Abstract
The plant cell wall is a complex network of polysaccharides and proteins that provides strength and structural integrity to plant cells, as well as playing a vital role in growth, development, and defense response. Cell wall polysaccharides can be broadly grouped into three categories: cellulose, pectins, and hemicelluloses. Dynamic interactions between polysaccharides and cell wall-associated proteins contribute to regions of flexibility and rigidity within the cell wall, allowing for remodeling when necessary during growth, environmental adaptation, or stress response activation. These polysaccharide interactions are vital to plant growth, however they also contribute to the level of difficulty encountered when attempting to analyze cell wall structure and composition. In the past, lengthy protocols to quantify cell wall monosaccharides contributing to cellulose as well as neutral and acidic cell wall polysaccharides have been used. Recently, a streamlined approach for monosaccharide quantification was described. This protocol combines a simplified hydrolysis method followed by several runs of high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD). Here, we present an updated version of this protocol in which we can analyze all nine cell wall monosaccharides in a single high-performance liquid chromatography HPAEC-PAD gradient profile. The inclusion of an enzymatic starch degradation, as well as alternate internal standards for added quantification accuracy, and a ready-to-use Python script facilitating data analysis adds a broadened scope of utility to this protocol. This protocol was used to analyze Arabidopsis light-grown seedlings and dark-grown hypocotyls, but is suitable for any plant tissues.
Keywords: Cell wall, Cellulose, HPLC, Monosaccharides, Saeman hydrolysis, Arabidopsis
Background
Understanding plant cell wall structure and composition have been at the forefront of both academic and industrial plant research for many years. Polysaccharide and cell wall protein interactions provide structure to plant cells, while also actively playing important biological roles during growth and adaptation to diverse external conditions (Cosgrove, 2005 and 2016; Kesten et al., 2017). The plant cell wall acts as an important physical barrier during biotic or abiotic interactions, but can also provide an endogenous source of signaling molecules released during stress response (Cosgrove, 2005 and 2016; Kesten et al., 2017 and 2019). These cell wall-derived molecules can activate signaling cascades to alert the plant host of an invading pathogen or a need to redirect growth resources. Cell wall polysaccharides are also important for industrial uses–pectins are widely used in the food and cosmetics industries, while cellulose is important for the food, paper, and textile industries (Pettolino et al., 2012). As such, efficient and informative methods for characterizing plant cell wall polysaccharides are vital in both academia and industry to further understand the biological role of the plant cell wall and improve ease of polymer extraction while reducing waste during industrial applications.
Experiments from the 1940s conducted in wood established the use of dilute acid hydrolysis at high temperature to study sugar decomposition into monomers (Saeman, 1945). Such studies have provided a foundation for many current protocols, including the one presented here. Streamlining this analysis has allowed us to improve both the accuracy and efficiency of our measurements. Other recently described protocols have established the combination of the widely-used trifluoroacetic acid (TFA) hydrolysis approach with a single HPLC run, or a dual sulfuric acid-based h ydrolysis method, referred to as a “one-step two-step sulfuric acid hydrolysis” approach, with multiple HPAEC-PAD runs (Zhang et al., 2012; Voiniciuc et al., 2016; Yeats et al., 2016a and 2016b). This “one-step two-step” approach makes use of a Saeman hydrolysis procedure followed by what has been previously referred to as a “matrix hydrolysis” (Yeats et al., 2016a). Saeman hydrolysis, the “two-step” portion of this approach, is essentially the addition of concentrated sulfuric acid hydrolysis (72% sulfuric acid) directly to plant material in order to swell the sample. After one hour at room temperature, water is added to the sample to dilute the sulfuric acid concentration to 4%. This portion of the hydrolysis is then performed at 121 °C for one hour. The use of a dilute acid hydrolysis immediately following a strong acid swelling hydrolysis allows the release of glucose from all possible sources, including heavily cross-linked and rigidly packed cell wall components, such as crystalline cellulose. The “one-step” portion of the hydrolysis, also called the “matrix hydrolysis”, refers to the hydrolysis of samples only at 121 °C with 4% sulfuric acid. By simply treating samples with a “matrix hydrolysis”, only monosaccharides from easily hydrolysable sources will be released. Therefore, the difference between these two hydrolyses allows the quantification of crystalline cellulose in what is referred to as a “one-step two-step” hydrolysis approach. In our protocol, we combine the efficient “one-step two-step” sulfuric acid hydrolysis described by Yeats et al. (2016a) with the inclusion of an optional enzymatic starch degradation and the use of alternate internal standards to elute all cell wall sugars and quantify glucose derived from crystalline cellulose in a single HPAEC-PAD run. Our additions to the previously mentioned protocols contribute significant improvements in both ease and accuracy of monosaccharide measurement that will greatly benefit the field of plant cell wall research.
