Abstract
Circadian rhythms have been extensively studied in Drosophila; however, still little is known about how the electrical properties of clock neurons are specified. We have performed a behavioral genetic screen through the downregulation of candidate ion channels in the lateral ventral neurons (LNvs) and show that the hyperpolarization-activated cation current Ih is important for the behaviors that the LNvs influence: temporal organization of locomotor activity, analyzed in males, and sleep, analyzed in females. Using whole-cell patch clamp electrophysiology we demonstrate that small LNvs (sLNvs) are bursting neurons, and that Ih is necessary to achieve the high-frequency bursting firing pattern characteristic of both types of LNvs in females. Since firing in bursts has been associated to neuropeptide release, we hypothesized that Ih would be important for LNvs communication. Indeed, herein we demonstrate that Ih is fundamental for the recruitment of pigment dispersing factor (PDF) filled dense core vesicles (DCVs) to the terminals at the dorsal protocerebrum and for their timed release, and hence for the temporal coordination of circadian behaviors.
SIGNIFICANCE STATEMENT Ion channels are transmembrane proteins with selective permeability to specific charged particles. The rich repertoire of parameters that may gate their opening state, such as voltage-sensitivity, modulation by second messengers and specific kinetics, make this protein family a determinant of neuronal identity. Ion channel structure is evolutionary conserved between vertebrates and invertebrates, making any discovery easily translatable. Through a screen to uncover ion channels with roles in circadian rhythms, we have identified the Ih channel as an important player in a subset of clock neurons of the fruit fly. We show that lateral ventral neurons (LNvs) need Ih to fire action potentials in a high-frequency bursting mode and that this is important for peptide transport and the control of behavior.
Keywords: bursting neuron, Drosophila melanogaster, HCN, Ih, ion channel, pigment dispersing factor
Introduction
Circadian (circa: around, diem: day) rhythms are biological rhythms with a period of ∼24 h that have evolved in essentially all organisms. They confer an important adaptive value by allowing the anticipation to the daily changes in environmental conditions associated to the rotation of our planet. The “around the clock” coordination of behavior and physiology in Drosophila is regulated by ∼150 neurons grouped in different clusters and named after their anatomic localization (Yoshii et al., 2012). Among them, the small lateral ventral neurons (sLNvs) have been identified as a fundamental group in the control of behavioral rhythms under free running conditions, communicating via the release of the neuropeptide pigment dispersing factor (PDF; Renn et al., 1999; Peng et al., 2003; Lin et al., 2004; Yoshii et al., 2009) and glycine (Frenkel et al., 2017). The large LNvs (lLNvs), on the other hand, are highly relevant for arousal and the PDF they release provides wake promoting functions (Parisky et al., 2008; Shang et al., 2008; Sheeba et al., 2008a).
Although the mechanisms that give rise to the cell-autonomous cycling of gene and protein expression and comprise the core of the molecular clock have been described thoroughly (Top and Young, 2018), one of the challenges of the field now is to understand how different clock neurons communicate to each other. It is indeed the emerging properties of these clock neuronal circuits acting concertedly that provide the system with plasticity and adaptability (Beckwith and Ceriani, 2015). But to understand the communication taking place within clock neurons, it is paramount to examine the physiological properties of the different neuronal groups. The type, amount, and distribution of ion channels present in the membrane of a neuron determine features such as excitability and action potential firing pattern. In particular, clock neurons change their electrical activity on a daily basis, with higher action potential firing during the day than at night, a phenomenon that has been described both in mammals and flies (for review, see Allen et al., 2017).
In Drosophila, several ion channels have already been found to play roles in different aspects of circadian function, such as the calcium dependent voltage-gated potassium channel slowpoke (slo; Ceriani et al., 2002; Fernandez et al., 2007) and its binding protein (slob; Ceriani et al., 2002; Jaramillo et al., 2004; Tabuchi et al., 2018), the cation channel narrow abdomen (na; Nash et al., 2002; Lear et al., 2005; Flourakis et al., 2015), the voltage-gated potassium channel Shaw (Hodge and Stanewsky, 2008; Buhl et al., 2016; Smith et al., 2019), the inward rectifying potassium channel Ir (Ruben et al., 2012), the temperature sensitive trpA1 channel (Lee and Montell, 2013), the potassium channel hyperkinetic (hk; Fogle et al., 2015) and the voltage-gated potassium channel Shal (Feng et al., 2018; Smith et al., 2019). Under the hypothesis that additional ion channels are involved in determining the characteristic physiological properties of the LNvs that ensure circadian organization of locomotor activity, we performed a behavioral genetic screen downregulating candidate ion channels using RNA interference (RNAi) specifically in LNvs. Following this strategy, we have been able to identify several ion channels that, when knocked down, alter circadian locomotor behavior under free running conditions. Of those, we have first focused our attention on the hyperpolarization-activated cation current Ih, since, as it has been described in mammalian neurons (Luthi and McCormick, 1998), its biophysical properties make it particularly suitable to mediate the organization of action potential firing in bursts, a firing mode that characterizes lLNvs (Cao and Nitabach, 2008; Sheeba et al., 2008b; Muraro and Ceriani, 2015) and, we show here, also sLNvs. Consistently, we demonstrate that perturbing Ih causes a decrease in the frequency of LNvs bursting that is accompanied by a reduction in PDF immunoreactivity and in the complexity of sLNv axonal termini. Moreover, we have found that the disruption of Ih is accompanied by an increase in sleep. Altogether, our results reveal a novel function of Ih in determining LNvs physiology and the behaviors they command, and uncover several additional ion channels with putative roles in these important clock clusters, for future exploration.
Materials and Methods
Fly strains
All fly strains used in this study are detailed in Table 1. UAS lines for RNAi downregulation of candidate ion channels were obtained from the Bloomington Stock Center [the ones associated to the Drosophila RNAi Screening Center (DRSC)], from the Vienna Drosophila Resource Center (VDRC), and from the National Institute of Genetics Fly Stock Center (NIG). Information for each of these lines is also available in Tables 2, 3 (for the positive hits of the genetic screen), 4 (for the negative hits). Flies were grown and maintained at 25°C in standard cornmeal medium under 12/12 h light/dark cycles (LD) unless stated otherwise. For experiments involving the adult-specific GeneSwitch (GS) expression system, 2- to 4-d-old adult males raised in normal cornmeal food were transferred to food containing RU486 (mifepristone, Sigma) in 80% ethanol to a final concentration 200 μg/ml or with the same amount of ethanol (vehicle) in control treatments. These experiments were done with a line that includes a UAS-CD8::GFP transgene on the II chromosome. For experiments involving the TARGET system (McGuire et al., 2004a) flies were raised at 21°C, induction of the expression system was achieved by increasing the temperature to 30°C. Newly eclosed males were used for all circadian rhythmicity experiments, 3- to 7-d-old non-virgin females were used for sleep and electrophysiology experiments, a mix of males and females was used for immunofluorescence determination.
Table 1.
Reagents and resources used for this work
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Rat polyclonal anti-PDF | Depetris-Chauvin et al. (2011) | N/A |
| Chicken polyclonal anti-GFP | Aves Lab | Catalog #GFP-1020, RRID:AB_10000240 |
| Alexa Fluor 647-AffiniPure Donkey Anti-Rat | Jackson ImmunoResearch | catalog #712-605-150, RRID:AB_2340693 |
| Cy2-AffiniPure Donkey Anti-Chicken | Jackson ImmunoResearch | catalog #703-225-155, RRID:AB_2340370 |
| Chemicals | ||
| NaCl, sodium chloride | Sigma-Aldrich | S7653; CAS: 7647-14-5 (BioXtra, ≥99.5%; AT) |
| KCl, potassium chloride | Sigma-Aldrich | P3911; CAS: 7447-40-7 (ACS reagent, 99.0–100.5%) |
| CaCl2.2H2O, calcium chloride dihydrate | Sigma-Aldrich | 223506; CAS: 10035-04-8 (ACS reagent, ≥99%) |
| MgCl2.6H2O, magnesium chloride hexahydrate | Sigma-Aldrich | M2670; CAS: 7791-18-6 (BioXtra, ≥99.0%) |
| NaH2PO4, sodium phosphate monobasic | Sigma-Aldrich | S8282; CAS: 7558-80-7 (BioXtra, ≥99.0%) |
| NaHCO3, sodium bicarbonate | Sigma-Aldrich | S6297; CAS: 144-55-8 (BioXtra, 99.5–100.5%) |
| D-(+)-glucose | Sigma-Aldrich | G8270; CAS: 50-99-7 (≥99.5%; GC) |
| Protease from Streptomyces griseus | Sigma-Aldrich | P5147; CAS: 9036-06-0 (Type XIV, ≥3.5 units/mg solid, powder) |
| Potassium D-gluconate | Sigma-Aldrich | G4500; CAS: 299-27-4 (≥99%) |
| EGTA | Sigma-Aldrich | E3889; CAS: 67-42-5 (for molecular biology, ≥97.0%) |
| HEPES | Sigma-Aldrich | H3375; CAS: 7365-45-9 (≥99.5%; titration) |
| RU-486, mifepristone | Sigma-Aldrich | M8046, CAS: 84371-65-3 (≥98%) |
| Paraformaldehyde | Sigma-Aldrich | 441244; CAS: 30525-89-4 |
| NaCl, sodium chloride (for PBS solution) | Cicarelli Laboratorios | 750; CAS: 7647-14-5 |
| Na2HPO4, sodium phosphate dibasic | Sigma-Aldrich | S3264; CAS: 7558-79-4 (for molecular biology, ≥98.5%; titration) |
| Triton X-100 | Sigma-Aldrich | T9284; CAS: 9002-93-1 (BioXtra) |
| Goat serum | Natocor | 734 |
| Vetbond tissue adhesive | 3 M | 1469SB |
| Organisms/strains | ||
| D. melanogaster: w1118 | Bloomington Drosophila Stock Center | BDSC: 5905; FlyBase: FBal0018186 |
| D. melanogaster: pdf-GAL: y[1] w[*]; P{w[+mC]=Pdf-GAL4.P2.4}2 | Bloomington Drosophila Stock Center | BDSC: previously 6900, now available as part of 25031; FlyBase: FBtp0011844 |
| D. melanogaster: UAS-CD8::GFP: y[1] w[*]; P{w[+mC]=UAS-mCD8::GFP.L}LL5, P{UAS-mCD8::GFP.L}2 | Bloomington Drosophila Stock Center | BDSC: 5137; FlyBase: FBst0005137 |
| D. melanogaster: c929-Gal4: w[*]; P{w[+mW. h]=GawB}dimm[929] crc[929] | Bloomington Drosophila Stock Center | BDSC:25373; FlyBase: FBst0025373 |
| D. melanogaster: tub-Gal80TS: w[*]; P{w[+mC]=tubP-GAL80[ts]}2/TM2 | Bloomington Drosophila Stock Center | BDSC: 7017; FlyBase: FBst0007017 |
| D. melanogaster: pdfGS: w*; P{UAS-mCD8::GFP.L}LL5; P{Pdf-GS}3/TM3, Sb1 | Bloomington Drosophila Stock Center Depetris-Chauvin et al. (2011) |
BDSC: 80956 FlyBase: FBst0080956 |
| D. melanogaster: UAS-dicer2: w[1118]; P{UAS-dicer2, w[+]} | VDRC | VDRC ID: 60008 |
| D. melanogaster: R6-Gal4: P{?GawB}crcR6 | Helfrich-Forster et al. (2007) | FlyBase: FBti0016844 |
| D. melanogaster: pdf-RFP: Pdf-RFP transgene has 0.6 kb of Pdf regulatory genomic DNA 0.5 kb upstream the start site of transcription and 0.1 kb downstream) fused to DNA encoding mRFP1, a monomeric soluble red fluorescent protein (Shaner et al., 2004). Injected into y w flies. | Ruben et al. (2012) | FlyBase: FBrf0219602 |
| D. melanogaster: Ihf01485: PBac{WH}Ihf01485 | Exelixis at Harvard Medical School | FlyBase: FBst1017022 |
| D. melanogaster: Ihf03355: PBac{WH}Ihf03355 | Exelixis at Harvard Medical School | FlyBase: FBst1018427 |
| D. melanogaster: RNAi of cac: P{KK101478}VIE-260B | VDRC | VDRC ID: 104168 FlyBase: FBst0476026 |
| D. melanogaster: RNAi of cac: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02572}attP2 | Bloomington Drosophila Stock Center | BDSC: 27244; FlyBase: FBst0027244 |
| D. melanogaster: RNAi of Ca-α1T: P{KK100082}VIE-260B | VDRC | VDRC ID: 108827 FlyBase: FBst0480621 |
| D. melanogaster: RNAi of ClC-a: P{KK101247}VIE-260B | VDRC | VDRC ID: 110394 FlyBase: FBst0481966 |
| D. melanogaster: RNAi of CngA: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02039}attP2 | Bloomington Drosophila Stock Center | BDSC: 26014; FlyBase: FBst0026014 |
| D. melanogaster: RNAi of CngA: P{KK108314}VIE-260B | VDRC | VDRC ID: 101745 FlyBase: FBst0473618 |
| D. melanogaster: RNAi of Ih: P{KK100190}VIE-260B | VDRC | VDRC ID: 110274 FlyBase: FBst0481852 |
| D. melanogaster: RNAi of Ih: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF03253}attP2 | Bloomington Drosophila Stock Center | BDSC:29574; FlyBase: FBst0029574 |
| D. melanogaster: RNAi of Ork1: P{KK107843}VIE-260B | VDRC | VDRC ID: 104883 FlyBase: FBst0476711 |
| D. melanogaster: RNAi of Ork1: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF01926}attP2 | Bloomington Drosophila Stock Center | BDSC:25855; FlyBase: FBst0025885 |
| D. melanogaster: RNAi of Shal on the III chromosome | NIG Fly Stock Center | NIG Stock ID: 9262R-3 |
| D. melanogaster: RNAi of tipE on the III chromosome | NIG Fly Stock Center | NIG Stock ID: 1232R-3 |
| D. melanogaster: RNAi of tipE: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02148}attP2/TM3, Sb[1] | Bloomington Drosophila Stock Center | BDSC:26249; FlyBase: FBst0026249 |
| D. melanogaster: RNAi of Atpα: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02910}attP2 | Bloomington Drosophila Stock Center | BDSC: 28073; FlyBase: FBst0028073 |
| D. melanogaster: RNAi of Atpα: P{KK108782}VIE-260B | VDRC | VDRC ID: 100619 FlyBase: FBst0472492 |
| D. melanogaster: RNAi of Calx: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02937}attP2 | Bloomington Drosophila Stock Center | BDSC: 28306; FlyBase: FBst0028306 |
| D. melanogaster: RNAi of Calx: P{KK109144}VIE-260B | VDRC | VDRC ID: 104789 FlyBase: FBst0476622 |
| D. melanogaster: RNAi of Ca-α1D: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF01848}attP2 | Bloomington Drosophila Stock Center | BDSC: 25830; FlyBase: FBst0025830 |
| D. melanogaster: RNAi of Ca-α1D: w[1118]; P{GD1737}v51490/CyO | VDRC | VDRC ID: 51490 FlyBase: FBst0469449 |
| D. melanogaster: RNAi of Ca-α1T: w[1118]; P{GD7754}v31963 | VDRC | VDRC ID: 31963 FlyBase: FBst0459316 |
| D. melanogaster: RNAi of Ca-α1T: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02150}attP2 | Bloomington Drosophila Stock Center | BDSC: 26251; FlyBase: FBst0026251 |
| D. melanogaster: RNAi of eag: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF01471}attP2 | Bloomington Drosophila Stock Center | BDSC: 31678; FlyBase: FBst0031678 |
| D. melanogaster: RNAi of eag: P{KK107309}VIE-260B | VDRC | VDRC ID: 100260 FlyBase: FBst0472134 |
| D. melanogaster: RNAi of Hk: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02965}attP2/TM3, Sb[1] | Bloomington Drosophila Stock Center | BDSC: 28330; FlyBase: FBst0028330 |
| D. melanogaster: RNAi of Hk: P{KK109058}VIE-260B | VDRC | VDRC ID: 101402 FlyBase: FBst0473275 |
| D. melanogaster: RNAi of inx2: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02446}attP2 | Bloomington Drosophila Stock Center | BDSC: 29603; FlyBase: FBst0029306 |
| D. melanogaster: RNAi of inx2: P{KK111067}VIE-260B | VDRC | VDRC ID: 102194 FlyBase: FBst0474063 |
| D. melanogaster: RNAi of Ir: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF01841}attP2 | Bloomington Drosophila Stock Center | BDSC: 25823; FlyBase: FBst0025823 |
