Abstract
Background:
Pathologic fibrosis is characterized by dysregulation of gene expression with excessive extracellular matrix production. The genetic basis for solid organ fibrosis is well described in the literature. However, there is a paucity of evidence for similar processes in the musculoskeletal (MSK) system. The purpose of this review is to provide an overview of existing evidence of genetic predisposition to pathologic fibrosis in the cardiac, pulmonary, and MSK systems, and to describe common genetic variants associated with these processes.
Methods:
A comprehensive search of several databases from 2000 to 2019 was conducted using relevant keywords in the English language. Genes reported as involved in idiopathic fibrotic processes in the heart, lung, hand, shoulder, and knee were recorded by 2 independent authors.
Results:
Among 2373 eligible studies, 52 studies investigated genetic predisposition in terms of variant analysis with the following organ system distribution: 36 pulmonary studies (69%), 15 hand studies (29%), and 1 knee study (2%). Twenty-two percent of gene variants identified were associated with both pulmonary and MSK fibrosis (ie, ADAM, HLA, CARD, EIF, TGF, WNT, and ZNF genes). Genetic variants known to be involved in the MSK tissue development or contractility properties in muscle were identified in the pulmonary fibrosis.
Conclusion:
Despite shared genetic variations in both the lung and hand, there remains limited information about genetic variants associated with fibrosis in other MSK regions. This finding establishes the necessity of further studies to elucidate the genetic determinants involved in the knee, shoulder, and other joint fibrotic pathways.
Level of evidence:
Level III.
Keywords: arthrofibrosis, genetic predisposition, theragnostic, Dupuytren disease, musculoskeletal, total knee arthroplasty
Fibrotic diseases represent a major source of morbidity and mortality in contemporary healthcare [1–5]. It has been estimated that up to 45% of all deaths are related to fibroproliferative processes [6]. Fibrogenesis is a complex, dynamic molecular process involving numerous pathways and cell types [6]. In simple terms, fibrogenesis begins as a primary insult to an organ system that elicits activation of effector cells followed subsequently by deposition of extracellular matrix (ECM) to stabilize and repair the compromised tissue. In the nonpathologic state, this matrix is resorbed and replaced by healthy host tissue. In the pathologic state—characterized by continued injury stimulus—there is ongoing deposition of ECM coupled with insufficient resorption, resulting in a fibrotic phenotype. Fibrotic phenotypes alter the form of the tissue with resultant dysfunction. Tissue dysfunction is tolerated to varying degrees depending on the organ system involved, and likewise different organ systems exhibit varying abilities to recover from the fibrotic insult. Interestingly, there is substantial evidence that profibrotic molecular pathways of disease pathogenesis remain conserved across different organ systems, including the heart, lung, liver, pancreas, kidney, bone marrow, and skin, among others [6,7].
Understanding the molecular mechanisms underlying fibrogenesis has led to emerging therapies targeted at halting and reversing the course of disease with variable success [8,9]. Despite these advancements, the efficacy of such therapies is limited by the ability to identify and target at-risk cohorts. More recently, genome-wide association studies have been used to identify genetic variants associated with various fibrotic diseases. Genetic variants have been used to predict disease onset and severity, therefore facilitating risk stratification and early targeted intervention [10].
A subclass of fibroproliferative diseases represents a unique clinical and therapeutic challenge, as the pathologic state arises in the absence of continued injury stimulus. These processes are designated idiopathic and are the focus of this review. Of great interest is arthrofibrosis after total knee arthroplasty (TKA), an idiopathic fibrotic process in patients with well-fixed and well-aligned components and no known modifiable or nonmodifiable risk factors [11]. Despite the success of contemporary TKAs, arthrofibrosis remains a major failure mode affecting 4%−10% of patients undergoing primary TKA [11]. Risk factors such as limited preoperative range of motion, delayed physical therapy, and complexity of initial surgery have been described. However, it is unclear why these factors lead to debilitating arthrofibrosis in some patients, yet result in acceptable joint range of motion and clinical outcomes in others [12].