Materials and Reagents
2 ml screw-cap tubes (Sarstedt, catalog number: 72.694.006)
50 ml conical tubes (Greiner Bio-One, catalog number: 7.210 261)
500 μl autosampler Snap Ring vials (Sigma, catalog number: 27422)
Autosampler vial lids (Sigma, catalog number: 24757)
Stainless steel 25 ml grinding jars (Retsch, catalog number: 02.462.0119)
Stainless steel 12 mm grinding balls (Retsch, catalog number: 05.368.0032)
Tin weigh boats, 5 x 9 mm (Santis Analytical AG, catalog number: SA76981103)
0.22 μm sterile PES-membrane filter (Life Systems Design AG, catalog number: 99255)
Metal spatula
Plastic weighing papers (HuberLab, catalog number: 12.9702.080)
Conical tubes (Grenier Bio, catalog number: 7.210 261)
Nylon mesh, 60 μm pore size (Sefar Nitex, catalog number: 3A03-0060-110-00)
Square Petri plates (Greiner Bio-One, catalog number: 7.688.102)
-
Homogenizing beads (depending on amount of material):
Small glass beads (2.85-3.45 mm beads, Roth, catalog number: A557.1)
Large metal balls (12 mm beads, Retsch, catalog number: 05.368.0032)
CarboPac PA20 column (3 x 150 mm, Thermo Fisher Scientific, catalog number: 060142)
CarboPac PA20 guard column (3 x 30 mm, Thermo Fisher Scientific, catalog number: 060144)
Arabidopsis thaliana
Ethanol (HCI Shop, ETH Zurich, catalog number: 02000107)
Chloroform (Sigma, catalog number: 25693)
Methanol (HCI Shop, ETH Zurich, catalog number: 02000342)
Acetone (Sigma, catalog number: 24201-4X2.5L-GL-R)
-
(Optional) Enzymatic starch degradation
Amyloglucosidase (Sigma, catalog number: 10102857001)
α-Amylase (Sigma, catalog number: 10102814001)
Sulfuric acid (Sigma, catalog number: 258105)
Ultrapure water (Milli-Q or equivalent)
-
Monosaccharide analysis standards:
L-fucose (Sigma, catalog number: F2252-5G)
D-glucose (Sigma, catalog number: G7528-1KG)
D-galactose (Sigma, catalog number: 48260)
D-xylose (Sigma, catalog number: W360600-SAMPLE)
D-mannose (Sigma, catalog number: 63579)
L-arabinose (Roth, catalog number: 5118.2)
L-rhamnose (Sigma, catalog number: W373011-SAMPLE-K)
D-galacturonic acid monohydrate (Sigma, catalog number: 48280)
D-glucuronic acid (Sigma, catalog number: G5269-10G)
-
(Optional) Alternate internal standards
D-sedoheptulose (CarboSynth, catalog number: MS139006)
D-ribose (Sigma, catalog number: R7500-5G)
Sodium hydroxide, 50% solution in water (Sigma, catalog number: 415413)
Sodium acetate, anhydrous (Sigma, catalog number: 32319-1KG-R)
Lugol solution (Sigma, catalog number: 32922-6X1L)
Liquid nitrogen
Equipment
Freeze-dryer (Christ, Alpha 2-4)
Microcentrifuge (Eppendorf, model: 5424 R)
Sample concentrator (Stuart, model: SBHCONC/1)
Autoclave-compatible rack (Karter Scientific, catalog number: 125A7)
Microbalance (Mettler Toledo MX5)
Tissue homogenizer (Retsch MM200)
Micro-centrifuge tube shaker (Eppendorf ThermoMixer F1.5)
Tube rotator (Labinco LD79, catalog number: 79000)
Speed-vacuum centrifuge (Eppendorf Concentrator Plus)
Autoclave (Thermo Fisher Scientific, Sterico, Varioklav)
Heating block (Stuart, model: SBH130D)
Autosampler (Dionex, model: AS-1)
-
Dionex ICS-5000 (Dionex, model: DC-5)
ED Electrochemical Detector (without cell, product number: 072042)
ED Cell (no reference or working electrode, product number: 072044)
Gold (Au) on Polytetrafluororethylene (PTFE) Disposable Electrode (product number: 066480)
Software
-
Chromeleon 8 (Thermo Fisher Scientific)
Available for a fee at https://www.thermofisher.com/order/catalog/product/CHROMELEON7
-
Microsoft Excel
Available for a fee at https://www.office.com
-
GraphPad Prism
Available for a fee at https://www.graphpad.com
-
Spyder5 (Anaconda3)
Freely available at https://www.anaconda.com
-
Python 3.6
Freely available at https://www.python.org
Procedure
-
Generating and preparing plant material
-
Grow plants in desired conditions
Note: Plant samples can be collected from any desired growth method; we suggest the following in vitro method for Arabidopsis material generated from either light-grown (any day cycle) seedlings or dark-grown hypocotyls.
