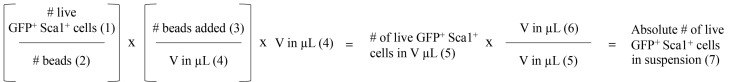Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological malignancy that arises from transformation of T-cell primed hematopoietic progenitors. Although T-ALL is a heterogenous and molecularly complex disease, more than 65% of T-ALL patients carry activating mutations in the NOTCH1 gene. The majority of T-ALL–associated NOTCH1 mutations either disrupt the negative regulatory region, allowing signal activation in the absence of ligand binding, or result in truncation of the C-terminal PEST domain involved in the termination of NOTCH1 signaling by proteasomal degradation. To date, retroviral transduction models have relied heavily on the overexpression of aggressively truncated variants of NOTCH1 (such as ICN1 or ΔE-NOTCH1), which result in supraphysiological levels of signaling activity and are rarely found in human T-ALL. The current protocol describes the method for mouse bone marrow isolation, hematopoietic stem and progenitor cell (HSC) enrichment, followed by retroviral transduction with an oncogenic mutant form of the NOTCH1 receptor (NOTCH1-L1601P-ΔP) that closely resembles the gain-of-function mutations most commonly found in patient samples. A hallmark of this forced expression of constitutively active NOTCH1 is a transient wave of extrathymic immature T-cell development, which precedes oncogenic transformation to T-ALL. Furthermore, this approach models leukemic transformation and progression in vivo by allowing for crosstalk between leukemia cells and the microenvironment, an aspect unaccounted for in cell-line based in vitro studies. Thus, the HSC transduction and transplantation model more faithfully recapitulates development of the human disease, providing a highly comprehensive and versatile tool for further in vivo and ex vivo functional studies.
Keywords: Mouse model, In vivo, T-ALL, NOTCH1 signaling, NOTCH1 mutations , Leukemia development, Retrovirus, Transformation, Hematopoietic stem cell, Bone marrow, Transplantation
Background
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological tumor arising from the malignant transformation of hematopoietic progenitor cells primed towards T-cell development. T-ALL accounts for up to 15% of pediatric and 25% of adult ALL (acute lymphoblastic leukemia) and occurs more frequently in males than females ( Goldberg et al., 2003 ). Despite being characterized by a high level of heterogeneity and molecular complexity ( Liu et al., 2017 ), more than 65% of T-ALL patients carry gain-of-function mutations in the NOTCH1 gene ( Weng et al., 2004 ). The resulting constitutive activation of NOTCH1 signaling is therefore the most prominent oncogenic pathway in T-cell transformation.
NOTCH1 proteins are developmentally conserved type I transmembrane receptors that play a prominent instructive role in T-cell lineage commitment and cell growth and proliferation during thymocyte development ( Radtke et al., 1999 ; Defto and Bevan, 2000). NOTCH1 signaling is initiated upon binding of transmembrane ligands expressed on the surface of neighboring cells. This interaction prompts cleavage of the extracellular domain of the receptor by the ADAM10 metalloprotease followed by γ-secretase cleavage in the transmembrane domain of the receptor. Subsequent release from the membrane results in translocation of the intracellular domain (ICN1) to the nucleus and transcriptional activation of target genes (Kopan and Ilagan, 2009; Andersson et al., 2011 ). Finally, termination of NOTCH1 signaling is regulated by phosphorylation of the C-terminal PEST domain, which targets ICN1 for ubiquitination and FBXW7-mediated proteasomal degradation ( O'Neil et al., 2007 ; Thompson et al., 2007 ). Constitutive activation of NOTCH1 in human T-ALL results from disruption of the mechanisms stringently regulating this multi-step process. The majority of T-ALL-associated mutations result either in disruption of the negative regulatory region, resulting in NOTCH1-signaling activation in the absence of ligand binding, or truncation of the C-terminal PEST domain which allows ICN1 to evade proteasomal degradation, thus impairing termination of signaling activity ( Weng et al., 2004 ). In the past decade, the introduction of intensive combination chemotherapy approaches has increased the cure rate of pediatric T-ALL to nearly 90%. Nonetheless, the prognosis for patients with primary refractory or relapse T-ALL remains very poor ( Goldberg et al., 2003 ; Litzow and Ferrando, 2015), underscoring the need to further decipher the molecular pathology underlying T-ALL transformation, identification of more specific therapeutic targets, and development of more effective and less toxic multi-drug therapies.
To date, in vitro human T-ALL-derived cell-line cultures remain the most commonly employed experimental approach to the study of T-ALL. Notably, this model does not correct for genetic drift associated with decade-long periods of culture and fails to account for the crosstalk between leukemia cells and the tumor microenvironment ( Passaro et al., 2015 ; Pitt et al., 2015 ). Animal models therefore provide an alternative approach to functional studies that more accurately recapitulate human T-ALL in vivo. The mouse hematopoietic stem cell (HSC) retroviral transduction and transplantation model has been extensively applied to mimic initiation and progression of NOTCH1-driven T-ALL ( Wendorff et al., 2010 ; Medyouf et al., 2011 ; Gachet et al., 2013 ). It has been shown that retroviral expression of ICN1 or ΔE-NOTCH1 (the extracellular domain-truncated form of the receptor) in murine bone marrow progenitors drives thymus-independent T-cell development and rapidly induces T-ALL development ( Pear et al., 1996 ; Perez- Garcia et al., 2013 ). However, it should be noted that these aggressively truncated variants of NOTCH1, which result in supraphysiological levels of signaling activity, are rarely found in human T-ALL ( Chiang et al., 2008 ). In fact, ΔE-NOTCH1 truncated alleles are expressed in only a small subset of patients carrying chromosomal translocations (1%) ( Ellisen et al., 1991 ; Palomero et al., 2006 ). Much more commonly, human T-ALLs harbor acquired gain-of-function point mutations, which occur predominantly within the extracellular HD and the C-terminal PEST domains ( Weng et al., 2004 ). Of note, 20% of T-ALL patients exhibit co-occurrence of these two mutation-driven activating mechanisms, which generate levels of activity well below those of ΔE-NOTCH1 ( Chiang et al., 2008 ). As the outcome of NOTCH signaling is highly dose dependent in many developmental settings (Artavanis- Tsakonas et al., 1999 ; Bray, 2016), we hypothesized that the level of activity has a similar impact on the process of leukemogenesis. The current protocol describes the method for mouse hematopoietic stem cell (HSC) retroviral transduction with an oncogenic mutant NOTCH1 receptor (NOTCH1-L1601P-ΔP) that closely resembles the gain-of-function mutations found in patient samples. The L1601P-ΔP mutant contains a common HD point mutation (L1601P) that enables ligand-independent activation ( Malecki et al., 2006 ), together with a frameshift mutation that results in truncation of amino acids 2,473-2,555, which comprise the negative regulatory PEST domain (ΔP). We outline the method for isolation of bone marrow cells from mice, enrichment for hematopoietic stem and progenitor cells, followed by retroviral transduction and transplantation into isogenic recipients (Figure 1). In this model, forced expression of constitutively active mutant NOTCH1 typically results in a wave of extrathymic T-cell lymphopoiesis 3 weeks after transplantation, which manifests as the transient appearance of GFP+ preleukemic CD4+ CD8+ double-positive (DP) immature T cells in peripheral blood (Figure 4). Transformation resulting in lethal leukemia progression occurs 12-15 weeks following transplantation ( Wendorff et al., 2019 ) (Figure 4). When combined with constitutive- or inducible-knockout transgenic models, the protocol can be easily modified to address the role of candidate genes in NOTCH1-driven T-ALL transformation, tumor initiation and tumor progression ( Wendorff et al., 2019 ). Furthermore, this approach provides a rapid and efficient model system for pre-clinical assessment of potential novel anti-leukemic drugs ( Herranz et al., 2015 ; Sanchez- Martin et al., 2017 ).
