Abstract
Cytoplasmic localization of mRNAs is common to all organisms and serves the spatial expression of genes. Cis-acting RNA signals (mostly found in the mRNA’s 3'-UTR), called zipcodes recruit trans-acting RNA-binding proteins that facilitate the localization of the mRNA. UV-cross-linking or affinity purification has been applied to identify such proteins but suffer from the need for stable RNA-protein binding or direct contact of protein and RNA. To identify stably or transiently interacting proteins that directly or indirectly associated with the localization elements and the body of the mRNA, we developed an in vivo proximity labeling method we call RNA-BioID. In RNA-BioID, we tether a fusion of the BirA* biotin ligase and the MS2 coat protein (MCP) at the 3'-UTR of MS2-tagged β-actin mRNA in vivo. Exposing BirA* expressing cells to biotin in the media and induces biotinylation of β-actin mRNA-associated proteins that can be isolated with streptavidin beads. This technique allowed us to identify by mass spectrometry analysis the β-actin mRNA 3'-UTR-interacting proteome in fibroblasts. The protocol can be useful to identify the interacting proteome of any mRNA in mammalian cells.
Keywords: RNA-BioID, Proximity labeling, Biotinylation, RNA-binding protein, RNA tagging
Background
Localization of mRNAs to specific subcellular sites is a widespread phenomenon and has been observed from bacteria to humans (Bovaird et al., 2018; Fei and Sharma, 2018). Diverse human pathologies of the neural system are linked to defects in mRNA localization (Tolino et al., 2012; Bovaird et al., 2018). Additionally, this mechanism is essential for developmental processes including mesoderm formation in the clawfrog Xenopus laevis or the determination of the embryonic body axes of the fruit fly Drosophila melanogaster (Jansen, 2001). To achieve intracellular asymmetry of transcripts, mRNAs are incorporated into motor-protein containing particles that can move along the cytoskeleton to distinct cellular sites, where the transcripts are locally translated (Marchand et al., 2012; Buxbaum et al., 2015). Such particles containing mRNA and RNA-binding proteins (RBPs) are beginning to assemble during transcription and undergo several changes upon maturation in the cytoplasm (Holt et al., 2019). Localized mRNAs differ from non-localized ones by containing cis-acting motifs termed localization elements or zip codes that are recognized by specific RBPs (Bovaird et al., 2018). Zipcodes can be positioned in the coding sequence but are mainly found in the 3' untranslated region (3'-UTR; Bovaird et al., 2018).
A well-studied example is the 54-nt long zip code in the 3'-UTR of β-actin mRNA that is essential for targeting the mRNA to the leading edge of fibroblasts as well as to growth cones and dendrites in neurons (Ross et al., 1997; Eom et al., 2003; Buxbaum et al., 2015). The zip code is recognized by the RBP ZBP1 (aka IGF1 or IGF2BP1) that is not only essential for the localization of β-actin mRNA (Ross et al., 1997; Oleynikov and Singer, 2003) but also represses the translation of the mRNA until it has arrived at the target site (Hüttelmaier et al., 2005). However, additional RBPs and supplementary factors are involved in splicing, translational regulation, stabilization, transport, and decay of β-actin mRNA.
Obtaining a complete set of the proteins that associate with any mRNA remains challenging, despite a large range of high-throughput methods available so far that include diverse pulldown approaches as well as cross-linking and immunoprecipitation (CLIP) (for an overview see Ramanathan et al., 2019). A specific challenge that remains is the identification of transient interactors that are not directly contacting the RNA. To solve this, we recently introduced a proximity biotinylation method-based on tethering the BioID biotin ligase BirA (BirA*) via MS2 coat binding protein (MCP) at the 3'-UTR of MS2-tagged β-actin mRNA (Park et al., 2014). In the presence of biotin, BirA* generates AMP-biotin (‘activated biotin’), which reacts with accessible lysine residues in its vicinity (10-20 nm). After lysis, biotinylated proteins can be isolated via streptavidin affinity purification and identified using standard mass spectrometry techniques. The RNA-BioID method can in principle be applied to any MS2-tagged mRNA and allows the identification of its associated proteins as well as probing its environment by indirect or transient interactions.