Gas or liquid sterilize seeds and stratify at 4 °C for 2-3 days.
Plates: prepare MS + 1% sucrose + 0.9% agar square Petri plates; sterilize nylon mesh and lay on top of agar plates; sow approximately 100-120 seeds at top of plates; collect material at desired time point in 50 ml conical tubes; remove seeds before processing.
For light-grown plants, grow plants in light for 14 days under long-day conditions (16 h light, 8 h dark) with light optimized for Arabidopsis (130-150 μE m-2 s-1) at 20-22 °C.
For dark-grown hypocotyls, sow multiple lines of seed per plate (without nylon); leave seeds sown on plates in light for 2-4 h; cover in several layers of aluminum foil and allow to grow in climate-controlled chamber for 5 days at 20-22 °C.
Leave plants in the dark for 24-48 h prior to harvesting to deplete starch reserves; if not possible, an enzymatic starch degradation can be performed after homogenization of plant material (proceed to Step A5).
-
If analyzing aerial plant tissues (containing chlorophyll), harvest material in enough 70% ethanol to fully submerge plant material, and continue exchanging ethanol until chlorophyll is depleted and the liquid no longer has any trace of green colour.
If non-chlorophyll-containing plant portions are to be analyzed, harvest and flash freeze samples with liquid nitrogen and proceed to grinding in Step A5.
Remove ethanol and use freeze-dryer to dry material over 2 days (or longer if necessary).
-
Grind all plant material using tissue homogenizer with 12 mm diameter metal balls and metal containers.
Notes:
If generating only a small amount of material, it is possible to harvest roots into microcentrifuge tubes and use a tissue homogenizer with racks for microcentrifuge tubes and glass beads for grinding. In this case, aliquot ≤ 200 mg plant material to ensure thorough homogenization.
When collecting plant material, it is imperative to avoid collection of agar, soil, or any growth media containing sugars with samples (N.B. agar will contribute to galactose quantification). Nylon mesh can be used as described above to prevent agar adhesion to roots.
-
-
Starch degradation conducted as previously described (Hostettler et al., 2011) with modifications as follows:
Aliquot ground plant material up to ~0.5 ml in a 2 ml micro-centrifuge tube.
Add 1 ml 80% ethanol (v/v) and heat samples at 95 °C for 10 min. Mix well by vortexing each sample for 10-15 s.
-
Centrifuge at room temperature (RT) at 3,000 × g for 5 min; discard supernatant.
Note: All centrifugation steps should be performed at RT unless otherwise indicated.
-
Continue with the following washing steps: shaking in a ThermoMixer for 10 min during each wash, centrifuging at 3,000 × g for 5 min, vortexing to re-suspend pellet between washes, discard supernatant, and add the next washing solution: 1 ml 50% (v/v) ethanol, 1 ml 20% (v/v) ethanol, 1 ml water, and finally 1 ml 80% (v/v) ethanol.
Note: After Step B4, the final wash should be mostly clear, but pellet may still be green.
Dry the pellet at room temperature, using a speed-vacuum centrifuge, or in an oven at 60 °C for at least 30 min or over-night until completely dry. Re-suspend in 400 μl water and vortex to mix.
Boil at 95 °C for 10-15 min–do not cool on ice.
-
Prepare a digestion mixture of 9 parts amyloglucosidase and 1 part α-amylase.
Note: Calculate volume of digestion mixture needed based on total number of samples, keeping in mind that 20 μl of the mixture is required per sample. One sample requires 18 μl of amyloglucosiades and 2 μl α-amylase.
-
Add 380 μl 0.22 M sodium acetate to 20 μl of the prepared 9:1 amyloglucosidase: α-amylase mixture; combine with 400 μl sample.
