Abstract
Superoxide dismutases (SODs) act as a primary defence against reactive oxygen species (ROS) by converting superoxide anion radicals (O2-) into molecular oxygen (O2) and hydrogen peroxide (H2O2). Members of this enzyme family include CuZnSODs, MnSODs, FeSODs, and NiSODs, depending on the nature of the cofactor that is required for proper activity. Most eukaryotes, including yeast, possess CuZnSOD and MnSOD. This protocol aims at assessing the activity of the yeast Saccharomyces cerevisiae MnSOD Sod2p from cellular extracts using nitroblue tetrazolium staining. This method can be used to estimate the cellular bioavailability of Mn2+ as well as to evaluate the redox state of the cell.
Keywords: Superoxide dismutase, Yeast, Manganese, Sod2p, Nitroblue tetrazolium, Redox state
Background
SODs are defined as metal-containing antioxidant enzymes that reduce harmful free radicals of oxygen formed during normal aerobic metabolism to oxygen and hydrogen peroxide. These enzymes are classified based on the metal required as cofactor for proper enzymatic activity: CuZnSODs, MnSODs, FeSODs, and NiSODs. In the yeast Saccharomyces cerevisiae, there are two SODs: the CuZn-Sod1p and the Mn-Sod2p (Abreu and Cabelli, 2010). This protocol focuses on the determination of the enzymatic activity of the Mn-Sod2p, found in the yeast mitochondrial matrix. In this protocol, activity of Sod2p is visualized through nitroblue tetrazolium staining. According to this method, the excitation of riboflavin by light, catalyzed by tetramethylethylenediamine (TEMED), generates superoxide radicals, which convert the yellow nitroblue tetrazolium into blue formazan. In the regions in which Sod2p is present, the superoxide radicals are rapidly removed and formazan formation is prevented. Sod2p is thereby revealed in clear bands on a blue background (Packer, 2002). The method described here includes inhibition of the CuZn-Sod1p by potassium cyanide and thereby enables to determine specifically for the enzymatic activity of the Mn-Sod2p. Apart from providing a method to quickly determine the enzymatic activity of Sod2p, this protocol can be used to correlate the activity of the mitochondrial Sod2p to the bioavailability of manganese cations required for proper activity, a decreased manganese content in the close vicinity of Sod2p resulting in a lower enzymatic activity ( Thines et al., 2018 ). Besides, due to the implication of both Sod2p and manganese cations in resistance against oxidative stress, this protocol can be used to assess the redox state of yeast cells, a decreased enzymatic activity reflecting a reduced ability of the cell to neutralize free radicals.
Materials and Reagents
425-600 µm diameter acid-washed glass beads (Sigma-Aldrich, catalog number: G8772)
Petri dishes (Sigma-Aldrich, catalog number: P5606-400EA)
50 ml Falcon® tubes (Dutscher, catalog number: 352070)
Eppendorf tubes (VWR, catalog number: 89000-028)
Bovine serum albumin standard, 2 mg/ml (Thermo Scientific, catalog number: 23210)
Protease inhibitor cocktail [4 mg/ml of leupeptin (Roth, catalog number: CN33.2), aprotinin (Roth, catalog number: A162.3), antipain (Roth, catalog number: 2933.2), pepstatin (Roth, catalog number: 2936.3), and chymostatin (Sigma-Aldrich, catalog number: EI6)]
Yeast extract KAT (Ohly, catalog number: OHLY® KAT)
Glucose (Merck, catalog number: 1083469029)
Nitroblue tetrazolium (Sigma-Aldrich, catalog member: N6876)
4-15% Mini-PROTEAN® TGXTM Precast Protein Gels, 10-well, 50 µl (Bio-Rad, catalog number: 4561084)
MilliQ water
Ethylenediaminetetraacetic acid (EDTA) disodium salt dihydrate (Sigma-Aldrich, catalog number: E4884)
Ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) (Sigma-Aldrich, catalog number: E3889)
NaCl (Sigma-Aldrich, catalog number: S9888)
Triton X-100 (Sigma-Aldrich, catalog number: X100)
Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: 10837091001)
Bicinchoninic acid (Supelco, catalog number: B9643)
CuSO4·5H2O (Sigma-Aldrich, catalog number: 209198)
Trizma base (Sigma-Aldrich, catalog number: 93362)
HCl (Sigma-Aldrich, catalog number: H1758)
NaOH (Sigma-Aldrich, catalog number: 795429)
Glycerol (Sigma-Aldrich, catalog number: G5516)
Bromophenol blue (Sigma-Aldrich, catalog number: B0126)
Glycine (Sigma-Aldrich, catalog number: 50046)
TEMED (Sigma-Aldrich, catalog number: T9281)
Riboflavin (Sigma-Aldrich, catalog number: 47861)
KCN (Sigma-Aldrich, catalog number: 60178)
Na2HPO4 (Sigma-Aldrich, catalog number: S7907)
NaH2PO4 (Sigma, Aldrich, catalog number: S3139)
K2HPO4 (Sigma-Aldrich, catalog number: 1551128)
KH2PO4 (Sigma-Aldrich, catalog number: 1551139)
Liquid nitrogen
YD plates (see Recipes)
NaPO4 buffer (0.1 M, pH 7.8) (see Recipes)
Tris buffer (1 M, pH 6.8) (see Recipes)
Potassium phosphate buffer (1 M, pH 7.8) (see Recipes)
EDTA (100 mM, pH 8.0) (see Recipes)
EGTA (100 mM, pH 8.0) (see Recipes)
Lysis buffer (see Recipes)
CuSO4·5H2O (4%) (see Recipes)
2x cc. sample buffer for native gels (see Recipes)
Native gel running buffer (see Recipes)
Staining solution (see Recipes)
Equipment
Centrifuge for Eppendorf tubes (Hettich Zentrifugen, model: MIKRO 20)
Vortex (VWR, model: 444-0996)
Autoclave (Systec, model: VX/VE)
Protein gel cassette (Bio-Rad, model: 1645052)
Gel scanner (Amersham, model: 29083461)
Incubator shaker for yeast growth in liquid culture (Edmund Bühler GmbH, model: VKS-75 control)
Procedure
-
S. cerevisiae cellular extracts preparation
Streak the S. cerevisiae strains to be analyzed from corresponding glycerol stocks on YD plates (Recipe 1). Incubate for two days at 28 °C.
Using a sterile toothpick, select individual colonies.
-
Grow yeast cells at 28 °C under agitation (120 rpm) in 50 ml YD medium to an OD600 of 3 (OD600 = 1 corresponds to a density of 1.25 x 107 cells/ml).
Note: From this step, keep your samples on ice as much as possible.
Centrifuge the yeast culture in 50 ml Falcon® tubes at 3,500 × g at 4 °C for 5 min.
Discard the supernatant and resuspend the pellet in 2 ml ice-cold water.
Centrifuge at 16,000 × g at 4 °C for 20 s.
Discard the supernatant and resuspend the pellet in 200 µl ice-cold lysis buffer (Recipe 7).
Add 200 µg acid-washed 425-600 µm diameter glass beads to the cell suspension.
Perform cell lysis by vortexing 8 x 30 s at 3,000 rpm, with a 15 s break on ice between each vortexing step.
Centrifuge at 2,400 × g at 4 °C for 20 s.
Transfer the supernatant (cell lysate) in a new Eppendorf tube and store it at -80 °C if not used directly, with prior snap freezing in liquid nitrogen.
-
Protein concentration quantification
Note: This protocol includes protein quantification using the bicinchoninic acid assay. If familiar with any other method for protein concentration quantification like Bradford assay, this can be used as well.
Prepare standard solutions of bovine serum albumin (BSA) from a 2 mg/ml stock solution according to the Table 1 for establishment of a calibration curve:
Dilute the cell lysates so that their concentration is covered by the calibration curve. If starting from a 50 ml culture harvested at an OD600 of 3, samples can be diluted 10 times (10 µl of cell lysate in 90 µl MilliQ water).
Mix 49 ml bicinchoninic acid with 1 ml CuSO4·5H2O (4%) (Recipe 8).
Mix 20 µl of each standard/sample with 200 µl of the mix bicinchoninic acid/ CuSO4·5H2O.
Incubate for 30 min at 37 °C.
Read the absorbance at 562 nm.
Determine a calibration curve using the standards and deduce the concentration of the samples to be analyzed using this calibration curve. Figure 1 illustrates a typical linear regression that could be obtained with the BSA standards prepared as described above.