Materials and Reagents
Beaker (500 ml, 1 L)
50 ml conical tube
21 G needle
T175 flasks (Corning, catalog number: CLS431080)
10 cm culture dishes (Corning, catalog number: 430167)
6-well dishes (Corning, catalog number: CLS3516)
FACS tubes (Falcon, catalog number: 352235)
Reprosil-Pur C18-AQ, 1.9 μm resin (Dr. Maisch GmbH)
25 cm fused silica emitters with an inner diameter of 75 μm (DNU-MS GbR)
Bench cleaner (terralin® PAA, catalog number: 126203, store at room temperature)
1.5 ml safe seal pyrogen-free tubes (Sarstedt, catalog number: 72.706.201)
0.45 μm filter (Sartorius, catalog number: 10109180)
pH strip 0.0-6.0; 6.0-10.0 (MN, Part No: 1138922; 1178895)
PVDF membrane (GE Healthcare AmershamTM HybondTM-P Membranes, catalog number:10471085, store at room temperature)
Phage-ubc-nls-2xmcp-egfp-BirA* (Addgene Plasmid, catalog number: 131132) or phage-ubc-nls-2xmcp-egfp-cMyc BirA* (Addgene Plasmid, catalog number: 131133) or phage-ubc-nls-2xmcp-mCherry-BirA* (Addgene Plasmid, catalog number: 131136)
pCEP4-tat (Addgene Plasmid, catalog number: 22502)
pRSV-Rev (Addgene Plasmid, catalog number: 12253)
pMDLg/pRRE (Addgene Plasmid, catalog number: 12251)
pMD2.G (Addgene Plasmid, catalog number: 12259)
Hek293 FT cells (Thermo Scientific, catalog number: R70007)
Wildtype mouse embryonic fibroblasts (MEFs, e.g., Park et al., 2014)
Magnetic sepharose streptavidin Beads (GE, catalog number: 28985799, store at 4 °C)
MBS MEFs (MEFs with 24 MS2 binding sites (MBS) inserted in the 3’-UTR of β-actin gene loci; see Park et al., 2014)
Fugene 6 (Promega, catalog number: E2691, store at 4 °C)
Trypsin (Sigma-Aldrich, catalog number: T4049, store at -20 °C)
Fetal calf serum (FBS; Sigma-Aldrich, catalog number: F4135, store at -20 °C)
Alkaline phosphatase-conjugated streptavidin
Trypsin, sequencing grade, modified, 0.5 μg/μl (Promega, catalog number: V5113)
LentiX concentrator (Takara, catalog number: 631231, store at 4 °C)
Lysyl endopeptidase Lys-C (Waco, catalog number: 129-02541), stocks: 0.5 μg/μl in H2O
OptiMEM (Thermo scientific, catalog number: 31985062, store at 4 °C)
DMEM high glucose media (Sigma-Aldrich, catalog number: D6429, store at 4 °C)
1x PBS (Sigma-Aldrich, catalog number: D6429, store at 4 °C)
Penicillin-Streptomycin solution (Sigma-Aldrich, catalog number: P4333, store at -20 °C)
Bleach powder (Schülke perform® disinfectant, catalog number: 107912, store at room temperature)
Hand disinfectant (STERILLIUM Virugard solution, catalog number: 126064, store at room temperature)
Biotin (Sigma-Aldrich, catalog number: B4501, store at 4 °C)
Complete protease inhibitor, EDTA free (Roche, catalog number: 11697498001, store at 4 °C)
5x Roti Quant Bradford reagent (Roth, catalog number: K015.3, store at 4 °C)
SDS (sodium dodecyl sulfate) (Roth, catalog number: CN30.1, store at room temperature)
Bromophenol blue (Roth, catalog number: A512.1, store at room temperature)
Glycerol (Sigma-Aldrich, catalog number: G6279, store at room temperature)
Nu-PAGE SDS-PAGE gel 4-12% (Invitrogen, catalog number: WG1402BOX, store at 4 °C)
NuPAGE MOPS SDS Running Buffer (20x) (Invitrogen, catalog number: NP0001, store at 4 °C)
β-mercaptoethanol (Roth, catalog number: 42273, store at room temperature)
p-Nitrotetrazoliumblauchlorid (NBT) (Roth, catalog number: 4421.2, store at -20 °C)
