Abstract
Bacteriocins are small ribosomally synthesized antimicrobial peptides produced by some microorganisms including lactic acid bacteria (LAB), a group of Gram-positive bacteria (cocci, rods) expressing high tolerance for low pH. Bacteriocins kill bacteria rapidly and are biologically active at very low concentrations. Bacteriocins produced by LAB are primarily active against closely related bacterial species. Many bacteriocins have been investigated with respect to their potential use in promoting human, plant, and animal health, and as food biopreservatives. Bacteriocins produced by LAB are particularly interesting since several LAB have been granted GRAS (Generally Recognized as Safe) status. Because it is not always possible to extract active bacteriocins secreted from cells grown in liquid medium, we developed a simple and inexpensive peptide extraction procedure using a semi-solid nutrient-rich agar medium. We hereby present a detailed procedure that leads to the rapid extraction of secreted bioactive bacteriocin peptides from the oral species Streptococcus mutans, a prolific bacteriocin-producing species, and its potential application for bacteriocin extraction from other LAB (e.g., Streptococcus, Lactococcus, Enterococcus). We also present a simple method for the detection of bacteriocin activity from the purified extracellular peptide extract.
Keywords: Bacteriocin, Extracellular peptide extraction, Lactic acid bacteria, Oral streptococci, Streptococcus mutans, Spot-on-Lawn assay, Bacteriocin activity
Background
Most bacteria in nature do not exist independently but persist in complex multispecies biofilm communities ( López et al., 2010 ). Numerous physical and nutritional interactions exist between bacteria contributing to the biofilm growth and survival. The production and secretion of bacteriocins to the extracellular space confer a distinct ecological advantage to the producer in competing against other bacteria that are present in the same ecological niche (Donia and Fischbach, 2015). Therefore, bacteria tightly regulate expression of bacteriocins to foster survival of the species within the biofilm.
The cariogenic organism Streptococcus mutans is found in the dental plaque, a biofilm community that forms on the tooth surface ( Kolenbrander et al., 2010 ). Under normal circumstances, the dental plaque remains relatively stable (microbial homeostasis), contributing to dental health. However, a change in local environmental conditions can disrupt this natural homeostasis, leading to an ecological pressure that can cause disease. S. mutans is one of the most prolific producers of bacteriocins (referred to as mutacins) and bacteriocin production is considered to be an important factor in the colonization and establishment of S. mutans in the dental biofilm (Merritt and Qi, 2012). In our previous publications, we highlighted the role of S. mutans bacteriocins in the antagonistic interactions among oral streptococci (Dufour and Lévesque, 2013; Dufour et al., 2020 ). We also showed that DNA uptake and bacteriocin production were controlled by a tighty regulated quorum sensing system suggesting that DNA exchange could be enhanced by bacteriocin secretion during the state of genetic competence ( Perry et al., 2009a and 2009b; Dufour et al., 2011 ).
Most strains of S. mutans encode bacteriocins. Identification of putative bacteriocins can be easily done in silico by screening of genomic DNA sequences. For instance, the freely available software BAGEL4 (http://bagel4.molgenrug.nl) can be used for the detection of putative bacteriocin genes. It also analyzes the surrounding region on the bacterial genome for genes possibly encoding proteins involved in transport, immunity, and regulation. However, the putative bacteriocin loci identified in silico do not necessarily represent functional bacteriocin production. Consequently, bacteriocins must be extracted and purified in order to study them. Only few bacteriocins produced by S. mutans have been purified and characterized ( Nicolas et al., 2007 ). This may be due to the many challenges associated with the production and extraction of these extracellular peptides. High variability of peptide recovery from one extraction to another is also usually observed. Since the volumetric bacteriocin production is dependent on the total biomass production, a nutrient-rich medium (e.g., animal tissue extracts) is also suggested for efficient production. In this protocol, we developed an extraction method from submerged cultures using a commercial medium supplemented with mineral salts and yeast extract ( Dufour et al., 2020 ). This procedure has been successfully used by our group to extract biologically active bacteriocin peptides from oral streptococci (S. mutans, S. salivarius) isolated from the dental plaque biofilm of caries-free and caries-active subjects.
