Abstract
Exploring the structure and function of protein complexes requires their isolation in the native state–a task that is made challenging when studying labile and/or low abundant complexes. The difficulties in preparing membrane-protein complexes are especially notorious. The cyanobacterium Synechocystis sp. PCC 6803 is a widely used model organism for the physiology of oxygenic phototrophs, and the biogenesis of membrane-bound photosynthetic complexes has traditionally been studied using this cyanobacterium. In a typical approach, the protein complexes are purified with a combination of His-affinity chromatography and a size-based fractionation method such as gradient ultracentrifugation and/or native electrophoresis. However, His-affinity purification harbors prominent contaminants and the levels of many proteins are too low for a feasible multi-step purification. Here, we have developed a purification method for the isolation of 3x FLAG-tagged proteins from the membrane and soluble fractions of Synechocystis. Soluble proteins or solubilized thylakoids are subjected to a single affinity purification step that utilizes the highly specific binding of FLAG-affinity resin. After an intensive wash, the captured proteins are released from the resin under native conditions using an excess of synthetic 3x FLAG peptide. The protocol allows fast isolation of low abundant protein complexes with a superb purity.
Keywords: Protein purification, Membrane protein complexes, Synechocystis 6803 , Photosystems, FLAG-tag, Affinity chromatography
Background
Cyanobacteria have been used as the preferred model systems to study the biogenesis and function of photosynthetic protein complexes for decades. The photosynthetic apparatus in cyanobacteria is very similar to eukaryotic systems (algae and plants), but cyanobacteria have all the advantages of prokaryotic models, such as fast growth and small genomes that allow easy genetic manipulation and the use of standard tools of bacterial genetics. Particularly, Synechocystis sp. PCC 6803 (hereafter Synechocystis) is established as a favorite model organism in this field, mainly because it is transformable with high efficiency and integrates DNA by functional homologous recombination. Moreover, a glucose tolerant substrain of Synechocystis, which can proliferate using carbohydrate supplement, enables studying the function of photosynthetic genes by reverse genetics (Williams, 1988).
The biogenesis of photosystem II (PSII), a large membrane embedded pigment-protein complex responsible for photosynthetic water oxidation, has been studied intensively in Synechocystis ( Nixon et al., 2010 ). The assembly of the PSII complex from individual subunits requires complicated machinery of many auxiliary (assembly) factors, which are typically involved in a certain assembly step ( Komenda et al., 2012 ). A conventional approach for the analysis of PSII assembly complexes is using Synechocystis mutants lacking one or several PSII subunits. PSII biogenesis is then blocked at a particular step where the missing subunit would bind to an intermediate complex of the assembly process. In such mutants, the otherwise transient PSII pre-complexes can accumulate to a level that allows their detection and even purification, which has traditionally been achieved via His-tagged PSII (core) subunits ( Dobáková et al., 2007 and 2009; Boehm et al., 2011 and 2012).
His-tag has several advantages: it is small enough to be easily introduced via a simple primer design, and it is relatively inexpensive to use. However, after His-affinity purification the preparations from Synechocystis often contain prominent contaminating proteins ( Boehm et al., 2011 and 2012; Liu et al., 2011 ); therefore, additional purification steps are usually required to obtain the satisfactory quality for functional and structural studies. Additional purification steps may, however, compromise the yield and nativity of the complex of interest. Moreover, the levels of many protein complexes in the cell are simply too low for feasible multi-step purification protocols, PSII assembly intermediates are such an example. For these reasons, a fast, gentle, single-step purification method is highly desirable.
Affinity tags, based on biomolecular interactions offer superior specificity as compared to the general metal affinity of His-tag ( Lichty et al., 2005 ). Large, proteinaceous tags such as small ubiquitin-like modifier (SUMO) or maltose binding protein (MBP) are well suited for the isolation of small soluble proteins due to their stable nature, which can even promote the solubility of the protein (Esposito and Chatterjee, 2006). However, for studying larger and potentially labile membrane-protein complexes, a less bulky tag with minimal interference on protein-protein interactions is preferable. Short peptide tags with high-affinity to proteins or antibodies seem to be the best suited for this purpose. One such tag is the short octapeptide called FLAG (DYKDDDDK; Hopp et al., 1988 ). Compared to His-, and several other tags, using FLAG-tag results in superior purity with little compromise in yield ( Lichty et al., 2005 ). Moreover, because the elution of FLAG-tagged proteins is possible using a synthetic FLAG-peptide, the usage of disrupting chemicals, high ion concentrations or extreme pH can be avoided. Even though the price per mg purified protein of FLAG-affinity resin is approximately 50 times higher than that of Ni-NTA resin ( Lichty et al., 2005 ); for the purification of labile and/or low-abundant complexes it can well worth the time and costs saved compared to optimizing multi-step purification procedures. Recently, FLAG-tag based approaches have been employed to purify pathogenic variants of the human Huntingtin protein from mammalian and insect cell lines ( Harding et al., 2009 ), as well as the integral membrane Vo-part of the Pichia pastoris V-type ATPase ( Li et al., 2017 ).