5-Bromo-4-chloro-3-indolyl-phosphate Disodium Salt (BCIP) (Roth, catalog number: A155.2, store at -20 °C)
Ammonium Bicarbonate (Sigma-Aldrich, catalog number: 09830, store at room temperature)
NuPage SDS-PAGE gels (Thermo scientific, catalog number: NP0315BOX, store at 4 °C)
PHOENIX Peptide Cleanup Kit (PreOmics, catalog number: P.O.00023)
Dithiothreitol (VWR, catalog number: 0281-5G, store at room temperature)
Urea (Merck, catalog number: 1.08488.1000, store at room temperature)
Thiourea (Merck, catalog number: 1.07979.0250, store at room temperature)
Iodoacetamide (Merck, catalog number: 8.04744.0025, store at room temperature)
Acetonitrile (Merck, catalog number: 1.00030.2500)
Formic acid (Sigma-Aldrich, catalog number: 33015-500ML)
Acetic acid (Roth, catalog number: 3738.5)
Trifluoracetic acid (TFA) (Merck, catalog number: 1.08262.0100)
NaCl
MgCl2
Tris-Cl
TBS
Tween 20
Deoxycholate
HEPES
EDTA
LiCl
Complete media (500 ml) (see Recipes)
Serum-free media (100 ml) (see Recipes)
Biotin Media (100 ml) (see Recipes)
50 mM Biotin solution (see Recipes)
BCIP (5-bromo-4-chloro-3-indolyl phosphate) (see Recipes)
NBT (Nitro blue tetrazolium) (see Recipes)
AP (Alkaline phosphatase) substrate buffer (see Recipes)
1x TBS (Tris-buffered saline) (see Recipes)
1x TBST (Tris-buffered saline-Tween) (see Recipes)
Lysis Buffer (see Recipes)
Wash Buffer 1 (see Recipes)
Wash Buffer 2 (see Recipes)
Wash Buffer 3 (see Recipes)
Wash Buffer 4 (see Recipes)
20 mM ABC (Ammonium bicarbonate) buffer (see Recipes)
2x SDS (Sodium dodecyl sulfate) sample buffer (see Recipes)
Denaturation buffer (see Recipes)
Reduction buffer (see Recipes)
Alkylation buffer (see Recipes)
Solvent A* (see Recipes)
Solvent A (see Recipes)
HPLC solvent A (see Recipes)
HPLC solvent B (see Recipes)
Equipment
37 °C water bath (Fisher Scientific, Isotemp 205 Water Bath)
Cell culture incubator, set to 5% CO2 (Thermo Scientific, HeracellTM 150i CO2 Incubators)
Centrifuge (Eppendorf, model: 5430)
Cell sorter (BD, model: FACS Aria I)
End-to-end rotator (Heidolph, model: REAX 2)
Thermomixer (Eppendorf, catalog number: 5350 000.013)
Magnetic stand (Bio-Rad, catalog number: 1614916)
Centrifuge (Heraeus Fresco 17, Thermo Fisher Scientific, catalog number: 75002420)
Western blot SDS-PAGE running apparatus (Bio-Rad, Mini Trans-Blot® Cell)
Western blot SDS-PAGE wet transfer apparatus (Bio-Rad, Mini Trans-Blot Module, catalog number: 1703935)
Concentrator plus vacuum centrifuge (Eppendorf, catalog number: 5305 000.304)
EASY-nLC 1200 (Thermo Fisher Scientific) or any (U) HPLC system compatible with the MS system of choice
LTQ Orbitrap or Q Exactive mass spectrometer (Thermo Fisher Scientific) or any state-of-the-art high-resolution mass spectrometer.
Fluorescence microscope (Zeiss CellObserver, 40x, 1.4 NA oil immersion objective, GFP filterset 38HE)
Software
MaxQuant software suite (www.maxquant.org, Max-Planck-Institute of Biochemistry, Martinsried), or other programs that can be used for downstream analysis
Perseus (http://www.coxdocs.org/doku.php?id=perseus:common:download_and_installation, Max-Planck-Institute of Biochemistry, Martinsried)
Procedure
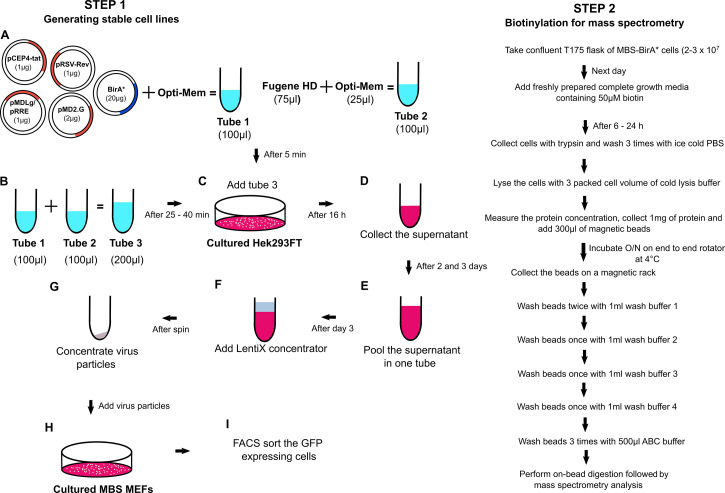
A flowchart demonstrating the general outline of the experimental procedure is shown in Figure 1.
Figure 1. Flowchart of the RNA-BioID procedure.
