Abstract
The study of host-pathogen interactions has improved our understanding of both pathogenesis and the response of the host to infection, including both innate and adaptive responses. Neutrophils and macrophages represent the first line of innate host defense against any infection. The zebrafish is an ideal model to study the response of these cells to a variety of pathogens. Zebrafish possess both neutrophils and macrophages exhibiting similar defense mechanisms to their human counterparts. The transparency of zebrafish embryos greatly facilitates in vivo tracking of infection dynamics in a non-invasive manner at high-resolution using labelled pathogens, while immune cells can also be labelled transgenically to enable even more in-depth analysis. Here we describe a procedure for performing a bacterial infection assay in zebrafish embryos using fluorescently-labelled E. coli bacteria and demonstrate the monitoring and quantification of the infection kinetics. Of note, this procedure helps in understanding the functional role of genes that are important in driving the innate immune response.
Keywords: Infection, Fluorescent bacteria, Innate immunity, Zebrafish, Imaging
Background
Host-pathogen interaction studies are important to understand disease pathogenesis and also for the development of effective treatments. Multiple aspects of both the host and pathogen need to be considered to determine potential risk factors in the host and key virulence factors in the pathogen. Innate immunity represents the first line of defense against infections, triggering a cascade of responses, including inflammation, neutralization and recruitment of components of the adaptive immune system (Akira et al., 2006). Neutrophils are rapidly recruited to the site of bacterial infection in response to chemoattractant gradients of host chemokines released from damaged cells as well as bacterial products themselves, where they initiate the immune response by phagocytosis and netolysis (Renshaw and Trede, 2012; Deng et al., 2013). Macrophage recruitment follows, with these cells focused on removing dead cells, remodeling injured tissue and coordinating adaptive immune cells (Weiss and Schaible, 2015).
The development and function of innate immune cells are controlled by a range of specific genes, the dysregulation of which can lead to a number of pathological states, including enhanced susceptibility to infection and chronic inflammation. Being short-lived, neutrophils are depleted rapidly in response to exposure to a pathogen, with so-called ‘emergency granulopoiesis’ being initiated to generate additional neutrophils (Hall et al., 2012; Manz and Boettcher, 2014). Granulocyte colony-stimulating factor (G-CSF) acting via its receptor, G-CSFR, is a key mediator of emergency granulopoiesis through its action promoting the proliferation and differentiation of relevant hematopoietic progenitor cells (Panopoulos and Watowich, 2008; Liongue et al., 2009). Understanding the role of factors that regulate the innate immune response during bacterial infection in an appropriate in vivo model can provide unique insights into infection and immunity.
Zebrafish represents a very attractive in vivo model to perform host-pathogen studies, with its transparent embryos allowing in vivo imaging at high-resolution to track infection in real-time, by tagging the pathogens with fluorescent markers (Basheer et al., 2019). In addition, using transgenic approaches in zebrafish, neutrophils and macrophages can also be monitored via fluorescent tags (Ellett et al., 2011; Gray et al., 2011). Importantly, these cells along with other components of the immune system share remarkable similarity with those of humans (Meeker and Trede, 2008). The zebrafish innate immune system forms early during their development with the generation of macrophages at 24 h post fertilization (hpf) and neutrophils by 32-48 hpf (Herbomel et al., 1999; Willett et al., 1999; Bennett et al., 2001). The zebrafish adaptive immune system develops later, allowing the innate immune system to be studied independently. Zebrafish infection models of different pathogen–bacterial, viral and fungal–have been established (Gratacap and Wheeler 2014; Masud et al., 2017; Varela et al., 2017).
Our research has employed a zebrafish bacterial infection model to understand the role of key genes involved in innate immunity in defense against infection. Escherichia coli (E. coli) bacteria expressing green fluorescent protein (GFP) were microinjected into 72 hpf larvae derived from either wild-type zebrafish or those mutant for the gene encoding G-CSFR (Figure 2). These were monitored by fluorescence microscopy over a time-course to determine the relative infection kinetics (Basheer et al., 2019). Overall, this procedure helps in understanding different host-pathogen interactions and unravel the functions of key immune genes involved in the process using zebrafish as an animal model.