Materials and Reagents
Sterile 13 ml culture tube with ventilation cap (any type, e.g., Sarstedt, catalog number: 62.515.006)
Screw cap tube 50 ml (any type, e.g., Sarstedt, catalog number: 62.547.205)
Disposable glass Pasteur pipets 22.9 cm (9”) (e.g., VWR, catalog number: 89061-526)
Disposable syringe, 50 ml (any type, e.g., Thermo Fisher Scientific, Fisherbrand, catalog number: 14-823-43)
Acrodisc® syringe filters with Supor® membrane 0.2 µm, 25 mm (Pall Corporation, catalog number: 4506)
Petri dishes with clear lid 100 x 15 mm (any type, e.g., Thermo Fisher Scientific, Fisherbrand, catalog number: FB0875712)
Aluminum foil (any product)
Bacteriocin-producing LAB (e.g., Streptococcus, Lactococcus, Enterococcus)
Bacterial target strain: Micrococcus luteus (e.g., strain ATCC 4698)
Dehydrated Todd-Hewitt broth (Thermo Fisher Scientific, BD Difco, catalog number: DF0492-17-6)
Dehydrated Columbia broth (Thermo Fisher Scientific, BD Difco, catalog number: DF0944-17-0)
Yeast extract (BioShop Life Science Products, catalog number: YEX401)
Agar (BioShop Life Science Products, catalog number: AGR001)
NaH2PO4 (e.g., Millipore Sigma, catalog number: S8282)
Na2HPO4 (e.g., Millipore Sigma, catalog number: S9763)
MgSO4 (e.g., Millipore Sigma, catalog number: M7506)
FeSO4 (e.g., Millipore Sigma, catalog number: F7002)
CaCO3 (e.g., Millipore Sigma, Sigma-Aldrich, catalog number: C4830)
Chloroform ACS Reagent Grade (Caledon Laboratory Chemicals, catalog number: 3000-1-40)
Acetonitrile (for HPLC) (Thermo Fisher Scientific, Fisher Chemical, catalog number: A998)
Trifluoroacetic acid (Peptide and Protein Analysis/Certified) (Thermo Fisher Scientific, Fisher Chemical, catalog number: O4902)
Methanol (for HPLC) (Thermo Fisher Scientific, Fisher Chemical, catalog number: A452SK-4)
Hydrochloric acid solution, 6 N ACS (Certified) (Thermo Fisher Scientific, Fisher Chemical, catalog number: SA56-4)
Milli-Q water or any filtered ultra-pure water
THYE broth (500 ml) (see Recipes)
BAC semi-solid agar medium (2 L) (see Recipes)
Columbia-CaCO3 agar plates (500 ml) (see Recipes)
Columbia soft agar (500 ml) (see Recipes)
ACN-TFA solvent (500 ml) (see Recipes)
Methanol:water, 95:5 v/v [pH 2] (500 ml) (see Recipes)
Methanol:water v/v gradient (500 ml) (see Recipes)
Equipment
Heavy-duty low form beaker 4 L (any type, e.g., VWR, catalog number: 10536-520)
Heavy-duty low form beaker 250 ml (any type, e.g., VWR, catalog number: 10536-390)
Scoop type spatula (any type, e.g., VWR International, catalog number: 470149-438)
Centrifuge bottles 250 ml (Thermo Fisher Scientific, Nalgene PPCO, catalog number: 3120-0250)