The FLAG-affinity purification protocol presented here is designed for the purification of proteins tagged with 3x FLAG epitope (DYKDDDDKDYKDDDDKDYKDDDDK). The protocol has been successfully used to resolve the roles of the PSII assembly factors Ycf39 ( Knoppová et al., 2014 ), CyanoP ( Knoppová et al., 2016 ), Pam68 ( Bučinská et al., 2018 ) and RubA ( Kiss et al., 2019 ); as well as pigment-protein complexes containing the chlorophyll-synthase ( Chidgey et al., 2014 ), protoporphyrinogen oxidase ( Skotnicová et al., 2018 ) and ferrochelatase enzymes ( Pazderník et al., 2019 ). Although the method was originally developed for membrane-bound protein complexes, with minor modifications it can also be applied to soluble proteins.
Materials and Reagents
Disposable semi-micro cuvettes (BRAND, catalog number: 759015)
50 ml conical tubes (JET biofil, catalog number: CFT011500)
Round paint brush, ø 6 mm
-
Pipette tips:
10 µl (Neptune, catalog number: 2340)
200 µl (Eppendorf, catalog number: 0030000870)
1,000 µl (Eppendorf, catalog number: 0030000927)
1.5 ml microtubes (Deltalabs, catalog number: 4092.3N)
7 ml screw cap vials for cell lysis (BioSpec Products, catalog number: 3205)
Plastic chromatography column (Bio-Rad, catalog number: 7311550)
Synechocystis cells expressing a 3x FLAG-tagged protein
BG-11 liquid medium and agar plates ( Rippka et al., 1979 )
100-200 µm glass microbeads (PRECIOSA ORNELA, crystal microbeads B 134)
cOmplete protease inhibitor cocktail tablets (Sigma-Aldrich, catalog number: 11836145001)
(Optional) Microconcentrators with suitable cut-off filter (e.g., Amicon Ultra-0.5, Sigma-Aldrich)
Liquid nitrogen
Crushed ice
Styrofoam box or equivalent for keeping ice
Reverse osmosis (RO) water
Deionized water (Merck, catalog number: 1167545000)
Methyl alcohol (PENTA, catalog number: 21240-11000)
n-dodecyl-β-D-maltoside (DDM; BioChemica, catalog number: A0819,0005)
ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich, catalog number: A2220)
Proteus Clarification Mini Spin Column (Generon, catalog number: GEN-MSF500)
-
3x FLAG peptide, 20 mg, > 85% purity (custom synthesis, Genscript, USA)
Notes:
An alternative is to purchase 3x FLAG peptide (Sigma-Aldrich, catalog number: F4799-4MG), however, the custom synthesis is roughly 6 times cheaper.
Prepare a 3 mg/ml solution in deionized water and store at -20 °C as 100-200 µl aliquots. Thawed peptide can be stored at 4 °C until use.
