
Sanger sequence the resulting amplicon with the primers 519R and 926F (Table 2).
BLASTn query each sequence against the NCBI 16S rRNA gene database to confirm identity.
Confirm the presence of the plasmid in putative transconjugants by amplifying a nucleotide sequence common to all of the modular plasmids but absent in the recipient's chromosome. The primers PS#MTL-for and PS#MTL-rev (Table 2) (annealing temp 60 °C) amplify a 514 bp region incorporating sections of catP gene and ColE1 Gram-negative replicon.
Expected results and comments are described in Table 4.
-
Verification of autonomous plasmid replication by Southern blotting
Extract DNA from transconjugants using the Wizard Genomic DNA Purification kit, following the manufacturer's instructions.
-
Digest 1 µg of extracted DNA with HindIII for 3 h at 37 °C to produce restriction fragments of various sizes.
Note: HindIII only cuts the plasmid once, resulting in a single linear fragment, whereas restriction of the genomic DNA results in fragments of various sizes.
Separate restriction fragments by size by gel electrophoresis (0.8% agarose, TBE).
Photograph gel image with transilluminator to facilitate size inference in the final blot.
Gently rock gel in depurination solution (0.25 M HCl) for 7 min and rinse in ddH2O (double distilled water).
Gently rock the gel in denaturation solution (0.5 M NaOH, 1.5 M NaCl) for 30 min three times and rinse in ddH2O.
Gently rock the gel in neutralisation solution (0.5 M Tris-HCl [pH 7.4], 3 M NaCl) for 30 min three times and rinse in ddH2O.
Transfer DNA from gel to nylon membrane using the Hybaid blotter (1.5 h) and UV-crosslinked.
Create probe by digesting pMTL83151 with HindIII and ApaLI. This produces two fragments (~3,000 bp and ~1,500 bp), the smaller of which is specific to a region common to all of the plasmids, but not present in the recipient chromosome. Gel purify the smaller fragment and use as probe template DNA.
Perform Southern blotting with DIG High Prime DNA Labelling and Detection Starter Kit II, following the manufacturer's instructions.
-
Visualise hybridizing bands by exposing membrane to x-ray film.
This protocol enables the establishment and verification of autonomously replicating plasmids transferred into Roseburia inulinivorans and Eubacterium rectale and will provide an essential tool in investigating the biochemistry and physiology of these important organisms by genetic manipulation.
Figure 1. Culturing of strictly anaerobic bacteria using the Hungate methodology (growth in tubes) and anaerobic workstation (growth on Petri plates).
Figure 2. Detailed diagrammatic representation of the optimised conjugation protocol for Roseburia inulinivorans and Eubacterium rectale.
Supplemented with 5 or 7.5 µg/ml chloramphenicol (Cm5 or Cm7.5).
Table 3. PCR protocol.
| Temperature | Time | |
| Initial denaturation | 94 °C | 5 min |
| 25 cycles | 95 °C | 30 s |
| Depending on primers | 30 s | |
| 72 °C | 2 min | |
| Final Extension | 72 °C | 8 min |
| Hold | 4 °C |
Table 4. Verification procedures for putative transconjugants.