Note: If processing larger sample volumes, sample can be resuspended in larger volume of water. For digestion, simply combine equal parts sample with digestion mixture and proceed as indicated.
Digest at 37 °C for a minimum of 2 h.
-
Check if there is remaining starch by staining a small portion of the plant material with Lugol solution.
Note: Lugol staining suggested procedure:
Mix 20 μl sample with 80 μl 100% ethanol.
Boil for 5 min at 90 °C.
Centrifuge for 5 min at 5,000 x g. Discard supernatant.
-
Add 25 μl Lugol solution; check for colour change after 5 min.
Lugol solution can be used at full concentration without dilution of purchased solution.
Colour change to a deep blue/black colour indicates presence of starch.
If colour change is observed, continue digestion overnight and check with Lugol again. Continue digestion until colour change is no longer observed (Figure 1).
Once no colour change is observed, proceed to Step A11.
Centrifuge samples at 5,000 × g for 5 min and proceed with the insoluble fraction remaining.
-
Cleaning and production of final cell wall-derived alcohol insoluble residue (AIR) preparation
Aliquot de-starched insoluble fraction up to 0.5 ml in a 2 ml micro-centrifuge tube.
Add 1.5 ml of a 1:1 methanol:chloroform (v/v) mixture to sample and vortex to mix thoroughly. Mix samples for 2 h using the tube rotator set to 15 rpm, or any standard tube mixer.
Centrifuge at 10,000 × g for 5-10 min at room temperature and remove supernatant.
Add 1.5 ml RT acetone and vortex to mix thoroughly. Mix for 30 min using a tube rotator set to 15 rpm or any standard tube mixer.
Centrifuge at 10,000 × g for 5-10 min at room temperature and remove supernatant.
Dry the final pellet at room temperature, using a speed-vacuum centrifuge, or in an oven at 60 °C for at least 30 min or overnight until completely dry. The final product is the cell wall-derived alcohol insoluble residue (AIR).
-
Sample and standard hydrolysis conducted as described in Yeats et al. (2016a) (see Figure 2 in this manuscript). Briefly, the method is summarized as follows:
-
Weigh 1 ± 0.1 mg AIR per technical replicate into 2 ml screw-cap tubes using a microbalance and tin weigh boats; record final weight (required for analysis steps).
Notes:
If spatula used for weighing AIR needs to be cleaned in between samples, do not use any kind of tissue/paper towel. Use the plastic weighing papers to wipe and rinse thoroughly with 100% ethanol in between samples.
We recommend a minimum of two technical replicates per sample per hydrolysis (meaning, 2 technical replicates for samples subjected only to matrix hydrolysis, and 2 technical replicates subjected to Saeman hydrolysis + matrix hydrolysis) and three biological replicates for each experimental analysis.
-
Prior to sample hydrolysis, internal standards (sedoheptulose or ribose) can be added directly to weighed AIR aliquots. Allow internal standards to completely dry for 30 min or longer as necessary, either at room temperature or using a sample concentrator.
For our assays, we added 150 μg of sedoheptulose to AIR material; however, this amount must be optimized based on the sample dilution that will be measured considering the working range of the instrument of choice.
Addition of the standards (ribose or sedoheptulose) is not mandatory. However, they provide an added measure of certainty to ensure consistency of hydrolysis and quantification.
Sedoheptulose worked better for this analysis as we observed a “background” peak with the same retention time as ribose. However, with a slightly different instrument or sample, ribose may be used. This should be tested prior to hydrolysis of all samples and standards.
-
One sample set will be subjected to Saeman as well as matrix hydrolysis, while a second sample set will be subjected only to matrix hydrolysis.