2-Morpholinoethanesulfonic acid (MES; AppliChem, catalog number: A0689,0250)
MgCl2·6H2O (PENTA, catalog number: 16330-31000)
Glycerol (PENTA, catalog number: 14550-11000)
CaCl2·2H2O (PENTA, catalog number: 16790-31000)
α-FLAG antibody (Merck, catalog number: F7425)
MES buffer (1 L) (see Recipes)
0.5 M MES-NaOH, pH 6.5 (250 ml) (see Recipes)
1 M MgCl2 (100 ml) (see Recipes)
1 M CaCl2 (100 ml) (see Recipes)
Equipment
Temperature controlled growth facility equipped with an orbital shaker and a light source
Laminar flow hood
Metallic inoculation loop
Gas burner
500 ml Erlenmeyer flasks
Round culture flask (10 L)
CimarecTM i Maxi magnetic stirrer (Thermo Fisher Scientific, catalog number: 50088143) and a magnetic stir bar
Spectrophotometer (WPA S1200 Spectrawave)
-
Centrifuges and rotors:
Centrifuge (Sigma Laborzentrifugen GmbH, model: Sigma 8KS, rotor: 12505-H)
Centrifuge (Sigma Laborzentrifugen GmbH, model: Sigma 3K30, rotors: 12155-H [+15 ml conical tube adapters, 13081] and 12158-H)
Centrifuge (Eppendorf, model: 5418)
Centrifuge (Eppendorf, model: 5415 R)
-
Centrifuge bottles:
500 ml (Nalgene, catalog number: 3141-0500)
80 ml (Sigma Laborzentrifugen GmbH, catalog number: 15080)
27 ml (Sigma Laborzentrifugen GmbH, catalog number: 15032)
Deep freezer
Regular freezer
Mini-Beadbeater-16 (BioSpec Products, catalog number: 607) equipped with a 7 ml vial adapter (BioSpec Products, catalog number: 607TC8)
Vortex V-1 Plus (Biosan, catalog number: BS-010203-AAG)
1 ml Hamilton syringe (Hamilton, catalog number: 81330)
-
Automatic pipettes:
0.5-10 µl (Eppendorf, catalog number: 3120000020)
10-100 µl (Eppendorf, catalog number: 3120000046)
100-1,000 µl (Eppendorf, catalog number: 3120000062)
Multi-Bio RS-24 rotator (Biosan, catalog number: BS-010117-AAG)
Cold hood or cold room, 10 °C
A retort stand and a ring clamp
25 ml glass beakers
Procedure
-
Cell culture and harvesting
-
Working in a laminar flow hood, inoculate Synechocystis cells using a metallic inoculation loop into five, 500 ml Erlenmeyer flasks, each containing 200 ml BG-11 medium. Grow the cells at 28 °C and 40 µmol/m2/s photosynthetic photon flux density on a rotary shaker set to 150 rpm until the culture reaches mid/end of logarithmic growth phase.
Note: When the optical density of the culture at 750 nm reaches 0.6-0.8, which corresponds to 9-12 x 107 Synechocystis cells/ml.
Collect your cultures (OD750 ~0.8) into a 10 L flask and dilute it to 4 L final volume to gain OD750 ~0.2 density. Grow the culture until mid/end of logarithmic growth phase with magnetic stirrer (240 rpm) and air bubbling in the same conditions as above.
Divide the cell culture into 500 ml centrifuge bottles and pellet the cells by centrifuging at 10,000 × g in a cooling centrifuge (Sigma 8KS, rotor 12505-H) at 4 °C for 20 min.
Suspend the cell pellets into a total of 30 ml of MES buffer.
Notes:
All media and equipment used for the preparation of the cell culture should be sterilized. Use the gas burner to sterilize the metal inoculation loop, and autoclave all glassware and growth media prior to use.
Cyanobacterial cultures have negligible absorbance at 750 nm; therefore, the OD at this wavelength is primarily dependent on light scattering (turbidity). Different spectrophotometers differ greatly in their various optical properties, and hence the light scattering value by OD750 may vary. Therefore, it is important to establish the OD at which your culture is at the mid/end of logarithmic growth phase by measuring a growth curve a priori. Under our standard conditions, the doubling time of Synechocystis is approximately 12 h; hence, a culture with OD750 = 0.2 will reach desirable OD750 in about a day.
In our experience, larger volumes of cyanobacterial cultures (i.e., 4 L) tend to grow slower, probably due to limitations in gas exchange. Therefore, an intensive bubbling of the culture by air is desirable.
At Step A4 you may freeze the cells in liquid nitrogen and store them at -80 °C until use.
-
-
Cell lysis and thylakoid isolation
Pellet the cells in 80 ml centrifuge tubes at 10,000 × g (Sigma 3K30, rotor 12155-H) at 4 °C for 10 min.
Discard the supernatant and resuspend the cells into 5 ml of MES buffer using a wet paint brush. Add 30 ml MES buffer to the centrifuge tube and pellet the cells as above.
Resuspend the cells into 12 ml of MES buffer and add protease inhibitor: dissolve 1 tablet into 1 ml of deionized water to obtain a 50x stock solution and pipet accordingly to obtain 1x working concentration. You may store the rest of the inhibitor at -20 °C for later use.