| Confirmation | Expected result | Comment | |
|---|---|---|---|
| Gram-staining | Absence of contamination | Pink or purple bacilli | Common contaminants, such as coccus species are easily distinguished from Roseburia and Eubacterium by Gram staining. |
| 16S rRNA gene sequencing | Absence of contamination | Match to recipient DNA | |
| Plasmid-specific PCR | Presence of plasmid | 514 bp amplicon | Recipient chromosomes lack the amplified region of DNA |
| Anaerobic incubation | Absence of donor | No aerobic growth | Recipient anerobes are incapable of aerobic growth |
| Southern blotting | Autonomous replication | Single band of same size as linear plasmid | Chromosome insertion would change the size of the probe hybridising band |
Recipes
-
Resazurin solution
Add 100 mg of powdered resazurin to 100 ml ddH2O
-
Anaerobic phosphate buffered saline (PBS)
Dissolve PBS tablets in 1 L of ddH2O and add 1 ml resazurin solution
Place solution in boiling waterbath for 15 min
Bubble solution with 100% CO2 until liquid turns from purple to clear
Dispense in 100 ml aliquots in Wheaton bottles
-
Mineral solution 1
K2HPO4 3.0 g
ddH2O to 1 L
Store at 4 °C
-
Mineral solution 2
KH2PO4 3.0 g
(NH4)2SO4 6.0 g
NaCl 6.0 g
MgSO4 0.6 g
CaCl2 20.6 g
ddH2O to 1 L
Store at 4 °C
-
Short chain fatty acid solution
Acetic acid 17 ml
Propionic acid 6 ml
n-Valeric acid 1 ml
Iso-Valeric acid 1 ml
Iso-Butyric acid 1 ml
Store at 4 °C
-
Vitamin solution 1
Biotin 1 mg
Cobalamin 1 mg
p-Aminobenzoic acid 3 mg
Folic acid 5 mg
Pyridoxamine 15 mg
ddH2O to 100 ml
Store at -20 °C
-
Vitamin solution 2
Thiamin 5.0 mg
Riboflavin 5.0 mg
ddH2O to 100 ml
-
Haemin solution
KOH 0.28 g
Ethanol 95% 25 ml
Haemin 100 mg
ddH2O to 100 ml
Store at 4 °C
-
SOC
Bacto tryptone 2 g
Yeast Extract 0.5 g
NaCl 200 μl of 5 M
KCl 250 μl of 1 M
ddH2O to 100 ml
Stir to dissolve, autoclave, then cool to room temperature
Add 1 ml of filter sterile 2 M Mg stock solution (1 M MgCl2·6H2O and 1 M MgSO4·7H2O) to give a final conc of 20 mM
Before use add 20 µl of 1 M sterile glucose per 1 ml of SOC
-
LB
Bacto tryptone 1 g
Yeast extract 0.5 g
NaCl 1 g
Deionized H2O to 100 ml
-
LA
Bacto tryptone 1 g
Yeast extract 0.5 g
NaCl 1 g
Agar 1.5 g
Deionized H2O to 100 ml
-
YCFAGSC and AMM
Bacto casitone 10.0 g
Yeast extract 2.5 g
NaHCO3 4.0g
Glucose 2.0 g
Soluble starch 2.0 g
Cellobiose 2.0 g
Mineral solution 1 150.0 ml
Mineral solution 2 150.0 ml
Haemin solution 10.0 ml
Vitamin solution 1 (before autoclaving) 1.0 ml
Vitamin solution 2 (after autoclaving) 1.0 ml
Resazurine 1.0 ml
L-cysteine 1.0 g
Acetic acid (0.7 ml) to make AMM or short chain fatty acid solution (3.1 ml) to make YCFAGSC
Add 20 g of agar to these recipes to make AMM or YCFAGSC agar
Distilled water up to 1 L
Acknowledgments
The Rowett Institute (University of Aberdeen) receives financial support from the Scottish Government Rural and Environmental Sciences and Analytical Services (RESAS). The protocol is derived from work published in “Heterologous gene expression in the human gut bacteria Eubacterium rectale and Roseburia inulinivorans by means of conjugative plasmids” Anaerobe 59: 131-140 (2019) ( Sheridan et al., 2019 ).
Competing interests
The authors state that there are no competing interests
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Aquino de Muro M. and Priest F. G.(2000). Construction of chromosomal integrants of Bacillus sphaericus 2362 by conjugation with Escherichia coli . Res Microbiol 151(7): 547-555. [DOI] [PubMed] [Google Scholar]
- 2. Barcenilla A., Pryde S. E., Martin J. C., Duncan S. H., Stewart C. S., Henderson C. and Flint H. J.(2000). Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol 66(4): 1654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bryant M. P.(1972). Commentary on the Hungate technique for culture of anaerobic bacteria. Am J Clin Nutr 25(12): 1324-1328. [DOI] [PubMed] [Google Scholar]
- 4. Cockburn D. W., Orlovsky N. I., Foley M. H., Kwiatkowski K. J., Bahr C. M., Maynard M., Demeler B. and Koropatkin N. M.(2015). Molecular details of a starch utilization pathway in the human gut symbiont Eubacterium rectale . Mol Microbiol 95(2): 209-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duncan S. H., Aminov R. I., Scott K. P., Louis P., Stanton T. B. and Flint H. J.(2006). Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces . Int J Syst Evol Microbiol 10): 2437-2441. [DOI] [PubMed] [Google Scholar]