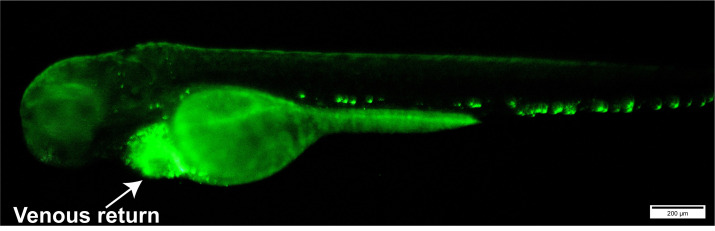
Figure 2. Image of a 72 hpf embryo with E. coli-GFP bacteria injected into their venous return (shown by arrow head).
Scale bars = 200 μm.
Materials and Reagents
Transfer pipette, 3 ml (Heinz Herenz, catalog number: 1131303)
Mesh strainer, 12-15 cm in diameter (pore size should be less than zebrafish embryo with chorion diameter, approximately 0.5 mm mesh)
Petri plate (PLP, catalog number: S9014UV20)
Parafilm
Thin-wall borosilicate with filament, 1.0 mm x 0.78 mm x 15 cm (SDR Scientific, catalog number: 30-0039 GC100TF-15)
Microloader pipette tips (Point of Care Diagnostics Pty Ltd, catalog number: EPP5242 956.003)
Corning square dish, 245 mm Non-Treated (Corning, catalog number: 38020)
Paper towel
Multiwell tissue culture plates, 6-well and 12-well (Interpath, Greiner, catalog numbers: 657160, 665180)
Cuvettes (Bio-Rad, catalog number: 2239950)
Breeding/mating tanks, 3 L (Techniplast)
Zebrafish (Danio rerio), wild-type
E. coli-GFP bacteria (ATCC, catalog number: 25922GFP)
Agar bacteriological grade (Astral Scientific, catalog number: J637-500)
N-phenylthiourea, PTU (Sigma-Aldrich, catalog number: P7629-10G)
Benzocaine (Sigma-Aldrich, catalog number: E1501-100G)
NaCl (Astral Scientific, catalog number: 0241-5kg)
KCl (Sigma-Aldrich, catalog number: P9541-500G)
CaCl2·2H2O (Sigma-Aldrich, catalog number: C3306-250G)
MgSO4·7H2O (Sigma-Aldrich, catalog number: 63138-1KG)
Methylene blue (Sigma-Aldrich, catalog number: M9140-25G)
NaOH
Phosphate buffered saline-PBS tablets (Astral Scientific, catalog number: AME404-200TABS)
Triton X-100 (Sigma-Aldrich, catalog number: X100-500ML)
Blu-tack
BD BactoTM Tryptic Soy Broth (Soybean-Casein Digest Medium) (Becton Dickinson, catalog number: 211825)
Ampicillin, 100 mg/ml, ready-made solution (Sigma-Aldrich, catalog number: A5354-10ML)
Milli-Q or distilled water
Phenol red, 0.5%, liquid, sterile-filtered (Sigma-Aldrich, catalog number: P0290)
Mineral oil (Sigma-Aldrich, catalog number: M5904-5ML)
60x E3 media (see Recipes)
E3 media (see Recipes)
Methylene blue stock solution (see Recipes)
E3 containing 0.3 mg/L methylene blue (see Recipes)
PTU stock, 0.3% (w/v) (see Recipes)
E3 containing 0.003% (w/v) PTU (see Recipes)
Benzocaine stock solution (see Recipes)
10x PBS stock solution (see Recipes)
PBS (see Recipes)
1% Triton X-100 in PBS (see Recipes)
Equipment
Glass bottles, 1 L and 2 L
Magnetic stir bar–PTFE Cylindrical 15 mm length x 6 mm diameter (PLP, Cowie, catalog number: 001.115.6)
Eppendorf tube racks
Pipettes, 100-1,000 μl, 20-200 μl, 2-10 μl, 0.5-2 μl
Watch makers forceps, pointed tweezers, style 5, 114 mm (ProSciTech, catalog number: T65-SA)
Incubator, 28 °C (Sanyo, catalog number: MIR-162)
Incubator, 37 °C (Thermo Scientific, Hereaus, catalog number: B6030)
Shaking incubator (200-225 rpm) (Infors HT Multitron Standard incubator)
Spectrophotometer (Bio-Rad SmartSpec Plus Spectrophotometer, catalog number: 273BR03695)