A potential explanation is that there may be a genetic predisposition to arthrofibrosis in a subset of patients. So far, however, there is no evidence to directly substantiate this hypothesis. Based on previous work demonstrating conserved molecular processes of fibrogenesis in different solid organs, the goal of the current review is to establish if there are identifiable overlapping genetic determinants of fibrosis across multiple organ systems-namely the cardiac, pulmonary, and musculoskeletal (MSK) systems. We focused on cardiac and pulmonary processes because of their synovial and encapsulated organization analogous to the joint organization. The current review aims to (1) identify the presence of a genetic predisposition in different organ systems by evaluating the idiopathic fibrotic disease processes in each system, (2) evaluate the type and function of genes implicated in each organ system, and (3) identify overlapping genetic predisposition between systems.
Materials and Methods
A protocol was designed for this systematic review to identify shared genetic determinants of human idiopathic fibrotic diseases in the cardiac, pulmonary, and MSK systems. This study was reported according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses methodology.
Data Sources and Search Strategies
A comprehensive search of several databases from January 1, 2000, to October 7, 2019, was conducted in the English language. The databases included Ovid MEDLINE(R) and Epub Ahead of Print, In-Process & Other Non-Indexed Citations, and Daily, Ovid EMBASE, Ovid Cochrane Central Register of Controlled Trials, Ovid Cochrane Database of Systematic Reviews, and Scopus. The search strategy was designed and conducted by an experienced librarian. Controlled vocabulary supplemented with keywords was used to search for the genomics of fibrosis in the cardiac, pulmonary, and MSK systems in humans. Pulmonary processes included idiopathic pulmonary fibrosis (IPF). MSK processes emphasized in our review included Dupuytren disease, adhesive capsulitis of the shoulder, and arthrofibrosis of the knee, as these idiopathic conditions are most clinically relevant. The research strategy, including all search terms and combinations used, is available in Appendix A.
Study Selection
Two authors (LD and ARO) independently reviewed all titles and abstracts using the Covidence systematic review software (Veritas Health Innovation, Melbourne, Australia). The quality of the studies was assessed according to the Newcastle—Ottawa quality assessment scale (Appendix B) [13]. Discordances in study inclusion were resolved by evaluation discussion and consensus. Studies about fibrosis in anatomic locations other than the selected organs were excluded, as were studies about true hereditary or chronic diseases (nonidiopathic or sporadic fibrosis such as cystic fibrosis and scleroderma), studies emphasizing confounding factors such as smoking, nonspecific fibrotic processes (eg, atrial fibrillation, sarcoidosis, myositis), nongenetic studies (genomic expression without gene variant discovery), imaging or therapeutic articles, and animal studies. Editorials, reviews, symposia, case reports, and case series also were excluded.