| D. melanogaster: RNAi of Ir: P{KK102249}VIE-260B | VDRC | VDRC ID: 107389 FlyBase: FBst0479211 |
| D. melanogaster: RNAi of Irk2: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF01838}attP2 | Bloomington Drosophila Stock Center | BDSC: 25820; FlyBase: FBst0025820 |
| D. melanogaster: RNAi of Irk2: w[1118]; P{GD203}v4341 | VDRC | VDRC ID: 4341 FlyBase: FBst0465076 |
| D. melanogaster: RNAi of KCNQ: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02562}attP2 | Bloomington Drosophila Stock Center | BDSC: 27252; FlyBase: FBst0027252 |
| D. melanogaster: RNAi of KCNQ: P{KK109039}VIE-260B | VDRC | VDRC ID: 106655 FlyBase: FBst0478479 |
| D. melanogaster: RNAi of Ncc69: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF03097}attP2 | Bloomington Drosophila Stock Center | BDSC: 28682; FlyBase: FBst0028682 |
| D. melanogaster: RNAi of Ncc69: P{KK108763}VIE-260B | VDRC | VDRC ID: 106499 FlyBase: FBst0478323 |
| D. melanogaster: RNAi of Nckx30C: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02574}attP2 | Bloomington Drosophila Stock Center | BDSC: 27246; FlyBase: FBst0027246 |
| D. melanogaster: RNAi of nrv1: w[1118] P{GD959}v46542 | VDRC | VDRC ID: 46542 FlyBase: FBst0466759 |
| D. melanogaster: RNAi of nrv1: P{KK100406}VIE-260B | VDRC | VDRC ID: 103702 FlyBase: FBst0475560 |
| D. melanogaster: RNAi of nrv2: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF03081}attP2 | Bloomington Drosophila Stock Center | BDSC: 28666; FlyBase: FBst0028666 |
| D. melanogaster: RNAi of nrv2: w[1118]; P{GD960}v2660 | VDRC | VDRC ID: 2660 FlyBase: FBst0456497 |
| D. melanogaster: RNAi of para: w[1118]; P{GD3392}v6131 | VDRC | VDRC ID: 6131 FlyBase: FBst0470199 |
| D. melanogaster: RNAi of para: P{KK108534}VIE-260B | VDRC | VDRC ID: 104775 FlyBase: FBst0476611 |
| D. melanogaster: RNAi of picot: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF01940}attP2 | Bloomington Drosophila Stock Center | BDSC: 25920; FlyBase: FBst0025920 |
| D. melanogaster: RNAi of picot: P{KK106848}VIE-260B | VDRC | VDRC ID: 101082 FlyBase: FBst0472955 |
| D. melanogaster: RNAi of ppk: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF03250}attP2 | Bloomington Drosophila Stock Center | BDSC: 29571; FlyBase: FBst0029571 |
| D. melanogaster: RNAi of ppk: P{KK104185}VIE-260B | VDRC | VDRC ID: 108683 FlyBase: FBst0480493 |
| D. melanogaster: RNAi of ppk12: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02027}attP2 | Bloomington Drosophila Stock Center | BDSC: 27092; FlyBase: FBst0027092 |
| D. melanogaster: RNAi of ppk12: P{KK101805}VIE-260B | VDRC | VDRC ID: 105131 FlyBase: FBst0476959 |
| D. melanogaster: RNAi of ppk25: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02434}attP2 | Bloomington Drosophila Stock Center | BDSC: 27088; FlyBase: FBst0027088 |
| D. melanogaster: RNAi of ppk25: P{KK109736}VIE-260B | VDRC | VDRC ID: 101808 FlyBase: FBst0473681 |
| D. melanogaster: RNAi of ppk28: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02153}attP2 | Bloomington Drosophila Stock Center | BDSC: 31878; FlyBase: FBst0031878 |
| D. melanogaster: RNAi of ppk28: P{KK106316}VIE-260B | VDRC | VDRC ID: 100946 FlyBase: FBst0472819 |
| D. melanogaster: RNAi of sei: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF01474}attP2/TM3, Ser[1] | Bloomington Drosophila Stock Center | BDSC: 31681; FlyBase: FBst0031681 |
| D. melanogaster: RNAi of sei: P{KK105733}VIE-260B | VDRC | VDRC ID: 104698 FlyBase: FBst0476547 |
| D. melanogaster: RNAi of Sh: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF01473}attP2/TM3, Ser[1] | Bloomington Drosophila Stock Center | BDSC: 31680; FlyBase: FBst0031680 |
| D. melanogaster: RNAi of Sh: P{KK109112}VIE-260B | VDRC | VDRC ID: 104474 FlyBase: FBst0476332 |
| D. melanogaster: RNAi of Shal: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02154}attP2 | Bloomington Drosophila Stock Center | BDSC: 31879; FlyBase: FBst0031879 |
| D. melanogaster: RNAi of Shaw: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02982}attP2 | Bloomington Drosophila Stock Center | BDSC: 28346; FlyBase: FBst0028346 |
| D. melanogaster: RNAi of Shaw: P{KK108371}VIE-260B | VDRC | VDRC ID: 110589 FlyBase: FBst0482154 |
| D. melanogaster: RNAi of SK: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02571}attP2 | Bloomington Drosophila Stock Center | BDSC: 27238; FlyBase: FBst0027238 |
| D. melanogaster: RNAi of SK: P{KK107699}VIE-260B | VDRC | VDRC ID: 103985 FlyBase: FBst0475843 |
| D. melanogaster: RNAi of SLO2: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF03426}attP2 | Bloomington Drosophila Stock Center | BDSC: 32034; FlyBase: FBst0032034 |
| D. melanogaster: RNAi of stj: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF01825}attP2 | Bloomington Drosophila Stock Center | BDSC: 25807; FlyBase: FBst0025807 |
| D. melanogaster: RNAi of stj: P{KK101267}VIE-260B | VDRC | VDRC ID: 108569 FlyBase: FBst0480379 |
| D. melanogaster: RNAi of Teh2: w[1118]; P{GD3839}v9037 | VDRC | VDRC ID: 9037 FlyBase: FBst0471346 |
| D. melanogaster: RNAi of Teh2: P{KK112449}VIE-260B | VDRC | VDRC ID: 104951 FlyBase: FBst0476779 |
| D. melanogaster: RNAi of Teh4: w[1118]; P{GD3578}v11621/CyO | VDRC | VDRC ID: 11621 FlyBase: FBst0450303 |
| D. melanogaster: RNAi of Teh4: P{KK110985}VIE-260B | VDRC | VDRC ID: 102161 FlyBase: FBst0474030 |
| D. melanogaster: RNAi of trp: y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF01441}attP2 | Bloomington Drosophila Stock Center | BDSC: 31649; FlyBase: FBst0031649 |
| D. melanogaster: RNAi of trp: w[1118]; P{GD372}v1366 | VDRC | VDRC ID: 1366 FlyBase: FBst0451102 |
| D. melanogaster: UAS-pdf on the 2nd Chromosome | Renn et al. (1999) | N/A |
| Software | ||
| ImageJ | Schneider et al. (2012) | https://imagej.nih.gov/ij/ |
| Infostat | Di Rienzo et al. (2018) | https://www.infostat.com.ar/ |
| ClockLab | Actimetrics | https://www.actimetrics.com/products/clocklab/ |
| Rethomics | Geissmann et al. (2019) | https://rethomics.github.io/ |
| R | R Core Team (2014) | https://www.r-project.org/ |
| Micro Manager | Edelstein et al. (2014) | https://micro-manager.org/wiki/Download_Micro-Manager_Latest_Release |
| pClamp 9 | Molecular Devices | https://moleculardevices.app.box.com/s/d93nukl3chbo206t33cw5fpabsph6wh4 |
| Clampfit 10 | Molecular Devices | https://moleculardevices.app.box.com/s/l8h8odzbdikalbje1iwj85x88004f588 |
| Origin 8 | OriginLab | https://www.originlab.com/ |
| GraphPad | Prism8 | https://www.graphpad.com/ |
Table 2.
Positive hits of the ion channel downregulation behavioral screen
| Gene symbol | CG | RNAi info | Channel type | Tau (h) | Rhythm (%) | n | N |
|---|---|---|---|---|---|---|---|
| Pdf,dcr>cac | CG1522 | DRSC 27244 + VDRC KK 104168 | VG Ca++ channel | 24.48 ± 0.21* | 57 ± 11* | 75 | 5 |
| +>cac | 23.79 ± 0.07 | 97 ± 2 | 93 | 5 | |||
| Pdf,dcr>+ | 23.93 ± 0.10 | 95 ± 2 | 104 | 5 | |||
| Pdf,dcr>Ca-α1T | CG15899 | VDRC KK 108827 | VG Ca++ channel | 24.85 ± 0.14* | 30 ± 9* | 70 | 3 |
| +>Ca-α1T | 23.93 ± 011 | 84 ± 10 | 82 | 3 | |||
| Pdf,dcr>+ | 23.82 ± 0.67 | 76 ± 11 | 59 | 3 | |||
| Pdf,dcr>ClC-a | CG1116 | VDRC KK 110394 | VG Cl- channel | 21.78 ± 0.13* and 24.25 ± 0.20 | 4 ± 4* | 37 | 2 |
| +>ClC-a | 24.00 ± 0.01 | 97 ± 3 | 41 | 2 | |||
| Pdf,dcr>+ | 24.17 ± 0.06 | 76 ± 15 | 43 | 2 | |||
| Pdf,dcr>CngA | CG42701 | DRSC 26014 + VDRC KK 101745 | Cyclic-nucleotide G channel | 23.55 ± 0.06 | 65 ± 2* | 27 | 2 |
| +>CngA | 23,73 ± 0,29 | 91 ± 9 | 25 | 2 | |||
| Pdf,dcr>+ | 24,07 ± 0,11 | 98 ± 1 | 37 | 2 | |||
| Pdf,dcr>Ork1 | CG1615 | DRSC 25885 + VDRC KK 104883 | K+ leak channel | 25.00 ± 0.46* | 48 ± 12* | 33 | 2 |
| +>Ork1 | 24.28 ± 0.29 | 100 ± 0 | 34 | 2 | |||
| Pdf,dcr>+ | 24.23 ± 0,05 | 98 ± 1 | 30 | 2 | |||
| Pdf,dcr>Shal | CG9262 | NIG 9262R-3 | VG K+ channel | 24.59 ± 0.11* | 89 ± 6 | 59 | 4 |
| +>Shal | 23.97 ± 0.20 | 97 ± 3 | 39 | 3 | |||
| Pdf,dcr>+ | 24.01 ± 0.05 | 98 ± 2 | 67 | 4 | |||
| Pdf,dcr>tipE | CG1232 | NIG 1232R-3 | VG Na+ auxiliary subunit | 25.73 ± 0.40* | 85 ± 12 | 46 | 3 |
| +>tipE | 23.90 ± 0.01 | 96 ± 4 | 27 | 2 | |||
| Pdf,dcr>+ | 24.12 ± 0.04 | 98 ± 2 | 55 | 3 | |||
| Pdf,dcr>tipE | CG1232 | DRSC 26249 | VG Na+ auxiliary subunit | 25.60 ± 0.38* | 48 ± 16* | 47 | 3 |
| +>tipE | 23.73 ± 0.04 | 96 ± 5 | 30 | 2 | |||
| Pdf,dcr>+ | 24.12 ± 0.04 | 94 ± 4 | 55 | 3 |
This table includes a list of genes that, when downregulated exclusively in LNvs using these particular RNAi constructs, produced statistically significant alterations in free running period and/or percentage of rhythmicity. Values represent the average of mean values of N independent experiments ± SEM, n indicates total number of individuals tested; * indicates statistically significant difference (p < 0.05) after a one-way ANOVA comparing pdf,dicer>RNAi to control genotypes pdf,dicer/+ and RNAi/+. Tukey's test was used for means comparison and Levene's test for checking ANOVA assumption of homogeneity of variance. In the case where information for two RNAi constructs is given, it means that each RNAi on its own did not show significant differences compared with controls, but did show a trend toward an altered phenotype. For that reason, two different RNAis for the same gene were genetically combined to achieve added downregulation strength. In the case of ClC-a, the reduction of rhythmicity was because of the appearance of complex rhythms and not to the deconsolidation of locomotor activity organization; the tau of each component of complex rhythms is given. V: voltage, G: gated, DRSC: Drosophila RNAi Screening Center, VDRC: Vienna Drosophila Resource Center, NIG: National Institute of Genetics. Significant differences (p < 0.05) compared with genetic controls are displayed in boldface.
Table 3.
Ih genetic manipulations disrupt circadian locomotor activity organization
| Genotype | DD analysis |
LD analysis |
||||||
|---|---|---|---|---|---|---|---|---|
| Tau (h) | Rhythm (%) | n | N | MAI | EAI | n | N | |
| pdf,dicer/+ | 24.00 ± 0.07 | 95 ± 3 | 75 | 3 | 0.73 ± 0.02 | 0.88 ±0.02 | 55 | 2 |
| UAS-IhRNAi/+ | 23.71 ± 0.02 | 92 ± 6 | 39 | 0.77 ± 0.03 | 0.80 ± 0.03 | 21 | ||
| pdf,dicer>UAS-IhRNAi | 23.95 ± 0.03 | 74 ± 8* | 40 | 0.67 ± 0.03 | 0.80 ± 0.02 | 23 | ||
| pdfGS/+, RU | 24.59 ± 0.41 | 97 ± 2 | 64 | 3 | 0.64 ± 0.01 | 0.66 ± 0.02* | 76 | 3 |
| UAS-IhRNAi/+, RU | 23.75 ± 0.06 | 100 ± 0 | 41 | 0.72 ± 0.02 | 0.58 ± 0.01 | 50 | ||
| pdfGS>UAS-IhRNAi, RU | 24.81 ± 0.75 | 57 ± 12* | 67 | 0.59 ± 0.01* | 0.60 ± 0.01 | 74 | ||
| control | 24.02 ± 0.05 | 96 ± 2 | 86 | 4 | 0.68 ± 0.03 | 0.84 ± 0.02 | 62 | 2 |
| Ihf01485/+ | 23.87 ± 0.03 | 96 ± 3 | 80 | 0.71 ± 0.04 | 0.89 ± 0.02 | 32 | ||
| Ihf01485 | 23.56 ± 0.05 | 60 ± 3* | 98 | 0.55 ± 0.05 | 0.65 ± 0.03* | 34 | ||
| Ihf03355/+ | 23.86 ± 0.05 | 99 ± 1 | 96 | 0.73 ± 0.03 | 0.92 ± 0.02 | 30 | ||
| Ihf03355 | 23.88 ± 0.08 | 39 ± 12* | 104 | 0.51 ± 0.03* | 0.67 ± 0.03* | 30 | ||
DD analysis (left): the average free running period and percentage of rhythmicity of populations of male flies of the indicated genotypes are shown. Values represent the average of N independent experiments ± SEM, n indicates total number of individuals tested; * indicates statistically significant difference (p < 0.05) after a one-way ANOVA comparing experimental genotypes to genetic controls. UAS-IhRNAi refers to the genetic combination of two UAS-IhRNAi constructs: DRSC 29574 + VDRC KK 110274). In the case of the Ih null mutants, Ihf01485 and Ihf03355, homozygotes were compared with a w1118 control and to heterozygotes (Ihf03355 crossed by w1118). RU refers to the presence of the steroid RU486 (200 μg/ml), the activator of the GS system, in the food media. LD analysis (right): morning anticipation index (MAI) and evening anticipation index (EAI) were calculated for the same genotypes. *Indicates statistically significant difference (p < 0.05) after Kruskal–Wallis statistical analysis with Dunn's multiple comparisons test. Significant differences (p – 0.05) compared with genetic controls are displayed in boldface.
Locomotor behavior analysis
Male flies were entrained to LD during their entire development, and newly eclosed adult males were placed in 65 × 5 mm glass tubes and monitored for activity with infrared detectors and a computerized data collection system (Trikinetics). For experiments involving the GS expression system, newly eclosed adult males were placed in glass tubes containing standard food (supplemented with 200 mg/ml RU486 or vehicle, as indicated) and monitored for activity. Activity was monitored in light-dark conditions (LD) for 3–4 d, followed by constant darkness for at least 9 d [constant dark conditions (DD)1–DD9]. Period and rhythmicity parameters as FFT and power were estimated using ClockLab software (Actimetrics). Flies with a single peak over the significance line (p < 0.05) in a χ2 analysis were scored as rhythmic, which was confirmed by visual inspection of the actograms. For LD anticipatory analysis, the last day before switching to DD was used. Average activity plots at the population level were produced using the Clocklab average activity function for each animal, relativized to its own activity, integrated in 30-min bins and then the population average for each genotype was calculated. Morning anticipation index (MAI) was calculated as follow, the sum of relativized activity from zeitgeber time (ZT)21.5 from the previous day to ZT0 was divided by the sum of relativized activity from ZT19 from the previous day to ZT0 for each animal. Since data were not normally distributed a non-parametric ANOVA analysis, Kruskal–Wallis test followed by Dunn's multiple was used to test statistically significant differences. An equivalent procedure was performed for the evening anticipation index (EAI) using data from ZT7 to ZT12.
Sleep behavior analysis
Female flies were socially housed in vials from eclosion at 25°C under LD until they were 4–6 d old and afterward transferred to 65 × 5 mm glass tubes (Trikinetics) containing normal cornmeal food. Tubes were loaded onto Drosophila activity monitors (DAM) and locomotor activity was assessed using the DAM system under LD. Sleep data were calculated on the second day after fly loading into tubes to allow them to recover from anesthesia and to acclimate to the new environment. For experiments using the TARGET system (McGuire et al., 2004b) flies were raised at 21°C, socially housed in vials from eclosion until they were 6 d old and afterward transferred to 65 × 5 mm glass tubes. Monitors were kept for 2 d at 21°C to measure sleep under the restrictive temperature at which the RNAi is not expressed (which in all cases produced no effect), and then the incubator temperature was raised to 30°C for two more days to allow RNAi expression, always under LD. Sleep data were calculated on the second day at 30°C. The DAM System binning time was set to 1 min. Sleep was defined as no movement for 5 min (Hendricks et al., 2000; Shaw et al., 2000). Rethomics, a collection of packages running in R language (Geissmann et al., 2019), was used to infer sleep from locomotor activity data, to build graphs of sleep for 30 min as a function of the time of day, to get measurements of total sleep, day sleep, night sleep, sleep bout duration, sleep bout number, latencies to lights on and off and to get an activity index (defined as the average activity counts in the active minutes) of each individual fly. Behavioral experiments were conducted at least two to three times, with 15–30 individuals per genotype.