-
Generating stable cell lines expressing an MCP-GFP-BirA* fusion protein
The MCP-GFP-BirA* gene fusion under control of the UBC promoter was integrated into a third-generation lentiviral vector and is available in three different versions via Addgene (Addgene 131132/131133/131136). The three different versions can be used in similar ways.
For transduction, use the MCP-GFP-BirA* plasmid Gag/Pol in combination with Env and VSV-G plasmids to assemble and generate the complete viral particles in Hek293FT cells.
On the day before transfection, seed 4 x 106 Hek293FT cells in complete growth media in a 10 cm dish.
On the day of transfection, set up two 1.5 ml centrifuge tubes, each containing 500 μl of OptiMEM media.
-
To one tube, add the corresponding amounts of the following plasmids:
MCP-GFP-BirA* backbone plasmid: 20 µg
pCEP4-tat 1 μg
pRSV-Rev 1 μg
pMDLg/pRRE 1 μg
pMD2.G (Insert name VSV-G) 2 μg
In total 25 μg of plasmid DNA
To the other OptiMEM-containing centrifuge tube, add 75 µl Fugene HD transfection reagent (3 μl per μg plasmid).
Incubate both tubes separately for 5 min at room temperature.
Add the OptiMEM media containing plasmid DNA to the tube containing the OptiMEM-Fugene HD mix, mix briefly by pipetting 2-3 times, and incubate for at least 25 min (up to 45 min) at room temperature. This results in the formation of the DNA-Fugene complex.
Exchange the media of the 10 cm dish containing Hek293FT cells with fresh 10 ml of complete growth media.
-
Add the DNA-Fugene complex on top of the cells in a dropwise manner. Swirl the dish gently to mix the Fugene-DNA mix with the medium and incubate the dishes at 37 °C for at least 16 h.
Notes:
For biosafety reasons, keep the dishes on a tray inside the incubator to avoid spilling of the media in the incubator due to handling errors.
At this stage, the lentivirus is being produced, so strictly follow the biosafety class II regulations. After the 16 h incubation period, all dishes and pipettes that have and will come in contact with virus-containing solutions have to be treated with bleach prior to disposal. Always wear a lab coat and wipe the entire hood with ethanol. When you leave the working space treat your gloves with bleach as well.
Before the end of the 16 h incubation, prepare 500 ml of 10% bleach and keep it in a large beaker in the tissue culture hood for treating surfaces (prepare fresh 19% bleach every day, it remains effective only for 24 h).
Once the bleach solution is ready under the hood, carefully remove 8 ml of the media from the 10 cm dish and transfer it to a 50 ml conical tube. Keep it at 4 °C overnight.
Add 8 ml of fresh complete growth media to the dish and return it to the incubator for another 24 h.
At this time, seed wildtype (WT) MEFs and MBS MEFs (3 x 106/10 cm dish) and incubate at 37 °C in a cell culture incubator (5% CO2).
Repeat Steps A11-A13 twice more during the consecutive days. Collect the media from each day and pool it in the same 50 ml conical tube from Step A12, keep at 4 °C. After collecting all supernatants, spin the conical tube for 10 min at 500 × g and 4 °C to pellet large debris.
Filter the supernatant from this centrifugation through a 0.45 μm filter.
Concentrate the lentiviral particles in the supernatant with a LentiX concentrator according to manufacturer’s instructions.
After concentrating the viral particles, add 1 ml of serum-free media (Recipe 2) and carefully resuspend the pellet.
Remove the dishes with WT MEFs and MBS-MEFs from the incubator for transduction, replace the old media with fresh complete medium (Recipe 1) and add 100 μl of the viral particle solution per 10 cm dish with 10 ml of media. Store the rest of the virus particles at -80 °C. Place the MEFs back into the cell culture incubator and leave for 24 h.
Check the viral transduction efficiency under a fluorescence microscope (filter setting for GFP). If no GFP fluorescence is visible, repeat Step A19 with another aliquot of the stored viral particles.
Fluorescent cells should stably express the MCP-GFP-BirA* fusion protein.
On the last day of collection, bleach all the dishes that came in contact with the viral particles.
-
Isolating cells expressing a functional MCP-GFP-BirA* fusion protein
After generation of stable cell lines (WT-BirA*: WT MEFs expressing MCP-GFP-BirA*; MBS-BirA*: MEFs with 24x MBS in the 3'-UTR of β-actin and expressing MCP-GFP-BirA*), sort positive, GFP-expressing cells by fluorescence-activated cell sorting against GFP, in FACS (fluorescence-activated cell sorting) tube containing 1 ml of complete growth media. This can be done by a core facility at your institution.