Micropipette needle puller (Sutter Instrument, catalog number: P-97)
Bunsen burner
Atherton Chipmunk Autoclave Sterilizer
Pipette pump
Mini centrifuge (Thermo Fisher Scientific Pty Ltd, catalog number: 75004061)
Microcentrifuge (Thermo Fisher Scientific Pty Ltd, catalog number: THR75002410)
Magnetic stirrer (Ratek Instruments Pty Ltd, catalog number: MS10)
Refrigerator, 4 °C
Microinjector set up (SDR Clinical Technology, Nanoject II Nanoliter In Jecto, catalog number: 690131, Footswitch, catalog number: 690140, Support Base, catalog number: 690141, Micropipette Holder Kit Option, catalog number: M-PIP-Kit)
Stereo dissecting microscope with light base (Nikon, catalog number: SMZ745)
Fluorescence microscope (Olympus, model: MVX10 with DP72 camera)
Tissue Lyser II (Qiagen, model: 85300)
Fume hood
Software
GraphPad Prism 8 (http://www.graphpad.com)
ImageJ (https://imagej.nih.gov/ij/index.html)
Procedure
-
Preparing the embryos
Transfer adult zebrafish (one male and two females) to a 3 L breeding tank in the evening to allow them to acclimatize before stimulation of mating the next morning using light.
Collect embryos from the tanks using a mesh strainer and transfer them to a 10 cm Petri dish containing 25 ml E3 media with 0.3 mg/L methylene blue.
Remove any dead or unfertilized eggs (embryos turn black as they die and need to be removed by using suction from a transfer pipette and then discarded) from the dish and transfer the remaining fertilized eggs to an incubator held at 28 °C.
At 24 hpf, transfer embryos to E3 media with 0.3 mg/L methylene blue and 0.003% (w/v) 1-phenyl-2-thiourea (PTU) to prevent pigmentation.
Replace E3 media in the dish daily until 72 hpf.
Any embryos unhatched by 72 hpf (injection day) may be dissected out of the chorion membrane using pointed watch makers forceps.
-
Tryptic soy broth preparation
Weigh 15 g tryptic soy broth (TSB) powder and transfer to 1 L glass bottle.
Add 500 ml of Milli-Q water and a magnetic stir bar and place the bottle on a magnetic stirrer to allow dissolution.
Autoclave the media at 121 °C, pressure 400-500 kPa in an autoclave (Atherton Chipmunk Sterilizer) for 1 h 30 min to sterilize (make sure the lid of the glass bottle is loosened before autoclaving).
Once sterilization is complete, remove the media and close the bottle lid tightly.
Allow the media to completely cool down at room temperature before transferring to 4 °C for long term storage (3-4 weeks).
-
Tryptic soy-agar-ampicillin plate preparation
Weigh 6 g Agar-bacteriological grade and 15 g TSB powder to a 1 L glass bottle.
Add 500 ml of Milli-Q water and a magnetic stir bar and place the bottle on a magnetic stirrer to allow dissolution.
Autoclave the media at 121 °C in an autoclave, pressure 400-500 KPa (Atherton Chipmunk Sterilizer) for 1 h and 30 min to sterilize (make sure the lid of the glass bottle is loosened before autoclaving).
Once sterilized, remove the media and close the bottle lid tightly.