Electrophysiology
Three to 7-d-old female flies were anesthetized with a brief incubation of the vial on ice, brain dissection was performed on external recording solution which consisted of the following: 101 mm NaCl, 3 mm KCl, 1 mm CaCl2, 4 mm MgCl2, 1.25 mm NaH2PO4, 5 mm glucose, and 20.7 mm NaHCO3, pH 7.2, with an osmolarity of 250 mmol/kg (based on solution used by Cao and Nitabach, 2008). After removal of the proboscis, air sacks and head cuticle, the brain was routinely glued ventral side up to a Sylgard-coated coverslip using a few microliters of tissue adhesive 3 M Vetbond. The time from anesthesia to the establishment of the first successful recording was ∼15–19 min spent as following: 5–6 min for the dissection, 4–5 min for the protease treatment to remove the brain's superficial glia, and 6–8 min to fill and load the recording electrode onto the pipette holder, approach the cell, achieve the gigaohm seal and open the cell into whole-cell configuration to start recording. LNvs were visualized by red fluorescence in pdf-RFP using a Leica DM LFS upright microscope with 63× water-immersion lens and TK-LED illumination system (TOLKET S.R.L.). Once the fluorescent cells were identified, cells were visualized under IR-DIC using a Hamamatsu ORCA-ER camera and Micro Manager software. lLNvs were distinguished from sLNvs by their size and anatomic position. To allow the access of the recording electrode, the superficial glia directly adjacent to LNvs somas was locally digested with Protease XIV solution (10 mg/ml, Sigma-Aldrich P5147) dissolved in external recording solution. This was achieved using a large opened tip (∼20 µm) glass capillary (pulled from glass of the type GC100TF-10; Harvard Apparatus) and gentle massage of the superficial glia with mouth suction to render the underling cell bodies accessible for the recording electrode with minimum disruption of the neuronal circuits. After this procedure, protease solution was quickly washed by perfusion of external solution. Recordings were performed using thick-walled borosilicate glass pipettes (GC100F-10; Harvard Apparatus) pulled to 6–7 MΩ using a horizontal puller P-1000 (Sutter Instruments) and fire polished to 9–12 MΩ. Recordings were made using an Axopatch 200B amplifier controlled by pClamp 9.0 software via a Digidata 1322A analog-to-digital converter (Molecular Devices). Recording pipettes were filled with internal solution containing the following: 102 mm potassium gluconate, 17 mm NaCl, 0.085 mm CaCl2, 0.94 mm EGTA, and 8.5 mm HEPES, pH 7.2 with an osmolarity of 235 mmol/kg (based on the solution employed by Cao and Nitabach, 2008). Cell-attached configuration was achieved by gentle suction and recordings were performed in voltage-clamp mode with no hold. For whole-cell configuration, gigaohm seals were accomplished using minimal suction followed by break-in into whole-cell configuration using gentle suction in voltage-clamp mode with a holding voltage of –60 mV. Gain of the amplifier was set to 1 during recordings and a 5-kHz lowpass Bessel filter was applied throughout. Spontaneous firing was recorded in current clamp (I = 0) mode. Analysis of traces was conducted using Clampfit 10.4 software. Bursting frequency was calculated as the number of bursts in a minute of recording. For comparisons, all recordings were quantified at the same time postdissection as specified in the text and figure legends. For Action Potential (AP) firing rate calculation, the event detection tool of Clampfit 10.4 was used. In many cases, we were able to see the two different AP sizes reported previously (Cao and Nitabach, 2008); however, for AP firing rate calculation, only the large APs were taken into account. Traces shown in figures were filtered offline using a lowpass boxcar filter with smoothing points set to 9. Perfusion of external saline in the recording chamber was achieved using a peristaltic pump (MasterFlex C/L). All recordings were performed during the light phase, between ZT1 and ZT10.
Immunofluorescence detection
Heads of male and female flies were cut at ZT2 and ZT14, fixed in paraformaldehyde 4% in 0.1 m PB for 35–45 min at room temperature (RT) and brains were dissected afterward, washed five times in PBS-Triton X-100 0.1%, blocked with 7% normal goat serum for 2 h at RT and incubated with primary antibody (see antibodies information in Table 1), ON at 4°C. After five 15-min washes in PBS-Triton X-100 0.1%, brains were incubated with the secondary antibody. Confocal images were obtained in a Zeiss 710 Confocal Microscope or Pascal Confocal Microscope. All the photographs within the same experiment were taken with the same confocal parameters. In the pdf overexpression experiments, data were relativized to the average of intensities for each experiment because two different microscopes were used. The acquisition of sLNv soma images required different confocal parameters (laser intensity, gain, zoom).
PDF quantitation
For the quantitation of PDF intensity at the sLNv projections, we assembled a maximum intensity z-stack that contains the whole projection (approximate 10 images) and constructed a threshold image to create a ROI for measure immunoreactivity intensity using ImageJ (NIH). Data were analyzed with InfoStat software (Universidad Nacional de Córdoba, Argentina) and GraphPad. For quantitation of PDF intensity at the sLNv somas, we used a unique 1-µm image per cell, which was the one where the PDF cytoplasm immunoreactivity signal could be clearly differentiated from the empty nucleus. The draw tool from ImageJ (NIH) enabled to measure only the PDF signal at the cytoplasm, and this procedure was repeated for each cell (three to four) in each brain (only one brain hemisphere). Background intensity was subtracted for each brain and average intensity was calculated. Data were normalized using the average intensity for the whole population of brains of the experiment. This way of quantifying PDF in the sLNvs somas allowed a more precise assessment of neuropeptide levels and it may be the reason why we were able to detect circadian cycling of PDF levels, unlike previous reports that were unable to detect them (Park et al., 2000). Statistics analysis was done using the GraphPad program, after testing data normality one-way ANOVA and Sidak's multiple comparisons tests were performed to determine time-of-day genotype differences.
Analysis of structural plasticity
To assess the degree of complexity within the sLNvs dorsal projections we performed immunofluorescence against a membrane version of GFP. The maximum intensity z-stack image was transformed into a threshold image and Sholl analysis was performed with ImageJ (NIH) software. Each picture was corroborated by visual inspection to confirm the number of crosses in every 10-µm concentric Sholl ring. Data were analyzed by means of InfoStat software (Universidad Nacional de Córdoba).
Statistical analysis
The following statistical analyses were used in this study: one-way ANOVA and two-way ANOVA with post hoc Tukey's HSD test for multiple comparisons of parametric data, and non-parametric Kruskal–Wallis statistical analysis with multiple comparisons (p adjustment method = BH) as specified in figure legends. Parametric tests were used when data were normally distributed and showed homogeneity of variance, tested by Kolmogorov–Smirnov test and Levene's test, respectively. Sidak's and Dunn's multiple comparisons tests were performed after parametric and non-parametric ANOVA when GraphPad software was used. Sleep data tended to not show a normal distribution, hence non-parametric statistics were used. Statistical analyses were performed using Infostat for circadian rhythmicity and immunofluorescence experiments, R-based Rethomics package for sleep data and Origin software for electrophysiological parameters; p < 0.05 was considered statistically significant. Throughout the manuscript, n represents the total number of measurements compared in each experimental group (behavior of an individual, brain morphology, or neuronal recordings, depending of the experiment), and N represents the number of independent times an experiment was repeated. Boxes in box and whisker plots for sleep and electrophysiological parameters represent the median and interquartile range (the distance between the first and third quartiles). In all tables, parameters represent the mean value ± SEM. In dot plots for circadian power and tau and in fluorescence and structural plasticity, quantification lines represent the mean value; error bars depict the SEM.
Results
LNvs targeted behavioral screen for ion channels
To shed light onto how LNvs achieve the physiological properties that allow them to play a key role in the circadian organization of locomotor activity, we performed an ion channel downregulation behavioral screen. The pdf-Gal4 driver, in the presence of UAS-dicer2 (from here on pdf,dicer) was used to drive expression of UAS-RNAis to knock down the expression of candidate ion conductances solely in LNvs. The RNAis were chosen to target ion channel genes, genes coding for ion channel auxiliary subunits or genes coding for ion channel transporters which had not been reported to be involved in LNvs-driven circadian phenotypes before. The locomotor activity of pdf,dicer>RNAi male flies was recorded using DAM (Trikinetics) for 9 d in DD after 3 d of LD entrainment. Each RNAi was initially tested once and, in the case of showing a differential phenotype in DD, corresponding to either a change of circadian period or deconsolidation of locomotor activity, experiments were repeated. In some cases, a non-significant trend toward a phenotype was detected; and therefore, two RNAis that targeted different regions of the same gene were genetically combined to achieve a more potent downregulation. Tables 2, 3 show the positive hits of our screen, revealing novel ion channels or ion channel auxiliary subunits likely to play roles in LNvs circadian function, namely: cacophony (cac, CG1522), Ca-α1T (Ca2+-channel protein α1 subunit T, CG15899), ClC-a (Chloride channel-a, CG31116), CngA (Cyclic nucleotide-gated ion channel subunit A, CG42701), Ih (Ih channel, CG8585), Ork1 (Open rectifier K+ channel 1, CG1615), Shal (Shaker cognate l, CG9262), and tipE (temperature-induced paralytic E, CG1232). The RNAis that did not show altered circadian phenotypes in our screen are listed in Table 4. In total, 70 RNAis aimed at 36 different genes were tested.
Table 4.
Negative hits of the ion channel downregulation behavioral screen
| Gene | CG | RNAi info | Channel type | n | N |
|---|---|---|---|---|---|
| Atpα | CG5670 | DRSC 28073 | Na+/K+ ATPase α subunit | 16 | 1 |
| Atpα | CG5670 | VDRC KK 100619 | Na+/K+ ATPase α subunit | 45 | 3 |
| Calx | CG5685 | DRSC 28306 | Ca++ Na+ antiporter | 8 | 1 |
| Calx | CG5685 | VDRC KK 104789 | Ca++ Na+ antiporter | 13 | 1 |
| Ca-α1D | CG4894 | DRSC 25830 | VG Ca++ channel | 16 | 1 |
| Ca-α1D | CG4894 | VDRC GD 51490 | VG Ca++ channel | 16 | 1 |
| Ca-α1T | CG15899 | VDRC GD 31963 | VG Ca++ channel | 70 | 3 |
| Ca-α1T | CG15899 | DRSC 26251 | VG Ca++ channel | 50 | 2 |
| eag | CG10952 | DRSC 31678 | VG cation channel | 16 | 1 |
| eag | CG10952 | VDRC KK 100260 | VG cation channel | 16 | 1 |
| Hk | CG43388 | DRSC 28330 | VG K+ channel β subunit | 16 | 1 |
| Hk | CG43388 | VDRC KK 101402 | VG K+ channel β subunit | 14 | 1 |
| inx2 | CG4590 | DRSC 29306 | Gap junction channel | 13 | 1 |
| inx2 | CG4590 | VDRC KK 102194 | Gap junction channel | 9 | 1 |
| Ir | CG6747 | DRSC 25823 | VG K+ channel | 32 | 2 |
| Ir | CG6747 | VDRC KK 107389 | VG K+ channel | 15 | 1 |
| Irk2 | CG4370 | DRSC 25820 | Inwardly rectifying K+ channel | 31 | 2 |
| Irk2 | CG4370 | VDRC GD 4341 | Inwardly rectifying K+ channel | 13 | 1 |
| KCNQ | CG33135 | DRSC 27252 | VG K+ channel | 65 | 4 |
| KCNQ | CG33135 | VDRC KK 106655 | VG K+ channel | 15 | 1 |
| Ncc69 | CG4357 | DRSC 28682 | Na+ K+ Cl- symporter | 16 | 1 |
| Ncc69 | CG4357 | VDRC KK 106499 | Na+ K+ Cl- symporter | 16 | 1 |
| Nckx30C | CG18660 | DRSC 27246 | Na+ K+ Ca++ exchanger | 15 | 1 |
| nrv1 | CG9258 | VDRC GD 46542 | Na+/K+ ATPase β subunit | 14 | 1 |
| nrv1 | CG9258 | VDRC KK 103702 | Na+/K+ ATPase β subunit | 15 | 1 |
| nrv2 | CG9261 | DRSC 28666 | Na+/K+ ATPase β subunit | 15 | 1 |
| nrv2 | CG9261 | VDRC GD 2660 | Na+/K+ ATPase β subunit | 23 | 2 |
| para | CG9907 | VDRC GD 6131 | VG Na+ channel | 16 | 1 |
| para | CG9907 | VDRC KK 104775 | VG Na+ channel | 28 | 2 |
| picot | CG8098 | DRSC 25920 | Phosphate Na+ symporter | 15 | 1 |
| picot | CG8098 | VDRC KK 101082 | Phosphate Na+ symporter | 14 | 1 |
| ppk | CG3478 | DRSC 29571 | Amiloride-sensitive Na+ channel | 16 | 1 |
| ppk | CG3478 | VDRC KK 108683 | Amiloride-sensitive Na+ channel | 34 | 3 |
| ppk12 | CG10972 | DRSC 27092 | Amiloride-sensitive Na+ channel | 16 | 1 |
| ppk12 | CG10972 | VDRC KK 105131 | Amiloride-sensitive Na+ channel | 15 | 1 |
| ppk25 | CG33349 | DRSC 27088 | Amiloride-sensitive Na+ channel | 16 | 1 |
| ppk25 | CG33349 | VDRC KK 101808 | Amiloride-sensitive Na+ channel | 16 | 1 |
| ppk28 | CG4805 | DRSC 31878 | Amiloride-sensitive Na+ channel | 16 | 1 |
| ppk28 | CG4805 | VDRC KK 100946 | Amiloride-sensitive Na+ channel | 12 | 1 |
| sei | CG3182 | DRSC 31681 | VG K+ channel | 15 | 1 |
| sei | CG3182 | VDRC KK 104698 | VG K+ channel | 16 | 1 |
| Sh | CG12348 | DRSC 31680 | VG K+ channel | 16 | 1 |
| Sh | CG12348 | VDRC KK 104474 | VG K+ channel | 31 | 2 |
| Shal | CG9262 | DRSC 31879 | VG K+ channel | 15 | 1 |
| Shaw | CG2822 | DRSC 28346 | VG K+ channel | 16 | 1 |
| Shaw | CG2822 | VDRC KK 110589 | VG K+ channel | 16 | 1 |
| SK | CG10706 | DRSC 27238 | Ca++-activated K+ channel | 16 | 1 |
| SK | CG10706 | VDRC KK 103985 | Ca++-activated K+ channel | 16 | 1 |
| SLO2 | CG42732 | DRSC 32034 | Na+ activated K+ channel | 16 | 1 |
| stj | CG12295 | DRSC 25807 | VG Ca++ channel | 15 | 1 |
| stj | CG12295 | VDRC KK 108569 | VG Ca++ channel | 15 | 1 |
| Teh2 | CG15004 | VDRC GD 9037 | VG Na+ auxiliary subunit | 16 | 1 |
| Teh2 | CG15004 | VDRC KK 104951 | VG Na+ auxiliary subunit | 16 | 1 |
| Teh4 | CG15003 | VDRC GD 11621 | VG Na+ auxiliary subunit | 13 | 1 |
| Teh4 | CG15003 | VDRC KK 102161 | VG Na+ auxiliary subunit | 16 | 1 |
| trp | CG7875 | DRSC 31649 | Light-activated Ca++ channel | 10 | 1 |
| trp | CG7875 | VDRC GD 1366 | Light-activated Ca++ channel | 14 | 1 |
This table includes the list of UAS-RNAi transgenic lines that, when driven exclusively in LNvs, did not produced statistically significant alterations in free running period and/or percentage of rhythmicity compared with pdf,dicer/+ control genotype (after Student's t test analysis). N indicates number of independent experiments performed; n indicates number of individuals tested. The appearance of a gene in this table suggests that it may not be involved in the circadian function of LNvs. However, most of these RNAi constructs have not been individually tested for their actual performance on ion channel knock-down. Moreover, it should be noticed that for some genes, such as Shal and Ca-α1T, one RNAi construct was able to affect behavior, while others were not. Further investigations are necessary to determine the roles of these channels in LNvs function. Besides the efficiency of the particular RNAi transgenic line, another phenomenon that should be taken into account is that, in some cases, a homeostatic compensation of ion channel downregulation might have taken place. For instance, it is surprising that targeting the gene coding for the only classical voltage-gated sodium channel paralytic (para) in LNvs has not resulted in a behavioral phenotype. Most likely, this genetic manipulation has produced compensation, as it has been reported to happen for such an important and therefore highly regulated ion conductance (Lin and Baines, 2015). Interestingly, downregulation of para accessory subunit tipE does affect circadian behavior (see Table 2), indicating that less compensatory mechanisms may be in place to counterbalance such genetic manipulation, and that affecting para in this indirect way is probably having a detrimental effect on LNvs ability to fire action potentials. For all these reasons, this table only provides the information that the specific RNAi transgenic lines shown, in the particular conditions we have used, are not able to affect circadian behavior when driven in LNvs. Further analysis is necessary to make stronger statements in all cases. V: voltage, G: gated, DRSC: Drosophila RNAi Screening Center, VDRC: Vienna Drosophila Resource Center.
Although all of the positive hits of our behavioral screen are worth of further assessment, we focused our attention on the hyperpolarization-activated cation current Ih. Little is known about this channel in Drosophila, but its homologues in mammals have been implicated in diverse functions such as the generation of pacemaker potentials and the determination of neuronal excitability, among others (Luthi and McCormick, 1998). RNAi-mediated downregulation of Ih in LNvs produced a subtle but consistent decrease in locomotor rhythmicity without altering free running period (Table 3; Fig. 1A).
Figure 1.