Data Extraction
The studies included were reviewed and separated by organ systems, namely the cardiac, pulmonary, and MSK-related studies. Data were extracted manually to provide an overview of the evidence of a genetic predisposition in idiopathic fibrotic processes in the organ systems of interest.
Results
There were 2373 potentially eligible articles with 2134 (90%) being unrelated to the study question. These studies investigated genomic expression without genetic variant determination and were thus excluded. The full texts of 69 studies were reviewed, and 17 studies were subsequently excluded secondary to the above exclusion criteria (Fig. 1).
Fig. 1.
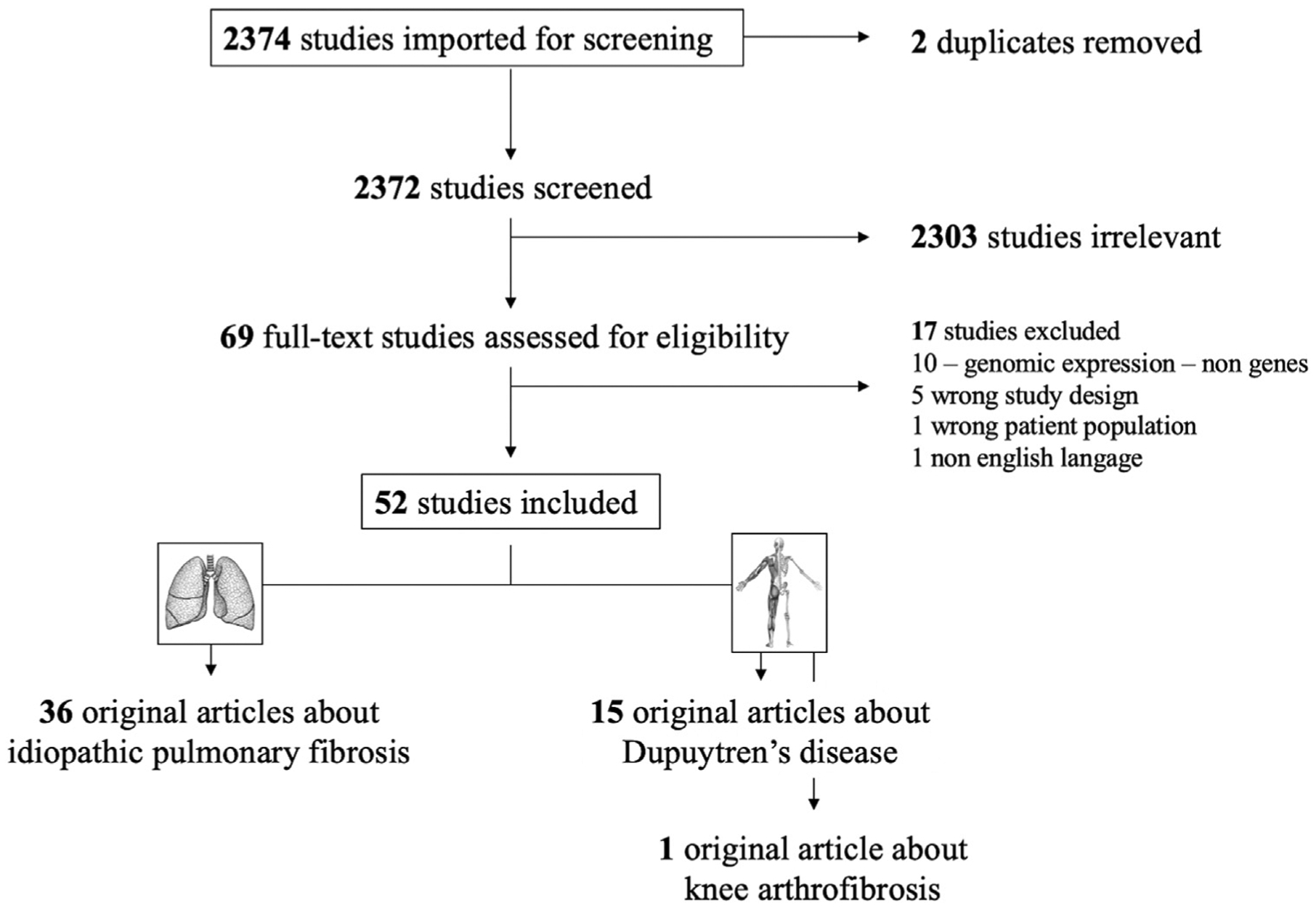
Flow diagram of the systematic research method for detecting genes predisposition in the heart, lung, hand, shoulder, and knee.
Ultimately, 52 studies (2.1%) were selected for inclusion in the systematic review. The 52 studies selected for this investigation contained data about genetic discovery and predisposition with the following organ system distribution: 36 pulmonary studies (69%), 15 hand studies (29%), and one knee study (2%) [14]. No evidence was found in this review for cardiac or shoulder idiopathic fibrosis according to our inclusion and exclusion criteria. Seven studies (13%) evaluated the genetic determinants in relation to disease severity (6 studies in the pulmonary literature [15–20] and 1 study in the hand literature [21]).
The genes involved in the fibrotic processes were summarized and divided into 2 categories: organ-specific genes (Table 1) and fibroproliferative genes (Table 2). The function of each fibroproliferative gene was identified and reported as follows: (1) ECM remodeling and fibroblast regulation, (2) immune system dysfunction, and (3) profibrotic mediators. MUC gene variants (mucosal tissue) were identified as a lung-specific entity predictive of IPF risk and disease severity [17,19,22–26]. Some MSK-specific genes were also identified in this review: ACAN gene (encodes an ECM protein in cartilaginous tissue) and EBF2 gene (a transcriptional regulator of osteoclast differentiation) [27].
Table 1.
Organ-Specific Genes Involved in Idiopathic Fibrosis in the Lung and Musculoskeletal Tissue From Our Review.
| Lung-specific genes | |
| Alveolar epithelial dysfunction | MUC2, MUC5AC, MUC5B, SFTPC, SFTPA2 |
| Musculoskeletal specific genes | |
| Extracellular matrix in cartilaginous tissue | ACAN |
| Osteoclast differentiation | EBF2 |
Table 2.