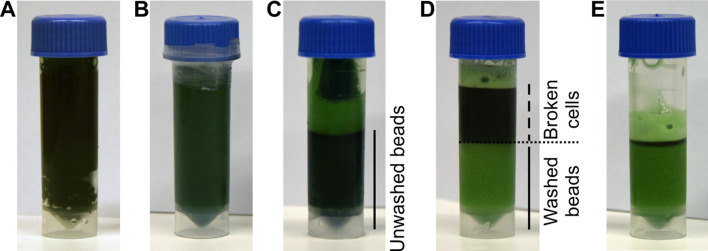
Add 3 ml of glass beads and 3 ml of cell suspension into four 7 ml screw cap vials (Figure 1A), close the lids and seal them with parafilm (Figure 1B). There should be approximately 1 cm of free space in the tube, you may add some buffer to reach the desired volume.
Break the cells for 60 s by Mini-Beadbeater-16. Allow the tubes to cool on ice protected from light for 5 min and repeat the breaking cycle 5 times.
After lysis, spin down the beads at 500 × g for 10 s at 4 °C, (Sigma 3K30, rotor 12155-H equipped with adapters for 15 ml conical tubes). Collect the supernatants into a 27 ml centrifuge tube. Wash the beads (Figure 1C) with one bed volume of MES buffer three-to-four times, until the supernatant and the beads appear light green (Figures 1D-1E).
Pool all supernatants from Step B6 and pellet the thylakoids by centrifuging at 35,000 × g (Sigma 3K30, rotor 12158-H) at 4 °C for 20 min.
At this point, if you are only interested in the isolation of membrane proteins, you may discard the supernatant. Otherwise, collect the supernatant into a new centrifuge tube (marked: SP, ‘soluble proteins’), and resuspend the pellet into 10 ml of MES buffer using a wet paint brush (mark the tube: TM, ‘thylakoid membranes’). Centrifuge the tubes as above.
Collect the supernatant from the ‘SP’-tube into a 50 ml conical tube and store it on ice. Discard the supernatants from the ‘TM’-tube and resuspend the pellets from both tubes (‘SP’ and ‘TM’) into 1/3 of soluble fraction volume of MES buffer. If you are continuing only with the membrane fraction use a total of 10 ml MES buffer and combine the pellets into a 50 ml conical tube.
Freeze a 300 µl aliquot of soluble proteins and a 100 µl aliquot of thylakoids in liquid nitrogen and store at -80 °C for further analysis by gel electrophoresis.
Notes:
After pelleting the cells, the supernatant may appear slightly bluish due to partial breaking of cells after freezing and thawing.
From Step B5 onwards it is important to keep the sample cooled (4-10 °C) and protect it from light to prevent the excitation of chlorophyll molecules and the consequent formation of reactive oxygen species. We recommend working under dim green light.
Thylakoids are easier to resuspend first into few ml volume, before adding the rest of the buffer.
At Step B10 you may also freeze the rest of the soluble protein and thylakoid samples in liquid nitrogen and store them at -80 °C until subsequent solubilization and purification.
-
Thylakoid solubilization
Adjust the chlorophyll concentration of the thylakoids to 0.5 µg/µl with MES buffer.
Add 1% n-dodecyl β-D-maltoside (DDM) to the thylakoid suspension and incubate the tube for 1 h at 10 °C in the dark (for example, you may cover the tube with aluminum foil) on a rotator mixer set to 10 rpm.
Transfer the suspension into a 27 ml centrifuge tube and pellet insoluble material at 46,000 × g (Sigma 3K30, rotor 12158-H) at 4 °C for 30 min.
Collect the supernatant into a 50 ml conical tube and discard the pellet.
Freeze 100 µl of the solubilized thylakoids in liquid nitrogen and store at -80 °C for further analysis, particularly for the identification of a potential problem with protein solubility.
-
FLAG-affinity purification
Wash the purification column with 1 ml of RO-water followed by 1 ml of deionized water.
Close the column, then add 1 ml of MES buffer and 600 µl of 1:1 Anti-FLAG-M2 agarose resin using a cut pipette tip (Video 1).
Open the column and allow the buffer to flow through. Wash the resin with an additional 1 ml of MES buffer (pipet the buffer gently on top of the resin and allow it to flow through; Figure 2A; Video 2).