Allow the media to completely cool down at room temperature before adding 500 μl ampicillin solution (100 mg/ml), for a final concentration of 100 μg/ml ampicillin.
Pour approximately 25 ml TSB-agar-ampicillin solution under flame into 10 cm Petri dishes without generating bubbles and allow them to completely cool down to room temperature (Figure 1).
Once cooled, close the lid of the Petri dish, seal with parafilm and store at 4 °C for later use.
-
Bacterial cultivation and enumeration
Inoculate E. coli-GFP bacteria into 10 ml of tryptic soy broth supplemented with 10 μl 100 mg/ml ampicillin.
Grow this bacterial culture overnight at 37 °C in a shaking incubator at 1.1-1.4 g.
Transfer 1 ml of bacterial culture into a cuvette and place in a spectrophotometer previously blanked against uninoculated TSB and use the absorbance as 600 nm to determine the colony forming units (CFU) according to Biorad SmartSpec Plus OD600 absorbance assay.
Perform serial dilutions of the bacterial culture (ten-fold serial dilutions between 10-6 and 10-10, 1 ml of bacterial culture in 9 ml of uninoculated TSB), plating 50-100 μl diluted broth onto TSB-agar-ampicillin plates and incubating the plates overnight at 37 °C incubator to determine the exact number of CFU in the broth.
Pellet the bacterial cells at 13,751 × g for 1-2 min and remove the supernatant.
Resuspend the bacterial cells in sterile PBS at a concentration of ~9 x 1010 CFU/ml based on the spectrophotometry.
-
Microinjection of bacterial cells into embryos
Form a thin-walled precision borosilicate glass capillary tube into a fine needle using a micropipette needle puller (After loading the borosilicate glass capillary tube, pull the needle with settings, heat–365, pull–45, velocity–80, time–150 and pressure–500, and press the pull button to make needles). These variable settings are increments that differ with each equipment (https://www.sutter.com/manuals/P-87_OpMan.pdf).
Place the needle onto a square plate/dish with Blu-tack to protect them from breakage before use.
Prepare the injection material by mixing 4 μl ~9 x 1010 CFU bacteria-PBS suspension with 1 μl 2% (w/v) phenol red to allow visualization of the injectate.
Using a microloader pipette, load the wide end of the needle with 3 μl injection material containing phenol red.
Shake the needle to bring the injection material to the tip of the needle and to remove any air bubbles.
Turn on the air source and the microinjector and transfer the needle into the microinjector source and secure tightly within the housing.
Adjust the micromanipulator to the correct position to allow for fine adjustments while injecting, bringing the needle close to the stage and area of injection.
Anesthesize 72 hpf embryos with 0.1 mg/ml of benzocaine in E3 media and transfer the Petri dish to the microinjection stage.
Break the tip of the microinjection needle using sharp watch makers forceps to an extent that allows the bacterial cells to expel easily, releasing consistent amounts and able to pierce the embryos with minimal damage.
The volume of injecting solution can be determined by injecting the material into a drop of mineral oil, measuring the diameter of the injected drop over a scale bar and calculating the volume. The volume can be adjusted by altering the injector pressure and/or the needle tip.
Once the microinjector is setup completely, align the embryos close to the needle tip.
-
Pierce the needle tip into the venous return of 72 hpf embryos (Figure 2) and use the foot pedal to inject 2-5 nano litres (nl) of injection solution into each embryo, repeating until the desired number is injected.
Note: The quality of the needle tip is crucial for performing consistent injections that do not unnecessarily damage the embryos.
Once injected, place the embryos in a Petri dish containing fresh E3 media with 0.3 mg/L of methylene blue and 0.003% PTU for 15 min to recover before they are transferred to a 28 °C incubator.