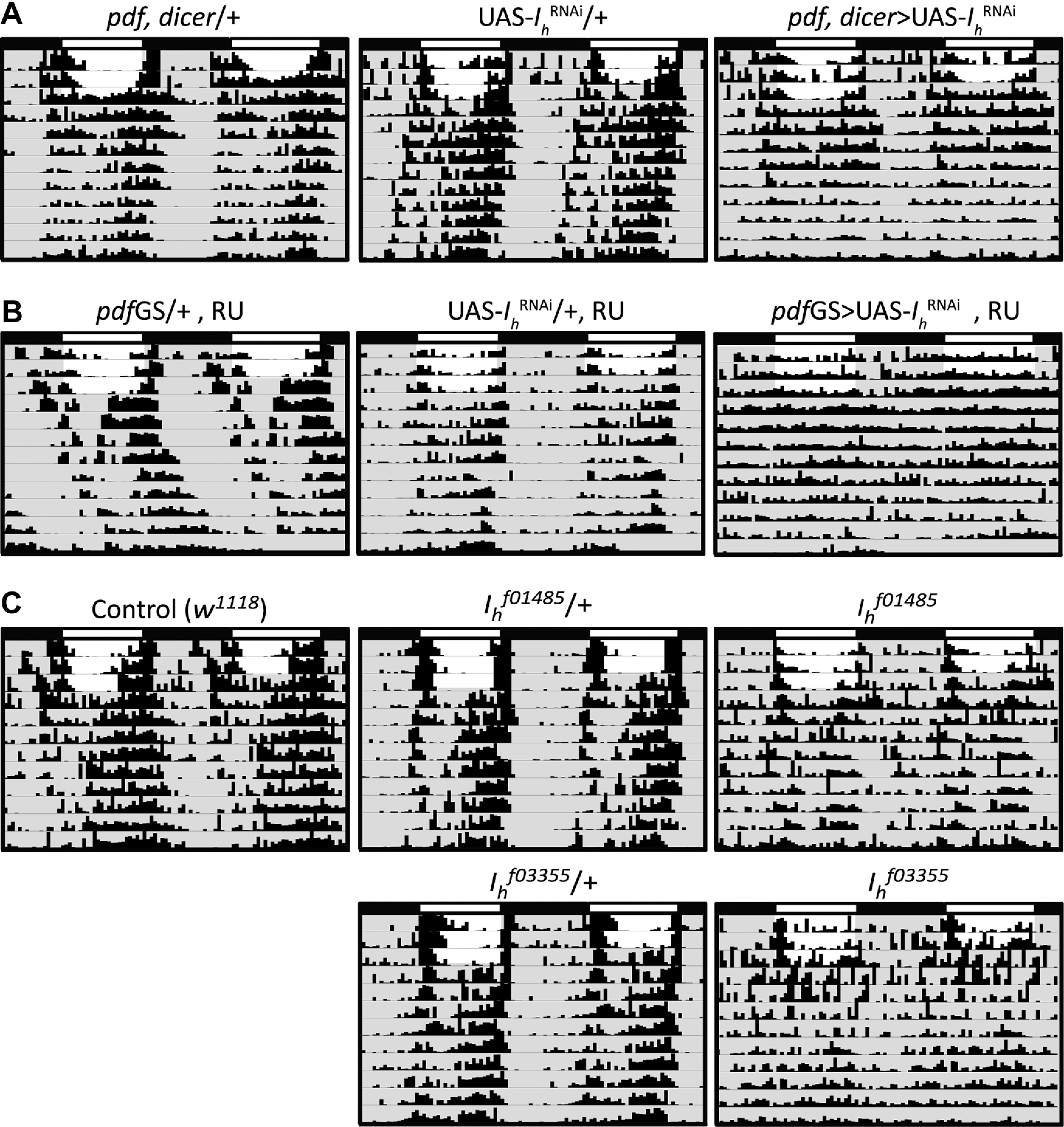
Ih genetic manipulations disrupt circadian locomotor activity organization. Representative double-plotted actograms of the different Ih genetic manipulations tested. A, LNvs constitutive downregulation of Ih using pdf,dicer and UAS-IhRNAi (in all cases, UAS-IhRNAi refers to the genetic combination of two UAS-IhRNAi constructs: DRSC 29574 + VDRC KK 110274) and genetic controls. B, LNvs acute downregulation of Ih using pdfGS and UAS-IhRNAi and genetic controls. RU refers to the presence of the steroid RU486, the activator of the GS system, in the food media. C, Homozygote Ih null mutants, Ihf01485 and Ihf03355, and controls (w1118 and heterozygote mutants, crossed by w1118). In the case of the experimental genotypes an actogram of an arrhythmic individual is shown, different genetic manipulations varied in the degree of arrhythmicity (see Table 3). No statistically significant alterations in free running period were found for these genetic manipulations.
The pdf-Gal4 driver used for the ion channel behavioral screen is active throughout development. Therefore, to dissect whether the behavioral phenotype observed was because of a developmental defect or to a postdevelopmental functional role, we downregulated Ih expression in LNvs in an adult-specific fashion using the GS-inducible system (Osterwalder et al., 2001). When the previously reported pdfGS driver (Depetris-Chauvin et al., 2011) was used to knock down Ih adult specifically in LNvs, we also observed a decrease in circadian rhythmicity (Table 3; Fig. 1B), indicating that the Ih channel is necessary postdevelopmentally in LNvs for the maintenance of circadian function. As a complementary approach, we assessed the circadian behavior of the Ih mutants, Ihf01485 and Ihf03355, which correspond to two independent transposon insertions previously characterized as null mutants because of the absence of Ih mRNA detectable by RT-PCR (Chen and Wang, 2012; Hu et al., 2015) and the lack of Ih protein by Western blotting (Hu et al., 2015). As expected, these mutants also showed reduced rhythmicity under free running conditions (Table 3; Fig. 1C). We found Ih mutants to be less rhythmic than any tissue-specific knock-down (LNvs-specific manipulations). This suggests a requirement for Ih not only in LNvs but also in other neuronal types for the rhythmic organization of locomotor activity under free running conditions. Another possibility, which does not exclude the one proposed, is that null Ih mutations simply produce more robust phenotypes than the RNAi mediated knock-down, which are normally not 100% efficient. All genetic manipulations did not, in any case, produce changes in free running period (Table 3). To assess whether Ih is important for circadian function also under entrained conditions, we analyzed morning and evening anticipatory behavior. Consistent with the strength of the phenotypes observed in DD, we detected a failure in both, morning and evening anticipation in Ihf03355 mutants, which is less pronounced in the Ihf01485 mutants and the adult-specific downregulation of Ih; no effects were detected under Ih constitutive knock-down, suggesting a less effective Ih downregulation by this genetic manipulation (Table 3). Taken together, these results suggest that the Ih channel contributes to define the firing properties of neurons controlling circadian behavior.
Ih is necessary for high-frequency bursting firing of LNvs
One of the main reasons why we decided to select Ih as the ion channel for in depth analysis is the association of Ih with the organization of action potential firing in bursts. It has been reported, mainly from mammalian thalamic relay and inferior olivary nucleus neurons, that a combination of a hyperpolarization-activated cation current such as Ih, together with a low-voltage activated T-type calcium current (a channel type also uncovered by our screen; Table 2), could mediate a bursting firing mode (Luthi and McCormick, 1998). This is because of Ih particular biophysical properties, which opens on hyperpolarization but carries a depolarizing current (mainly because of the influx of Na+). This current takes the membrane potential to the activation threshold of the T-type voltage-gated Ca2+ channel that depolarizes the membrane up to the action potential firing threshold, opening the classical voltage-gated Na+ channels. Because Ih is slow to close and does not inactivate, the membrane stays in a depolarized state for longer, generating a burst of action potentials. Once Ih closes, the classical voltage-gated K+ channels that repolarize the membrane, together with the leak K+ channels, produce the after-hyperpolarization that kick starts the following burst, activating Ih again (Luthi and McCormick, 1998).
Another relevant observation is that firing in bursts is an effective way of releasing neuropeptides, which are stored in dense core vesicles (DCVs). In contrast to small clear vesicles containing classical fast neurotransmitters, neuropeptide-filled DVCs require a larger amount of Ca2+ entering the cell to reach the more distantly located DVCs at a concentration that would allow the activation of the fusion and release mechanism (for review, see van den Pol, 2012; Nusbaum et al., 2017). Since lLNvs have been described to fire action potentials in a bursting mode (Cao and Nitabach, 2008; Sheeba et al., 2008b) and also to be neuropeptide-releasing neurons (Helfrich-Forster, 1995; Renn et al., 1999), the hypothesis we formulated is that Ih participates in the active bursting firing mode of LNvs and plays a role in the release of PDF.
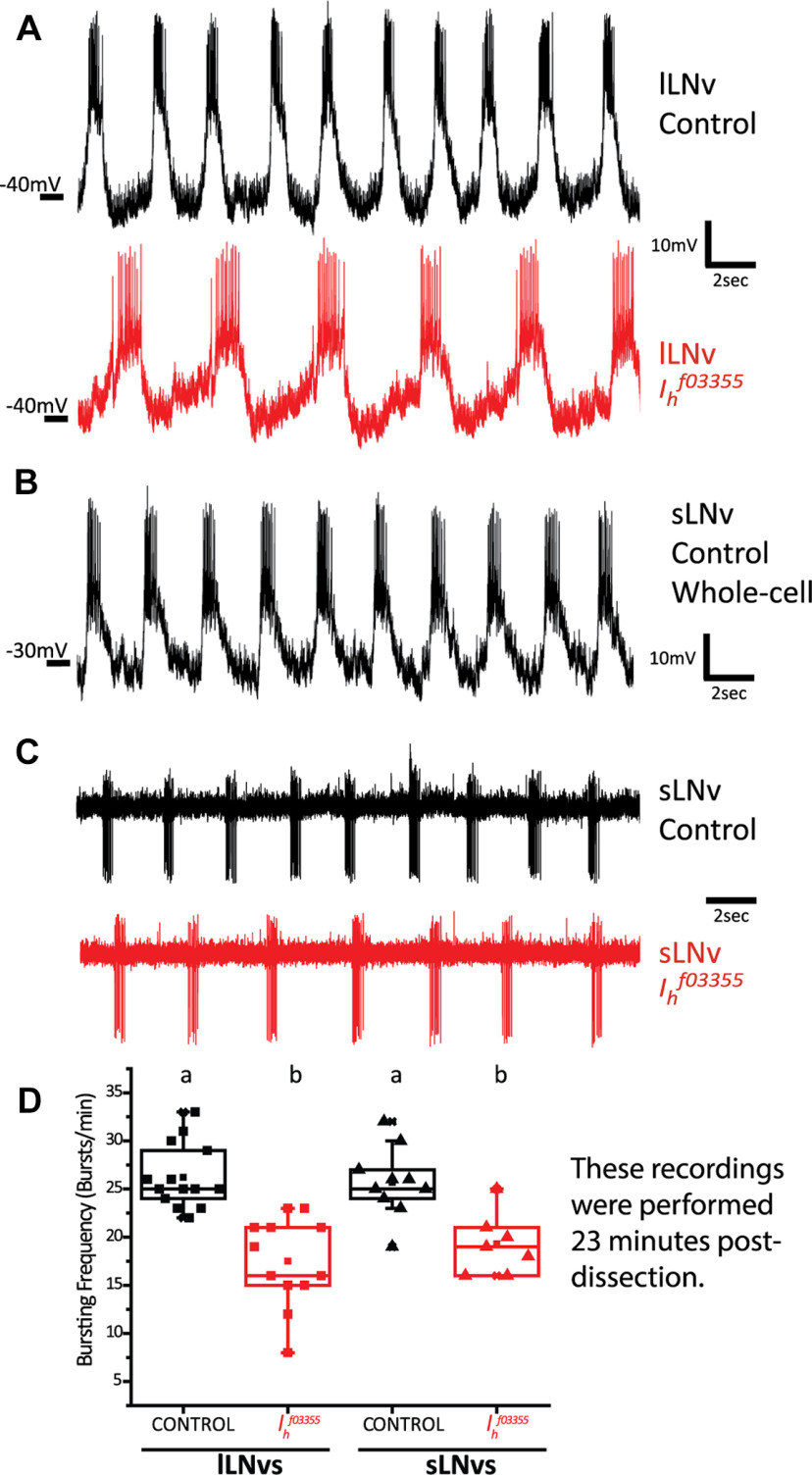
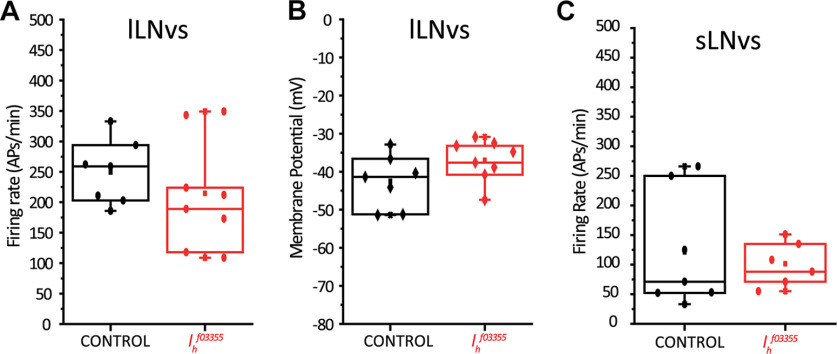
We first tested our hypothesis in the lLNvs, which have effectively been shown to be bursting neurons (Cao and Nitabach, 2008; Sheeba et al., 2008b). We performed ex vivo whole-cell current clamp recordings of control pdf-RFP (expressing a red fluorofore in the LNvs thus enabling the identification of the two neuronal types because of the difference in the size of their soma) lLNvs and compared their bursting frequency to Ihf03355 homozygote mutants. Since we have reported that lLNv bursting frequency also depends on synaptic inputs that are disrupted during the dissection protocol (Muraro and Ceriani, 2015) we compared the bursting frequency of control and Ihf03355 mutant lLNvs at exactly the same time postdissection (23 min). Figure 2A,D shows that although lLNvs from Ihf03355 homozygote mutants can still organize their action potential firing in bursts, they do so at a statistically significant lower frequency [mean bursting frequency ± SEM (bursts/minute) are lLNvsCONTROL = 26.2 ± 0.9 and lLNvsIhf03355 = 17.5 ± 1.4]. Other parameters, such as overall firing frequency and membrane potential, were not significantly affected in Ihf03355 mutants (Fig. 3A and B). The frequency of bursts is reduced without a significant decrease of the firing rate in the mutants; as a result, an increased number of spikes per burst is clearly visible (Fig. 2A).
Figure 2.
Ih is important for high-frequency bursting of LNvs. A, Representative traces of whole-cell patch clamp recordings of lLNvs of control (pdf-RFP, top) and Ih homozygote mutant genotypes (Ihf03355; pdf-RFP, bottom). B, Representative trace of a recording of a sLNv control (pdf-RFP) in whole-cell patch clamp configuration. C, Representative traces of cell-attached recordings of sLNvs of control (pdf-RFP, top) and Ih homozygote mutant genotypes (Ihf03355; pdf-RFP, bottom). D, Box plot showing the median and interquartile range of the bursting frequency quantification of lLNvs and sLNvs of control (pdf-RFP) and Ih homozygote mutant genotypes (Ihf03355; pdf-RFP). All quantifications were done at exactly 23 min postdissection. Different letters indicate significant differences (p < 0.05) after a one-way ANOVA with Tukey's test for means comparisons. n: lLNvsCONTROL = 14, lLNvsIhf03355 = 12, sLNvsCONTROL = 10, sLNvsIhf03355 = 7.
Figure 3.
Mutation of Ih does not significantly affect other electrophysiological parameters of LNvs. A, No statistical significant differences were found in action potential firing rate of lLNvs when comparing control (pdf-RFP) and Ih homozygote mutant genotypes (Ihf03355; pdf-RFP). B, No statistical significant differences were found in membrane potential (measured as the trough between bursts) of lLNvs when comparing control (pdf-RFP) and Ih homozygote mutant genotypes (Ihf03355; pdf-RFP). C, No statistical significant differences were found in action potential firing rate of sLNvs when comparing control (pdf-RFP) and Ih homozygote mutant genotypes (Ihf03355; pdf-RFP). Membrane potential was not quantified in sLNvs as recordings were made in cell-attached configuration, and it is not possible to measure this parameter under this configuration. All quantifications were done at exactly 23 min postdissection. In all cases, p > 0.05 after Student's t test. n: lLNvsCONTROL = 7, lLNvsIhf03355 = 8, sLNvsCONTROL = 7, sLNvsIhf03355 = 6.
Next, we tested our hypothesis in the sLNvs. Information regarding sLNv electrophysiological properties is scarce (Cao and Nitabach, 2008; Li et al., 2018), probably because of the technical challenge that their small soma size represents. However, given the important role that sLNvs play in the control of circadian behavior, we analyzed their firing properties in detail. We report here that the sLNvs also fire action potentials organized in bursts (Fig. 2B). Obtaining a large amount of recordings in whole-cell configuration was a difficult task to achieve; thus, we recorded action potential firing rate and bursting frequency in a cell-attached configuration of the sLNvs of control (pdf-RFP) and Ihf03355 homozygote mutants. We found that, similarly to lLNVs, sLNvs show a decreased bursting frequency in the absence of Ih [mean bursting frequency ± SEM (bursts/minute) are sLNvsCONTROL = 25.7 ± 1.2 and sLNvsIhf03355 = 19.3 ± 1.3; Fig. 2C,D], without significantly affecting overall firing frequency (Fig. 3C).