Load the protein suspension into the column and collect the flowthrough into a glass beaker (Figure 2B; Video 3).
Wash the column with 1 ml of MES buffer. Discard the flowthrough.
Load the flowthrough from Step D4 to the column for the second time and collect the flowthrough. Repeat the binding once more.
Take a 0.5 ml sample from the final flowthrough, freeze it in liquid nitrogen and store at -80 °C for further analysis [Figure 3, flowthrough (FT)].
Wash the column with 1 ml of MES buffer [Figure 2C; Figure 3, first wash (W1)].
Close the column and add 1 ml of MES buffer: pipet the buffer with high enough speed to mix it thoroughly with the resin (Figure 2D; Video 4) (you may mix the buffer with the resin gently by pipetting up and down with a cut pipette tip, but some resin may stick to the tip and be lost). Open the column and allow the buffer to drain out [Figure 3, second wash (W2)].
Wash the column with 1 ml of MES buffer without mixing the resin. Repeat this washing step 3 times. In total, the resin is washed by 20 column volumes of MES buffer (Figure 2E).
Resuspend the resin into one bed volume of MES buffer and transfer it into a Proteus Clarification Mini Spin Column.
Add 3x FLAG-peptide into a final concentration of 300 µg/ml. Seal the top of the tube with parafilm and cover the whole tube with aluminum foil. Mix in a rotator mixer set to 10 rpm for 30 min at 10 °C.
Spin the column at 600 × g (Eppendorf 5415 R) at 4 °C for 3 min. Collect elution 1 into a 1.5 ml microtube and store on ice.
Repeat the incubation with FLAG-peptide and centrifuge as above to yield elution 2.
The obtained elutions can be pooled and the total elution volume (typically ~800 µl) can be immediately concentrated using microconcetrators (e.g., Amicon Ultra-0.5, Sigma-Aldrich) with a desired cut-off to achieve higher concentration for downstream analysis (protein gel electrophoresis, immunoblotting [see Figure 3, eluate (E)], protein mass spectrometry, single-particle analysis, size-exclusion chromatography, etc.).
Figure 1. Breaking of Synechocystis cells.
A. Screwcap tubes are filled with 3 ml of glass beads and 3 ml of cyanobacterial suspension; B. The tubes are sealed with parafilm and cells lysed using a bead beater. C. After breaking cells, the beads are spun to the bottom and the supernatant is collected. D. The beads are washed with MES buffer; E. After each washing step, the supernatant is collected.
Video 1. Column preparing.

Figure 2. Isolation of 3x FLAG-tagged proteins from Synechocystis using affinity chromatography.
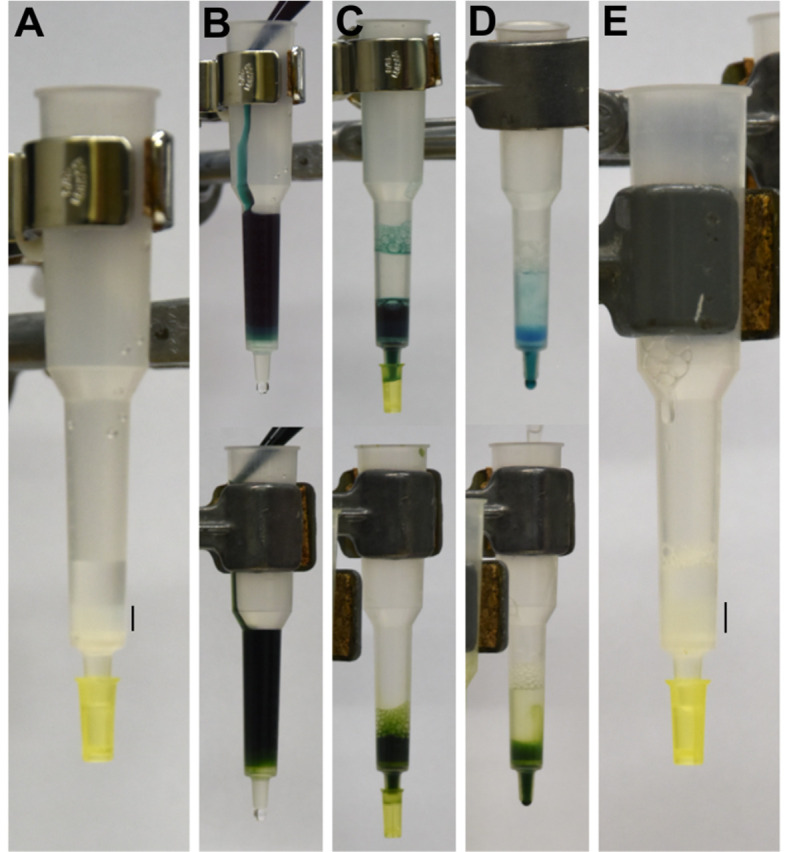
Individual purification steps for soluble proteins and detergent-solubilized membrane proteins are shown in upper panels and lower panels, respectively. A. Anti-FLAG resin is packed into a plastic chromatography column. B. Soluble protein fraction (upper panel) or detergent solubilized thylakoids (lower panel) are loaded to the column. C. Sample is allowed to flow through the column and the flowthrough is collected. Loading can be repeated several times. D. First wash (W1) is performed by gently pipetting buffer on top of the resin. E. Washing is continued as in panel D until the column is completely washed from excess pigment-proteins. FLAG resin is indicated with a black bar in panels A and E.