Fibrotic Genes Involved in the Lung (IPF) and Hand (DD) From Our Review.
| Fibrotic Pathway | Lung (IPF) | Hand (DD) |
|---|---|---|
| ECM remodeling and fibroblast regulation (cell-cell adhesion, apoptotic regulation, integrity and mechanotransduction) | ACTA-2, ACTG2, ATP1A2, BRSK2 (rs71469892), CARD10, CLPTM1L (rs4449583), DES, DPP9, DSP (rs72825054, rs2744373, rs2744371, rs17133512, rs145125286, rs2008756, rs2237105), DYM, EIF1AY, MCF2L (rs1278769), MDGA2 (rs7144383), MFSD2A, MGMT, MIR4457(rs4449583), MYH11, NLGN4Y, PIGA, RNPS1, RPS4Y1, RTEL1, SCN1A, SLFNL1, SLN, SPPL2C (rs17690703), SYNPO2, TAGLN, TP53 (rs12951053, rs12602273), TPM2, VDAC3 | AEN, CARD11, DAD1, DDR2, EIF3E (rs611744), EPDR1 (rs16879765), FBXL7 (rs6897647), FBXO45 (rs7426655), ITGA11, KRT15, KRT19, MAFB (rs8124695), MMP14, MMP21/22B, SAV1, TIMP2 (rs4789939), TMEM170A |
| Immune system dysfunction | DDX3Y, HLA (rs3117116), HLA-DRB1*15, IL-10, IL-1alpha, IL-4, IL-6 (intron 4G), IL1A, IL1B (rs1143627, rs1143634, rs1143643, rs1799916), IL1RN (rs11677397, rs2637988, rs397211, rs408392), IL3, IL4, IL8 (s4073T> A, rs2227307T> G, rs2227306C > T, IL8_ht3), KDM5D, R131H (FcgRIIa), UTY | CCL18, CFHR1, CFHR3, DGAT1, HCG27\\HLA-C (rs3132506, rs3130473), HCG4P6, HLA-A, HLA-DR4, HLA-DRB1*01, HLA-DRB1*15, ISG20, PRKX (rs17335275), SIRPB1, TRGC2, TRGV9 |
| Profibrotic mediators | ADAMTSL5, ALX4, CDKN1A (rs2395655, rs733590), CDKN2B, CNN1, FAM13A (rs2609260), FAM83D, FZD5, GBP4, HFE (C282Y, S65C, H63D), IFI44L, IFIT3, IGFBP5, IGFBP6, KAAG1, OAS-2, PCDH9, SLC1A2, SLC22A11, SLC39A8, SLC6A14, SLCO4C1, TF, TGF- β1, TNF-RII, TNF-α, TOLLIP(rs111521887, rs5743894, rs5743890), ZFY, ZNF385B | ADAM3A, FKBP1A, HSF1, LRRC33 (rs7426656), MAP4K5, NEDD4, RSPO2 (rs611744), SDK1, SERP2, SFRP4 (rs16879765), TAB2, TGFB1, TGFbRI-3’UTR, TGFbRII-3’UTR3, TGFbRII-e5, TGFbRIII-3’UTR, WNT2 (rs4730775), WNT4 (rs7524102), WNT5A, Wnt7B (rs8140558, rs6519955), X89654, ZBTB40, ZC3H12D, Zf9–3’, Zf9–3’2, Zf9-c, ZIC1, ZIM3, ZNF160, ZNF264 |
DD, Dupuytren disease; ECM, extracellular matrix; IPF, idiopathic pulmonary fibrosis.
We extracted 129 fibrotic gene variants for the pulmonary system and 100 fibrotic gene variants for the hand. Among these fibroproliferative genes, 51 gene variants (22%) were associated with both pulmonary and MSK fibrosis (Table 3). Overlapping gene families included the following: ADAM [28,29], HLA [14,21,29–32], CARD [27,28], EIF [27,33,34], TGF [20,35,36], WNT [27,34,35,37], and ZNF genes [27,29,33,34,38] (Table 3). The majority of overlap was shared between IPF and Dupuytren disease pathologic states. However, HLA variants did overlap across IPF, Dupuytren disease, and knee arthrofibrosis [14]. The subsets of genetic variants from genes known to be involved in the MSK tissue development [28] or contractility properties in muscle tissue [33] were identified in IPF studies; however, these genes were not even examined in available MSK investigations. Genes with unknown fibrotic function were summarized in Appendix C.