Reusable glass media bottle with cap 1 L (any type, e.g., Thermo Fisher Scientific, Fisherbran, catalog number: FB8001000)
Reusable glass media bottle with cap 500 ml (any type, e.g., Thermo Fisher Scientific, Fisherbrand, catalog number: FB800500)
Sep-Pak® C18 environmental cartridges, 55-105 µm (Waters, catalog number: WAT023635)
CO2 incubator (any type, e.g., Thermo Fisher Scientific, model: Hera Cell 150)
Water bath (any product)
Magnetic stirrer (any product)
Magnetic stir bar (any product)
Benchtop refrigerated centrifuge (any type, e.g., Eppendorf, model: 5810 R)
Floor refrigerated centrifuge (any type, e.g., Beckman Coulter, model: J2-21 M)
Ultra-low temperature freezer (any product, e.g., Thermo Fisher Scientific, model: Forma 900 Series)
SpeedVac vacuum concentrator (any product, e.g., Thermo Fisher Scientific, model: Savant SPD120)
Portable pump (any product, e.g., Bio-Rad, model: Econo Gradient Pump)
Bunsen burner
Cold (4 °C) lab chamber
Lab fume hood
Steam autoclave
Procedure
-
Preparation of bacteriocin production medium and bacterial growth
Prepare the bacteriocin production BAC semi-solid agar medium in a beaker and keep the sterilized medium in the cold room overnight (18-24 h) before inoculation.
Prepare a preculture of bacteriocin-producing LAB strain of interest into a sterile 13 ml culture tube with ventilation cap containing 10 ml of THYE broth, and incubate statically at 37 °C with 5% CO2 in a CO2 incubator for 18 h.
Using a sterile glass Pasteur pipet, stab the BAC semi-solid agar medium approximately 300 times under aseptic conditions (Figure 1).
Pour the overnight preculture of the bacteriocin-producing LAB strain on the surface of the BAC semi-solid agar medium and cover the beaker with the same aluminum foil used for sterilization of the medium. Use 5 ml of an overnight preculture at an OD600 of ~1.0 per liter of BAC semi-solid agar medium (Figure 2).
Incubate the mixture at 37 °C with 5% CO2 in a CO2 incubator for 72 h (Figure 3).
-
Extraction of secreted bacteriocins
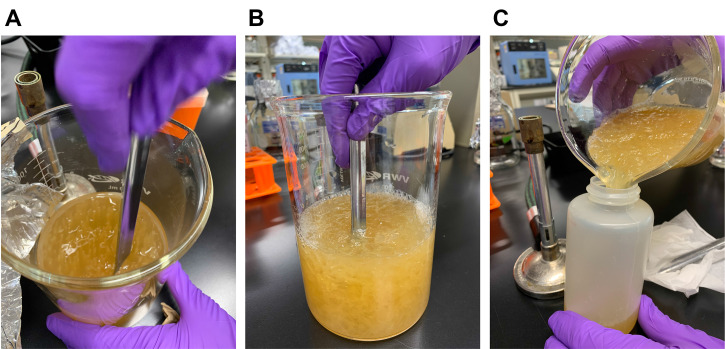
Mash the mixture using a spatula and transfer into 250 ml centrifuge bottles (Figure 4).
Store the bottles containing the mashed mixture in an ultra-low temperature freezer for 18 h.
Thaw the mixture by putting the bottles in a water bath at 65 °C for 1 h. The freeze/thaw cycle is necessary in order to disrupt the cell membrane of LAB.
Centrifuge at 15,000 × g for 40 min at 4 °C using a floor refrigerated centrifuge.
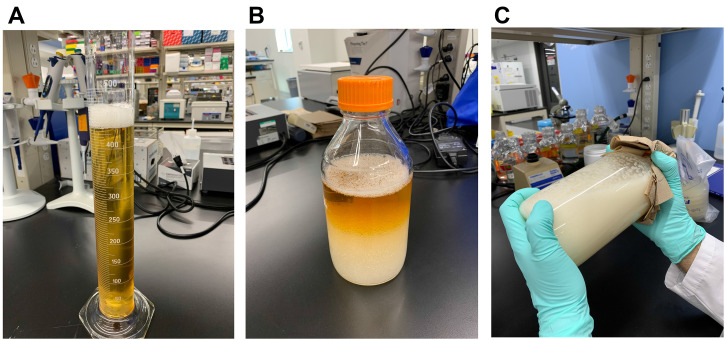
Carefully collect the supernatant (Figure 5A) and transfer into a 1 L glass bottle.