Video 2. Gentle washing of column.

Video 3. Sample loading.
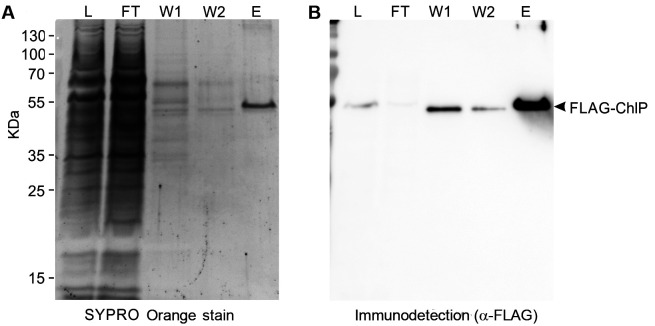
Figure 3. Immunoblot analysis following the purification of geranylgeranyl reductase enzyme tagged with 3x FLAG-tag (FLAG-ChlP).
The purification of FLAG-ChlP from Synechocystis was performed according to this protocol. Protein suspension that was loaded in Step D4 (L; solubilized thylakoid membranes of 0.5 µg chlorophyll), the first flowthrough in Step D4 (FT; equal volume to L), the first wash in Step D5 (W1; 1/25 volume of the wash fraction), the second wash in step 6 (W2; 1/25 volume of the wash fraction), and the eluate gained in Step D15 (E; 1/250 volume of total, non-concentrated eluate) were analyzed by SDS gel electrophoresis (A) and by a subsequent immunoblot (B). Prior to Western blotting on a PVDF membrane the gel was stained with SYPRO Orange (Sigma) to detect total proteins (A). Proteins blotted on the membrane were probed with the α-FLAG antibody (Merck) (A); the protein band, representing the isolated FLAG-ChlP, is indicated.
Video 4. Vigorous washing of the resin.
Notes:
When working with solubilized thylakoids, use buffers supplemented with 0.04% DDM at all steps. Use buffers without detergent when working with soluble proteins.
FLAG-resin should never be allowed to dry out during the purification.
Thaw the FLAG resin on ice and mix thoroughly before use.
The FLAG resin is stored in buffer as a 50% solution, which means that pipetting 600 µl of the well mixed solution will result in 300 µl resin bed volume.
For low abundant proteins, the number of binding steps (Step D6) can be increased.
The stringency of the purification can be altered by increasing the number of washing steps.
Notes
The MES buffer system pH 6.5 containing Mg2+ and Ca2+ ions used in this procedure has been optimized to maximize the stability of PSII complexes ( Dobáková et al., 2009 ). A different buffer composition may be required for the stability of different protein complexes.
The protein concentration in soluble fraction can be assessed e.g., by BCA assay or other standard methods. However, for assessing the concentration of the membrane fraction, it is more convenient to relate to its chlorophyll content [the protein concentration (µg/µl) in Synechocystis thylakoids is roughly ten times higher than that of chlorophyll].