Table 3.
Overview of Putative Functions of the Most Relevant Fibrotic Genes Overlapping in IPF and DD.
| Genes Family | Variants | Pathway | Samplesa | Agea | Malea | Racea | Tissue | Methods | Significanceb | Putative Function |
|---|---|---|---|---|---|---|---|---|---|---|
| ADAM genes | ADAMTSL5 [28] | IPF | 6/6 | 49(18) | 55 | NHWs | Lung biopsy | Sequencing | P = 7.68E-04 | Cell adhesion and protease, |
| ADAM3A [29] | DD | 13/11 | 69 (4) | 100 | NHWs | Palmar fasciectomy | Expression array | - | degradation of ECM | |
| X89654 [29] | ||||||||||
| HLA genes | HLA(rs3117116) [30] | IPF | 878/2017 | - | - | NHWs | Lung biopsy | GWAS | P = .02 | Immune system regulation |
| HLA-DRB1*15 [31] | IPF | 79/196 | 67(1) | 73 | - | Lung tissue/molecular | Molecular typing | P = .004 | ||
| HLA-A [29] | DD | 13/11 | 69 (4) | 100 | NHWs | Palmar fasciectomy | Expression array | - | ||
| HLADRBV01 [21] | DD | 121/51 | - | 100 | NHWs | Blood | PCR-SSPs | P = .02 | ||
| HCG27ǁHLA-C (rs3132506 [32], | DD | 40/40 | 60 (11) | 72 | Caucasians | Saliva | GWAS | P = 5.84E-05 | ||
| P = 5.31E-05 | ||||||||||
| rs3130473 [32]) | DD | 13/11 | 69 (4) | 100 | NHWs | Palmar fasciectomy | Expression array | - | ||
| HCG2P7 [29] | ||||||||||
| HCG4P6 [29] | ||||||||||
| CARD genes | CARD10 [28] | IPF | 6/6 | 49 (18) | 55 | NHWs | Lung biopsy | Sequencing | P = 6.73E-04 | Apoptosis signaling |
| CARD11 [27] | DD | 5408/9961 | - | - | - | Blood | GWAS | P = 2.89E-08 | ||
| EIF genes | EIF-1AY [33] | IPF | 22/11 | 62 (6) | 77 | - | Lung tissue | Expression array | P < .0001 | Apoptotic pathway in fibroblast |
| EIF-3E [27] | DD | 5408/9961 | - | - | - | Blood | GWAS | P = 3.7E-19 | ||
| TGF genes | TGF-B1 [20] | IPF | 128/140 | 66 (10) | 63 | Whites | - | - | P = .002 | Cell proliferation, stimulates sustained |
| TGFbRI-3’UTR [36] | DD | 184/181 | 63 (10) | 82 | Caucasians | Blood | PCR-RFLP | P = .048 | production of collagen | |
| TAB2 [27] | DD | 5408/9961 | - | - | - | Blood | GWAS | P = 2.03E-08 | ||
| ZNF genes | ZFY[33] | IPF | 22/11 | 62 (6) | 77 | - | Lung tissue | Expression array | P < .0001 | Transcription repressor and apoptosis |
| ZNF385B [33] | - | signaling | ||||||||
| DMRT1 [27] | DD | 5408/9961 | - | - | - | Blood | GWAS | P = .0237 | ||
| DMRT2 [27], (rs10809650 [37] | DD | 960/3117 | - | - | Caucasians | Blood/Saliva | GWAS | P = 6.2E-09 | ||
| rs1080964 [37]), | P = 1.2E-08 | |||||||||
| DMRT3 [27] | DD | 5408/9961 | - | - | - | Blood | GWAS | P = 9.76E-12 | ||
| ZNF160 [29] | DD | 4/10 | 69 (4) | 100 | Whites | Nodules | Expression array | - | ||
| ZNF264 [27] | DD | 5408/9961 | - | - | - | Blood | GWAS | P = 1.42E-13 | ||
| ZBTB40 [27] | P = 7.68E-12 | |||||||||
| ZC3H12D [27] | P = 2.03E-08 | |||||||||
| Zf9–3’ [38] | DD | 138/255 | 61 (11) | 86 | Caucasians | Blood | PCR-RFLP | P = .0025 | ||
| ZIM3 [27] | DD | 5408/9961 | - | - | - | Blood | GWAS | P = 1.42E-13 | ||
| WNT genes | FZD5 [33] | IPF | 22/11 | 62 (6) | 77 | - | Lung tissue | Expression array | P = .0237 | Wnt signaling regulating cell growth and differentiation in specific cell types, including regulation of cell fate and |
| WNT2 (rs4730775 [34]) | DD | 960/3317 | - | - | Caucasians | Blood/Saliva | GWAS | P = 3.0E-08 | ||
| WNT4 | ||||||||||
| (rs7524102 [34]) | P = 2.8E-09 | patterning during embryogenesis. | ||||||||
| WNT7B (rs8140558 [34], | P = 1.2E-22 | |||||||||
| rs6519955 [34]) | P = 3.2E-33 | |||||||||
| RSPO2 (rs611744 [34]) | P = 7.9E-15 | |||||||||