- 6. Duncan S. H., Hold G. L., Barcenilla A., Stewart C. S. and Flint H. J.(2002). Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces . Int J Syst Evol Microbiol 5): 1615-1620. [DOI] [PubMed] [Google Scholar]
- 7. Heap J. T., Pennington O. J., Cartman S. T., Carter G. P. and Minton N. P.(2007). The ClosTron: a universal gene knock-out system for the genus Clostridium . J Microbiol Methods 70(3): 452-464. [DOI] [PubMed] [Google Scholar]
- 8. Heap J. T., Pennington O. J., Cartman S. T. and Minton N. P.(2009). A modular system for Clostridium shuttle plasmids . J Microbiol Methods 78(1): 79-85. [DOI] [PubMed] [Google Scholar]
- 9. Muyzer G., Teske A., Wirsen C. O. and Jannasch H. W.(1995). Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments . Arch Microbiol 164(3): 165-172. [DOI] [PubMed] [Google Scholar]
- 10. Neville B. A., Sheridan P. O., Harris H. M., Coughlan S., Flint H. J., Duncan S. H., Jeffery I. B., Claesson M. J., Ross R. P., Scott K. P. and O'Toole P. W.(2013). Pro-inflammatory flagellin proteins of prevalent motile commensal bacteria are variably abundant in the intestinal microbiome of elderly humans. PLoS One 8(7): e68919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Purdy D., O'Keeffe T. A., Elmore M., Herbert M., McLeod A., Bokori-Brown M., Ostrowski A. and Minton N. P.(2002). Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier . Mol Microbiol 46(2): 439-452. [DOI] [PubMed] [Google Scholar]
- 12. Scott K. P., Martin J. C., Campbell G., Mayer C. D. and Flint H. J.(2006). Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium"Roseburia inulinivorans" . J Bacteriol 188(12): 4340-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scott K. P., Martin J. C., Chassard C., Clerget M., Potrykus J., Campbell G., Mayer C. D., Young P., Rucklidge G., Ramsay A. G. and Flint H. J.(2011). Substrate-driven gene expression in Roseburia inulinivorans: importance of inducible enzymes in the utilization of inulin and starch . Proc Natl Acad Sci U S A 1: 4672–4679.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scott K. P., Martin J. C., Mrazek J. and Flint H. J.(2008). Transfer of conjugative elements from rumen and human Firmicutes bacteria to Roseburia inulinivorans . Appl Environ Microbiol 74(12): 3915-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sheridan P. O., Martin J. C., Minton N. P., Flint H. J., O'Toole P. W. and Scott K. P.(2019). Heterologous gene expression in the human gut bacteria Eubacterium rectale and Roseburia inulinivorans by means of conjugative plasmids . Anaerobe 59: 131-140. [DOI] [PubMed] [Google Scholar]
- 16. Sheridan P. O., Martin J. C., Lawley T. D., Browne H. P., Harris H. M. B., Bernalier-Donadille A., Duncan S. H., O'Toole P. W., K, P. S. and H, J. F. (2016). Polysaccharide utilization loci and nutritional specialization in a dominant group of butyrate-producing human colonic Firmicutes. Microb Genom 2(2): e000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Turner S., Pryer K. M., Miao V. P. and Palmer J. D.(1999). Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol 46(4): 327-338. [DOI] [PubMed] [Google Scholar]
- 18. Williams D. R., Young D. I. and Young M.(1990). Conjugative plasmid transfer from Escherichia coli to Clostridium acetobutylicum . J Gen Microbiol 136(5): 819-826. [DOI] [PubMed] [Google Scholar]
- 19. Wood J., Scott K. P., Avgustin G., Newbold C. J. and Flint H. J.(1998). Estimation of the relative abundance of different Bacteroides and Prevotella ribotypes in gut samples by restriction enzyme profiling of PCR-amplified 16S rRNA gene sequences . Appl Environ Microbiol 64(10): 3683-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhernakova A., Kurilshikov A., Bonder M. J., Tigchelaar E. F., Schirmer M., Vatanen T., Mujagic Z., Vila A. V., Falony G., Vieira-Silva S., Wang J., Imhann F., Brandsma E., Jankipersadsing S. A., Joossens M., Cenit M. C., Deelen P., Swertz M. A., LifeLines Cohort S., Weersma R. K., Feskens E. J., Netea M. G., Gevers D., Jonkers D., Franke L., Aulchenko Y. S., Huttenhower C., Raes J., Hofker M. H., Xavier R. J., Wijmenga C. and Fu J.(2016). Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352(6285): 565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]