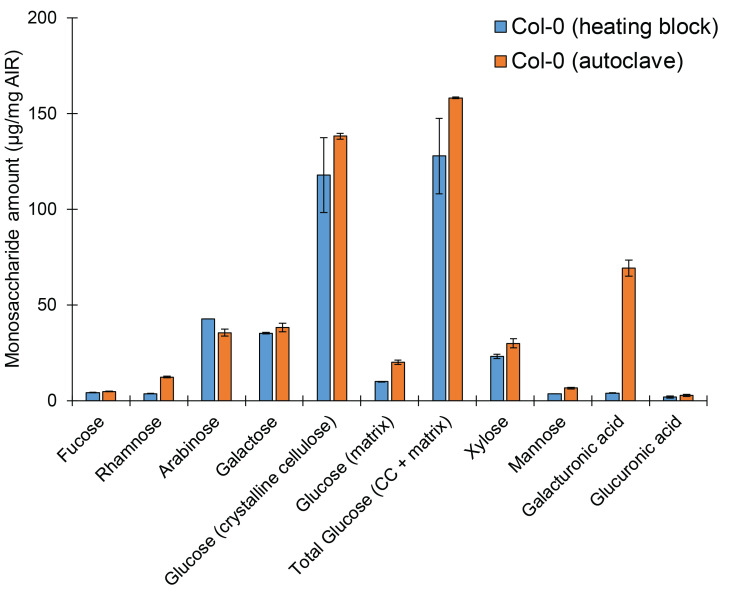
Note: Previously, this method was described using autoclaving as the hydrolysis method (Yeats et al., 2016a). We confirm this as an efficient method of hydrolyzing upwards of 50 samples at once. However, we also confirmed the validity of the HPLC analysis and quantification if hydrolysis is accomplished using a heating block at 121 °C for 1 h. However, data is more consistent when using autoclaving to accomplish hydrolysis (Figure 3), as is demonstrated by reduced error. Further, it appears hydrolysis of galacturonic acid may be incomplete using heating block hydrolysis, although all other monosaccharide values seem to be consistent across either method. It may also be possible to use a heating block at a lower temperature (80-100 °C) for a longer hydrolysis time, however this should be tested and optimized.
-
For standard curve analysis, make a 100 μg stock solution containing all monosaccharides for quantification as well as the appropriate internal standard. Dilute into appropriate standard concentrations based on assaying range.
Note: Recommended concentrations to generate the standard curve are as follows: 0.05 μg, 0.1 μg, 0.5 μg, 1 μg, 2 μg, 5 μg.
-
To consider sugar-specific losses during hydrolysis and calculate monosaccharide-specific correction factors, prepare two recovery standards by combining 500 μl of the 100 μg standard mixture with 900 μl water.
One recovery standard is subjected to the same conditions as the matrix hydrolysis samples (water + acid, hydrolysis for 1 h at 121 °C).
The second recovery standard is treated with acid, but is not subjected to heat hydrolysis.
After hydrolyses are complete, allow samples to cool at room temperature and centrifuge for 1 min at 20,000 × g to pellet any insoluble material. The supernatant is used in the next setp.
-
-
HPLC analysis
-
Dilute sample supernatants as required before injection (1:10, 1:20, 1:50, or 1:100 dilutions may be used depending on the starting material and detector sensitivity) and pipet into autosampler vials.
Notes:
Appropriate standards and dilutions may vary based on samples or detector sensitivity; it is recommended to test standards as well as sample dilutions thoroughly to optimize conditions before completing processing of all material.
It is strongly recommended to randomize order of sample analysis and run a standard after every 10-15 samples to ensure sensitivity and accuracy of measurement is consistent.
-
Make eluents; purge with and maintain under helium gas, or as directed by manufacturer (Rohrer, 2017). Any eluents containing sodium acetate must be filtered using a 0.22 μm PES filter. All eluents must be purged for a minimum of 10 min before addition of 50% sodium hydroxide solution, followed by additional purging of a minimum of 10 min after addition.
Eluent A = water
Eluent B = 50 mM sodium hydroxide
Eluent C = 100 mM sodium hydroxide, 100 mM sodium acetate
Eluent D = 200 mM sodium hydroxide
Inject 10 μl of each standard, recovery standard, and sample onto a 3 x 150 mm CarboPac PA20 column equipped with a 3 x 50 mm CarboPac PA20 guard column.
Maintain column temperature at 36 °C with a flow rate of 0.4 ml/min.
-
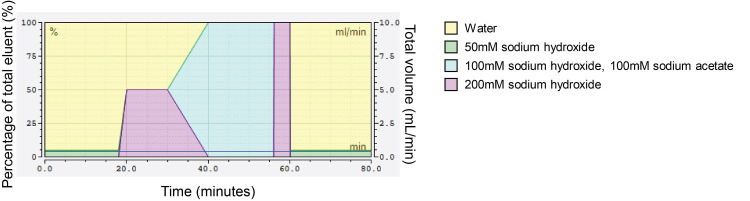
Use the following elution profile to elute all monosaccharides and standards (Figure 4, Table 1): 0-18 min 4.8% B, 95.2% A; 18-20 min linear gradient to next condition; 20-30 min 50% D, 50% A; 30-40 min linear gradient next condition; 40-56 min 100% C; 56-56.1 min linear gradient to 50% D; 56.1-60 min 50% D; 60-60.1 min change to next condition; 60.1-80 min 4.8% B, 95.2% A to equilibrate column back to starting conditions:
Notes:
Retention times of peaks may shift as more samples run, therefore regular “column flushing” and monitoring of column performance may be necessary.