After sorting, spin down the cells at 500 × g for 5 min at room temperature. Resuspend and, depending on the number of positive cells identified in cell sorting, culture them either on 10 cm dish or bigger dishes (10,000 cells/each 10 cm dish).
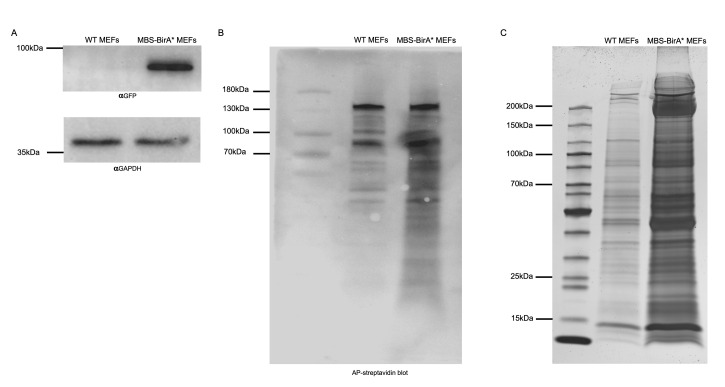
When at least 10,000 cells have been isolated, test for the GFP expression levels in both transduced cell types (WT-BirA*/MBS-BirA*). For checking GFP expression, prepare cell lysates in the lysis buffer (Recipe 10). Add 3 packed cell volume (PCV) of lysis buffer and and pass 10-12 times through a 21 G needle. Centrifuge the lysate at 12,000 × g for 10 min at 4 °C to remove cell debris. Take out the soluble fraction and keep in a new 1.5 ml centrifuge tube at 4 °C. Use at least 10 µg of total protein for a Western blot against GFP and a housekeeping gene (e.g., GAPDH). Use untransfected WT MEFs as negative control and GAPDH as loading control. This step is important to check whether the full NLS-HA-2XMCP-eGFP-BirA* construct is expressing giving a band at 91.4 kDa (Figure 2A).
To check for the activity of BirA* and biotinylation efficiency, use a 6-well dish. Seed duplicates of 3 x 104 cells of WT untransfected, WT-BirA* and MBS-BirA* and culture them in complete media for 24 h. On the next day, exchange the media. Use complete media containing 50 μM of biotin with only on one well of each duplicate. To the second set of duplicates just exchange the media with new complete media without biotin.
Culture the cells with the new media (with or without biotin) for at least 6 h and up to 24 h. Biotinylation efficiency might not increase after 6 h, depending on the cell line.
At the end of the incubation, wash the wells of the 6-well dishes two times with cold PBS and finally isolate the cells by adding trypsin.
Isolate the cells by adding 500 μl/well of warm trypsin solution and incubate the dish at 37 °C for 2-3 min. After 2 min, check under a cell culture microscope (at 10x magnification) if all cells are completely detached. If not, leave the cells in the incubator but do not incubate longer than 5 min.
Remove the solution with the cells and transfer them to a 15 ml conical tube. Add 6 ml of complete media containing FBS to deactivate the trypsin.
Spin down the cells for 5 min at 500 × g at room temperature.
Remove the media and add 1 ml of cold 1x PBS, resuspend the pellet with a 1 ml tip and pellet the cells again by centrifugation for 4 min at 500 × g at 4 °C.
Repeat Step B10 to completely remove the media.
Add 3 PCV lysis buffer containing 1x complete protease inhibitors to each pellet, resuspend cells and keep them on ice for 10 min. Lyse the cells by passing the pellet 10-15 times through a 21 G needle. After the lysis, spin the lysates for 10 min at 12,000 × g at 4 °C to remove the unbroken cells.
Determine the protein concentration (e.g., by a Bradford assay following the manufacturer’s protocol).
Mix 10 µg of total protein with SDS-PAGE loading buffer.
Boil the mixture for 5 min at 95 °C and afterward spin down debris for 3 min at 12,000 × g at room temperature.
Load the supernatant from step 15 on a Nu-PAGE SDS-PAGE gel in 1x MOPS running buffer and run at 200 V until the front dye exits the gel.
Transfer the protein from the gel onto an activated PVDF membrane by semi-dry Western blotting (2.5 h at 250 mA).
After the transfer is complete, wash the PVDF membrane in a small box with TBST (Recipe 9) twice for 5 min each at room temperature with rotation or tilting.
In order to probe for biotinylated proteins, incubate with alkaline phosphatase-conjugated streptavidin.
Incubate the membrane with AP substrate buffer (Recipe 7) for 5 min at room temperature.
Remove AP substrate buffer. In a 15 ml conical tube, mix 5 ml of freshly made AP substrate buffer together with NBT (Recipe 6) (final concentration 50 μg/ml) and BCIP (Recipe 5) (final concentration 50 μg/ml). Pour the mixture on top of the membrane and protect it from light.