Add an equal volume of chloroform (Figure 5B) and shake vigorously by hand for 20 min (Figure 5C). Store the mixture at 4 °C overnight (18-24 h).
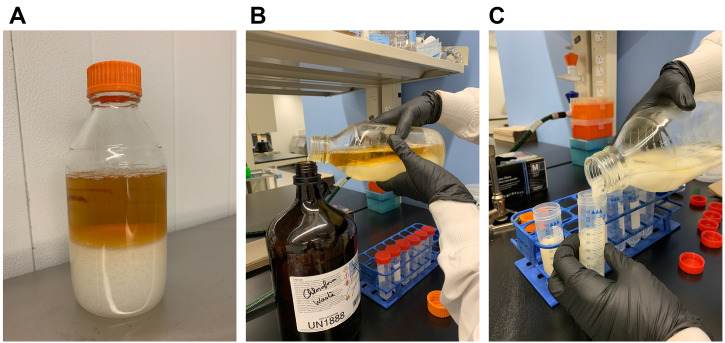
Remove the upper layer and transfer the white layer into screw cap tube 50 ml (Figure 6). Let the emulsion settle on its own (approx. 5-10 min).
Collect the floating white interfacial layer and transfer into a glass beaker (Figure 7).
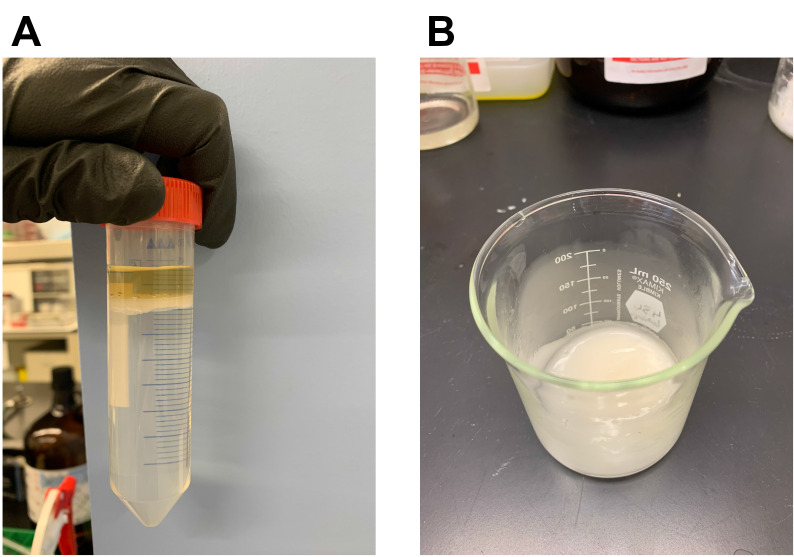
Keep the material in a fume hood until complete evaporation of the residual chloroform (approx. 2-3 days). A bacteriocin-containing powder of brownish color should be obtained (Figure 8).
Dissolve the extracellular peptide extract in 25 ml of ACN-TFA solvent.
Transfer the resuspended extracellular peptide extract in a screw cap tube 50 ml.
Centrifuge at 3,200 × g for 40 min at 4 °C using a benchtop refrigerated centrifuge to remove insoluble materials.
Transfer the clear supernatant into a clean screw cap tube 50 ml.
The supernatant is passed through a 0.22 µm syringe filter with low protein binding membrane.
Prewash a C18 cartridge with 100 ml of methanol:water, 80:20 v/v.