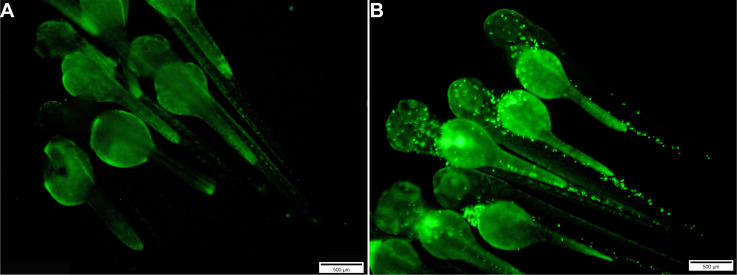
In performing the injection assay, each experimental group should have a minimum of 30-60 embryos, including both sterile PBS injected and uninjected controls (Figure 3).
After microinjection, remove any dead or deformed embryos and un-injected larvae from the plate.
To determine the actual number of CFU injected per embryo, five injected embryos should be homogenized and plated directly onto TSB-Agar-Amp plates, followed by overnight growth at 37 °C prior to CFU counting.
-
Imaging of injected embryos
Transfer the injected embryos to 12-well or 6-well plates for imaging.
Anesthetize the embryos with E3 media containing 0.1 mg/ml of benzocaine and orient them in the desired angle for imaging using Olympus Cell Sens Standard software with a Olympus MVX10 fluorescence microscope (Olympus, model: MVX10 with a LH100HG and a U-MGFPA/XL GFP mirror filter attached to a DP74 camera for photography).
-
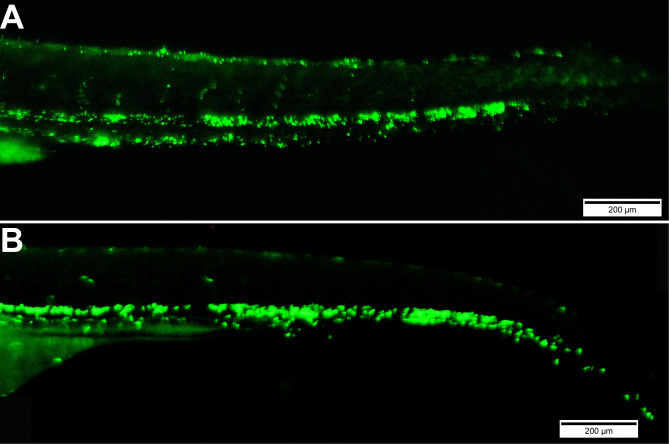
Repeat this at regular intervals, such as 0, 4, 8, 16, 24 and 48 h post infection (hpi), to monitor the rate of infection and pathogenesis of GFP positive E. coli bacteria (Figure 4).
To determine relative survival, visually inspect embryos to determine the number of dead embryos based on the absence of their heart beat.
-
Bacterial enumeration by plate counting
For bacterial enumeration, collect 10 live injected larvae at each time point (for example 0, 4, 8, 16, 24 and 48 hpi) separately in Eppendorf tubes using a transfer pipette post euthanasia with an overdose of 50 μg/ml benzocaine in E3 media.
Remove all the E3 media, rinse once in fresh E3 media to remove all the benzocaine.
Transfer 1,000 μl 1% Triton X-100 in sterile PBS solution to the tube.
Homogenize the samples in a Qiagen tissue lyser II for 5 min at a frequency of 30 s. If required, repeat homogenization for 5 min to ensure complete embryo disintegration.