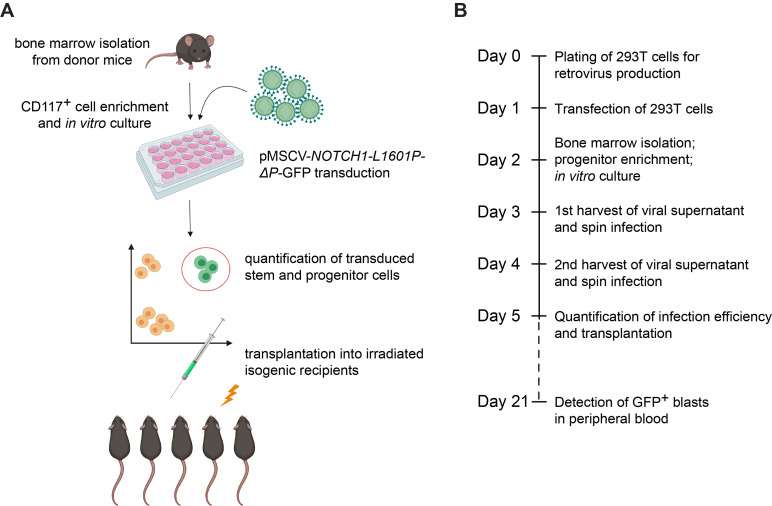
Figure 1. Experimental design for generation of oncogenic NOTCH1-driven T-ALL from mouse hematopoietic stem cells.
A. Cartoon representation of key experimental steps. B. Timeline of the experimental strategy.
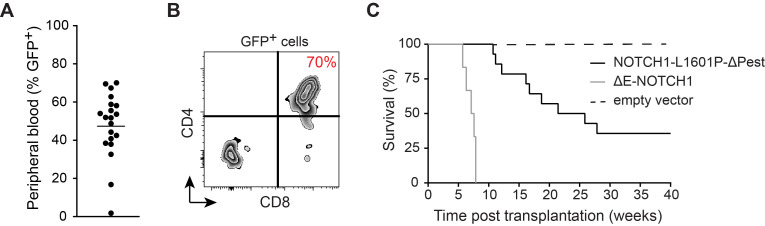
Figure 4. Monitoring development of NOTCH1-induced T-ALL.
A. Percentage of GFP+ pre-leukemia blasts in peripheral blood. B. Preleukemia blast-cell CD4/CD8 surface expression 21 days after transplantation of NOTCH1-L1601P-ΔP retrovirally transduced murine bone marrow stem and progenitor cells. C. Comparative Kaplan-Meier survival curves in weeks following transplantation of 100,000 NOTCH1-L1601P-ΔP (solid black), 100,000 pMSCV empty vector controls (dashed black) and 50,000 ΔE-NOTCH1-pMSCV-transduced murine hematopoietic stem and progenitor cells (grey).
Materials and Reagents
Pipette tips
Parafilm
Kimwipes Delicate Task Wipers, 1-Ply (Kimberly-Clark Professional, catalog number: 06-666)
150 mm TC-treated cell culture dishes (Corning, Falcon, catalog number: 353025)
24-well TC-treated microplates (Corning, Falcon, catalog number: 353847)
5 ml round-bottom polystyrene tubes (Corning, Falcon, catalog number: 352008)
15 ml Conical bottom Centrifuge Tubes (Corning, Falcon, catalog number: 352196)
50 ml Conical bottom Centrifuge Tubes (Corning, Falcon, catalog number: 352070)
3 ml BD Luer-Lok tip Syringes (Becton Dickinson, catalog number: 309657)
30 ml BD Luer-Lok tip Syringes (Becton Dickinson, catalog number: 302832)
Sterile 25 mm PES syringe filters, 0.45 µm pore size (Thermo Fischer Scientific, catalog number: 7252545)
40 µm cell strainer (Corning, Falcon, catalog number: 352340)
70 µm cell strainer (Thermo Fisher Scientific, catalog number: 22-363-548)
Amicon Ultra-15 Centrifugal 15 ml sample volume filter units NMWL 100KDa (EMD Millipore, catalog number: UFC910008)
U-100 Insulin syringes, 1 ml, 26 G (BD Biosciences, catalog number: 329652)
U-100 Insulin syringes, 0.5 ml, 29 G (Exelint international, catalog number: 26028)
Goldenrod Animal Lancet, 5 mm (Medipoint, catalog number: NC9891620)
LS Columns for negative cell selection (Miltenyi Biotec, catalog number: 130-042-401)
C57BL/6 bone marrow donor mice aged 6-12 weeks and isogenic C57BL/6 bone marrow recipient mice (min. 6 weeks of age) (The Jackson Laboratory, catalog number: 000664)
293T cells (ATCC, catalog number: CRL-3216)
pCL-Eco Retrovirus Packaging Vector (Addgene, catalog number: 12371) ( Naviaux et al., 1996 )
pMSCV-NOTCH1-L1601P-ΔP ( Chiang et al., 2008 )
jetPEI DNA Transfection Reagent provided with 150 mM NaCl solution (Polyplus, catalog number: 101-10N)
PBS: Cell Culture Phosphate Buffered Saline (1x) without calcium and magnesium (Corning, catalog number: 21040CV)
DMEM (Corning, catalog number: 10-013-CV)
Opti-MEM I Reduced Serum Medium, GlutaMAX Supplement (Gibco, catalog number: 51985)
Fetal Bovine Serum (FBS), heat inactivated and sterile (Sigma-Aldrich, catalog number: F4135)
Penicillin (10,000 IU) and Streptomycin (10,000 µg/ml) in a 100-fold working concentration (Corning, catalog number: 30002CI)
HEPES 1 M (Gibco, catalog number: 15630080)
0.5 M EDTA (Lonza, catalog number 51201)
β-mercaptoethanol, 55 mM (Gibco, catalog number: 21985023)
Trypan blue (Gibco, catalog number: 15250061)
ACK Lysing Buffer (Quality Biologicals, catalog number: 118-156-721)
CD117 Microbeads, mouse (Miltenyi Biotec, catalog number: 130-091-224)
Lineage Cell Depletion Kit, mouse (Miltenyi Biotec, catalog number: 130-090-858)
Polybrene Infection/Transfection Reagent (EMD Millipore, catalog number: TR-1003-G)
CountBright Absolute Counting Beads for flow cytometry (Invitrogen, catalog number: C36950)
Recombinant murine IL-3 (Peprotech, catalog number: 213-13)
Recombinant murine IL-6 (Peprotech, catalog number: 216-16)
Recombinant murine IL-7 (Peprotech, catalog number: 217-17)
Recombinant murine Flt3-L (Peprotech, catalog number: 250-31L)
Recombinant murine SCF (Peprotech, catalog number: 250-03)
Recombinant murine TPO (Peprotech, catalog number: 315-14)
Anti-mouse Sca-1 (Ly-6A/E) monoclonal antibody (clone D7), PE-Cyanine7 conjugate (eBioscience, catalog number: 25-5981)
(Optional) Anti-mouse CD4 monoclonal antibody (clone RM4-5), APC-conjugate (BD Biosciences, catalog number: 561091)
(Optional) Anti-mouse CD8 monoclonal antibody (clone 53-6.7), PE-Cyanine7 conjugate (BD Biosciences, catalog number: 561097)
DAPI (4',6-Diamidino-2-Phenylindole, Dilactate) (Invitrogen, catalog number: D3571)
Heparin, 1,000 USP units/ml (Sagent Pharmaceuticals, catalog number: 25021-400-30)
Ketamine HCl injection (100 mg/ml: controlled substance); consult institution’s veterinarian
Xylazine sterile solution (20 mg/ml: AKORN Animal Health; catalog number: 139-236); consult institution’s veterinarian
Staining medium (SM; see Recipes)
293T culture/transfection medium (see Recipes)
Growth-factor supplemented transduction medium (see Recipes)
Ketamine/xylazine anesthesia cocktail (see Recipes)
Equipment
1 ml/200 µl/20 µl pipettes
Mortar and pestle
A cell culture incubator, 37 °C/5% CO2
Dissection tools: scissors, forceps, scalpel blade (recommended source: Fine Science Tools www.finescience.com)
Hemocytometer (recommended) or automated cell counter
Laminar flow biosafety cabinet
Refrigerated centrifuge fitted with a swing-bucket rotor and plate adapters