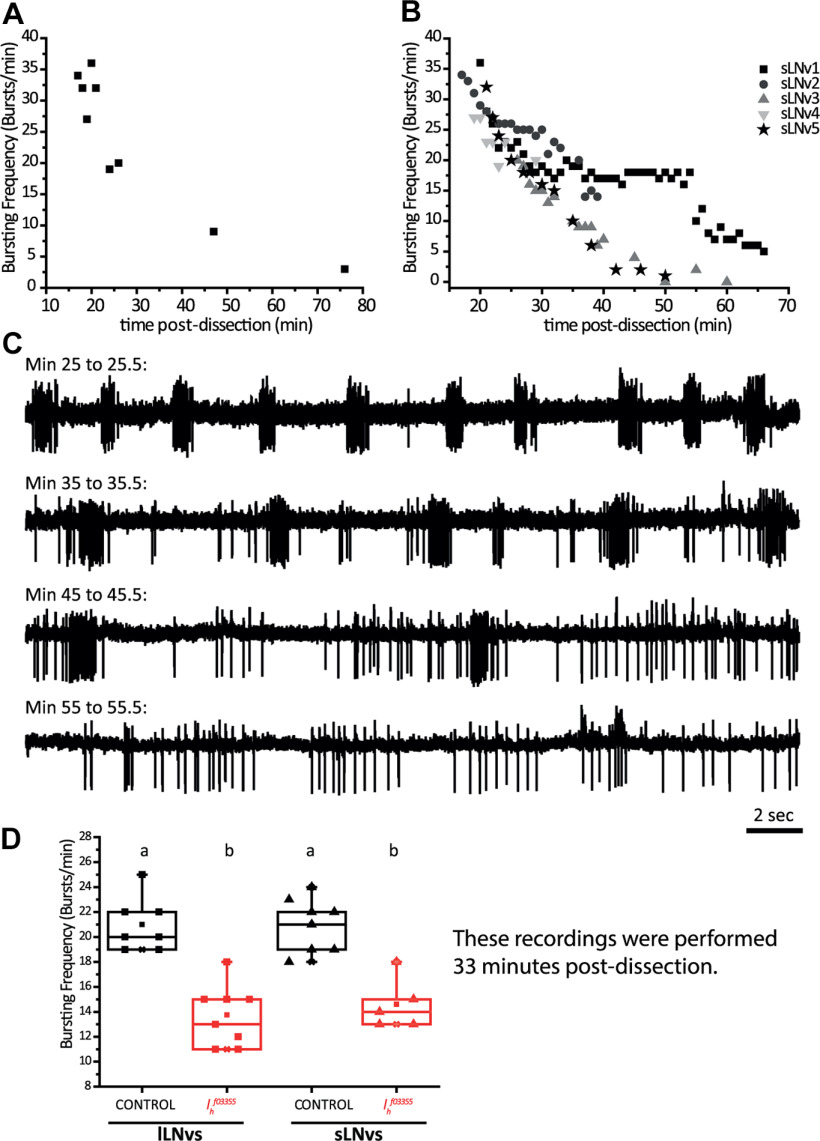
A feature that should be remarked is that both types of LNvs display equivalent basal bursting frequencies (Fig. 2D), suggesting that this parameter depends on common synaptic inputs and/or shared intrinsic mechanisms. We have previously reported that lLNvs bursting frequency relies to some extent on synaptic inputs coming from the visual neuropiles, which indirectly involve L2 lamina neurons and the neurotransmitter acetylcholine (Muraro and Ceriani, 2015). The dependence of lLNv bursting on these synaptic inputs is illustrated by the fact that this parameter decays as a function of the time elapsed since brain dissection that removes the retina (Muraro and Ceriani, 2015). We found that sLNv bursting frequency also decays with time ex vivo, which can be seen both at the population level (Fig. 4A) and also in individual cells (Fig. 4B,C), suggesting that both types of LNvs depend on synaptic inputs which are gradually lost after dissection. Alternatively, it might be that lLNvs rely on visual circuit inputs to burst, and sLNv bursting depends on lLNv bursting. Certainly, the neuronal processes of lLNvs are better localized, spanning all over the optic lobes, to integrate visual information. However, the sLNvs have been shown to receive direct input from the Hofbauer–Buchner (HB) eyelet extraretinal organ (Schlichting et al., 2016), whose integrity may also be compromised during dissection. Whether sLNvs and lLNvs rely on similar or different synaptic inputs to support bursting frequency, or whether one LNv group depends on the other to detect synaptic information from visual organs, will require further investigation.
Figure 4.
sLNvs bursting depends on synaptic inputs. As has been demonstrated before for lLNvs (Muraro and Ceriani, 2015), we show here that sLNvs bursting frequency also decays as a function of the time ex vivo. A, The number of bursts in the initial minute of recording of nine individual control (pdf-RFP) sLNvs recorded at different times postdissection is shown. For the late points the preparation was left in the chamber on purpose before establishing the recording. B, Shows the bursting frequency of five individual control (pdf-RFP) sLNvs where the recordings were long enough to appreciate the decay in this parameter as a function of time postdissection not only as a population as in A, but as individual cells. C, Shows 30-s windows of cell-attached recording of a representative sLNv (sLNv3 in B) at different times postdissection. From top to bottom, the 30 s starting at 25, 35, 45, and 55 min postdissection are shown. At the beginning of the recording, all action potentials are organized in bursts. As time passes, action potentials become less organized in bursts, going through a phase of bursting-tonic firing and becoming purely tonic toward the end. This figure shows that the fact that in A, B the neurons have a tendency toward the cero bursting frequency does not mean that the neurons are not firing, but that they are doing so in a tonic mode. D, Box plot showing the median and interquartile range of the bursting frequency quantification of lLNvs and sLNvs of control (pdf-RFP) and Ih homozygote mutant genotypes (Ihf03355; pdf-RFP), these quantifications were done at exactly 33 min postdissection. Different letters indicate significant differences (p < 0.05) after a one-way ANOVA with Tukey's test for means comparisons. n: lLNvsCONTROL = 7, lLNvsIhf03355 = 8, sLNvsCONTROL = 8, sLNvsIhf03355 = 5.
We also compared bursting frequency in the same recordings but at a different time postdissection; as expected, this analysis also showed that both lLNvs and sLNvs present equivalent bursting frequency, and the lack of Ih produces a significant reduction of this parameter, which is of the same magnitude in the two LNv groups [mean bursting frequency ± SEM (bursts/minute) are as following lLNvsCONTROL = 21.0 ± 0.9, lLNvsIhf03355 = 13.8 ± 0.9, sLNvsCONTROL = 21.1 ± 1.5, sLNvsIhf03355 = 14.6 ± 0.9; compare Figs. 4D, 2D]. Although we cannot be certain of the effects of Ih over the bursting frequency of LNvs in an intact animal, our ex vivo results suggest that both LNv clusters share common mechanisms to control their bursting firing frequency, which appear to be controlled intrinsically, likely involving the Ih current, as well as rely on synaptic inputs.
Ih channel and the sLNvs outputs
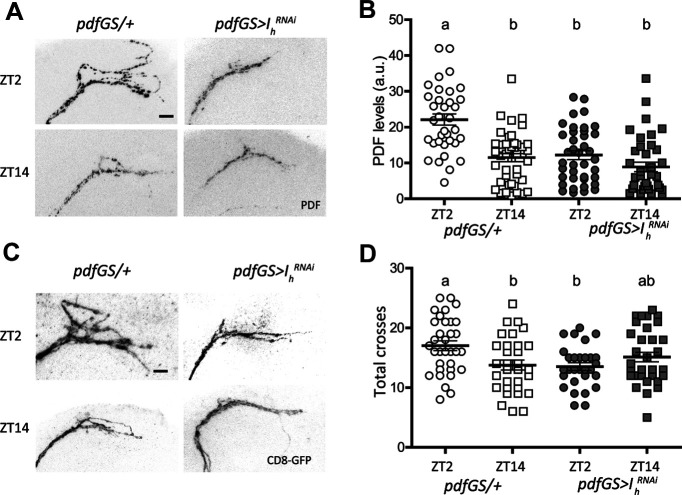
Over the years, it has been demonstrated that communication from the sLNvs to other clock clusters is crucial for coherent circadian behavior under free running conditions (Renn et al., 1999; Peng et al., 2003; Grima et al., 2004; Stoleru et al., 2004; Fernandez et al., 2007; Yoshii et al., 2009; Yao and Shafer, 2014; Frenkel et al., 2017). The rhythmic accumulation of PDF neuropeptide in sLNvs axonal termini has been implicated in this communication, with high immunoreactivity detected in the early morning and low immunoreactivity at night (Park et al., 2000). We hypothesized that release of DVCs containing PDF would be affected by the decrease in bursting activity that accompanies Ih downregulation. To test this, we performed anti-PDF immunofluorescence in whole brains of flies with adult-specific downregulation of Ih. Figure 5A,B shows that PDF immunoreactivity in controls (pdfGS/+) displays the normal cycling pattern; however, on downregulation of Ih (in pdfGS>IhRNAi) PDF levels at the axonal termini are constantly reduced and clamped in a night-like state.
Figure 5.
Ih downregulation affects PDF levels and structural plasticity. A, Confocal images of representative sLNvs dorsal projections of individual flies of control (pdfGS/+) and Ih downregulation (pdfGS>IhRNAi) at day (top) and night (bottom) showing their PDF content. Flies were kept in LD 12:12 at 25°C for 7 d in food containing RU486. Brains were dissected at ZT02 and ZT14 and standard anti-PDF immunofluorescence detection was performed. The bar indicates 10 µm. B, PDF quantitation of the sLNvs dorsal projections for the four conditions mentioned before. Circles represent day time, squares, night time; empty symbols are the control genotype (pdfGS/+) and filled symbols, the experimental one (pdfGS>IhRNAi). Different letters indicate significant differences, analysis included a two-way ANOVA (genotype and time of day; F(3,150) = 18.58 p < 0.0001 with Tukey's post hoc test, α = 0.05), n = 35–43 per group. C, Confocal images of sLNvs projections illustrating their complexity at ZT02 and ZT14 for both in the control and Ih downregulated genotypes. Procedure as in A but with immunofluorescence against GFP. The bar indicates 10 µm. D, Complexity quantitation was asses by Sholl analysis (ImageJ) corroborated by visual inspection of each picture. Symbols as in B, analysis included a two-way ANOVA (F(3,123) = 4.24 p < 0.01 with Tukey's post hoc test, α = 0.05). In B and D the mean ± SEM are shown. Different letters indicate significant differences. n = 34–38 per group.
In addition to PDF cyclic accumulation, sLNvs show circadian variation of the complexity of their axonal arborizations (Fernandez et al., 2008) to contact different synaptic targets at different times of the day (Gorostiza et al., 2014). This structural synaptic plasticity has been shown to be activity-dependent (Depetris-Chauvin et al., 2011; Sivachenko et al., 2013; Petsakou et al., 2015), therefore we wondered whether Ih downregulation would affect this property. Figure 5C,D shows that total axonal crosses measured by Sholl analysis in controls display the normal cycling pattern, where the terminals are maximally spread (and more complex) in the early morning and less complex at night, where axonal terminals are collapsed together. In contrast, Ih downregulation leads to axonal projections that display little complexity throughout the day, accompanying the reduced PDF levels. Our speculation on why Ih downregulation leaves both, PDF and terminal complexity at levels similar to ZT14 is that Ih underlies high activity bursting firing, a property that is functional during the day. Downregulation of this channel impairs this high-frequency bursting that would be associated to increased PDF levels and the spreading of sLNv axonal projections in the morning, both phenomena that have been described to be clock and activity-dependent (Depetris-Chauvin et al., 2011; Sivachenko et al., 2013; Petsakou et al., 2015). Moreover, we have previously described that structural plasticity depends on PDF levels (Depetris-Chauvin et al., 2014), so the collapsed state of the projections could be linked to PDF decrease as well. To corroborate whether the defects shown on Ih downregulation are linked to reduced PDF levels we used the GS system to express pdf in the context of Ih downregulation. Figure 6A,C,D shows that indeed, in the context of a surplus of PDF, cycling of this neuropeptide in the sLNvs axonal terminals is restored, while PDF expression in controls cycles with reduced (yet significant) amplitude.
Figure 6.
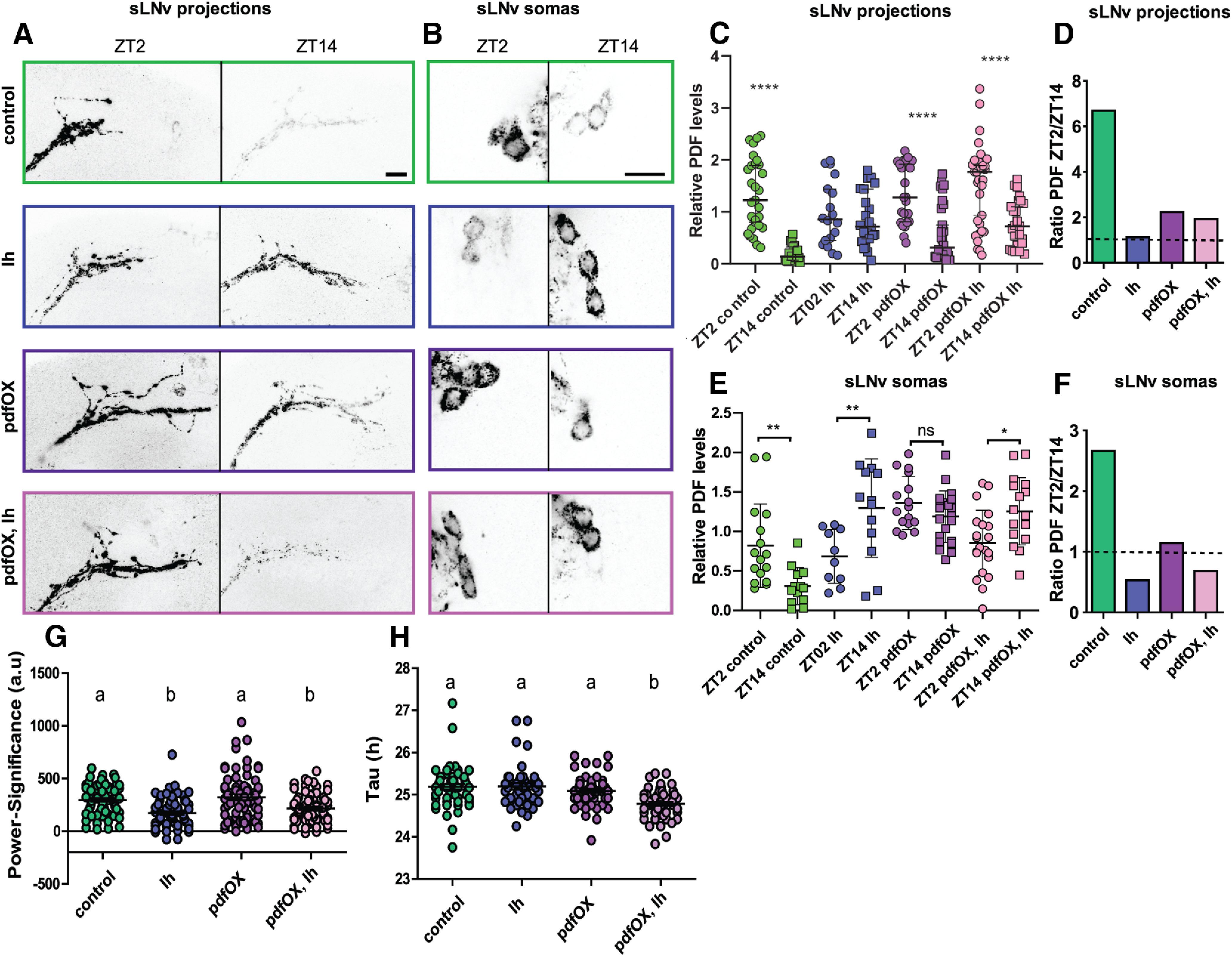
PDF transport is affected on Ih manipulation. A, B, Confocal images of representative sLNv projections (A) and somas (B) of individual flies of pdfGS/+ (control), pdfGS>IhRNAi (Ih), pdfGS>UAS-pdf (pdfOX) and pdfGS>UAS-pdf, IhRNAi (pdfOX, Ih) at day (left) and night (right) showing their PDF content. Flies were kept in LD 12:12 at 25°C for 7 d in food containing RU486. Brains were dissected at ZT02 and ZT14 and standard anti-PDF immunofluorescence detection was performed. Bars indicate 10 µm. C, E, PDF quantitation of the sLNv dorsal projections (C) or somas (E) for the four genotypes mentioned before. Circles represent day time, squares, night time; each color is a different genotype. Asterisks represent significant statistical differences. For the projections, a non-parametric ANOVA Kruskal–Wallis test and Dunn's comparisons test showed differences among the two time points in control, pdfOX and pdfOX, Ih groups but not in Ih group (Kruskal–Wallis statistic(8,196) = 71.95, p < 0.0001, n = 18–28). Immunoreactivity from somas was analyzed with one-way ANOVA and Sidak's multiple comparisons test and revealed differences between the two time points in every genotype except pdfOX, although in Ih and pdfOX, Ih showed differences in the anti-phase direction compared with the control, ANOVA F(7,120) = 10.95, p < 0.0001, n = 10–22 (each point is the average of three to four cell somas for one hemi-brain of an individual fly). D, F, Morning to evening PDF level ratios for axonal projections (D) or somas (F). G, Locomotor behavior under constant darkness of the same genotypes as before. Experiments were performed as in Figure 1 and Table 3. The rhythmicity measured as power-significance was analyzed by Kruskal–Wallis test followed by Dunn's comparisons test and showed a significant reduction of power-significance in Ih and pdfOX, Ih compared with control and pdfOX as indicated by different letters (Kruskal–Wallis statistic(4,31) = 31.40, p < 0.0001, n = 65–72). H, Free running period values were analyzed as well. The same type of analysis reveals a reduction of tau in pdfOX, Ih compared with all the other genotypes as indicated by a different letter (Kruskal–Wallis statistic(4,31) = 38.28, p < 0.0001, n = 45–58). In C, G and H the median ± confidence intervals are shown. In E the mean ± Standard Deviation is shown. ns, not significant.
To investigate whether the decreased PDF levels seen at the dorsal projections are because of decreased PDF production or to a failure to recruit PDF-loaded vesicles (i.e., transport) toward the axonal terminal, we measured PDF levels in the sLNv somas. We analyzed somatic PDF levels (see methods) and found that PDF immunoreactivity cycles in the sLNv somas in a way that resembles its cycling at the axonal terminals, with more PDF during the early morning and less PDF at the beginning of the night (Fig. 6B,E,F). Interestingly, in the context of Ih downregulation, somatic PDF shows an abnormal accumulation during the night, which could be because of a decreased daytime transport toward the axonal terminals that results in anti-phase cycling of somatic PDF levels. PDF overexpression per se increases overall levels, preventing PDF cycling in the somas, albeit not in the terminals. On the other hand, PDF overexpression in the context of Ih knock-down does not rescue the night-time abnormal PDF accumulation in the somas, however, it does rescue cycling in the projections (Fig. 6C).