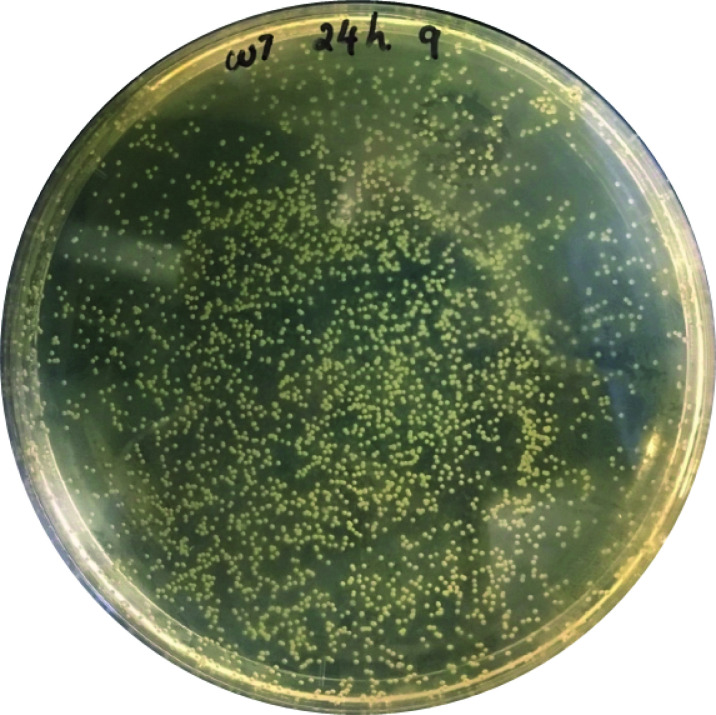
Plate 50-100 μl of the homogenate onto a TSB-agar-ampicillin plate and incubate at 37 °C for up to 48 h to determine the number of CFU per larvae (Figure 5) at each time point of infection to deduce the infection kinetics.
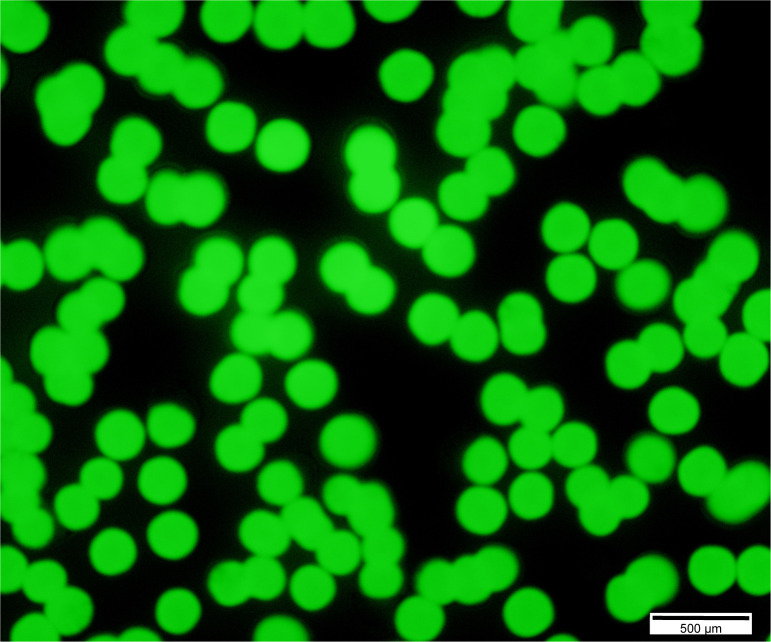
Confirm the growth of E. coli-GFP bacteria on TSB-agar-ampicillin plates by observing them under fluorescence microscopy (Figure 6).
Figure 1. Image of a Tryptic soy broth (TSB)-Agar-ampicillin (Amp) plate.
Figure 3. Images of groups of uninjected embryos (A) and those injected with E. coli-GFP bacteria (B).
Scale bars = 500 μm.
Figure 4. Images of embryos injected at 72 hpf with E. coli-GFP bacteria and visualized at 0.1 hpi (A) and 48 hpi (B).
Scale bars = 200 μm.
Figure 5. Image of a TSB-Agar-Amp plate used to quantify E. coli-GFP CFU in embryos.
Figure 6. Fluorescent image of E. coli-GFP bacterial colonies on a TSB-Agar-Amp plate.
Scale bars = 500 μm.
Data analysis
-
Bacterial colonies from the plate could be either counted manually or using ImageJ plugin “analyze particles” as follows:
Open the image of the bacterial plate with colonies.
Select the region of interest by either selecting oval or elliptical shape or use the free hand tool to draw the region of interest.