MACS MultiStand (Miltenyi Biotec, catalog number: 130-042-303)
MidiMACS Separator (Miltenyi Biotec, catalog number: 130-042-302)
A Fluorescence Microscope with green fluorescent protein (GFP) excitation/emission filters
A Flow Cytometer instrument with a Blue (488 nm), Violet (405 nm), Green (561 nm) and Red (633 nm) laser (recommended: LSRFortessa, BD Biosciences)
A gamma-source or alternative small-animal irradiator
Software
FlowJo (FlowJo LLC, Ashland, OR, USA)
GraphPad Prism Software (La Jolla California USA, www.graphpad.com)
Procedure
This approach to retroviral transduction and bone marrow transplantation can be used to overexpress any oncogene of interest to generate either lymphoid or myeloid leukemia in mice. Transformation efficiency, kinetics of disease progression and tumor penetrance will depend on both the tumorigenicity of the oncogene used and experimental variables, such as the number of transplanted transduced hematopoietic stem cells.
Note: Please refer to the Notes section of this protocol for additional information and see Figure 4 for a comparison of the pMSCV-NOTCH1-L1601P-ΔP and ΔE-NOTCH1 leukemia models (recommended cell numbers, kinetics of leukemia development and penetrance).
The procedure described here has been optimized for modeling T-ALL development driven specifically by the L1601P-ΔP mutant form of the NOTCH1 receptor. For example, transfection of 293T cells is modified to account for reduced transfection efficiency when using larger constructs, such as the pMSCV-NOTCH1-L1601P-ΔP-ires-GFP (> 15 kb) and counteract the resulting reduction in viral titer. Furthermore, the protocol recommends bone marrow isolation from the hip bones and spinal vertebrae, in addition to the tibiae and femurs. This approach greatly increases the total number of bone marrow stem and progenitor cells available for subsequent transduction, while simultaneously reducing the number of experimental animals needed. Although all mice listed in this protocol (both donor and recipient mice) are on the C57BL/6 background, other mouse strains may be used if bone marrow donor and recipient mice are on congenic genetic backgrounds. However, it is important to remember that each strain has unique background alleles that may interact with and modify the expression of a mutation, transgene or other genetic insert (https://www.jax.org/news-and-insights/2006/june/the-importance-of-genetic-background-in-mouse-based-biomedical-research).
Please note that the experimental procedure outlined in this protocol requires 6 consecutive days of hands-on activity (Days 0-5), with a subsequent follow-up on Day 21 (Figures 1A and 1B; Figure 4).
Important: All experimental procedures must be approved by the local animal care committee.
Notes:
Prior to beginning the protocol, calculate the number of bone marrow donor mice needed based on the desired number of recipient animals; as a “rule of thumb”, when harvesting marrow from only the 2 hind legs, n = 1 bone marrow donor is sufficient for transplantation into n = 2-3 recipient animals; when harvesting marrow from all bones (hind and fore limbs, pelvic bone and spinal column), n = 1 bone marrow donor is sufficient for transplantation into n = 6-8 recipient animals.
An additional untreated congenic wild-type mouse is needed as a source of helper bone-marrow cells on Day 5 of the protocol.
Transient transfection of 293T cells
Day 0:
Plate 293T cells 24-48 h before transformation at a density that will result in approximately 60-70% confluence at time of transfection (for 293T medium; see Recipes). Use 293T cells that have been passaged no less than 2x after thawing of a fresh vial of live-frozen cells. Do not exceed a total of 20 passages. Use only tested mycoplasma negative cells ( Young et al., 2010 ).
Note: Plating density should be optimized based on the size of the culture plate and the growth kinetics of the 293T cells being used for virus production. The values below have been optimized for 150 mm tissue culture plates, with a conservative ratio of 1:1 of 293T culture plates to the number of donor mice and total bones used for bone marrow isolation.
Day 1:
In the evening, perform transient transfection of 293T cells (Table 1); see https://www.polyplus-transfection.com/wp-content/uploads/2015/09/CPT_101_jetPEI_vU.pdf for general guidelines.
Table 1. Amount of transfection reagent and retroviral construct recommended per 1x 150 mm plate of 293T cells (scale up as needed).
| Tube #1 | Amount per 150 mm plate |
|---|---|
| jetPEI transfection reagent | 50 µl |
| NaCl | 450 µl |
| Total Volume/plate | 500 µl |
| Tube #2 | Amount per 150 mm plate |
| pMSCV-NOTCH1-L1601P-ΔP retroviral construct | 20 µg |
| pCL-Eco packaging construct | 15 µg |
| NaCl | X µl |
| Fill to Total Volume/plate | 500 µl |
Prepare the DNA and transfection mix in separate tubes in sterile conditions:
Add the contents of Tube #1 to Tube #2 (always add transfection reagent to DNA mix and not vice versa), vortex gently and allow to stand at room temperature for 20-30 min.
Add the mix to 293T cells drop-wise; 1 ml per 150 mm plate.
Incubate overnight (12-16 h).
Day 2:
-
Transient transfection of 293T cells, continued
In the morning, remove transfection medium and very gently replace with 13-15 ml fresh 293T medium. Place in incubator until the next day.