| SFRP4 | ||||||||||
| (rs16879765 [34]) | P = 5.6E-39 | |||||||||
| EBF2 [27] | DD | 5408/9961 | - | - | - | Blood | GWAS | P = 3.14E-11 |
ADAM, ADAM metallopeptidase; CARD, caspase recruitment domain; DD, Dupuytren disease; ECM, extracellular matrix; EIF, eukaryotic translation initiation factor; GWAS, genome-wide association study; HLA, major histocompatibility complex; IPF, idiopathic pulmonary fibrosis; NHW, non-Hispanic white; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; PCR-SSPs, polymerase chain reaction-sequence-specific primers; TGF, transforming growth factor; ZNF, zinc finger protein.
Samples: number of inclusions (fibrosis/control) and demographics from fibrosis group when noted. Age expressed as mean and standard deviation (SD), Male gender expressed in %.
Significance: P value after correcting population if available. In cases of genes significance involved in several articles, the lowest P value is reported.
Discussion
The identification of genetic determinants of fibroproliferative processes is required for a more robust understanding of how fibrosis in multiple tissues is caused at the cellular level in humans. Advancing our understanding of genetically controlled cellular mechanisms that mediate fibrosis will be key to guide patient-specific strategies to predict, diagnose, and control the risk of pathologic fibrogenesis [39]. Previous investigations have highlighted the “two-hit disease” as the hallmark of fibrotic diseases that are not hereditary—a patient’s genetic predisposition is the first hit, but is not activated until a second hit occurs—that is an environmental stressor that ultimately tips the balance from normal homeostasis to pathologic fibrosis [40]. Largely, these studies have focused on the solid organ fibrosis [8,9] and so far, no literature review has investigated the possibility of an overlapping genetic predisposition between cardiac, pulmonary, and MSK systems. The present systematic review provides a comprehensive evaluation of the existing literature and important information about shared genetic variations among pulmonary and hand systems.
Evidence of Genetic Predisposition
To evaluate evidence of genetic predisposition, the present investigation sought to identify genetic variation at the DNA level. Contemporary studies most often evaluate these differences via a genome-wide association study and report findings in terms of associated variants [19,23,26,27,30,32,35,41–43]. With these specific criteria, there was a large mismatch between the studies included for screening (n = 2373 articles) and those included for final analysis (n = 52 articles). Despite our narrow focus, this review was able to demonstrate shared genetic variation between the pulmonary and MSK systems. Genomic research is more prevalent and advanced for pulmonary fibrotic diseases compared to MSK disorders. The skewed representation of pulmonary fibrosis studies is secondary to the catastrophic clinical course and high mortality associated with this disease [8]. Nevertheless, the findings of the present study suggest that the current understanding of the genetic variation of fibroproliferative processes among the included systems is limited to data on IPF and Dupuytren disease. Despite clinical evidence and established molecular pathways, the present review identified only a single study related to knee fibrosis [14] and no studies related to cardiac or shoulder fibrosis.