Incubate the membrane on a shaker protected from light and check at 1 min intervals until you can see purple bands from the biotinylated proteins (Figure 2B).
Once you tested the biotinylation is working properly, continue with large scale biotinylation experiment for purification of biotinylated proteins and mass spectrometry.
-
RNA-BioID
To capture and analyze biotinylated proteins in RNA-BioID, you need a confluent T175 flask of cells (2-3 x 107) for each round of mass spectrometry analysis.
On the day before the streptavidin pulldown, confluency should have reached 80%. Exchange media of one T175 flask, containing WT-BirA* or MBS-BirA* cells biotin-containing medium and culture for 16-24 h.
Wash the cells twice with PBS at room temperature to dispose of dead cells and serum-containing medium. Trypsinize for 2-3 min in the incubator. After checking cell detachment under a microscope, transfer cells into a conical tube and add 5 ml of serum-containing media to inactivate the trypsin.
Collect cells by centrifugation for 5 min at 500 × g for at 4 °C.
Wash cells twice with ice-cold PBS to completely remove the media.
Add 3 PCV of lysis buffer (including CompleteTM protease inhibitor mix) to each pellet and resuspend.
Lyse cells, collect lysate and determine protein concentration as mentioned above (Steps B12-B13).
For the isolation of biotinylated proteins, use at least 1 mg of total protein per lysate and 300 μl of magnetic streptavidin beads per 1 mg of protein.
To prepare the beads, mix the bead solution in the original vendor’s tube by quick vortexing, remove 200 μl of bead slurry to a fresh 1.5 ml tube and separate the beads from its storage buffer by capturing the beads on a magnetic stand for 5 min. Remove storage solution and wash the beads three times with 1 ml of lysis buffer. Rotate tubes with the bead slurry on an end-to-end rotator for 5 min.
Collect the beads on a magnetic stand and discard the wash buffer. Add 1 mg of total cell lysate onto the beads, remove from magnetic stand and leave mixture overnight with end to end rotation at 4 °C.
On the next day, collect the beads on a magnetic stand and wash twice with 1 ml of wash buffer 1 (Recipe 11), once with 1 ml of wash buffer 2 (Recipe 12), once with 1 ml of wash buffer 3 (Recipe 13) and once with 1 ml of wash buffer 4 (Recipe 14), each time by end-to-end rotation for 5 min at room temperature.
During the final washing with 1 ml of wash buffer 4, take 100 μl into a new 1.5 ml centrifuge tube and collect the beads via a magnetic stand and store the beads on ice until Step C14 to check enrichment of proteins with silver staining. Collect the beads from the remaining 900 μl on a magnetic stand.
Wash the beads 3 times with 50 mM of ammonium bicarbonate buffer each for 5 min at room temperature with rotation. Collect the beads and proceed immediately to Procedure D or store beads at -80 °C (safe stopping point).
From the remaining beads from step C12, add 50 μl of 1x SDS sample buffer (Recipe 16), boil the mixture for 5 min at 95 °C and afterward spin for 1 min at 12,000 × g at room temperature.
Keep the centrifuge tube on a magnetic stand for 2 min at room temperature and take out the supernatant to a new 1.5 ml centrifuge tube. Load 20 μl of the supernatant on a Nu-PAGE SDS-PAGE gel in 1x MOPS running buffer and run at 200 V until the front dye exits the gel.
Perform silver staining (Aboulaich, 2011) on this gel to compare proteins from WT-BirA* (shows proteins which are background biotinylated like mitochondria carboxylases and proteins which are unspecifically bound to the beads) and MBS-BirA* cells after treatment with biotin (Figure 2C).
-
On-bead tryptic digestion
Add an appropriate volume of denaturation buffer (Recipe 17) to the beads (usually 30-80 μl), such that they are well covered and can float in the suspension during incubation.
Add reduction buffer (Recipe 18) to a final concentration of 1 mM, incubate for 1 h at room temperature in a Thermomixer at 1,000 rpm.
Add alkylation buffer (Recipe 19) to a final concentration of 5.5 mM, incubate for 1 h at room temperature in a Thermomixer at 1,000 rpm.
Add 1 μg LysC for predigestion and check pH on a pH 6-10 pH strip. The pH needs to be around pH 8, otherwise, adjust with 1 M Tris-base pH 8. Incubate for 3 h at room temperature in the Thermomixer at 1,000 rpm.
Add 4 volumes of 20 mM ammonium bicarbonate buffer to dilute the urea in the buffer.