Notes:
293T cells will begin to produce GFP within 12 h of transfection. To confirm successful transfection and obtain a qualitative estimate of transfection efficiency, it is recommended to verify GFP production using a fluorescence microscope 16-24 h after transfection; transfected virus-producing cells with appear brightly fluorescent.
Perform all media changes with extreme care as cells have been sensitized by the transfection reagent and detach from the plate more easily with continued culture.
From this point onwards observe all safety requirements when handling BSL-2 hazardous materials.
-
Isolation of bone marrow cells and CD117+ cell enrichment
Notes:
This protocol recommends bone marrow isolation by crushing of bones using a mortar and pestle, an approach that results in higher and more consistent cell yields, while maintaining high cell viability. While isolation of bone marrow from hip bones and spinal vertebrae appears more cumbersome to novice researchers, it greatly increases the cell yield per mouse and decreases the number of experimental animals required for bone marrow extraction, a pivotal consideration when using transgenic mouse models. While dissection of tibiae and femurs from experimental mice is well established in the field, obtaining the hip bones and spinal vertebrae is less common. We highly recommend viewing the video tutorial published by Lo Celso and Scadden (2007) prior to undertaking this approach.
Retroviral transduction of mouse bone marrow cells is most commonly performed using negative selection of total lineage-negative bone marrow cells. Positive selection of CD117+ cells, as described here, tips the scales towards transduction of hematopoietic stem cells, which results in increased transformation efficiency of the target population with self-renewal potential.
Keep staining medium (SM) and the bone marrow cell suspension (BM) on ice at all times. Perform all centrifugation steps at 4 °C.
The values provided below are based on total BM isolated from all bones of n = 1 adult mouse (approx. 8-10 weeks of age) and should be scaled up appropriately based on the target number of bone marrow donor animals.
Sacrifice donor mice: CO2 asphyxiation followed by cervical dislocation is recommended.
Dissect the tibia, femurs and pelvic bones from each donor mouse; the total expected bone-marrow cell yield is approximately 120 x 106/animal, but varies with age of the donor animals. If needed, the total cell yield can be increased to 160 x 106-200 x 106/animal by additionally isolating the spinal vertebrae.
Clean the bones by scraping off muscle tissue using a scalpel blade. Remove remaining tissue by rubbing the bones between your fingers using Kimwipes/paper towel. It is recommended to gently break off the cartilage of the kneecaps and other joints by dislocation rather than cutting the bone, as this preserves the integrity of the bone, results in higher yields and reduces the risk of contamination. If isolating spinal vertebrae, separate the vertebrae into 3-4 individual segments and remove the spinal cord. Clean thoroughly by rubbing with Kimwipes. Keep cleaned bones in cold SM on ice.
Although not absolutely necessary, it is recommended to perform all subsequent steps in sterile conditions in a laminar flow biosafety cabinet to prevent contamination. Crush the bones in 5 ml cold SM using a mortar and pestle (crush, rather than grind, to retain high cell viability). Repeatedly aspirate and eject the cell suspension using a 3 ml syringe to generate a single-cell suspension and pass through a 40 µm cell strainer into a 50 ml collection tube placed on ice. Add 5 ml SM to the mortar and repeat this sequence approximately 3 times, until the bones are thoroughly crushed and appear translucent/white in color.
After the final round of crushing, wash the strainer with 5 ml SM; this results in approximately 40 ml of single-cell bone-marrow suspension per donor mouse.
Pellet cells at 500 rcf at 4 °C for 7 min.
Aspirate/pour off the SM (the pellet will appear very RED).
Perform red blood cell lysis by resuspending the pellet in 5 ml ACK buffer and incubate at room temperature for 5 min. Pass the cell suspension through a 70 µm cell strainer into a 50 ml conical tube placed on ice containing at least 20 ml SM to stop the lysis reaction, then fill the tube to capacity with SM to further dilute the ACK lysis buffer.
Pellet cells at 400 rcf at 4 °C for 10 min.
Aspirate/pour off the supernatant (the pellet should now appear near WHITE) and carefully resuspend the cell pellet in 5 ml cold SM. Add 15 ml cold SM and gently pulse-vortex to obtain 20 ml of a uniform cell suspension.
-
Determine the number of viable cells (Trypan blue exclusion) either by manual cell counting using a hemocytometer or an automated cell counter.
Note: Due to the heterogeneity of total bone marrow cells, the former approach results in more accurate quantification of cell number.
Continue with CD117+ positive-cell selection using anti-CD117 Microbeads following the manufacturer’s instructions (https://www.miltenyibiotec.com/US-en/resources/technical-documents/data-sheets.html). In the final step, elute into a 15 ml Eppendorf tube and add 10 ml SM. Pellet cells at 4 °C for 5 min at 500 rcf.
Resuspend the CD117+ cell-enriched pellet in 2 ml cold SM and proceed with cell quantification. Keep cells on ice.
Manual cell counting using a hemocytometer and Trypan blue exclusion of dead cells is recommended at this step of the protocol to ensure accurate cell counts. When harvesting cells from ALL bones (as above), the total expected yield is approximately 5 x 106 CD117+ cells/donor animal.
Plate CD117+-enriched cells in a 24-well tissue culture plate in 2 ml/well growth-factor supplemented medium (for GF medium, see Recipes) at a density of 0.5 x 106 cells/ml and culture overnight.
Day 3: First round of virus collection and bone-marrow progenitor cell transduction
-
First collection and concentration of the viral supernatant from 293T cultures
Note: Perform the following steps at room temperature in a Level 2 biosafety cabinet and observe all safety requirements of handling BSL-2 hazardous materials.
-
Collection and filtration the viral supernatant:
Collect the viral supernatant (VSN) from 293T cultures and remove large cell debris by passing through a 70 µm strainer into a 50 ml conical collection tube.
Carefully replenish cultures with 13-15 ml fresh 293T medium for the second collection of VSN on Day 4.
Remove small debris and aggregates by passing the VSN through a 0.45 µm PES syringe filter (this is most easily performed by aspirating the VSN from the 50 ml conical collection tube into a 30-60 ml syringe fitted with a large 18 G gauge needle, turning the syringe upwards to prevent spillage and contamination, and replacing the needle with a syringe filter prior to ejecting the VSN into a fresh 50 ml conical-bottom collection tube).
Note: Two consecutive filtration steps remove all particles that may result in clogging of the Amicon Ultra concentration column in the next step.
-
Concentration of the viral supernatant
Transfer VSN into an Amicon filtration column tube to maximum capacity (Amicon Ultra = 15 ml).
Centrifuge at 2,500 rcf at room temperature until approximately 80% of the VSN has passed through the filtration unit (typical final concentrate volume/15mL VSN volume = 200µl). If VSN is collected from > 1 x 150 mm 293T virus-producing dish, repeat Steps A2a-A2b using the same filtration unit until the desired final volume of VSN is reached. Transfer the final volume of concentrated VSN to an Eppendorf tube. Discard the Amicon filtration unit.