Although PDF overexpression rescues some of the Ih-related phenotypes at the cellular level, it fails to rescue free running behavior (Fig. 6G,H). A plausible explanation for this may be that PDF cycling in the terminals, although rescued, still shows reduced amplitude (Fig. 6D) and may not be enough to synchronize the remaining clusters. Alternatively, Ih downregulation and the associated reduction of bursting frequency may be affecting the release of other neuropeptides or neurotransmitters besides PDF, which might also contribute to the neuronal communication needed to maintain rhythmicity under constant conditions. PDF expression in the context of Ih downregulation subtly shortens the free-running period (Fig. 6H), which is reminiscent of reduced PDF levels (Renn et al., 1999), although the underlying mechanisms remain to be explored.
Overall, these results indicate that Ih defines an essential property of the sLNvs that ensures proper regulation of neuropeptide levels and structural plasticity and provide a causal link between the alteration of electrical activity and the disruption of circadian behavior. Moreover, the careful determination of PDF levels in the sLNv somas suggest that in the context of Ih downregulation there is defective PDF transport toward the axonal projections, underscoring that action potential firing in bursts is responsible for an active recruitment of DVCs to the terminals. Although the aberrant transport is a parsimonious hypothesis, caution is called for when interpreting dynamic events by interval sampling from fixed tissues. Further work will be necessary to specifically dissect the role of Ih in these complex processes and to investigate other possibilities such as aberrant turnover and altered peptide release.
Sleep and the Ih channel
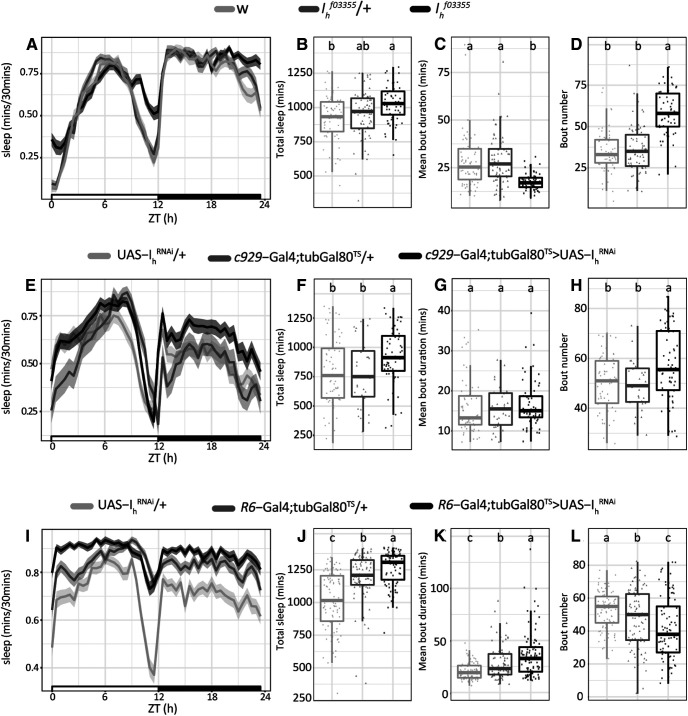
We then examined whether reduction in bursting firing frequency and hence, neuropeptide release, could affect sleep behavior. We first quantified sleep behavior in Ihf03355 mutants and found that homozygotes displayed an increase in total sleep, mainly because of a significant rise in the number of sleep bouts, which were shorter in duration but still resulted in an increase in total sleep during nighttime. Notably, the increase in sleep was more conspicuous toward the end of the night (Fig. 7A–D; Table 5). Given the ubiquitous nature of this genetic manipulation we reasoned that the deconsolidated sleep phenotype could arise from the lack of Ih in a plethora of neurons. To narrow down the cells where Ih is required for sleep regulation we continued the analysis using Ih RNAi-mediated downregulation in circumscribed neuronal groups.
Figure 7.
Genetic manipulations of Ih increase sleep. A, E, I, Sleep ethograms for the indicated genotypes, quantification of the relative amount of sleep every 30 min as a function of the time of the day (starting at ZT = 0, when lights are turned on) and its standard deviation (shadowed area). Black and white bars at the bottom represent daytime (white) and nighttime (black). B, F, J, Boxplots showing the total amount of sleep minutes for each genotype. C, G, K, Boxplots showing the average duration of sleep episodes for each genotype. D, H, L, Boxplots showing the total amount of sleep episodes for each genotype. For all the boxplots, different letters indicate significant differences (p < 0.05) after non parametric Kruskal–Wallis statistical analysis with multiple comparisons (p adjustment method = BH). Box represents the median and interquartile range of each parameter. For more information on sleep parameters see Table 5.
Table 5.
Sleep parameters after genetic manipulation ofIh
| Genotype | Total sleep (min) | Daytime sleep (min) | Nighttime sleep (min) | Bout duration (min) | Sleep bout number | Latency lights on (min) | Latency lights off (min) | Activity index | n | N | Temperature (°C) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ihf03355 | 1026.6 ± 17.8a | 430.0 ± 14.3a | 599.9 ± 8.4a | 17.6 ± 0.6b | 58.5 ± 1.8a | 25.0 ± 5.8b | 15.3 ± 1.9b | 4.24 ± 0.18a | 56 | 3 | 25 |
| Ihf03355/+ | 956.9 ± 21.6a,b | 394.6 ± 11.8a | 560.5 ± 15.0a,b | 29.0 ± 1.6a | 36.4 ± 1.7b | 71.5 ± 6.3a | 27.1 ± 3.7a | 2.18 ± 0.08b | 69 | ||
| w1118 | 922.2 ± 20.8b | 380.5 ± 12.6a | 533.4 ± 17.4b | 28.4 ± 1.7a | 35.3 ± 1.6b | 78.7 ± 7.3a | 27.6 ± 3.0a | 2.02 ± 0.05b | 66 | ||
| c929-Gal4;tub-Gal80TS>UAS-IhRNAi | 921.7 ± 31.6a | 464.1 ± 16.1a | 457.7 ± 21.6a | 17.0 ± 0.9a | 57.5 ± 1.9a | 18.1 ± 3.1a | 15.9 ± 2.7b | 2.16 ± 0.07b | 58 | 2 | 30 |
| c929-Gal4;tub-Gal80TS/+ | 773.0 ± 50.7b | 432.6 ± 25.5a | 340.4 ± 30.3b | 15.7 ± 0.9a | 49.4 ± 1.8b | 51.0 ± 12.0a | 45.7 ± 8.1a | 1.88 ± 0.05c | 31 | ||
| UAS-IhRNAi/+ | 793.5 ± 38.1b | 424.8 ± 21.5a | 368.7 ± 21.1b | 16.0 ± 0.8a | 50.6 ± 1.4b | 25.5 ± 7.5a | 17.4 ± 2.9b | 2.52 ± 0.09a | 61 | ||
| R6-Gal4;tub-Gal80TS>UAS-IhRNAi | 1264.9 ± 14.2a | 636.7 ± 7.6a | 630.7 ± 21.6a | 37.1 ± 2.5a | 41.7 ± 2.0c | 2.2 ± 0.7c | 16.8 ± 3.7b | 2.63 ± 0.08a | 85 | 3 | 30 |
| R6-Gal4;tub-Gal80TS/+ | 1195.6 ± 19.6b | 605.9 ± 10.8b | 589.6 ± 30.3b | 31.2 ± 3.1b | 47.3 ± 1.9b | 5.4 ± 1.1b | 24.7 ± 6.2b | 2.16 ± 0.06b | 87 | ||
| UAS-IhRNAi/+ | 1015.7 ± 23.7c | 515.0 ± 12.8c | 504.6 ± 21.1c | 20.5 ± 0.8c | 52.5 ± 1.2a | 13.6 ± 2.8a | 18.2 ± 2.4a | 2.21 ± 0.08b | 89 | ||
| pdf-Gal4, UAS-dicer2;tub-Gal80TS> UAS-IhRNAi | 955.9 ± 27.8a | 603.5 ± 12.3ab | 357.7 ± 20.5a | 25.8 ± 1.1b | 39.2 ± 1.6a | 17.2 ± 3.3b | 19.4 ± 4.6b | 1.82 ± 0.03a | 61 | 2 | 30 |
| pdf-Gal4, UAS-dicer2;tub-Gal80TS/+ | 847.9 ± 15.7b | 632.4 ± 7.4a | 215.5 ± 12.5c | 30.1 ± 1.0a | 30.0 ± 1.8b | 22.5 ± 1.7a | 36.5 ± 5.6a | 1.64 ± 0.03b | 63 | ||
| UAS-IhRNAi/+ | 848.7 ± 20.0b | 573.1 ± 12.9b | 274.9 ± 15.6b | 22.4 ± 1.1c | 41.9 ± 1.7a | 14.8 ± 1.9b | 24.5 ± 3.8a | 1.81 ± 0.03a | 61 | ||
| c929-Gal4>UAS-IhRNAi | 980.5 ± 29.9a | 496.1 ± 15.0a | 484.1 ± 17.2a | 18.2 ± 1.3a | 60.3 ± 2.1a | 8.6 ± 2.4c | 21.7 ± 2.6b | 1.98 ± 0.04a | 72 | 3 | 25 |
| c929-Gal4/+ | 793.1 ± 26.8b | 394.4 ± 13.5c | 401.4 ± 17.1b | 15.5 ± 0.9a | 52.7 ± 1.5b | 56.5 ± 7.7a | 32.3 ± 3.0a | 1.89 ± 0.03a,b | 71 | ||
| UAS-IhRNAi/+ | 919.0 ± 29.5a | 444.5 ± 15.9b | 474.9 ± 17.3a | 16.9 ± 1.1a | 58.7 ± 1.6a | 19.9 ± 4.0b | 26.5 ± 5.1b | 1.80 ± 0.03b | 70 | ||
| R6-Gal4>UAS-IhRNAi | 1111.7 ± 27.7a | 545.9 ± 15.7a | 551.2 ± 14.9a | 27.2 ± 0.6a | 53.3 ± 2.9a | 6.8 ± 2.6b | 30.9 ± 7.2a | 2.21 ± 0.09a | 65 | 3 | 25 |
| R6-Gal4/+ | 854.5 ± 35.9b | 409.7 ± 18.0b | 440.9 ± 20.8b | 15.6 ± 1.2b | 58.2 ± 2.3a | 19.0 ± 3.0a | 26.6 ± 4.4a | 2.06 ± 0.03a | 71 | ||
| UAS-IhRNAi/+ | 826.9 ± 28.1b | 400.6 ± 16.4b | 423.6 ± 15.5b | 14.1 ± 2.9b | 59.9 ± 1.4a | 32.9 ± 5.4a | 24.2 ± 2.5a | 1.81 ± 0.03b | 70 | ||
| pdf-Gal4, UAS-dicer2>UAS-IhRNAi | 715.9 ± 37.8b | 330.6 ± 20.7b | 385.3 ± 21.6b | 14.3 ± 0.9b | 52.2 ± 1.9a | 27.5 ± 6.2b | 36.5 ± 6.2a | 1.75 ± 0.04b | 64 | 2 | 25 |
| pdf-Gal4, UAS-dicer2/+ | 714.0 ± 29.3b | 320.8 ± 15.3b | 393.3 ± 18.3b | 14.0 ± 0.7b | 52.4 ± 1.6a | 70.0 ± 7.1a | 42.4 ± 6.3a | 1.73 ± 0.03b | 56 | ||
| UAS-IhRNAi/+ | 937.5 ± 28.1a | 414.3 ± 18.4a | 525.5 ± 15.2a | 18.7 ± 0.9a | 52.6 ± 1.7a | 39.0 ± 6.5b | 21.3 ± 4.6b | 1.89 ± 0.03a | 64 |
The following sleep parameters on the different genetic manipulations presented in the first column are shown: total sleep, daytime sleep, nighttime sleep, sleep bout duration, bout amount, latency to lights on, latency to lights off, and activity index (defined as the average activity counts in the active minutes). The last column shows the temperature at which each experiment was performed. Average ± SEM (Standard Error of the Mean) of N experiments using a final n number of individuals are displayed. Different letters indicate significant differences (p < 0.05) after non parametric Kruskal–Wallis statistical analysis with multiple comparisons (p adjustment method = BH). Sleep parameters where the experimental genotype showed statistically significant differences compared with genetic controls are displayed in bold.
It has previously been demonstrated a significant role of the lLNvs in arousal, as the PDF released by these neurons works as a strong arousal signal (Parisky et al., 2008; Shang et al., 2008; Sheeba et al., 2008a). We therefore analyzed sleep after acute downregulation of the Ih channel in the lLNvs along with other non-circadian peptidergic neurons (combining the c929-Gal4 driver with the TARGET system; McGuire et al., 2004a). Similar to the Ih mutants, these flies exhibited an increase in the number of sleep episodes that resulted in a significant rise in nighttime sleep (Figs. 7E–H, 8; Table 5); however, the duration of the sleep bouts remains unchanged, indicating that the short sleep bout phenotype observed in Ih mutants must derive from the lack of Ih in neurons not covered by the c929-Gal4 driver or to the lack of Ih in c929-Gal4 positive neurons during development (see Table 5).
Figure 8.
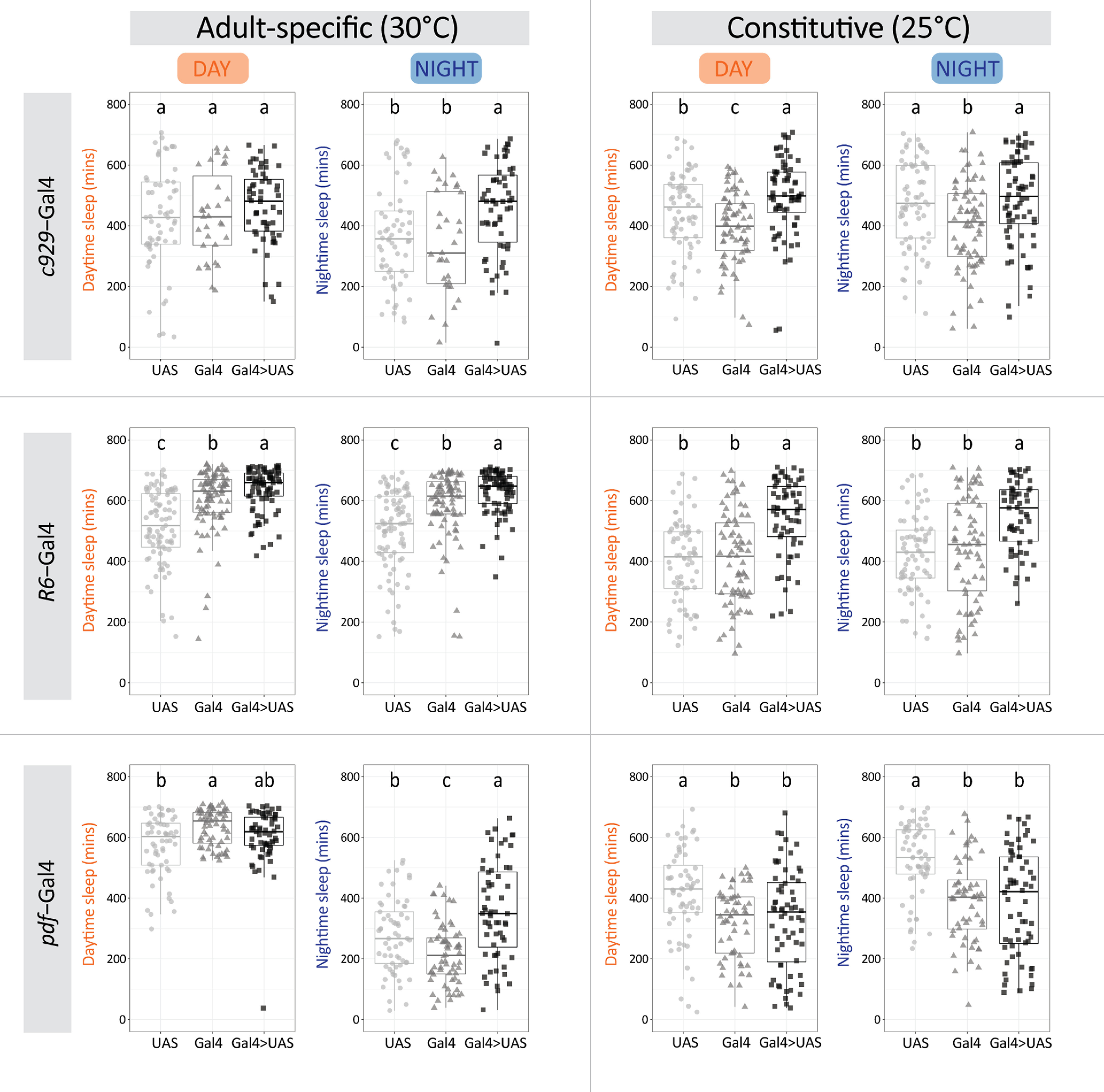
Relative contribution of specific clusters to sleep control. Boxplots show daytime and nighttime sleep duration on RNAi-mediated Ih downregulation using c929-Gal4 (top panel), R6-Gal4 (middle panel), and pdf-Gal4 (bottom panel), along with their genetic controls. Adult-specific manipulations performed using the TARGET system are shown in the left and correspond to the second day at the permissive temperature of 30°C. Constitutive genetic manipulations are shown on the right, all performed at the standard temperature of 25°C. Different letters indicate significant differences (p < 0.05) after non parametric Kruskal–Wallis statistical analysis with multiple comparisons (p adjustment method = BH). Box represents the median and interquartile range of each parameter. For more detailed information on sleep parameters see Table 5.
Although PDF released from the sLNvs has not been shown to play an arousal role as the one released by the lLNvs, diverse lines of evidence lend support to the notion that sLNvs can have an impact on sleep behavior (Chen et al., 2016; Guo et al., 2016). To test whether Ih from sLNvs had any influence on sleep behavior, we resorted to the sLNv driver R6-Gal4. This Gal4 line drives expression almost exclusively in the sLNv and very little (if at all) in the lLNv, with occasional expression in a few other non-clock neurons (Helfrich-Forster et al., 2007). As a consequence of acute downregulation of the Ih channel with R6-Gal4, flies showed a robust increase in the amount of sleep, both at daytime and nighttime (Figs. 7I–L, 8; Table 5). Surprisingly, this rise in sleep was because of a more consolidated sleep, as the sleep bout number was reduced but episodes lasted longer in the experimental flies compared with the genetic controls. Ih downregulation experiments were also performed constitutively, showing similar tendencies to the acute ones (Table 5).