Add 1 μg trypsin for digestion and check pH on a pH 6-10 pH strip (pH should be 8), incubate overnight at room temperature in a Thermomixer at 1,000 rpm.
Separate beads and supernatant by using a magnetic stand and transfer the supernatant to a new tube.
Acidify samples with TFA to pH ≤ 2.
-
Peptide clean-up
Load sample onto a PHOENIX Peptide Clean-up Kit device.
Perform clean-up procedure by following the kit protocol (washing and elution steps).
After elution, reduce the sample volume to ≤ 9 μl in a vacuum concentrator (30-40 min at 30 °C).
Add 1 μl solvent A* (Recipe 20) and adjust the sample volume to 10 μl with solvent A (Recipe 21).
-
Mass spectrometry analysis
Peptide samples are analyzed using online nanoflow liquid chromatography tandem mass spectrometry (LC-MS/MS). Note that this is a routine analysis in proteomics that can be done on any high-performance liquid chromatography system coupled to a state-of-the-art high-resolution mass spectrometer.
Here, nanoLC-MS/MS analyses are done on an EASY-nLC 1200 system connected to an LTQ-Orbitrap Elite through an electrospray Ionization source.
Pack a 25 cm long 75 μm-inner diameter analytical column with reversed-phase C18-AQ Reprosil-Pur 1.9 μm particles at a constant pressure of 600 bar, and cut the column to a final length of 20 cm.
Load peptides directly onto the column at a flow rate of 500 nl/min.
Reduce flow rate to 200 nl/min after loading, and separate peptides with a segmented linear gradient of HPLC solvent B (Recipe 23) (5-33-50-90%) for 87 min.
Operate the mass spectrometer in the positive ion mode, with the following acquisition cycle: a full scan is measured using the Orbitrap cell at a resolution R 120,000 followed by MS/MS of the top 15 most intense peptide ions in the linear ion trap using collision-induced dissociation (CID). Acquire precursor ions in the mass range from m/z 300 to 2000 and use the target values 1E6 charges for the full scans (Orbitrap analyzer), and 5E3 charges for the collision-induced dissociation (CID) in the Linear Ion Trap. Sequenced precursor masses need to be excluded from further selection for 60 s.
Figure 2. Functional validation of the integrated BirA* fusion construct.
A. Expression check of full-length BirA* construct via Western blot. The full-length fusion protein from the nls-HA-2x mcp-egfp-BirA* transgene, in total 91.4 KDa, is detectable by Western blot against the GFP part only in MBS MEF cells. GAPDH can be used as a negative control protein since it can be detected in lysates from WT and MBS-BirA* MEFs. B. Testing for BirA* activity in MEFs using AP coupled streptavidin. After incubation of MEFs with 50 μM biotin for 24 h, cells were lysed and 10 μg of total cell lysates from WT and MBS-BirA* MEF cells were loaded per lane. Biotinylated proteins were detected in a colorimetric assay by Western blot using AP-streptavidin. C. A pattern of biotinylated proteins in RNA-BioID obtained after purification via streptavidin-coupled magnetic beads. 1/10th of the amount of magnetic beads that are normally used for mass spectrometry analysis were boiled in 100 μl of SDS sample buffer. Twenty microliter of these were loaded on a 10% SDS-PAGE, separated and detected by silver staining. The number and intensity of protein bands from the lysate of MBS-BirA* MEFs are much larger than that from WT-MEFs, indicating successful RNA-dependent biotinylation and capturing of associated proteins.