-
Sod2p activity staining
Mix a volume of cell lysate corresponding to 200 µg proteins (about 15 µl if starting from a 50 ml culture harvested at an OD600 of 3) with the same volume of 2x cc. sample buffer for native gels (Recipe 9).
Load the resulting mixture on a Mini-PROTEAN® TGXTM precast protein gel.
Run the gel for 4 h at 100 V at 4 °C in native gel running buffer (Recipe 10).
After gel migration, immerge it in 20 ml of a 1 mg/ml nitroblue tetrazolium solution for 15 min under agitation (40 rpm) and in the dark.
Rince the gel with MilliQ water.
Immerge the gel in 25 ml of the staining solution (Recipe 11) for 15 min under agitation (40 rpm) and in the dark.
Rince the gel with MilliQ water.
Expose to light for 15-30 min and scan the gel. The unstained region of the gel corresponds to Sod2p. Figure 2 illustrates the gel obtained for the wild-type yeast strain and for the strains deleted for the genes coding for the CuZn-Sod1p (sod1Δ) or for the Mn-Sod2p (sod2Δ).
Table 1. Preparation of the BSA standards.
| Tube | BSA concentration (mg/ml) | H2O volume (µl) | BSA volume (µl) |
|---|---|---|---|
| A | 2.000 | 0 | 300 from stock solution 2 mg/ml |
| B | 1.500 | 125 | 375 from stock solution 2 mg/ml |
| C | 1.000 | 325 | 325 from stock solution 2 mg/ml |
| D | 0.750 | 175 | 175 from tube B |
| E | 0.500 | 325 | 325 from tube C |
| F | 0.250 | 325 | 325 from tube E |
| G | 0.125 | 325 | 325 from tube F |
| H | 0.025 | 400 | 100 from tube G |
| I | 0 | 400 | 0 |
Figure 1. Example of calibration curve obtained for the bicinchoninic acid assay from BSA standards prepared as described here.
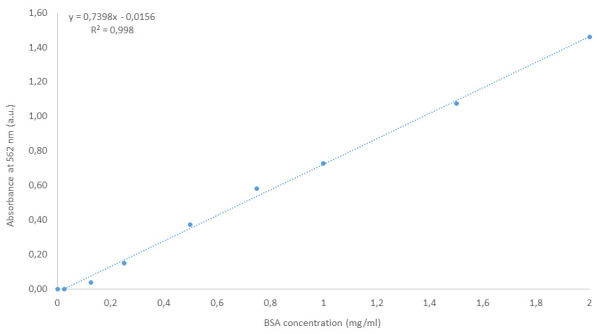
The equation of the linear regression as well as the corresponding R2 are mentioned on the graph.
Figure 2. Activity of Sod2p assessed in-gel for the wild-type, sod1Δ, and sod2Δ yeast strains.
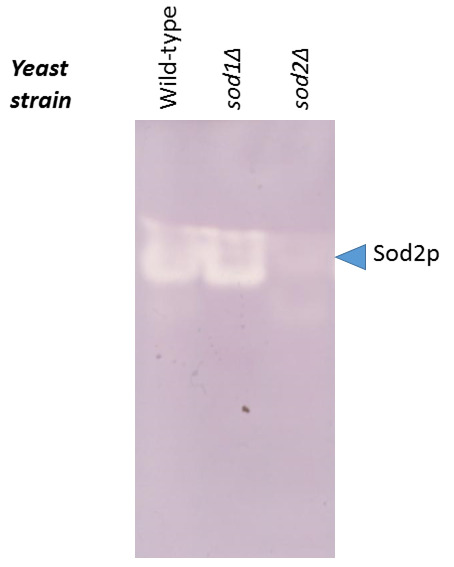
The intensity of the white band at the level of the arrow correlates with the level of activity of Sod2p (the whiter this region, the higher the activity of Sod2p).
Notes
The intensity of the signal corresponding to the activity of Sod2p can be correlated to the availability of Mn2+ and to the redox status of the cell. In this perspective, a less intense white band on the blue background, reflecting a decreased activity of Sod2p, might be correlated to (i) a decreased bioavailability of Mn2+ in the close vicinity of Sod2p due to its action as cofactor, or to (ii) a reduced cellular ability to resist to oxidative stress due to the implication of Sod2p and manganese cations in neutralizing free radicals. A more quantitative approach can be carried out by quantifying the signal corresponding to Sod2p using any software that is routinely used to quantify Western blotting signals.