Note: Generate 100 µl of VSN (20x concentration) for each well of the 24-well culture plate containing CD117+-enriched bone marrow in a volume of 2 ml. i.e., if CD117+-enriched bone marrow cells were plated in 12 wells on Day 2 of the protocol, concentrate the VSN to a final volume of 1,200 µl. Adjust volume as needed with fresh OptiMEM cell culture medium. Remember to add the appropriate amount of Polybrene ( Step B1 below).
-
-
First transduction of CD117+-enriched bone marrow progenitor cells
Note: Pre-warm the centrifuge for spin-infection to 28 °C. Set the deceleration speed (break) to 0.
Add Polybrene reagent to the Eppendorf tube containing concentrated VSN to a final concentration of 80 µg/ml (20x).
Add 100 µl of the 20x VSN containing 20x Polybrene to each well of the 24-well plate containing CD117+-enriched bone marrow cells in 2 ml GF-supplemented media.
Seal the plate with parafilm to prevent contamination of the centrifuge during spin-infection.
Centrifuge at 1,100 rcf at 28 °C for 90 min.
Carefully remove parafilm and return the plate to a TC incubator. Allow the cells to recover for at least 6 h.
After 6-8 h, gently remove of the media, ~1 ml (gently tilt the plate; use a 1 ml pipettor and keep the pipette tip level with the surface of the media while aspirating to prevent aspiration of bone marrow cells).
Add 1 ml of fresh GF-supplemented transduction medium and allow cells to recover overnight (12-16 h).
Day 4: Second round of virus collection and bone-marrow progenitor cell transduction
-
Second collection and concentration of the viral supernatant from 293T cultures
Repeat Steps A1a-A1c and A2a-A2b as on Day 3 of the protocol.
-
Second transduction of CD117+-enriched bone marrow progenitor cells, previously transduced on Day 3 and cultured overnight in GF-supplemented medium.
Repeat Steps B1-B7 as on Day 3 of the protocol.
-
Irradiation of recipient mice
Notes:
It is recommended to use 8-week-old mice as recipients, although a 6-12-week range is acceptable. As recipient mice should be irradiated at least 8 h prior to transplantation, this protocol recommends irradiation on the evening of the day before transplantation. Deliver a total body dose (TBI) of 900 Rads in a single dose, which qualifies as reduced exposure (lethality < 100%). The lethal dose varies between strains of mice, and 900 Rads TBI is optimal for the adult C57BL/6 strain, as used in this protocolF.
Following irradiation, recipient animals must be placed on an antibiotic/analgesic-supplemented drinking water regimen for 1 week; this must be outlined in your laboratory’s approved animal experimentation protocol.
Day 5: Quantification and transplantation of transduced bone marrow cells
-
Isolation and lineage depletion of helper bone marrow cells
-
Isolate bone marrow from 2 hind limbs of a wild-type adult mouse of the same strain/genetic background (congenic) as the animals used throughout the protocol. Follow Steps B1-B11 of the bone marrow isolation protocol outlined on Day 2.
Notes:
Isolation of bone marrow from 2 hind limbs (tibia + humeri only) of an adult mouse (approximately 8 weeks of age) after red blood cell lysis yields 50 x 106-70 x 106 cells.
Many laboratories chose to bypass this step altogether, while others employ the complement-mediated T-cell depletion approach (when donor and recipient mice are 100% syngeneic). Due to the transfer of mice between laboratories and varyingly stringent approaches to backcrossing, we recommend the slightly more cumbersome approach of using lineage-depleted helper cells to avoid any possible immunological responses in the donor-recipient transplantation setting.
Perform absolute cell quantification using the method of choice (manual cell quantification using a hemocytometer and Trypan blue exclusion or an automated cell counter; the former being the preferred method).
Perform magnetic bead-based lineage depletion of helper bone marrow following the manufacturer’s instructions (https://www.miltenyibiotec.com/US-en/resources/technical-documents/data-sheets.html). This approach relies on negative selection, i.e., unlike positive CD117-cell enrichment (Day 2 Section B) collect the flow-through from the magnetic column into a 15 ml conical bottom tube placed on ice. The total expected yield of the Lin- fraction from 2 hind limbs of one adult mouse is 5 x 106 cells.
Pellet the cells at 500 rcf at 4 °C for 5 min.
Resuspend the Lin- cells in 1 ml SM and quantify the absolute live cell number by trypan blue exclusion using a hemocytometer (see Step A2). Keep cells on ice until ready to proceed with the bone marrow transplantation step below.
Gently pulse-vortex the helper bone marrow to obtain a uniform single-cell suspension. Transfer the appropriate number of cells to a fresh tube 15 ml tube: you will need 2 x 105-3 x 105 helper bone marrow cells per recipient mouse (i.e., if 10 recipient mice are being used for the transplant, collect a volume containing the equivalent of 2 x 106-3 x 106 cells).
Proceed to Step B13 of Day 2 of the protocol.
-
-
Harvesting and quantification of transduced bone marrow stem and progenitor cells
In a laminar flow cabinet, resuspend pMSCV-NOTCH1-L1601P-ΔP transduced bone marrow progenitor cells using a 1 ml pipettor (Day 3 Step B7). Pool cell suspensions from the 24-well plate in a 15 ml/50 ml conical tube.
Pellet cells by centrifugation at 500 rcf at room temperature for 5 min.
Resuspend cells in a known volume of OptiMEM transduction medium without growth factors and quantify the absolute number of live cells by manual counting using a hemocytometer and Trypan blue exclusion of dead cells. The resuspension volume is guided by the size of the cell pellet (approximately 5 ml if cells were initially harvested from all bones).
-
Transfer two individual 100 µl aliquots of the cell suspension to 1.5 ml Eppendorf tubes for staining and quantification (this aliquot should contain no fewer than 50,000-100,000 cells to ensure accurate quantification in the next step). Keep the suspension of transduced cells in a 37 °C TC incubator until quantification is complete.
Note: Loosen the screwcap of the 15 ml/50 ml conical collection tube to allow free air flow.
-
When using the recommended PE-Cyanine7-conjugated anti-Sca-1 antibody, to a 100 µl aliquot of cells add 500 µl of SM and 1 µl of the anti-Sca-1 antibody. To the second aliquot of cells, add 500 µl of SM only (this negative control allows more accurate discrimination of the Sca-1-positive versus the Sca-1-negative population when performing subsequent flow cytometry analysis).
Note: When using alternative fluorochrome conjugates, all antibodies should be titrated prior to use, although a 1:200 dilution works well for the majority of commercially available anti-Sca-1 conjugates. The 1:600 dilution in this step has been empirically established for the specific antibody listed in this protocol (see Materials and Reagents).
Stain cells on ice for 45-60 min in the dark.
Add 1 ml of SM to wash cells and centrifuge at 500 rcf for 5 min at 4 °C. Be very careful when aspirating the supernatant as the pellet may be quite small.
-
Resuspend the pellet in 500 µl SM + 25 µl CountBright quantification beads. Add 5 µl of DAPI (pre-diluted to 0.5 mg/ml in H2O) for dead cell exclusion.