We then tested the effect of Ih downregulation in all PDF-positive neurons and found that this manipulation results in an increase in nighttime sleep when performed acutely using the TARGET system (Fig. 8). However, in the case of constitutive downregulation, no significant differences were identified (Fig. 8; Table 5). It should be noted that the pdf-Gal4, UAS-dicer2 control line used for these experiments shows abnormally short sleep bouts, likely underscoring some genetic background effects. This short sleep time of the parental line may conceal a subtle sleep phenotype associated to Ih constitutive downregulation, perhaps resulting in an underestimation of the effect in the acute manipulation too. Alternatively, both groups could be contributing to sleep regulation through different mechanisms/signals that, when impaired at the same time, result in a nonlinear combination of effects. Acute and truly cell-type-specific manipulations of LNvs are therefore required to further dissect their role on the control of sleep behavior.
Collectively, our work demonstrates that Ih certainly plays a role in the control of sleep behavior, both on the overall levels and the timing of sleep. Alterations in the timing of sleep is particularly prevalent, highlighted by a recurrent decrease in the latency to the first sleep episode after lights-on observed in the majority of the Ih genetic manipulations (Table 5). Further work will be necessary to pinpoint how different neurons recruit Ih to regulate various aspects of their physiology. In particular, the role of neuropeptides in sleep control is widely recognized and involves many neurons throughout the brain. We have initiated here an analysis that includes heterogeneous neuronal clusters including the sLNvs and the lLNvs, but it is likely that Ih manipulation will impair neuropeptide trafficking in other sleep-related neurons as well.
Discussion
The physiology of a particular neuron is not regulated by a single ion channel type but by a complex array of different players: they go from the leak conductances that determine input resistance and resting membrane potential which influence dendritic processes, including summation and propagation of synaptic inputs, to the abundance and quality of voltage-gated ion channels that determine the dynamics of action potential firing, and ultimately dictate the release of classical neurotransmitters and neuropeptides. If we add to this picture the channels that are directly or indirectly activated by ligands, we will be able to comprehend, and model, neuronal physiology. We have performed a downregulation screen to describe novel ion channels playing roles in establishing the electrical properties of the LNvs, with the aim of advancing the understanding of LNvs physiology. We focused our attention on Ih, a poorly studied ionic current in Drosophila.
Since the discovery of the first hyperpolarization-gated current in cardiac function (Noma and Irisawa, 1976) a great deal of information has been gained about the role of this type of channels in determining the physiology of the mammalian heart and brain (Luthi and McCormick, 1998). An interesting feature of Ih channels is that they are not only sensitive to hyperpolarization, but are also modulated by cyclic nucleotides, hence the name of the channel family hyperpolarization cyclic nucleotide-gated (HCN). The mammalian genome contains four HCN channel genes HCN1–HCN4, each with specific activation characteristics, distinct but in some cases partially overlapping expression patterns, and different roles in neuronal physiology (He et al., 2014). Drosophila's Ih channel is the sole member of the HCN family in its genome (Marx et al., 1999), but up to 12 different splice variants can be generated, providing diverse channels with particular biophysical properties (Gisselmann et al., 2005). A phylogenetic analysis indicates that Drosophila Ih (also referred in the literature as DMIH) diverged from a common ancestor before the emergence of the four vertebrate subtypes (Jackson et al., 2007). Interestingly, the domain organization of Ih is similar to its vertebrate counterparts, and the interaction between domains is conserved to the point that domain swapping between Drosophila Ih and vertebrate HCN channels produce similar biophysical results (Ishii et al., 2007).
Drosophila Ih has not been explored in depth yet but it has been reported in the visual system where it regulates the release of glutamate from amacrine cells (Hu et al., 2015), and at the larval neuromuscular junction where it affects neurotransmitter release (Hegle et al., 2017). An analysis of Ih mutants shows that this channel controls a variety of behaviors (Chen and Wang, 2012). Particularly relevant for our work is the fact that Ih has been reported to control circadian rhythms and sleep in Drosophila by acting on dopaminergic neurons (Gonzalo-Gomez et al., 2012). Although Gonzalo-Gomez et al. (2012) did not find any PDF disruptions in the sLNvs of the Ih mutants they generated, our current analysis of adult-specific downregulation of Ih shows that it does affect PDF levels and the structural plasticity at the sLNvs dorsal projections, as well as the accumulation of PDF in the somas albeit with altered circadian dynamics. This highlights the importance of using strategies where a genetic manipulation is performed acutely, to avoid homeostatic compensations that may conceal a phenotype. Together, the collective evidence indicates that Ih may be modulating circadian rhythms and sleep by exerting its role in more than one neuronal type. Whether the molecular mechanisms that regulate and are regulated by Ih in LNvs and in dopaminergic neurons are similar will require further examination.
Sleep behavior has been previously reported in Ih mutant flies, and, taking into account our contribution, the accumulated evidence raises some controversy that deserves special attention. Using an independently generated Ih null mutant, Gonzalo-Gomez et al. (2012) reported that total sleep was unchanged; but they showed, similar to our results, a deconsolidation of sleep resulting from an increase in sleep bout number of shorter duration (Gonzalo-Gomez et al., 2012). On the other hand, during the initial characterization of the mutants used in our study, Chen et al., reported the opposite sleep phenotype, that is, a decrease in total sleep, with no changes in bout number (Chen and Wang, 2012). One important element to take into account is that Ih mutant flies are hyperactive (Table 5) and therefore inferring sleep from activity data should be approached with caution. Since Ih mutants display an increase in sleep, and hyperactivity would result in an underestimation of sleep, our results are validated. It is not uncommon to come across published fly sleep data inferred from activity monitoring where basal activity is not reported (Chen and Wang, 2012), a practice that warrants further attention.
Perhaps more informative than the mutants is our sleep analysis following acute and cluster-specific downregulation of Ih. We show here that c929+ peptidergic neurons use Ih to promote arousal, which in the case of the lLNvs would likely be mediated by PDF release following high-frequency neuronal bursting. However, the release of other neuropeptides could also be affected by Ih downregulation; in fact additional neurons besides lLNvs contribute to the waking state within the c929-Gal4 driven group (Parisky et al., 2008). Moreover, specific downregulation of Ih in the sLNvs also results in an increase of sleep, however, this increase corresponds to a more consolidated sleep and therefore, although an increase in total sleep is produced on Ih downregulation with both R6-Gal4 and c929-Gal4, the properties of these sleep increases are different. This may indicate that, either the neuropeptide/s these neuronal clusters are releasing with the help of Ih, or the effects these signals are conveying to their particular downstream targets, are probably different. These findings should be thoroughly characterized in the future, as they suggest a participation of the sLNvs in the neuronal circuits governing sleep. Interestingly, in a recent genome-wide association study, ion channels were one of the two main pathways associated to sleep duration both in humans and flies, indicating an evolutionarily conserved function of ion channels in regulating a complex behavior such as sleep (Allebrandt et al., 2017).
Although no role has been directly demonstrated for Ih in Drosophila clock neurons before our work, the HCN family has been proposed to contribute to the circadian variations in neuronal excitability in the mammalian suprachiasmatic nucleus (SCN; Colwell, 2011). HCN channels have been reported to be expressed in the SCN (Notomi and Shigemoto, 2004) but their function has been difficult to discern because of a lack of strong and significant phenotypes following genetic and pharmacologic manipulations, which could be because of the heterogeneity of the SCN neuronal population and the genetic compensation that may arise from having several HCN channel genes (de Jeu and Pennartz, 1997; O'Neill et al., 2008; Atkinson et al., 2011). Relevant to our study, bursting firing in thalamic intergeniculate leaflet neurons, which act to regulate circadian rhythms generated by the SCN, is decreased when Ih is inhibited (Ying et al., 2011). Also in accordance to our findings are the results from HCN genetic manipulation in the heart sinoatral node cells where the lack of Ih (called If in the context of the heart) produces bradicardia (a decreased heart rate equivalent to a decreased bursting frequency in neurons; for review, see Herrmann et al., 2011). Taking advantage of the fact that Drosophila clock neuron clusters are well identifiable, and that there is only one member of the HCN channel family, we were able to show that Ih is a crucial player defining the high activity bursting physiology of LNvs, and that this regulates neuronal outputs and behavior.
The importance of cyclic nucleotide cascades in LNvs has been recognized before (Shang et al., 2011; Duvall and Taghert, 2013; Sabado et al., 2017). Our screen has uncovered two cyclic nucleotide-modulated channels (Ih and CngA), suggesting that the integration of information signaled by cyclic nucleotides is crucial for circadian function at rapid time frames, a hypothesis that has already been proposed (Seluzicki et al., 2014; Sabado et al., 2017). The case of Ih, being modulated by both hyperpolarizing voltage and cyclic nucleotides provides additional complexity, as they could serve as coincidence detectors (Fig. 9, scheme). The biophysics of Ih, and therefore the firing properties of LNvs, are affected by both the membrane voltage and the levels of cyclic nucleotides, therefore it is likely that the timing of arrival of these signals may significantly affect the LNvs neuronal output. Albeit purely speculative for the LNvs, the Ih current has been proposed to work as a coincidence detector in other systems (Yamada et al., 2005; Pavlov et al., 2011; Baumann et al., 2013). Further research will be necessary to reveal which are the neuronal inputs that contribute to the hyperpolarization and to the variations of cyclic nucleotide levels. Interestingly, HCN channels have been reported to be activated by vasoactive intestinal peptide (Sun et al., 2003), which is considered a functional homolog of PDF. Therefore, activation of the PDFR signaling cascade could result in an increase in cyclic nucleotide (i.e., cAMP), thus modulating Ih, adding players to the already complicated integration of synaptic and cell-autonomous cues coordinated at the sLNvs.
Figure 9.
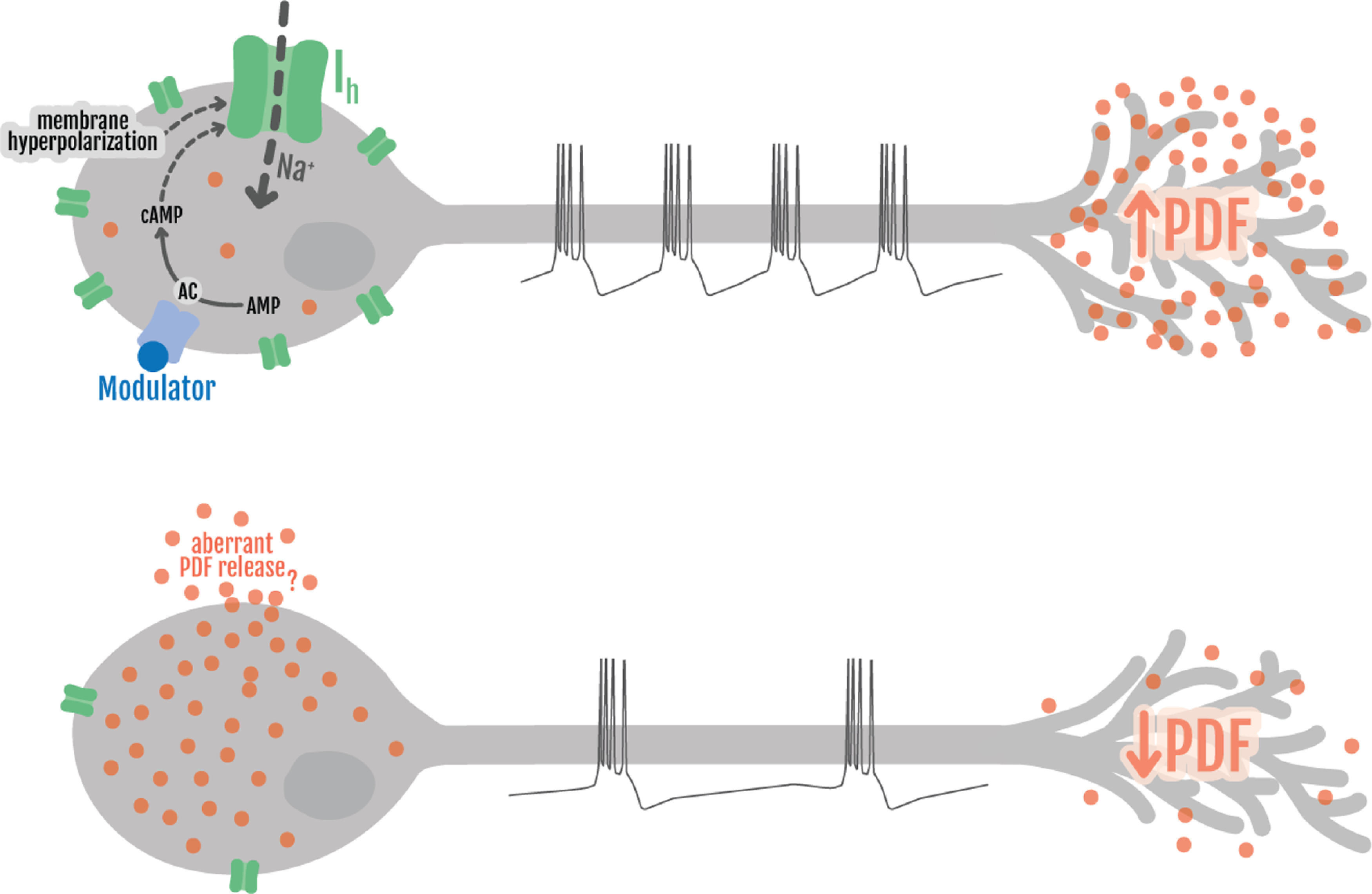
Model summarizing the findings reported and the hypotheses raised by this work. Ih channel (in green) responds to membrane hyperpolarization and is modulated by cyclic nucleotides, serving as coincidence detector for electrical and chemical signals mediated by ligands that activate G-protein-coupled receptors (such as PDF, dopamine, or other neuropeptides, symbolized by “modulator,” in blue). Ih function is necessary to allow LNvs to fire action potentials in a high-frequency bursting mode, which permits the release of PDF (in orange) at high levels and in a timely manner (top neuron). In the absence or on Ih knock-down (bottom neuron), bursting does not reach such high frequency, and PDF levels at the axonal projections are reduced. Associated to the decreased bursting frequency, large quantities of PDF accumulate at the soma, likely because of a failure in DVCs transport. This may give rise to a hypothetical aberrant PDF release from the overloaded soma, likely overriding the internal temporal control. All in all, these cellular disruptions result in anomalies at the behavioral level, such as disorganization of circadian locomotor activity and an increase in sleep. At the level of the axonal projections, the model represents an early daytime situation, where in control animals PDF levels are high and the axonal terminal are spread out. However, the accumulation and possible aberrant release of PDF from the soma is more likely to happen during the night (Fig. 6).
One question that remains to be answered is whether the circadian clock is directly regulating Ih function in LNvs. Although our work has not focused on this issue, a plausible hypothesis is that Ih expression levels may be changing at different times of the day. This is suggested by the work by Abruzzi et al. (2011) where they performed chromatin immunoprecipitation tiling array assays with a number of circadian proteins, and showed that the circadian transcription factor CLOCK cycles in its binding to Ih regulatory sequences in Drosophila heads (Abruzzi et al., 2011). Among all the positive hits of our screen, Ih is the only one that appears to be directly controlled by the circadian clock according to Abruzzi et al. (2011).
Our experiments demonstrate that the sLNvs, considered a central piece of the clock neuron circuitry puzzle, organize their action potential firing in bursts. This bursting frequency depends on synaptic inputs, as it has been shown for the lLNvs (Muraro and Ceriani, 2015). LNvs bursting frequency seems to be also influenced by cell autonomous mechanisms since, as we demonstrate here, a null mutation in Ih produces a decrease in this parameter. Interestingly, the mutation produces a decrease that is of the same magnitude in sLNvs and lLNvs, suggesting that this ion channel regulates bursting frequency in comparable ways in both neuronal types. Remarkably, the DN1 clock neuron cluster has recently been shown to fire action potentials in bursts and that this temporal coding, i.e., the timing of spiking, is relevant for the control of sleep behavior (Tabuchi et al., 2018).
The importance of gaining as much information as possible about the Ih current is underscored by the discovery of several Ih channelopathies. Information from both, patients and genetic animal models, has brought to light the asociation of mutations on HCN channel genes, or accessory subunits, to different conditions such as epilepsies, autism spectrum disorders, neuropathic pain, Parkinson's disease, depression and cardiac dysfunction among others (DiFrancesco and DiFrancesco, 2015; Brennan et al., 2016; Ku and Han, 2017). In this context, learning about Drosophila Ih helps understanding the basic characteristics of this current and Drosophila, with its less complex genome and fantastic genetic amenability, could serve in the future as a model organism to discover interacting proteins and pathways, to ultimately unravel the underlying pathologic mechanisms of Ih channelopathies.
Footnotes
This work was supported by Agencia Nacional de Promoción Científica y Tecnológica Grants PICT-2011-2185 (to M.F.C.) and PICT-2011-2364 and PICT-2015-2557 (to N.I.M.), the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) Grant PIP-11220130100378 (to N.I.M.) and the Fondo de Convergencia Estructural del MERCOSUR Grant COF 03/11 to Instituto de Investigación en Biomedicina de Buenos Aires (IBioBA). M.F.C., N.I.M., and L.F. are members of the CONICET. Stocks obtained from the Bloomington Drosophila Stock Center (National Institutes of Health Grant P40OD018537), the NIG-Fly Stock Center, and the Vienna Drosophila Resource Center (Dietzl et al., 2007) were used in this study. We thank Dr. Zuo Ren Wang for Ih mutant fly stocks. We also thank Esteban Beckwith and Quentin Geissmann for indispensable help with sleep quantification software. We thank Andres Liceri for fly maintenance.
The authors declare no competing financial interests.