The resultant liquid obtained at Step A18 is passed through a Sep-Pak® C18 cartridge using 50 ml of increasing concentration of methanol:water v/v (50%, 60%, 70%, 80%, and 95% [pH 2]) using a portable pump at a flow rate of 1 ml/min. All fractions are tested for bacteriocin activity using a spot-on-lawn assay.
To obtain a more concentrated peptide extract, dry the active fractions using a SpeedVac vacuum concentrator at room temperature and dissolve using ACN-TFA solvent to acquire desirable concentrations.
Keep an aliquot of the extracellular peptide extract at 4 °C for immediate use while the rest is stored in a freezer (-20 °C).
-
Detection of bacteriocin activity using spot-on-lawn
Prepare a preculture of a target strain into a sterile 13 ml culture tube with ventilation cap containing 3 ml of THYE broth, and incubate statically at 37 °C with 5% CO2 in a CO2 incubator for 18 h.
Measure the absorbance (OD600) of the preculture and dilute the target strain in 4 ml of Columbia soft-agar pre-warmed at 55 °C to obtain a concentration of approximately 107 CFU/ml using a plot standard curve (OD600 vs. CFU/ml).
Pour the inoculated soft-agar onto the surface of a Columbia-CaCO3 agar plate. Keep at room temperature until the top layer has solidified.
Prepare a series of two-fold serial dilutions (1/2, 1/4, 1/8, 1/16, 1/32, 1/64, 1/128) of the extracellular peptide extract in ACN-TFA solvent.
Drop 20 µl aliquots of two-fold serially diluted extracellular peptide extract on top of Columbia-CaCO3 agar plate overlay containing the sensitive target strain. Wait until the drops have been completely absorbed before incubating the plates. To accelerate the absorption, keep the plates next to the flame of a Bunsen burner with the lid slightly off for 10-15 min.
Incubate the plates at 37 °C with 5% CO2 in a CO2 incubator for 18 h.
Figure 1. Stabbing of the bacteriocin production medium.
Using a sterile glass Pasteur pipet of 22.9 cm, the semi-solid agar medium is stabbed multiple times. The depth of the stabs should correspond to the tip length of the pipet used (approx. 120 mm for a Pasteur pipet of 22.9 cm).
Figure 2. Inoculation of the bacteriocin production medium.
Figure 3. Culture of bacteriocin-producing LAB obtained after 72 h of incubation.
Bacterial growth of Streptococcus mutans LAB761, a strain encoding several bacteriocin loci, can be seen in stabbed BAC semi-solid agar medium.
Figure 4. The stabbed bacteriocin medium showing bacterial growth (A) is mashed into small pieces using a spatula (B). The mashed mixture is then transferred into centrifuge bottles (C).
Figure 5. Chloroform extraction of bacteriocins.
One volume of chloroform is added to one volume of collected supernatant (A). The mixture (B) is mixed thoroughly by hand for approx. 20 min (C).
Figure 6. Bacteriocin extraction following chloroform method.
The upper phase (A) corresponding to the “chloroform waste” is carefully removed (B) and the white material containing the secreted peptides is transferred into 50 ml tubes (C).
Figure 7. Extraction of bacteriocin following chloroform method.
A floating white interfacial layer should be clearly visible (A). The white interface corresponding to the bacteriocin fraction is transferred into a glass beaker (B).
Figure 8. Appearance of the extracellular peptide extract following complete evaporation.