A short “column flushing” period is incorporated into the gradient profile (~5 min of 100 mM NaOH at 56 min); however, it is also possible to run a flushing profile periodically as follows: 30 min 100% Eluent C followed by 30 min 100% Eluent D with a short equilibration step (approximately 10-15 min) back to starting conditions (95.2% Eluent A, 4.8% Eluent B).
-
If significant changes in column performance are observed, immediate action to fully flush the column as per the manufacturer’s instructions must be taken. In short, we accomplished column flushing by conducting the following:
Disconnect column from detector and general machine flow by turning the electrode/detector off and unscrewing the column outlet.
Wash column with 2 M NaOH (allowing flow-through to drip into a waste receptacle) for 1 h; adjust flow rate until pressure reaches similar level to running pressure during sample analysis (in this case, ~2,200 psi).
Re-equilibrate column with starting conditions (in the case of this protocol, 4.8% Eluent B, 95.2% Eluent A) for 30 min; again, adjust flow rate until pressure reaches similar level to running pressure during sample analysis, and collect flow-through in a waste receptacle.
Re-attach column to the system, and run a water sample followed by a standard before continuing with sample analysis using the normal gradient profile.
Technical notes from the manufacturer were consulted thoroughly for this analysis (Basumallick and Rohrer, 2017).
-
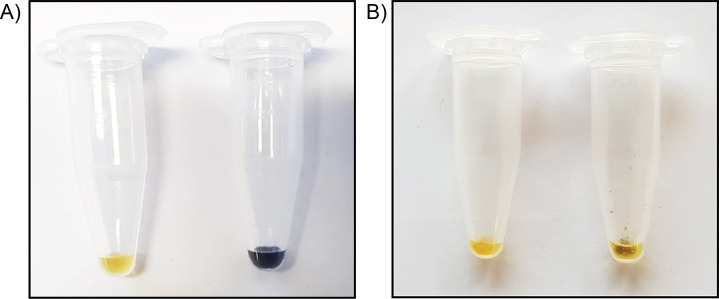
Figure 1. Lugol stain reveals presence or absence of starch.
A. Full concentration Lugol alone has a faint yellow colour (left tube), while Lugol added directly to starch extracted from corn shows a strong dark brown/purple colour change (right tube). B. Both tubes contain cell wall AIR material; the tube on the left has been subjected to starch degradation, while the tube on the right has not. Following the procedure described in Step A10, addition of 25 μl full concentration Lugol will stain AIR samples containing starch a dark brown/purple colour, as observed in the right tube. If the Lugol remains yellow when added to the sample, starch is not present, and one may proceed with Step A11.
Figure 2. Gradient profile of single HPLC run to elute all cell wall monosaccharides.
Each block of colour corresponds to a particular eluent, indicated in the legend on the right. The primary y-axis (left) features percentage, referring to what percentage each eluent comprises the final mixed eluent that flows through the column, with the total always equaling to 100%. The secondary y-axis (right) indicates the volume (ml/minute) used up per eluent, relating to the percentage of the total mixed eluent to which each individual eluent contributes. The x-axis indicates the amount of time (minutes) for which the indicated eluent composition should proceed.
Figure 3. Sample hydrolysis may be accomplished using either a heating block or autoclave, with minor differences.
Monosaccharide elution profile of cell wall AIR derived from light-grown wild-type Col-0 seedlings. A minimum of two technical replicates per sample per hydrolysis method were used. Bars represent average of 2 biological replicates ± standard error.
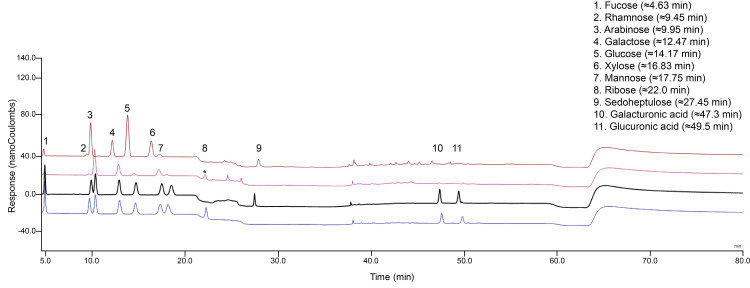
Figure 4. Elution profile of cell wall monosaccharides in a single HPAEC-PAD run.