Note: When using CountBright beads, use a pipettor to thoroughly resuspend the cells and make careful note of the final resuspension volume as this will be used in the next step to calculate the absolute number of transduced cells. Similarly, be sure to thoroughly vortex the CountBright beads and note the exact volume added for quantification and the number of beads/µl in the Lot # of CountBright beads being used.
-
Run samples on a flow cytometer to determine the frequency of transduced GFP+ Sca-1+ cells. Be sure to gate on live cells only (DAPI-) and use the “no anti-Sca-1 antibody” control to set the gate. Draw an additional gate on the population of CountBright beads (Figures 2A-2D).
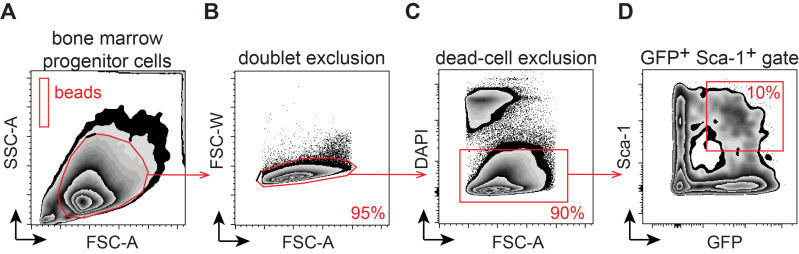
Note: The live cell population (DAPI-) will contain a mix of GFP- (untransduced) cells and GFP+ (transduced) cells. The expected percentage of successfully transduced bone marrow stem and progenitor cells (Sca-1+) should be no less than 10-15% (GFP+ Sca-1+ double-positive cells). See Notes section for additional comments.
Calculate the absolute number of GFP+ Sca-1+ double-positive cells based on the number of cells in the target population and the number of CountBright beads collected. Follow the manufacturer’s instructions for cell quantification (see Data analysis and Figure 5).
Remove cells from the TC incubator (Step B3 of Day 5) and resuspend by pipetting up and down/gently pulse-vortexing to obtain a uniform single cell-suspension.
Remove an aliquot of transduced cells based on the calculations performed in Steps B4-B10 of Day 5. You will need 100,000 GFP+ Sca-1+ cells/recipient mouse. We have previously established that transplantation of 100,000 transduced cells results in 70-80% tumor penetrance (it is not recommended to exceed 150,000 GFP+ Sca-1+ cells as this results in tumor development from multiple clones). For example: if 10% of transduced cells are GFP+ Sca-1+ and the total cell suspension contains 1.5 x 106 cells/ml, you will need 0.67 ml/recipient animal. Therefore, if transplanting 10 recipient animals, 6.7-10 ml of total cell suspension is required for transplantation.
Pool together the appropriate volume of transduced Sca-1+ GFP+ cells from the previous step and the required volume of helper bone marrow prepared in Step A6 of Day 5 above. The volume of helper bone marrow is dictated by the concentration of the cell suspension and the target number of recipient mice (use 2 x 105-3 x 105 helper bone marrow cells per recipient mouse).
Pellet cells at 500 rcf at 4 °C for 5 min.
-
Thoroughly resuspend the cells in sterile cold PBS (stored at 4 °C, or pre-chilled on ice) and pass through a 40 µm filter to remove debris/clumps. The total volume of PBS is dictated by the number of recipient animals, i.e., when performing transplantation by retro-orbital injection, resuspend cells in 100-50 µl PBS/recipient mouse.
Note: It is recommended to use a 20% excess of cells resuspended in an equivalent 20% excess volume (i.e., if transplanting n = 10 recipient mice, prepare a cell suspension sufficient for n = 12 recipient mice). Remember to collect any remaining cell suspension from the bottom side of the 40 µm filter using a 1 ml pipettor as the entire volume of cell suspension will not pass through the strainer by gravity pull alone when using such as small volume.
-
Transplantation of retrovirally transduced bone marrow cells
The result of the final step above is a single-cell suspension containing a mixture of transduced GFP+ Sca-1+ cells (100,000/recipient) + un-transduced progenitor cells + Lin- helper bone marrow cells.
-
Anesthetize irradiated recipient animals (Day 4; Section C) by intraperitoneal injection of 100 µl ketamine/xylazine cocktail using a 1 ml insulin syringe (see Recipes).
Note: Alternatively, inhalational anesthesia with isoflurane is commonly used, though this has a very short recovery period and is not recommended for users without extensive prior experience.
-
Once animals are fully sedated, inject 50-100 µl cell suspension into the retro-orbital sinus using a 0.5 ml 29 G insulin syringe ( Yardeni et al., 2011 ).
Note: Please refer to Yardeni et al. (2011) for a comprehensive technical overview of retro-orbital injections in mice.
Closely monitor animals and maintain on antibiotic-supplemented drinking water for 7 days.
-
Figure 2. Flow-cytometry based quantification of retrovirally transduced bone marrow progenitor cells.
The figure depicts the stepwise gating strategy for A. Total in vitro expanded CD117+-enriched cells and CountBright quantification beads; B. doublet exclusion; C. dead cell exclusion (DAPI-); and D. frequency of transduced hematopoietic stem and progenitor cells (HSPCs: GFP+ Sca-1+).
Figure 5. GFP+-Sca1+ absolute quantification.
CountBright bead-based approach to quantification of the absolute number of pMSCV-NOTCH1-L1601P-ΔP-GFP transduced hematopoietic stem and progenitor cells (see above and follow manufacturer’s instructions).
Day 21: Detection of transient CD4+ CD8+ double-positive preleukemia cells in peripheral blood
Notes:
Any method of blood collection must be approved by the local animal care committee and be outlined in your laboratory’s approved animal experimentation protocol. Only trained personnel may perform this procedure.
This protocol recommends blood sampling by submandibular puncture, which is a rapid technique that minimizes trauma to experimental animals (Figure 3).
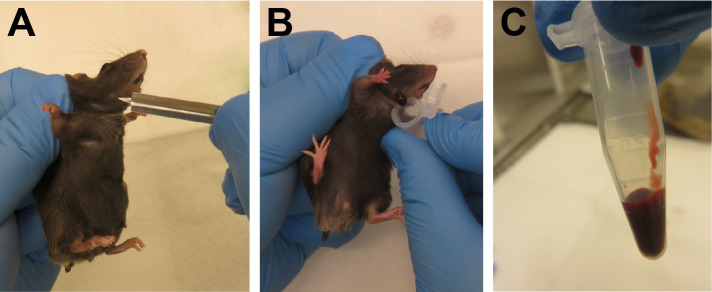
Figure 3. Blood sampling by submandibular puncture.
A. The “freckle” aids in correct localization of the submandibular vein. B. Collection of 2-4 drops of blood. C. Maximum recommended volume of peripheral blood obtained during this procedure.
If assessing CD4 and CD8 surface expression (optional), prepare 50 µl antibody cocktail/peripheral blood sample. When using the antibodies recommended in this protocol (see Materials and Reagents) dilute the antibodies in SM as follows: anti-CD4-APC 1:400 and anti-CD8-PeCy7 1:800. Keep the antibody cocktail in the dark at 4 °C until ready to proceed with staining.
Prepare 1.5 ml Eppendorf tubes containing 15 µl Heparin (n = number of recipient mice on Day 5 above).