Data analysis
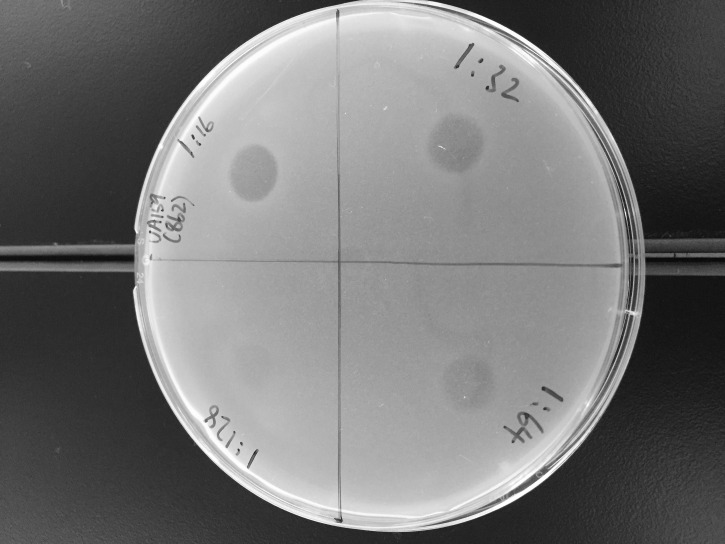
Plates are visually examined for evidence of growth inhibition (Figure 9).
Bacteriocin concentration is expressed as arbitrary unit (AU) per millimiter. AU is defined as the reciprocal of the highest dilution at which bacterial growth inhibition is visually detectable. Based on Figure 9, the bacteriocin concentration is estimated at 3,200 AU/ml (64 x [1,000 µl/20 µl]).
Figure 9. Antimicrobial activity of the extracellular peptide extract from S. mutans detected using the spot-on-lawn assay.
Aliquots (20 µl) of two-fold serially-diluted peptide extract (1/16, 1/32, 1/64 and 1/128 dilutions are shown) are tested.
Recipes
-
THYE broth (500 ml)
In an autoclavable glass bottle (1 L) with cap, add 15 g of dehydrated Todd-Hewitt broth, 1.5 g of Yeast extract, and 400 ml of Milli-Q water
Place a magnetic stir bar in the bottle and stir using a magnetic stirrer to completely dissolve
Adjust final volume to 500 ml with Milli-Q water
-
Autoclave on wet cycle at 121 °C for 30 min
Note: There is no need for pH adjustment. The cap must be loose to allow steam to escape. Broth can be stored at room temperature.
-
BAC semi-solid agar medium (2 L)
In a glass beaker (4 L), add 60 g of dehydrated Todd-Hewitt broth, 6 g of Yeast extract, 2 g of NaH2PO4, 0.4 g of Na2HPO4, 1.4 g of MgSO4, 0.01 g of FeSO4, 6 g of agar, and 1.6 L of Milli-Q water
Place a magnetic stir bar in the bottle and stir using a magnetic stirrer to completely dissolve
Adjust final volume to 2 L with Milli-Q water. Cover the beaker with aluminum foil
-
Autoclave on wet cycle at 121 °C for 55 min
Note: There is no need for pH adjustment. The medium is kept at room temperature for 18 h before inoculation.
-
Columbia-CaCO3 agar plates (500 ml)
In an autoclavable glass bottle (1 L) with cap, add 17.5 g of dehydrated Columbia broth, 0.5 g CaCO3, 7.5 g of agar, and 400 ml of Milli-Q water
Place a magnetic stir bar in the bottle and stir using a magnetic stirrer to dissolve
Adjust final volume to 500 ml with Milli-Q water
-
Autoclave on wet cycle at 121 °C for 30 min
Note: There is no need for pH adjustment. The cap must be loose to allow steam to escape. Although Columbia agar base powder can be purchased, we recommend to use the dehydrated Columbia broth and add agar separately. Lower bacteriocin activity was detected using Columbia agar base powder.
-
Pour agar plates under aseptic conditions
Note: Precipitate of CaCO3 will not dissolve completely. Agar plates are stored at 4 °C.
-
Columbia soft-agar (500 ml)
In an autoclavable glass bottle (1 L) with cap, add 17.5 g of dehydrated Columbia broth, 3 g of agar, and 320 ml of Milli-Q water
Place a magnetic stir bar in the bottle and stir using a magnetic stirrer to completely dissolve
Adjust final volume to 500 ml with Milli-Q water
-
Autoclave on wet cycle at 121 °C for 30 min
Note: There is no need for pH adjustment. The cap must be loose to allow steam to escape. Soft-agar is kept at 55 °C and used the same day.