Recipes
-
YD plates
2 g (2% w/v) yeast extract KAT
2 g (2% w/v) glucose
Adjust to 100 ml with MilliQ water and autoclave
Pour in Petri dishes
-
NaPO4 buffer (0.1 M, pH 7.8)
4.48 ml of 1 M Na2HPO4
0.52 ml of 1 M NaH2PO4
Adjust to 50 ml with MilliQ water and verify the pH
-
Tris buffer (1 M, pH 6.8)
121.14 g Trizma base
Dilute in approx. 800 ml MilliQ water
Adjust pH to 6.8 using HCl
Adjust to a final volume of 1 L with MilliQ water
-
Potassium phosphate buffer (1 M, pH 7.8)
14.894 g K2HPO4
1.972 g KH2PO4
Adjust to 100 ml with MilliQ water and verify the pH
-
EDTA (100 mM, pH 8.0)
3.7224 g EDTA
Dilute in approx. 80 ml MilliQ water
Adjust pH to 8.0 using NaOH
Adjust to a final volume of 100 ml with MilliQ water
-
EGTA (100 mM, pH 8.0)
3.8035 g EGTA
Dilute in approx. 80 ml MilliQ water
Adjust pH using NaOH
Adjust to a final volume of 100 ml with MilliQ water
-
Lysis buffer
10 ml (10 mM) NaPO4 buffer pH 7.8 (Recipe 2)
5 ml (5 mM) of 100 mM EDTA
5 ml (5 mM) of 100 mM EGTA
0.292 g (50 mM) NaCl
100 µl (0.1% v/v) Triton X-100
50 µl Protease inhibitor cocktail
1 ml (1 mM) of 100 mM phenylmethylsulfonyl fluoride (PMSF) (0.0174 g PMSF in 1 ml ethanol)
Adjust to a final volume of 100 ml with MilliQ water
-
CuSO4·5H2O (4%)
2 g CuSO4·5H2O
Dilute in 50 ml MilliQ water
-
2x cc. sample buffer for native gels
1.875 ml (62.5 mM) of 1 M Tris-HCl buffer pH 6.8 (Recipe 3)
12 ml (40% v/v) glycerol
0.3 ml (0.01% w/v) of 1% (w/v) bromophenol blue (0.5 g bromophenol blue in 50 ml MilliQ water)
Adjust to 30 ml with MilliQ water
-
Native gel running buffer
30.3 g (250 mM) Trizma base
144.1 g (1.9 M) glycine
Adjust to 1 L with MilliQ water
To be diluted 10 times before use
-
Staining solution
5 ml (100 mM) potassium phosphate buffer pH 7.8 (Recipe 4)
162.7 µl (28 mM) TEMED
0.0005 g (0.028 mM) riboflavin
0.0163 g (5 mM) KCN
Adjust to 50 ml with MilliQ water
Acknowledgments
This protocol was adapted from established published procedures (Luk and Culotta, 2001). The work was supported by grants from the Fonds National de la Recherche Scientifique (FNRS, grant PDR-T.0206.16). L.T. is a research fellow at the ‘Fonds pour le Formation à la Recherche dans l’Industrie et dans l’Agriculture’.
Competing interests
The authors declare that they have no conflicts of interest with the contents of this article.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Abreu I. A. and Cabelli D. E.(2010). Superoxide dismutases-a review of the metal-associated mechanistic variations. Biochim Biophys Acta 1804(2): 263-274. [DOI] [PubMed] [Google Scholar]
- 2. Luk E. E. and Culotta V. C.(2001). Manganese superoxide dismutase in Saccharomyces cerevisiae acquires its metal co-factor through a pathway involving the Nramp metal transporter, Smf2p . J Biol Chem 276(50): 47556-47562. [DOI] [PubMed] [Google Scholar]
- 3. Packer L.(2002). Superoxide dismutase. United States: Elsevier Science, 197. [Google Scholar]
- 4. Thines L., Deschamps A., Sengottaiyan P., Savel O., Stribny J. and Morsomme P.(2018). The yeast protein Gdt1p transports Mn2+ ions and thereby regulates manganese homeostasis in the Golgi . J Biol Chem 293(21): 8048-8055. [DOI] [PMC free article] [PubMed] [Google Scholar]