Collect 2-4 drops/mouse of peripheral blood by submandibular puncture using a 5 mm lancet into the previously prepared collection tube containing heparin (Figure 3).
Prepare 4 ml round-bottom FACS tubes containing 1 ml ACK lysis buffer.
Transfer approximately 75 µl of peripheral blood into ACK buffer and incubate at room temperature for 5 min.
Add 2 ml SM and pellet the cells at 500 rcf at room temperature for 5 min.
Resuspend the cell pellet in 1 ml ACK buffer and repeat the lysis and wash steps 3 times until red blood cells are thoroughly lysed (the final pellet may remain pink rather than white; however, it is not advisable to reduce the number of lysis steps as red blood cells are very “sticky” and often result in clogging of the flow cytometer).
After the final wash step, resuspend the pellet in 50 µl antibody cocktail and incubate on ice for 45 min in the dark.
Wash cells in 1 ml SM and pellet at 500 rcf for 5 min. Resuspend in 500 µl SM supplemented with DAPI (5 µg/ml final concentration).
Analyse samples using a flow cytometer instrument fitted with a Blue 488 nm (GFP), Green 561 nm laser (PE-Cy7), Violet 405 nm (DAPI) and Red 633 nm (APC) laser.
Data analysis
Quantification of the absolute number of GFP+ Sca-1+ double-positive cells, Day 5 (Figure 2 and Figure 5)
Quantification using CountBright Absolute Counting Beads for flow cytometry is performed according to the manufacturer’s instructions. The following values are required for this calculation:
https://www.thermofisher.com/order/catalog/product/C36950
The absolute number of acquired GFP+ Sca-1+ cells (Figure 2D; Step B9 of Day 5).
The absolute number of acquired beads (Figure 2A; Step B9 of Day 5).
The absolute # of CountBright beads added to the sample (Step B8 of Day 5).
The volume of SM in which cells were resuspended for flow cytometry analysis (Step B8 of Day 5).
The volume of the aliquot of bone-marrow cell suspension used for quantification (Step B4 of Day 5).
The absolute volume in which bone marrow cells were resuspended (Step B3 of Day 5).
Calculate the absolute number of GFP+ Sca-1+ double-positive cells in the culture.
Notes
In this protocol, replication-defective retrovirus encoding the human NOTCH1 oncogene and a traceable GFP marker (green fluorescent protein; this could alternatively be RFP, an antibiotic resistance gene, etc.) is produced by transient co-transfection of 293T cells with pCL-Eco (a plasmid that expresses the gag-pol-env packaging functions). The 293T cells produce an ecotropic retrovirus specific for mouse cells, but not entirely ineffective for human cells, necessitating stringent observation of BSL-2 biohazard conditions.
This protocol was optimized using the jetPEI transfection reagent. Alternative transfection systems may be used successfully but will require individual optimization.
Here we outline the generation of oncogenic NOTCH1-L1601P-ΔP-driven tumors. Due to the > 15kb size of this retroviral construct, it is difficult to achieve transduction efficiency of bone marrow progenitor cells that exceeds 10% (GFP+ Sca-1+). A robust 70-80% frequency of transformation is achieved upon transplantation of 100,000 transduced progenitor cells, which is preceded by 45 ± 20% GFP+ pre-leukemia blasts in peripheral blood on day 21 (Figures 4A and 4B). Tumor progression commonly results in mortality 12-25 weeks post-transplant, reflecting kinetically more variable tumor development when compared to ΔE-NOTCH1-driven T-ALL (Figure 4C). When using the highly oncogenic constitutively active ΔE-NOTCH1 allele, transduction efficiency of HSCs falls in the 30 ± 10% range, resulting in 100% transformation efficiency. Subsequent T-ALL development occurs 6-8 weeks after transplantation of 50,000 GFP+ Sca-1+ progenitor cells, with GFP+ pre-leukemia blasts in the peripheral blood 21 days post-transplant reaching 60-70% and consequently 0% survival exceeding 8 weeks (Figure 4C). This protocol has also been successfully employed to generate alternative mouse models of leukemia, such as ΔE-NOTCH1, c-Myc or Flt3-ITD overexpression-induced T-ALL and AML leukemia, with 100% penetrance upon transduction of Lin- C57BL/6-derived bone marrow cells and subsequent transplantation of 50, 000 transduced cells with 100% penetrance by 12 weeks (previously published data), illustrating it’s efficacy across multiple lineage-specific oncogenes.
Recipes
-
Staining Medium (SM)
PBS
2% FBS
1 mM Hepes
0.5 µM EDTA
-
293T growth/transfection medium
DMEM
10% FBS
100 U/ml penicillin G
100 µg/ml streptomycin
-
Growth-factor supplemented CD117+ cell culture/transduction medium
Opti-MEM I Reduced Serum Medium, GlutaMAX Supplement
10% FBS
100 U/ml penicillin
100 µg/ml streptomycin
55 µM 2-mercaptoethanol (β-mercaptoethanol)
10 ng/ml recombinant mouse IL-3
10 ng/ml recombinant mouse IL-6
25 ng/ml recombinant mouse IL-7
50 ng/ml recombinant mouse Flt3-L
50 ng/ml recombinant mouse SCF
25 ng/ml recombinant mouse TPO
-
Ketamine/xylazine anesthesia cocktail (1 ml)
150 µl Ketamine (100 mg/ml)
150 µl Xylazine (20 mg/ml)
700 µl PBS
Acknowledgments
This work was supported by the National Institute of Health grants R35 CA210065 (AF) and R01 CA155743 (AF). AAW was supported by a Rally Foundation fellowship. We thank D. Herranz for critical review of the protocol.
Competing interests
The authors declare no conflict of interest.