Clear the region outside of interest–Edit > Clear Outside.
Convert them to 16 bit grey scale image–Image > Type > 16-bit.
Adjust the image threshold–Image > Adjust > Threshold and click apply once the threshold is adjusted to the desired level.
If the colonies are crowded and not separated well enough, follow–Process > Binary > Watershed.
Count the colonies–Analyze > Analyze Particles, in the pop-up window select the size of the pixels (10-10,000) and circularity (0.00-1.00) depending on the size of the bacterial colonies, check the boxes “Display results”, “Clear results”, “Summarize” and “Add to manager”. This will provide the number of colonies and an overview of the colonies selected.
Analyse the bacterial load enumerated by plate counting and survival of the infected embryos using GraphPad Prism 8 software.
The bacterial load data can be analysed using non-parametric unpaired Student’s t-test and survival of the infected embryos can be displayed as a Kaplan-Meier curve, with statistical significance determined using a log-rank (Mantel-Cox) test.
Recipes
-
60x E3 media34.8 g NaCl
1.6 g KCl
5.8 g CaCl2·2H2O
9.78 g MgCl2·6H2O
To prepare a 60x stock, dissolve the ingredients in Milli-Q water, to a final volume of 2 L
Adjust the pH to 7.2 with NaOH and autoclave them (temperature: 121 °C, pressure: 400-500 KPa, duration: 1.5 h)
Store at room temperature for up to 1 year
-
E3 media
Dilute 16.5 ml of the 60x stock to 990 ml with Milli-Q water
Store at room temperature for up to 1 year
-
Methylene blue stock solution
Add 0.1 g of methylene blue powder to 100 ml of Milli-Q water and mix them
Store at room temperature for up to 1 year
-
E3 containing 0.3 mg/L of methylene blue
Add 600 μl methylene blue stock solution to 1 L E3 media
Store at room temperature for up to 1 year
-
PTU stock [0.3% (w/v)]
Add 0.3 g PTU to 100 ml E3 media
Dissolve at 65 °C for overnight
Store at room temperature for up to 1 year
-
E3 containing 0.003% (w/v) of PTU
Add 10 ml of 0.3% (w/v) PTU stock solution to 990 ml of E3 media
If crystals are present in 0.3% PTU stock, incubate solution at 65 °C in oven until dissolved
-
Benzocaine stock solution [10% (w/v)]
Add 10 g of benzocaine powder to 100 ml 100% ethanol (prepare in fume hood)
Store in the dark, wrap in foil
-
10x PBS stock
Dissolve 100 PBS tablets in 1 L Milli-Q water and autoclave (temperature: 121 °C, pressure: 400-500 KPa, duration: 1.5 h)
Store at room temperature for up to 1 year
-
PBS
Add 100 ml of 10x PBS to 900 ml Milli-Q water and autoclave (temperature: 121 °C, pressure: 400-500 KPa, duration: 1.5 h)
Store at room temperature for up to 1 year
-
1% Triton X-100 in PBS
Add 1 ml Triton X-100 to 99 ml sterile PBS
Store at 4 °C for up to 1 year
Acknowledgments
The authors recognize the support of a Deakin University International Research Scholarship (FB). The authors would like to thank the Deakin University Animal House staff for superb aquarium management that underpins the work described in this publication. This protocol was adapted from previous work (Fehr et al., 2015; Basheer et al., 2019).
Competing interests
The authors have no competing interests.