Four separate injections are presented: 1 μg/ml standards mixture using ribose as an internal standard (blue), 1 μg/ml standards mixture using sedoheptulose as an internal standard (black), and an example of a wild type Col-0 matrix hydrolysis profile (hydrolyzed using a heating block) (pink), and an example of a wild type Col-0 Saeman hydrolysis + matrix hydrolysis profile with sedoheptulose added as an internal standard (hydrolyzed using a heating block) (brown). After sample hydrolysis (pink), a background peak (*) appeared that clearly overlaps with the ribose peak (blue).
Table 1. Plant cell wall monosaccharide elution gradient steps.
| Time (min) | Eluent A (%) | Eluent B (%) | Eluent C (%) | Eluent D (%) |
| 0-18 | 95.2 | 4.8 | 0 | 0 |
| 20 | 50 | 0 | 0 | 50 |
| 30 | 50 | 0 | 0 | 50 |
| 40 | 0 | 0 | 100 | 0 |
| 56 | 0 | 0 | 100 | 0 |
| 56.1 | 0 | 0 | 0 | 100 |
| 60 | 0 | 0 | 0 | 100 |
| 60.1 | 95.2 | 4.8 | 0 | 0 |
| 80 | 95.2 | 4.8 | 0 | 0 |
Data analysis
All standard curve and sample peaks were integrated using Chromeleon 8.0 software and analyzed using Microsoft Excel as described in Yeats et al. (2016a). GraphPad Prism was used for statistical analyses and generating graphs.
-
In order to facilitate data entry into Microsoft Excel, a customized peak-calling analysis method in Chromeleon and a Python script can be used to copy integrated curve values into a sorted, transposed Excel spreadsheet. Values can subsequently be copied into a similar analysis sheet as the one described in Yeats et al. (2016a).
Raw data from Chromeleon runs must be saved as Excel spreadsheets within the same folder as the Python script.
Peak calling can be adjusted directly in Chromeleon to limit output to only sugars desired for analysis.
-
The Python script can be modified to analyze all cell wall sugars, as well as glucosamine if desired, or other additional sugars.
Note: If using the Python script for analysis, adjust the number of inputs in Line 9 of the script to reflect the number of peaks that will be called by the Chromeleon software based on the test parameters chosen in the software.
Conclusions:
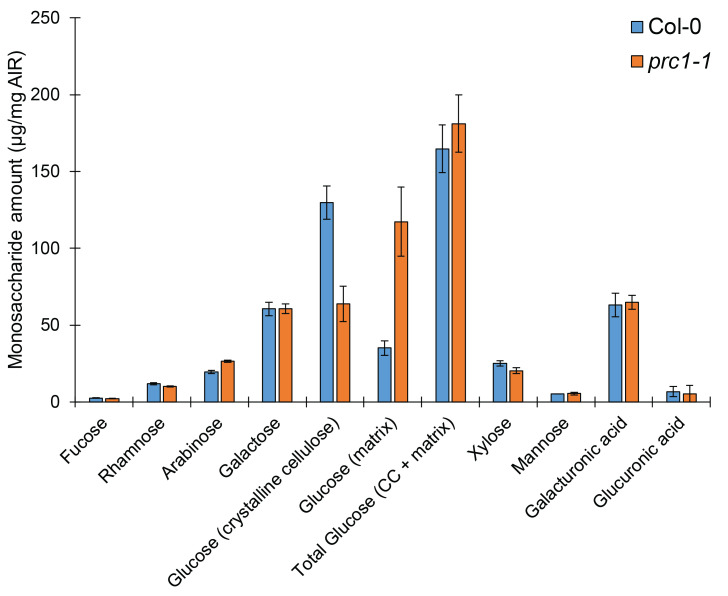
This protocol can be used to efficiently quantify all cell wall monosaccharides using a single HPAEC-PAD gradient profile. This quantification will allow the reliable characterization of monosaccharide composition, and allows for the determination of proportions of glucose that come from both the crystalline and non-crystalline portions of the plant cell wall. For example, using wildtype (Col-0), and the well-characterized cellulose-deficient mutant and prc1-1, we demonstrate that this method is sufficient for resolving differences between cell wall mutants (Figure 5). Additionally, this analysis has already been used in a recent study to quantify differences in glucose derived from crystalline cellulose (Kesten et al., 2019). This method is simple, reliable, and consistent, and can be used to better understand cell wall monosaccharide compositional changes in a biological context. Past methods rely on multiple hydrolyses approaches or HPLC gradients to quantify neutral cell wall monosaccharaides, uronic acids, and cellulose separately. Therefore, this analysis represents a streamlined alternative to total cell wall monosaccharide analysis.