Ethics
All animals used in the development of this protocol were housed in specific pathogen-free facilities at the Irving Cancer Research Center at the Columbia University Medical Center, New York, NY, in accordance with NIH guidelines for the Care and Use of Laboratory Animals. All animal procedures were approved by the Columbia University Institutional Animal Care and Use Committee (IACUC).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Andersson E. R., Sandberg R. and Lendahl U.(2011). Notch signaling: simplicity in design, versatility in function. Development 138(17): 3593-3612. [DOI] [PubMed] [Google Scholar]
- 2. Artavanis-Tsakonas S., Rand M. D. and Lake R. J.(1999). Notch signaling: cell fate control and signal integration in development. Science 284(5415): 770-776. [DOI] [PubMed] [Google Scholar]
- 3. Bray S. J.(2016). Notch signalling in context. Nat Rev Mol Cell Biol 17(11): 722-735. [DOI] [PubMed] [Google Scholar]
- 4. Chiang M. Y., Xu L., Shestova O., Histen G., L'Heureux S., Romany C., Childs M. E., Gimotty P. A., Aster J. C. and Pear W. S.(2008). Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest 118(9): 3181-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deftos M. L. and Bevan M. J.(2000). Notch signaling in T cell development. Curr Opin Immunol 12(2): 166-172. [DOI] [PubMed] [Google Scholar]
- 6. Ellisen L. W., Bird J., West D. C., Soreng A. L., Reynolds T. C., Smith S. D. and Sklar J.(1991). TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms . Cell 66(4): 649-661. [DOI] [PubMed] [Google Scholar]
- 7. Gachet S., Genesca E., Passaro D., Irigoyen M., Alcalde H., Clemenson C., Poglio S., Pflumio F., Janin A., Lasgi C., Dodier S., Soyer M., Dumenil G. and Ghysdael J.(2013). Leukemia-initiating cell activity requires calcineurin in T-cell acute lymphoblastic leukemia. Leukemia 27(12): 2289-2300. [DOI] [PubMed] [Google Scholar]
- 8. Goldberg J. M., Silverman L. B., Levy D. E., Dalton V. K., Gelber R. D., Lehmann L., Cohen H. J., Sallan S. E. and Asselin B. L.(2003). Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol 21(19): 3616-3622. [DOI] [PubMed] [Google Scholar]
- 9. Herranz D., Ambesi-Impiombato A., Sudderth J., Sanchez-Martin M., Belver L., Tosello V., Xu L., Wendorff A. A., Castillo M., Haydu J. E., Marquez J., Mates J. M., Kung A. L., Rayport S., Cordon-Cardo C., DeBerardinis R. J. and Ferrando A. A.(2015). Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat Med 21(10): 1182-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kopan R. and Ilagan M. X.(2009). The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137(2): 216-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Litzow M. R. and Ferrando A. A.(2015). How I treat T-cell acute lymphoblastic leukemia in adults . Blood 126(7): 833-41. [DOI] [PubMed] [Google Scholar]
- 12. Liu Y., Easton J., Shao Y., Maciaszek J., Wang Z., Wilkinson M. R., McCastlain K., Edmonson M., Pounds S. B., Shi L., Zhou X., Ma X., Sioson E., Li Y., Rusch M., Gupta P., Pei D., Cheng C., Smith M. A., Auvil J. G., Gerhard D. S., Relling M. V., Winick N. J., Carroll A. J., Heerema N. A., Raetz E., Devidas M., Willman C. L., Harvey R. C., Carroll W. L., Dunsmore K. P., Winter S. S., Wood B. L., Sorrentino B. P., Downing J. R., Loh M. L., Hunger S. P., Zhang J. and Mullighan C. G.(2017). The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 49(8): 1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lo Celso C. and Scadden D.(2007). Isolation and transplantation of hematopoietic stem cells(HSCs). J Vis Exp 2: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malecki M. J., Sanchez-Irizarry C., Mitchell J. L., Histen G., Xu M. L., Aster J. C. and Blacklow S. C.(2006). Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol 26(12): 4642-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Medyouf H., Gusscott S., Wang H., Tseng J. C., Wai C., Nemirovsky O., Trumpp A., Pflumio F., Carboni J., Gottardis M., Pollak M., Kung A. L., Aster J. C., Holzenberger M. and Weng A. P.(2011). High-level IGF1R expression is required for leukemia-initiating cell activity in T-ALL and is supported by Notch signaling. J Exp Med 208(9): 1809-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naviaux R. K., Costanzi E., Haas M. and Verma I. M.(1996). The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J Virol 70(8): 5701-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O'Neil J., Grim J., Strack P., Rao S., Tibbitts D., Winter C., Hardwick J., Welcker M., Meijerink J. P., Pieters R., Draetta G., Sears R., Clurman B. E. and Look A. T.(2007). FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med 204(8): 1813-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Palomero T., Barnes K. C., Real P. J., Glade Bender J. L., Sulis M. L., Murty V. V., Colovai A. I., Balbin M. and Ferrando A. A.(2006). CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia 20(7): 1279-1287. [DOI] [PubMed] [Google Scholar]
- 19. Passaro D., Irigoyen M., Catherinet C., Gachet S., C. Da Costa De Jesus, Lasgi C., Tran Quang C. and Ghysdael J.(2015). CXCR4 Is Required for Leukemia-Initiating Cell Activity in T Cell Acute Lymphoblastic Leukemia. Cancer Cell 27(6): 769-779. [DOI] [PubMed] [Google Scholar]
- 20. Pear W. S., Aster J. C., Scott M. L., Hasserjian R. P., Soffer B., Sklar J. and Baltimore D.(1996). Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med 183(5): 2283-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perez-Garcia A., Ambesi-Impiombato A., Hadler M., Rigo I., LeDuc C. A., Kelly K., Jalas C., Paietta E., Racevskis J., Rowe J. M., Tallman M. S., Paganin M., Basso G., Tong W., Chung W. K. and Ferrando A. A.(2013). Genetic loss of SH2B3 in acute lymphoblastic leukemia. Blood 122(14): 2425-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pitt L. A., Tikhonova A. N., Hu H., Trimarchi T., King B., Gong Y., Sanchez-Martin M., Tsirigos A., Littman D. R., Ferrando A. A., Morrison S. J., Fooksman D. R., Aifantis I. and Schwab S. R.(2015). CXCL12-Producing Vascular Endothelial Niches Control Acute T Cell Leukemia Maintenance. Cancer Cell 27(6): 755-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H. R. and Aguet M.(1999). Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10(5): 547-558. [DOI] [PubMed] [Google Scholar]
- 24. Sanchez-Martin M., Ambesi-Impiombato A., Qin Y., Herranz D., Bansal M., Girardi T., Paietta E., Tallman M. S., Rowe J. M., De Keersmaecker K., Califano A. and Ferrando A. A.(2017). Synergistic antileukemic therapies in NOTCH1-induced T-ALL. Proc Natl Acad Sci U S A 114(8): 2006-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thompson B. J., Buonamici S., Sulis M. L., Palomero T., Vilimas T., Basso G., Ferrando A. and Aifantis I.(2007). The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med 204(8): 1825-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wendorff A. A., Koch U., Wunderlich F. T., Wirth S., Dubey C., Bruning J. C., MacDonald H. R. and Radtke F.(2010). Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity 33(5): 671-684. [DOI] [PubMed] [Google Scholar]
- 27. Wendorff A. A., Quinn S. A., Rashkovan M., Madubata C. J., Ambesi-Impiombato A., Litzow M. R., Tallman M. S., Paietta E., Paganin M., Basso G., Gastier-Foster J. M., Loh M. L., Rabadan R., Van Vlierberghe P. and Ferrando A. A.(2019). Phf6 Loss Enhances HSC Self-Renewal Driving Tumor Initiation and Leukemia Stem Cell Activity in T-ALL . Cancer Discov 9(3): 436-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weng A. P., Ferrando A. A., Lee W., Morris J. P. t., Silverman L. B., Sanchez-Irizarry C., Blacklow S. C., Look A. T. and Aster J. C.(2004). Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia . Science 306(5694): 269-271. [DOI] [PubMed] [Google Scholar]
- 29. Yardeni T., Eckhaus M., Morris H. D., Huizing M. and Hoogstraten-Miller S.(2011). Retro-orbital injections in mice. Lab Anim(NY) 40(5): 155-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Young L., Sung J., Stacey G. and Masters J. R.(2010). Detection of Mycoplasma in cell cultures. Nat Protoc 5(5): 929-934. [DOI] [PubMed] [Google Scholar]