References
- Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M (2011) Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev 25:2374–2386. 10.1101/gad.178079.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allebrandt KV, Teder-Laving M, Cusumano P, Frishman G, Levandovski R, Ruepp A, Hidalgo MPL, Costa R, Metspalu A, Roenneberg T, De Pittà C (2017) Identifying pathways modulating sleep duration: from genomics to transcriptomics. Sci Rep 7:4555. 10.1038/s41598-017-04027-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen CN, Nitabach MN, Colwell CS (2017) Membrane currents, gene expression, and circadian clocks. Cold Spring Harb Perspect Biol 9:a027714 10.1101/cshperspect.a027714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson SE, Maywood ES, Chesham JE, Wozny C, Colwell CS, Hastings MH, Williams SR (2011) Cyclic AMP signaling control of action potential firing rate and molecular circadian pacemaking in the suprachiasmatic nucleus. J Biol Rhythms 26:210–220. 10.1177/0748730411402810 [DOI] [PubMed] [Google Scholar]
- Baumann VJ, Lehnert S, Leibold C, Koch U (2013) Tonotopic organization of the hyperpolarization-activated current (Ih) in the mammalian medial superior olive. Front Neural Circuits 7:117. 10.3389/fncir.2013.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith EJ, Ceriani MF (2015) Communication between circadian clusters: the key to a plastic network. FEBS Lett 589:3336–3342. 10.1016/j.febslet.2015.08.017 [DOI] [PubMed] [Google Scholar]
- Brennan GP, Baram TZ, Poolos NP (2016) Hyperpolarization-activated cyclic nucleotide-gated (HCN) channels in epilepsy. Cold Spring Harb Perspect Med 6:a022384. 10.1101/cshperspect.a022384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhl E, Bradlaugh A, Ogueta M, Chen KF, Stanewsky R, Hodge JJ (2016) Quasimodo mediates daily and acute light effects on Drosophila clock neuron excitability. Proc Natl Acad Sci USA 113:13486–13491. 10.1073/pnas.1606547113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Nitabach MN (2008) Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci 28:6493–6501. 10.1523/JNEUROSCI.1503-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA (2002) Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci 22:9305–9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS. (2011) Linking neural activity and molecular oscillations in the SCN. Nat Rev Neurosci 12:553–569. 10.1038/nrn3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Reiher W, Hermann-Luibl C, Sellami A, Cognigni P, Kondo S, Helfrich-Förster C, Veenstra JA, Wegener C (2016) Allatostatin A signalling in Drosophila regulates feeding and sleep and is modulated by PDF. PLoS Genet 12:e1006346. 10.1371/journal.pgen.1006346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Wang Z (2012) Functional study of hyperpolarization activated channel (Ih) in Drosophila behavior. Sci China Life Sci 55:2–7. 10.1007/s11427-012-4270-6 [DOI] [PubMed] [Google Scholar]
- De Jeu MT, Pennartz CM (1997) Functional characterization of the H-current in SCN neurons in subjective day and night: a whole-cell patch-clamp study in acutely prepared brain slices. Brain Res 767:72–80. 10.1016/s0006-8993(97)00632-x [DOI] [PubMed] [Google Scholar]
- Depetris-Chauvin A, Berni J, Aranovich EJ, Muraro NI, Beckwith EJ, Ceriani MF (2011) Adult-specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Curr Biol 21:1783–1793. 10.1016/j.cub.2011.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depetris-Chauvin A, Fernández-Gamba A, Gorostiza EA, Herrero A, Castaño EM, Ceriani MF (2014) Mmp1 processing of the PDF neuropeptide regulates circadian structural plasticity of pacemaker neurons. PLoS Genet 10:e1004700. 10.1371/journal.pgen.1004700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzo JA, Casanoves F, Balzarini MG, Gonzalez L, Tablada M, Robledo CW (2018) InfoStat versión 2018. Centro de Transferencia InfoStat, FCA, Universidad Nacional de Córdoba, Argentina: Available at http://www.infostat.com.ar. [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448:151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Difrancesco JC, Difrancesco D (2015) Dysfunctional HCN ion channels in neurological diseases. Front Cell Neurosci 6:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall LB, Taghert PH (2013) E and M circadian pacemaker neurons use different PDF receptor signalosome components in Drosophila. J Biol Rhythms 28:239–248. 10.1177/0748730413497179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N (2014) Advanced methods of microscope control using μManager software. J Biol Methods 1:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Zhang J, Li M, Shao L, Yang L, Song Q, Ping Y (2018) Control of sleep onset by Shal/Kv4 channels in Drosophila circadian neurons. J Neurosci 38:9059–9071. 10.1523/JNEUROSCI.0777-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez MP, Chu J, Villella A, Atkinson N, Kay SA, Ceriani MF (2007) Impaired clock output by altered connectivity in the circadian network. Proc Natl Acad Sci USA 104:5650–5655. 10.1073/pnas.0608260104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez MP, Berni J, Ceriani MF (2008) Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol 6:e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flourakis M, Kula-Eversole E, Hutchison AL, Han TH, Aranda K, Moose DL, White KP, Dinner AR, Lear BC, Ren D, Diekman CO, Raman IM, Allada R (2015) A conserved bicycle model for circadian clock control of membrane excitability. Cell 162:836–848. 10.1016/j.cell.2015.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogle KJ, Baik LS, Houl JH, Tran TT, Roberts L, Dahm NA, Cao Y, Zhou M, Holmes TC (2015) CRYPTOCHROME-mediated phototransduction by modulation of the potassium ion channel beta-subunit redox sensor. Proc Natl Acad Sci USA 112:2245–2250. 10.1073/pnas.1416586112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel L, Muraro NI, Beltran Gonzalez AN, Marcora MS, Bernabo G, Hermann-Luibl C, Romero JI, Helfrich-Forster C, Castano EM, Marino-Busjle C, Calvo DJ, Ceriani MF (2017) Organization of circadian behavior relies on glycinergic transmission. Cell Rep 19:72–85. [DOI] [PubMed] [Google Scholar]
- Geissmann Q, Garcia Rodriguez L, Beckwith EJ, Gilestro GF (2019) Rethomics: an R framework to analyse high-throughput behavioural data. PLoS One 14:e0209331 10.1371/journal.pone.0209331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselmann G, Gamerschlag B, Sonnenfeld R, Marx T, Neuhaus EM, Wetzel CH, Hatt H (2005) Variants of the Drosophila melanogaster Ih-channel are generated by different splicing. Insect Biochem Mol Biol 35:505–514. 10.1016/j.ibmb.2005.02.001 [DOI] [PubMed] [Google Scholar]
- Gonzalo-Gomez A, Turiegano E, Leon Y, Molina I, Torroja L, Canal I (2012) Ih current is necessary to maintain normal dopamine fluctuations and sleep consolidation in Drosophila. PLoS One 7:e36477. 10.1371/journal.pone.0036477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorostiza EA, Depetris-Chauvin A, Frenkel L, Pirez N, Ceriani MF (2014) Circadian pacemaker neurons change synaptic contacts across the day. Curr Biol 24:2161–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Chelot E, Xia R, Rouyer F (2004) Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431:869–873. 10.1038/nature02935 [DOI] [PubMed] [Google Scholar]
- Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, Rosbash M (2016) Circadian neuron feedback controls the Drosophila sleep–activity profile. Nature 536:292–297. 10.1038/nature19097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Chen F, Li B, Hu Z (2014) Neurophysiology of HCN channels: from cellular functions to multiple regulations. Prog Neurobiol 112:1–23. 10.1016/j.pneurobio.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Hegle AP, Frank CA, Berndt A, Klose M, Allan DW, Accili EA (2017) The Ih channel gene promotes synaptic transmission and coordinated movement in Drosophila melanogaster. Front Mol Neurosci 10:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C. (1995) The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc Natl Acad Sci USA 92:612–616. 10.1073/pnas.92.2.612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C, Shafer OT, Wulbeck C, Grieshaber E, Rieger D, Taghert P (2007) Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. J Comp Neurol 500:47–70. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI (2000) Rest in Drosophila is a sleep-like state. Neuron 25:129–138. 10.1016/s0896-6273(00)80877-6 [DOI] [PubMed] [Google Scholar]
- Herrmann S, Layh B, Ludwig A (2011) Novel insights into the distribution of cardiac HCN channels: an expression study in the mouse heart. J Mol Cell Cardiol 51:997–1006. 10.1016/j.yjmcc.2011.09.005 [DOI] [PubMed] [Google Scholar]
- Hodge JJ, Stanewsky R (2008) Function of the Shaw potassium channel within the Drosophila circadian clock. PLoS One 3:e2274. 10.1371/journal.pone.0002274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Wang T, Wang X, Han J (2015) Ih channels control feedback regulation from amacrine cells to photoreceptors. PLoS Biol 13:e1002115. 10.1371/journal.pbio.1002115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Tm, Nakashima N, Takatsuka K, Ohmori H (2007) Peripheral N- and C-terminal domains determine deactivation kinetics of HCN channels. Biochem Biophys Res Commun 359:592–598. 10.1016/j.bbrc.2007.05.127 [DOI] [PubMed] [Google Scholar]
- Jackson HA, Marshall CR, Accili EA (2007) Evolution and structural diversification of hyperpolarization-activated cyclic nucleotide-gated channel genes. Physiol Genomics 29:231–245. 10.1152/physiolgenomics.00142.2006 [DOI] [PubMed] [Google Scholar]
- Jaramillo AM, Zheng X, Zhou Y, Amado DA, Sheldon A, Sehgal A, Levitan IB (2004) Pattern of distribution and cycling of SLOB, Slowpoke channel binding protein, in Drosophila. BMC Neurosci 5:3. 10.1186/1471-2202-5-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku SM, Han MH (2017) HCN channel targets for novel antidepressant treatment. Neurotherapeutics 14:698–715. 10.1007/s13311-017-0538-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear BC, Lin JM, Keath JR, Mcgill JJ, Raman IM, Allada R (2005) The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron 48:965–976. 10.1016/j.neuron.2005.10.030 [DOI] [PubMed] [Google Scholar]
- Lee Y, Montell C (2013) Drosophila TRPA1 functions in temperature control of circadian rhythm in pacemaker neurons. J Neurosci 33:6716–6725. 10.1523/JNEUROSCI.4237-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MT, Cao LH, Xiao N, Tang M, Deng B, Yang T, Yoshii T, Luo DG (2018) Hub-organized parallel circuits of central circadian pacemaker neurons for visual photoentrainment in Drosophila. Nat Commun 9:4247. 10.1038/s41467-018-06506-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WH, Baines RA (2015) Regulation of membrane excitability: a convergence on voltage-gated sodium conductance. Mol Neurobiol 51:57–67. 10.1007/s12035-014-8674-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, Taghert PH (2004) The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci 24:7951–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthi A, Mccormick DA (1998) H-current: properties of a neuronal and network pacemaker. Neuron 21:9–12. [DOI] [PubMed] [Google Scholar]
- Marx T, Gisselmann G, Störtkuhl KF, Hovemann BT, Hatt H (1999) Molecular cloning of a putative voltage- and cyclic nucleotide-gated ion channel present in the antennae and eyes of Drosophila melanogaster. Invert Neurosci 4:55–63. 10.1007/s101580050007 [DOI] [PubMed] [Google Scholar]
- Mcguire SE, Mao Z, Davis RL (2004a) Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE 2004:pl6. [DOI] [PubMed] [Google Scholar]
- Mcguire SE, Roman G, Davis RL (2004b) Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet 20:384–391. 10.1016/j.tig.2004.06.012 [DOI] [PubMed] [Google Scholar]
- Muraro NI, Ceriani MF (2015) Acetylcholine from visual circuits modulates the activity of arousal neurons in Drosophila. J Neurosci 35:16315–16327. 10.1523/JNEUROSCI.1571-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash HA, Scott RL, Lear BC, Allada R (2002) An unusual cation channel mediates photic control of locomotion in Drosophila. Curr Biol 12:2152–2158. 10.1016/s0960-9822(02)01358-1 [DOI] [PubMed] [Google Scholar]
- Noma A, Irisawa H (1976) Membrane currents in the rabbit sinoatrial node cell as studied by the double microelectrode method. Pflugers Arch 364:45–52. 10.1007/BF01062910 [DOI] [PubMed] [Google Scholar]
- Notomi T, Shigemoto R (2004) Immunohistochemical localization of Ih channel subunits, HCN1-4, in the rat brain. J Comp Neurol 471:241–276. [DOI] [PubMed] [Google Scholar]
- Nusbaum MP, Blitz DM, Marder E (2017) Functional consequences of neuropeptide and small-molecule co-transmission. Nat Rev Neurosci 18:389–403. 10.1038/nrn.2017.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH (2008) cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320:949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H (2001) A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci USA 98:12596–12601. 10.1073/pnas.221303298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJL, Kang K, Kang K, Liu X, Garrity PA, Rosbash M, Griffith LC (2008) PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60:672–682. 10.1016/j.neuron.2008.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC (2000) Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci USA 97:3608–3613. 10.1073/pnas.070036197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov I, Scimemi A, Savtchenko L, Kullmann DM, Walker MC (2011) I(h)-mediated depolarization enhances the temporal precision of neuronal integration. Nat Commun 2:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M (2003) Drosophila free-running rhythms require intercellular communication. PLoS Biol 1:E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsakou A, Sapsis TP, Blau J (2015) Circadian rhythms in Rho1 activity regulate neuronal plasticity and network hierarchy. Cell 162:823–835. 10.1016/j.cell.2015.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2014) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: Available at https://www.R-project.org. [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH (1999) A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99:791–802. 10.1016/S0092-8674(00)81676-1 [DOI] [PubMed] [Google Scholar]
- Ruben M, Drapeau MD, Mizrak D, Blau J (2012) A mechanism for circadian control of pacemaker neuron excitability. J Biol Rhythms 27:353–364. 10.1177/0748730412455918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabado V, Vienne L, Nunes JM, Rosbash M, Nagoshi E (2017) Fluorescence circadian imaging reveals a PDF-dependent transcriptional regulation of the Drosophila molecular clock. Sci Rep 7:41560. 10.1038/srep41560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Rasband W, Eliceiri K (2012) NIH Image to ImageJ: 25 years of image analysis. J Nat Methods 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting M, Menegazzi P, Lelito KR, Yao Z, Buhl E, Dalla Benetta E, Bahle A, Denike J, Hodge JJ, Helfrich-Förster C, Shafer OT (2016) A neural network underlying circadian entrainment and photoperiodic adjustment of sleep and activity in Drosophila. J Neurosci 36:9084–9096. 10.1523/JNEUROSCI.0992-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluzicki A, Flourakis M, Kula-Eversole E, Zhang L, Kilman V, Allada R (2014) Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PLoS Biol 12:e1001810. 10.1371/journal.pbio.1001810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N, Campbell R, Steinbach P, Giepmans BNG, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572. [DOI] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, Rosbash M (2008) Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci USA 105:19587–19594. 10.1073/pnas.0809577105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Haynes P, Pirez N, Harrington Ki, Guo F, Pollack J, Hong P, Griffith LC, Rosbash M (2011) Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat Neurosci 14:889–895. 10.1038/nn.2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G (2000) Correlates of sleep and waking in Drosophila melanogaster. Science 287:1834–1837. 10.1126/science.287.5459.1834 [DOI] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC (2008a) Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol 18:1537–1545. 10.1016/j.cub.2008.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Gu H, Sharma VK, O'Dowd DK, Holmes TC (2008b) Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol 99:976–988. 10.1152/jn.00930.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivachenko A, Li Y, Abruzzi KC, Rosbash M (2013) The transcription factor Mef2 links the Drosophila core clock to Fas2, neuronal morphology, and circadian behavior. Neuron 79:281–292. 10.1016/j.neuron.2013.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P, Buhl E, Tsaneva-Atanasova K, Hodge JJL (2019) Shaw and Shal voltage-gated potassium channels mediate circadian changes in Drosophila clock neuron excitability. J Physiol 597:5707–5722. 10.1113/JP278826 [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M (2004) Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431:862–868. 10.1038/nature02926 [DOI] [PubMed] [Google Scholar]
- Sun Qq, Prince Da, Huguenard JR (2003) Vasoactive intestinal polypeptide and pituitary adenylate cyclase-activating polypeptide activate hyperpolarization-activated cationic current and depolarize thalamocortical neurons in vitro. J Neurosci 23:2751–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M, Monaco JD, Duan G, Bell B, Liu S, Liu Q, Zhang K, Wu MN (2018) Clock-generated temporal codes determine synaptic plasticity to control sleep. Cell 175:1213–1227.e18. 10.1016/j.cell.2018.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top D, Young MW (2018) Coordination between differentially regulated circadian clocks generates rhythmic behavior. Cold Spring Harb Perspect Biol 10:a033589 10.1101/cshperspect.a033589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Pol AN. (2012) Neuropeptide transmission in brain circuits. Neuron 76:98–115. 10.1016/j.neuron.2012.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R, Kuba H, Ishii Tm, Ohmori H (2005) Hyperpolarization-activated cyclic nucleotide-gated cation channels regulate auditory coincidence detection in nucleus laminaris of the chick. J Neurosci 25:8867–8877. 10.1523/JNEUROSCI.2541-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Shafer OT (2014) The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343:1516–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying SW, Tibbs GR, Picollo A, Abbas SY, Sanford RL, Accardi A, Hofmann F, Ludwig A, Goldstein PA (2011) PIP2-mediated HCN3 channel gating is crucial for rhythmic burst firing in thalamic intergeniculate leaflet neurons. J Neurosci 31:10412–10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Wülbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, Helfrich-Förster C (2009) The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila's clock. J Neurosci 29:2597–2610. 10.1523/JNEUROSCI.5439-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Rieger D, Helfrich-Förster C (2012) Two clocks in the brain: an update of the morning and evening oscillator model in Drosophila. Prog Brain Res 199:59–82. 10.1016/B978-0-444-59427-3.00027-7 [DOI] [PubMed] [Google Scholar]