-
ACN-TFA solvent (500 ml)
Prepare a solution of 35% acetonitrile-0.1% trifluoroacetic acid by diluting 175 ml of acetonitrile into 324.5 ml of Milli-Q water into a glass bottle (1 L) with cap
Add 0.5 ml of trifluoroacetic acid
-
Place a magnetic stir bar in the bottle and stir using a magnetic stirrer to mix
Note: The solvent is stored at room temperature.
-
Methanol:water, 95:5 v/v [pH 2] (500 ml)
Prepare a solution of 95% methanol by diluting 475 ml of methanol into 25 ml of Milli-Q water into a glass bottle (1 L) with cap
Place a magnetic stir bar in the bottle and stir using a magnetic stirrer to mix
-
Adjust the pH to 2.0 with HCl 6 N
Note: The solvent is stored at room temperature.
-
Methanol:water v/v gradient (500 ml)
Prepare a solution of 50%, 60%, 70% and 80% methanol by diluting 250 ml, 300 ml, 350 ml, or 400 ml of methanol into 250 ml, 200 ml, 150 ml, or 100 ml of Milli-Q water into a glass bottle (1 L) with cap, respectively
-
Place a magnetic stir bar in the bottle and stir using a magnetic stirrer to mix
Note: The solvents are stored at room temperature.
Acknowledgments
The authors acknowledge many helpful discussions with Aboud Barbour and Delphine Dufour. This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) grant RGPIN-2019-06454 (to C.M.L.) and by the Canadian Institutes of Health Research (CIHR) grant CMA-151711 (to C.M.L. and S.G.G.). C.M.L. is a recipient of a Canada Research Chair in Oral Microbial Genetics. This protocol was previously used in our reported work ( Dufour et al., 2020 J Bacteriol 202: e00762-19).
Competing interests
The authors declare no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Donia M. S. and Fischbach M. A.(2015). Small molecules from the human microbiota. Science 349(6246): 1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dufour D., Barbour A., Chan Y., Cheng M., Rahman T., Thorburn M., Stewart C., Finer Y., Gong S.-G. and Lévesque C. M.(2020). Genetic analysis of mutacin B-Ny266, a lantibiotic active against caries pathogens. J Bacteriol 202(12): e00762-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dufour D., Cordova M., Cvitkovitch D. G. and Lévesque C. M.(2011). Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J Bacteriol 193(23): 6552-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dufour D. and Lévesque C. M.(2013). Bacterial behaviors associated with the qorum-sensing peptide pheromone(‘alarmone’) in streptococci. Future Microbiol 8(5): 593-605. [DOI] [PubMed] [Google Scholar]
- 5. Kolenbrander P. E., Palmer R. J. Jr. Periasamy S. and Jakubovics N. S.(2010). Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol 8(7): 471-480. [DOI] [PubMed] [Google Scholar]
- 6. López D., Vlamakis H. and Kolter R.(2010). Biofilms. Cold Spring Harb Perspect Biol 2(7): a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Merritt J. and Qi F.(2012). The mutacins of Streptococcus mutans: regulation and ecology . Mol Oral Microbiol 27(2): 57-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nicolas G. G., Lavoie M. C. and LaPointe G.(2007). Molecular genetics, genomics and biochemistry of mutacins. G3: Genes, Genomes, Genomics 1(2): 193-208. [Google Scholar]
- 9. Perry J. A., Cvitkovitch D. G. and Lévesque C. M.(2009). Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA . FEMS Microbiol Lett 299(2): 261-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perry J. A. Jones M. B., Peterson S. N., Cvitkovitch D. G. and Lévesque C. M.(2009). Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol 72(4): 905-917. [DOI] [PMC free article] [PubMed] [Google Scholar]