Figure 5. Cell wall mutants can be clearly distinguished from wild-type using this hydrolysis and analysis method.
Monosaccharide elution profile of cell wall AIR derived from dark-grown Col-0 or cellulose-deficient prc1-1 hypocotyls. A minimum of two technical replicates per sample per hydrolysis method were used. Bars represent average of 2 or 3 biological replicates (for Col-0 or prc1-1, respectively) ± standard error.
Acknowledgments
We gratefully acknowledge S. Zeeman (ETH Zürich) and T. Yeats (Cornell University) for technical advice, C. Kesten (ETH Zürich) for scientific discussion, and S. Dora (ETH Zürich) and A. García Moreno (University of Málaga) for technical support during development and optimization of this protocol. As previously mentioned, this protocol was adapted and modified from Yeats, T., Vellosillo, T., Sorek, N., Ibáñez, A.B., and Bauser, S. (2016). Bio-protocol 6(20): e1978. This work has been financially supported by the Swiss National Foundation to C.S.-R. (SNF 31003A_163065/1; A.M.).
Competing interests
There are no conflicts of interest or competing interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Supplementary Data.
References
- 1.Basumallick L., Rohrer J.(2017). Determination of uronic acids and wood sugars in wood-based hydrolysates. Thermo Fisher Scientific. [Google Scholar]
- 2.Cosgrove D. J.(2005). Growth of the plant cell wall. Nat Rev Mol Cell Biol 6(11): 850-861. [DOI] [PubMed] [Google Scholar]
- 3.Cosgrove D. J.(2016). Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J Exp Bot 67(2): 463-476. [DOI] [PubMed] [Google Scholar]
- 4.Hostettler C., Kölling K., Santelia D., Streb S., Kotting O. and Zeeman S. C.(2011). Analysis of starch metabolism in chloroplasts. Methods Mol Biol 775: 387-410. [DOI] [PubMed] [Google Scholar]
- 5.Kesten C., Gámez-Arjona F.M., Menna A., Scholl S., Dora S., Huerta A.I., Huang H., Tintor N., Kinoshita T., Rep M., Krebs M., Schumacher K., Sánchez-Rodríguez C.(2019). Pathogen-induced pH changes regulate the growh-defense balance in plants. EMBO Journal: e101822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kesten C., Menna A. and Sanchez-Rodriguez C.(2017). Regulation of cellulose synthesis in response to stress. Curr Opin Plant Biol 40: 106-113. [DOI] [PubMed] [Google Scholar]
- 7.Pettolino F. A., Walsh C., Fincher G. B. and Bacic A.(2012). Determining the polysaccharide composition of plant cell walls. Nat Protoc 7(9): 1590-1607. [DOI] [PubMed] [Google Scholar]
- 8.Rohrer J.(2017). Eluent preparation for high-performance anion-exchange chromatography with pulsed amperometric detection. Thermo Fisher Scientific Technical: 1-7. [Google Scholar]
- 9.Saeman J. F.(1945). Kinetics of wood saccharification- hydrolysis of cellulose and decomposition of sugars in dilute acid at high temperature. Ind Eng Chem 37(1):43-52. [Google Scholar]
- 10.Voiniciuc C. and Günl M.(2016). Analysis of monosaccharides in total mucilage extractable from Arabidopsis seeds. Bio-protocol 6(9): e1801. [Google Scholar]
- 11.Yeats T. H., Sorek H., Wemmer D. E. and Somerville C. R.(2016). Cellulose deficiency is enhanced on hyper accumulation of sucrose by a H+-coupled sucrose symporter. Plant Physiol 171(1): 110-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeats T., Vellosillo T., Sorek N., Ibáñez A. B. and Bauer S.(2016). Rapid determination of cellulose, neutral sugars, and uronic acids from plant cell walls by one-step two-step hydrolysis and hpaec-pad. Bio-protocol 6(20): e1978. [Google Scholar]
- 13.Zhang Z., Khan N. M., Nunez K. M., Chess E. K. and Szabo C. M.(2012). Complete monosaccharide analysis by high-performance anion-exchange chromatography with pulsed amperometric detection. Anal Chem 84(9): 4104-4110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.