Recipes
-
Complete media (500 ml)
445 ml DMEM media
50 ml FBS
5 ml Penicillin-Streptomycin solution
-
Serum-free media (100 ml)
99 ml DMEM media
1 ml Penicillin-Streptomycin solution
-
Biotin Media (100 ml)
89 ml DMEM media
10 ml FBS
1 ml Penicillin-Streptomycin solution
50 µM biotin
-
50 mM Biotin solution
2.44 mg biotin
10 ml of serum-free DMEM
Pipetting is required to dissolve biotin completely. Sterilize by passing through a 0.22-μm syringe-driven filter unit
Active up to 8 weeks at 4 °C
-
BCIP
50 mg/ml BCIP
High-purity H2O
-
NBT
10 mg/ml NBT
High-purity H2O
-
AP substrate buffer
100 mM NaCl
5 mM MgCl2
100 mM Tris-Cl pH 9.5
-
1x TBS
50 mM Tris-Cl pH 7.5
150 mM NaCl
-
1x TBST
1x TBS
0.1% Tween 20
-
Lysis Buffer
50 mM Tris-Cl pH 7.5
150 mM NaCl
2.5 mM MgCl2
1 mM DTT
1% Triton X-100
1x proteinase inhibitor (Roche)
-
Wash Buffer 1
2% SDS
High-purity H2O
-
Wash Buffer 2
0.1% deoxycholate
1% Tween 20
500 mM NaCl
1 mM EDTA
50 mM HEPES pH 7.5
-
Wash Buffer 3
250 mM LiCl
0.5% Tween 20
0.5% deoxycholate
1 mM EDTA
10 mM Tris pH 8.1
-
Wash Buffer 4
50 mM Tris pH 7.4
50 mM NaCl
-
20 mM ABC buffer
79.06 mg ammonium bicarbonate
50 ml of RNase- and DNase-free water
-
2x SDS sample buffer
100 mM Tris-Cl pH 6.8
4% (w/v) SDS
0.2% (w/v) bromophenol blue
20% (v/v) glycerol
200 mM β-mercaptoethanol
Make 100 μl aliquots and store at -20 °C
-
Denaturation buffer
6 M urea
2 M thiourea
10 mM Tris buffer (pH 8.0)
-
Reduction buffer
1 M dithiothreitol
50 mM ammonium bicarbonate
-
Alkylation buffer
550 mM iodoacetamide
50 mM ammonium bicarbonate
-
Solvent A*
2% acetonitrile
1% formic acid
-
Solvent A
0.5% acetic acid
-
HPLC solvent A
0.1% formic acid
Milli-Q water
-
HPLC solvent B
0.1% formic acid
80% ACN in Milli-Q water
Acknowledgments
This protocol has been adapted from (Mukherjee et al., 2019). We thank Jeff Chao (FMI, Basel) and Julién Bethune (BZH, Heidelberg), for plasmids and cell lines. Silke Wahle (PCT Tübingen) is acknowledged for technical support. The project was funded as a project of the DFG Research Unit FOR2333 by a grant of the Deutsche Forschungsgemeinschaft (DFG JA696/11-1).
Competing interests
The authors declare no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Aboulaich N.(2011). Silver staining. Bio-101: e26. [Google Scholar]
- 2.Bovaird S., Patel D., Padilla J. A. and Lécuyer E.(2018). Biological functions, regulatory mechanisms, and disease relevance of RNA localization pathways. FEBS Lett 592(17): 2948-2972. [DOI] [PubMed] [Google Scholar]
- 3.Buxbaum A. R., Haimovich G. and Singer R. H.(2015). In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol 16(2): 95-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eom T., Antar L. N., Singer R. H. and Bassell G. J.(2003). Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J Neurosci 23(32): 10433-10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fei J. and Sharma C. M.(2018). RNA localization in bacteria. Microbiol Spectr 6(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holt C. E., Martin K. C. and Schuman E. M.(2019). Local translation in neurons: visualization and function. Nat Struct Mol Biol 26(7): 557-566. [DOI] [PubMed] [Google Scholar]
- 7.Hüttelmaier S., Zenklusen D., Lederer M., Dictenberg J., Lorenz M., Meng X., Bassell G. J., Condeelis J. and Singer R. H.(2005). Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438(7067): 512-515. [DOI] [PubMed] [Google Scholar]
- 8.Jansen R. P.(2001). mRNA localization: message on the move. Nat Rev Mol Cell Biol 2(4): 247-256. [DOI] [PubMed] [Google Scholar]
- 9.Marchand V., Gaspar I. and Ephrussi A.(2012). An intracellular transmission control protocol: assembly and transport of ribonucleoprotein complexes. Curr Opin Cell Biol 24(2): 202-210. [DOI] [PubMed] [Google Scholar]
- 10.Mukherjee J., Hermesh O., Eliscovich C., Nalpas N., Franz-Wachtel M., Maček B. and Jansen R. P.(2019). β-Actin mRNA interactome mapping by proximity biotinylation. Proc Natl Acad Sci U S A 116(26): 12863-12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oleynikov Y. and Singer R. H.(2003). Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr Biol 13(3): 199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park H. Y., Lim H., Yoon Y. J., Follenzi A., Nwokafor C., Lopez-Jones M., Meng X. and Singer R. H.(2014). Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science 343(6169): 422-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramanathan M., Porter D. F. and Khavari P. A.(2019). Methods to study RNA-protein interactions. Nat Methods 16(3): 225-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross A. F., Oleynikov Y., Kislauskis E. H., Taneja K. L. and Singer R. H.(1997). Characterization of a β-actin mRNA zipcode-binding protein. Mol Cell Biol 17(4): 2158-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tolino M., Köhrmann M. and Kiebler M. A.(2012). RNA-binding proteins involved in RNA localization and their implications in neuronal diseases. Eur J Neurosci 35(12): 1818-1836. [DOI] [PubMed] [Google Scholar]