Measurement of chlorophyll concentration: Pipet 10 µl of thylakoids into a 1.5 ml microtube and add 990 µl methanol using a Hamilton syringe. Vortex and incubate the tube in the dark at room temperature for 5 min. Pellet cell debris by centrifuging at 16,800 × g (Eppendorf 5418) at room temperature for 5 min. Measure the chlorophyll absorbance at 665.2 nm and sample turbidity at 750 nm, and calculate the chlorophyll a concentration (µg/µl) of the thylakoids: ((A665.2-A750)/α) x 100 x 1 cm-1 (α = 79.95 L g-1 cm-1; Porra et al., 1989 ).
Always perform a control purification with a wild-type strain to exclude contaminants from your analysis of results. For example, a small amount of Synechocystis trimeric photosystem I and tryptophanyl-tRNA synthetase are typical contaminants of our FLAG-tag purifications ( Knoppová et al., 2014 ; Bučinská et al., 2018 ; Skotnicová et al., 2018 ).
In case the purification fails or the protein yield is very low, make sure that your protein of interest is expressed and not degraded in vivo or during the preparation of cellular fractions (perform SDS-PAGE and immunodetection using commercially available anti-FLAG antibody (Merck). A representative SDS gel electrophoresis and subsequent immunoblot analysis of purification (Steps D7-D9) and elution (Step D15) are presented in Figure 3. In the case of membrane proteins, compare the unsolubilized and solubilized fractions to check protein solubility (a detergent with different properties than DDM may be required for the solubility or stability of the desired protein complex). In addition, compare your original SP and/or solubilized TM samples with the obtained flowthrough in Step D7 (Figure 3). The relative amount of your protein in flowthrough should significantly decrease when compared with the loaded material; otherwise it signifies a problem with binding to the resin. For the purification of proteins requiring metal cofactors an EDTA-free protease inhibitor should be used.
Recipes
-
MES buffer (1 L)
25 mM MES-NaOH, pH 6.5 (50 ml of 0.5 M stock)
10 mM MgCl2 (10 ml of 1 M stock)
10 mM CaCl2 (10 ml of 1 M stock)
25% glycerol (250 ml)
Adjust the volume to 1 L with deionized water in a measuring cylinder
After membrane protein solubilization, use buffer supplemented with 0.04% DDM for all purification steps
-
0.5 M MES-NaOH, pH 6.5 (250 ml)
24.41 g MES (2-Morpholinoethanesulfonic acid) (AppliChem)
Dissolve into ~200 ml of deionized water and set pH to 6.5 with 10 M NaOH
Adjust the volume to 250 ml in a measuring flask
-
1 M MgCl2 (100 ml)
20.33 g MgCl2·6H2O
Dissolve to deionized water and adjust the volume to 100 ml in a measuring flask
-
1 M CaCl2 (100 ml)
14.70 g CaCl2·2H2O
Dissolve to deionized water and adjust the volume to 100 ml in a measuring flask
Acknowledgments
This work was funded by the Czech Science Foundation (17‐08755S) and by the Czech Ministry of Education (project LO1416).
Competing interests
The authors declare no conflicts of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Boehm M., Romero E., Reisinger V., Yu J., Komenda J., Eichacker L. A., Dekker J. P. and Nixon P. J.(2011). Investigating the early stages of photosystem II assembly in Synechocystis sp. PCC 6803: isolation of CP47 and CP43 complexes . J Biol Chem 286(17): 14812-14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boehm M., Yu J., Reisinger V., Bečková M., Eichacker L. A., Schlodder E., Komenda J. and Nixon P. J.(2012). Subunit composition of CP43-less photosystem II complexes of Synechocystis sp. PCC 6803: implications for the assembly and repair of photosystem II . Philos Trans R Soc Lond B Biol Sci 367(1608): 3444-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bučinská L., Kiss É., Koník P., Knoppová J., Komenda J. and Sobotka R.(2018). The ribosome-bound protein Pam68 promotes insertion of chlorophyll into the CP47 subunit of photosystem II. Plant Physiol 176(4): 2931-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chidgey J. W., Linhartová M., Komenda J., Jackson P. J., Dickman M. J., Canniffe D. P., Konik P., Pilny J., Hunter C. N. and Sobotka R.(2014). A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell 26(3): 1267-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dobáková M., Sobotka R., Tichý M. and Komenda J.(2009). Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47(PsbB) in the cyanobacterium Synechocystis sp. PCC 6803 . Plant Physiol 149(2): 1076-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dobáková M., Tichý M. and Komenda J.(2007). Role of the PsbI protein in photosystem II assembly and repair in the cyanobacterium Synechocystis sp. PCC 6803 . Plant Physiol 145(4): 1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Esposito D. and Chatterjee D. K.(2006). Enhancement of soluble protein expression through the use of fusion tags. Curr Opin Biotechnol 17(4): 353-358. [DOI] [PubMed] [Google Scholar]