Ethics
All studies involving animals were approved by the Deakin University Animal Ethics Committee (G23/2016, 31/10/16-31/1/2020).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Akira S., Uematsu S. and Takeuchi O.(2006). Pathogen recognition and innate immunity. Cell 124(4): 783-801. [DOI] [PubMed] [Google Scholar]
- 2.Basheer F., Rasighaemi P., Liongue C. and Ward A. C.(2019). Zebrafish granulocyte colony-stimulating factor receptor maintains neutrophil number and function throughout the life span. Infect Immun 87:e00793-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett C. M., Kanki J. P., Rhodes J., Liu T. X., Paw B. H., Kieran M. W., Langenau D. M., Delahaye-Brown A., Zon L. I., Fleming M. D. and Look A. T.(2001). Myelopoiesis in the zebrafish, Danio rerio. Blood 98(3): 643-651. [DOI] [PubMed] [Google Scholar]
- 4.Deng Q., Sarris M., Bennin D. A., Green J. M., Herbomel P. and Huttenlocher A.(2013). Localized bacterial infection induces systemic activation of neutrophils through Cxcr2 signaling in zebrafish. J Leukoc Biol 93(5): 761-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellett F., Pase L., Hayman J. W., Andrianopoulos A. and Lieschke G. J.(2011). mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117(4): e49-e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fehr A., Eshwar A. K., Neuhauss S. C., Ruetten M., Lehner A. and Vaughan L.(2015). Evaluation of zebrafish as a model to study the pathogenesis of the opportunistic pathogen Cronobacter turicensis. Emerg Microbes Infect 4(5): e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gratacap R. L. and Wheeler R. T.(2014). Utilization of zebrafish for intravital study of eukaryotic pathogen-host interactions. Dev Comp Immunol 46(1): 108-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray C., Loynes C. A., Whyte M. K., Crossman D. C., Renshaw S. A. and Chico T. J.(2011). Simultaneous intravital imaging of macrophage and neutrophil behaviour during inflammation using a novel transgenic zebrafish. Thromb Haemost 105(05): 811-819. [DOI] [PubMed] [Google Scholar]
- 9.Hall C. J., Flores M. V., Oehlers S. H., Sanderson L. E., Lam E. Y., Crosier K. E. and Crosier P. S.(2012). Infection-responsive expansion of the hematopoietic stem and progenitor cell compartment in zebrafish is dependent upon inducible nitric oxide. Cell Stem Cell 10(2): 198-209. [DOI] [PubMed] [Google Scholar]
- 10.Herbomel P., Thisse B. and Thisse C.(1999). Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126(17): 3735-3745. [DOI] [PubMed] [Google Scholar]
- 11.Liongue C., Wright C., Russell A. P. and Ward A. C.(2009). Granulocyte colony-stimulating factor receptor: stimulating granulopoiesis and much more. Int J Biochem Cell Biol 41(12): 2372-2375. [DOI] [PubMed] [Google Scholar]
- 12.Manz M. G. and Boettcher S.(2014). Emergency granulopoiesis. Nat Rev Immunol 14(5): 302-314. [DOI] [PubMed] [Google Scholar]
- 13.Masud S., Torraca V. and Meijer A. H.(2017). Modeling infectious diseases in the context of a developing immune system. Curr Top Dev Biol 124: 277-329. [DOI] [PubMed] [Google Scholar]
- 14.Meeker N. D. and Trede N. S.(2008). Immunology and zebrafish: spawning new models of human disease. Dev Comp Immunol 32(7): 745-757. [DOI] [PubMed] [Google Scholar]
- 15.Panopoulos A. D. and Watowich S. S.(2008). Granulocyte colony-stimulating factor: molecular mechanisms of action during steady state and‘emergency’ hematopoiesis. Cytokine 42(3): 277-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renshaw S. A. and Trede N. S.(2012). A model 450 million years in the making: zebrafish and vertebrate immunity. Dis Model Mech 5(1): 38-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varela M., Figueras A. and Novoa B.(2017). Modelling viral infections using zebrafish: innate immune response and antiviral research. Antiviral Res 139: 59-68. [DOI] [PubMed] [Google Scholar]
- 18.Weiss G. and Schaible U. E.(2015). Macrophage defense mechanisms against intracellular bacteria. Immunol Rev 264(1): 182-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willett C. E., Cortes A., Zuasti A. and Zapata A. G.(1999). Early hematopoiesis and developing lymphoid organs in the zebrafish. Dev Dyn 214(4): 323-336. [DOI] [PubMed] [Google Scholar]