- 8. Harding R. J., Loppnau P., Ackloo S., Lemak A., Hutchinson A., Hunt B., Holehouse A. S., Ho J. C., Fan L., Toledo-Sherman L., Seitova A. and Arrowsmith C. H.(2019). Design and characterization of mutant and wildtype huntingtin proteins produced from a toolkit of scalable eukaryotic expression systems. J Biol Chem 294(17): 6986-7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hopp T. P., Prickett K. S., Price V. L., Libby R. T., March C. J., Cerretti D. P., Urdal D. L. and Conlon P. J.(1988). A short polypeptide marker sequence useful for recombinant protein identification and purification. Nature Biotechnology 6: 1204-1210. [Google Scholar]
- 10. Kiss É., Knoppová J., Aznar G. P., Pilný J., Yu J., Halada P., Nixon P. J., Sobotka R. and Komenda J.(2019). A Photosynthesis-Specific Rubredoxin-Like Protein Is Required for Efficient Association of the D1 and D2 Proteins during the Initial Steps of Photosystem II Assembly. Plant Cell 31(9): 2241-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Knoppová J., Sobotka R., Tichý M., Yu J., Konik P., Halada P., Nixon P. J. and Komenda J.(2014). Discovery of a chlorophyll binding protein complex involved in the early steps of photosystem II assembly in Synechocystis . Plant Cell 26(3): 1200-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Knoppová J., Yu J., Konik P., Nixon P. J. and Komenda J.(2016). CyanoP is involved in the early steps of Photosystem II assembly in the cyanobacterium Synechocystis sp. PCC 6803 . Plant Cell Physiol 57(9): 1921-1931. [DOI] [PubMed] [Google Scholar]
- 13. Komenda J., Sobotka R. and Nixon P. J.(2012). Assembling and maintaining the Photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol 15(3): 245-251. [DOI] [PubMed] [Google Scholar]
- 14. Li S., Hong T., Wang K., Lu Y. and Zhou M.(2017). Dissociation and purification of the endogenous membrane-bound Vo complex from Pichia pastoris. Protein Expr Purif 138: 76-80. [DOI] [PubMed] [Google Scholar]
- 15. Lichty J. J., Malecki J. L., Agnew H. D., Michelson-Horowitz D. J. and Tan S.(2005). Comparison of affinity tags for protein purification. Protein Expr Purif 41(1): 98-105. [DOI] [PubMed] [Google Scholar]
- 16. Liu H., Roose J. L., Cameron J. C. and Pakrasi H. B.(2011). A genetically tagged Psb27 protein allows purification of two consecutive photosystem II(PSII) assembly intermediates in Synechocystis 6803, a cyanobacterium . J Biol Chem 286(28): 24865-24871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nixon P. J., Michoux F., Yu J., Boehm M. and Komenda J.(2010). Recent advances in understanding the assembly and repair of photosystem II. Ann Bot 106(1): 1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pazderník M., Mareš J., Pilný J. and Sobotka R.(2019). The antenna-like domain of the cyanobacterial ferrochelatase can bind chlorophyll and carotenoids in an energy-dissipative configuration. J Biol Chem 294(29): 11131-11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Porra R. J., Thompson W. A. and Kriedemann P. E.(1989). Determination og accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy . Biochimica et Biophysica Acta 975: 384-394. [Google Scholar]
- 20. Rippka R., Deruelles J., Waterbury J. B., Herdman M. and Stanier R. Y.(1979). Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. Microbiology 111(1): 1-61. [Google Scholar]
- 21. Skotnicová P., Sobotka R., Shepherd M., Hajek J., Hrouzek P. and Tichy M.(2018). The cyanobacterial protoporphyrinogen oxidase HemJ is a new b-type heme protein functionally coupled with coproporphyrinogen III oxidase. J Biol Chem 293(32): 12394-12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Williams J. G. K.(1988). Construction of specific mutations in photosystem II photosynthetic reaction center by genetic-engineering methods in Synechocystis 6803 . Methods in Enzymology 167: 766-778. [Google Scholar]