It should be noted that failure to discover genes related to cardiac and shoulder diseases may be related to the study design. A dearth of information regarding cardiac fibrosis was in relation to ventricular fibrosis after chronic cardiomyopathy (ie, congestive heart failure, arrhythmia, or myocarditis). These disease states did not represent the idiopathic nature this review aimed to identify, and hence were excluded. There have been prior studies that have revealed an association between patients who are HLA-B27 positive and have adhesive capsulitis of the shoulder (when compared to controls) [44–48], but our current investigation and others did not identify the genetic predisposition in relation to adhesive capsulitis of the shoulder. These older studies were not included in the present investigation because they were published before the human genome sequence was established and used methodology that is no longer considered state of the art. Lastly, MeSH terms using genetic predisposition were associated with a different focus that included inheritance in family relatives, positive family history, and genomic expression that did not correspond to gene discovery.
Genes Involved in the Fibrotic Pathway
Pathways leading to idiopathic fibrosis have been largely overlooked in the literature. Excluding pulmonary-specific genes, this review highlighted a common pathway supported by genetic variants found in IPF and Dupuytren disease, including ECM remodeling and fibroblast regulation, immune dysfunction and profibrotic mediators. Although Friedman et al [9] highlighted core features shared by chronic pathology among multiple solid organs (including the lung, liver, skin, and kidney), this review demonstrated that the same core features are shared by the pulmonary and MSK systems. Several responses to injury summarize what is understood about the fibrotic process [9]. Firstly, the genes involved in cell death and apoptosis regulation were very common in the literature and consistent in the lung and hand in our review. CARD, EIF, and ZNF genes are involved in apoptotic signaling and similar genes appear to be involved in the different organ systems [20,27–29,33–36]. Secondly, cell stress causes dysregulation of the metabolic pathways. Although markers of endoplasmic reticulum stress have been identified in the alveolar epithelial cells (IPF), some genes involved in the endoplasmic reticulum signaling were expressed in the hand, including SERP2 [27], TMEM [27], SDR16C5 [33], and SLN variants [33]. Genes involved in changes in adenosine 5’ triphosphate generation were also associated with this metabolic dysregulation. Furthermore, TGFβ plays a central role in the epithelial to mesenchymal transition and interaction with integrins. Some overlapping profibrotic mediators were found in this study and TGFβ-related variants (ie, TGFβ1, TGFβ2, and TGFβ3) also were found in the lung and hand [20,35,36]. Lastly, immune system dysfunction is another common pathway identified in the literature: immune modulation via HLA complex [14,21,29–32] and inflammatory response via interleukin genes [15,22,49–54]. So far, only HLA genes were commonly reported in literature regarding the fibrosis in the lung, hand, shoulder, and knee.
Perspectives in MSK Research
Importantly, genetic variants associated with profibrotic processes of the knee and shoulder were underrepresented in this review when compared to the robust evidence observed in the hand. This lack of evidence does not mean genetic variants exist, and instead represents an opportunity for investigation in MSK research in humans [55]. In the knee, there is a body of molecular evidence demonstrating the differential expression of genes throughout the pathogenesis of arthrofibrosis in animal models [56–59]. Our laboratory has recently identified 380 genes most significantly upregulated or downregulated within the first 2 weeks after traumatic insult and immobilization leading to severe permanent contracture in the rabbit [57]. In the shoulder, the genomic expression of an intercellular adhesion model was found in adhesive capsulitis [60]. The same fibrotic pathways have been validated using microarray data and quantitative reverse transcription polymerase chain reaction analysis: cell-cell adhesion, apoptosis, protein kinase activity, or inflammatory response.
So far, there are no studies specifically addressing the genetic determinants of arthrofibrosis in the context of TKA. A few clinical studies indirectly acknowledged a predisposition to arthrofibrosis via reports of the bilateral occurrence of arthrofibrosis [61,62] or manipulation under anesthesia after primary TKA in a subset of patients [63]. As suggested by Clement et al, stiffness could be a predictor of stiffness for staged TKA [63] and genetic predisposition may lead to a cumulative incidence of manipulations under anesthesia for bilateral procedures [64].
Although the pathogenesis of arthrofibrosis after TKA is incompletely understood, translational work has been critical to identifying the importance of myofibroblast and related cellular pathways at play in the process [56]. Under pathologic conditions, myofibroblasts are activated by local growth factors (eg, TGFβ) resulting in increased collagenous and noncollagenous (eg, fibronectin, decorin) ECM production. This dysregulated accumulation of ECM elements results in tissue deformation and loss of function after TKA.
Despite an improved understanding of the fundamentals of arthrofibrosis over the past decade, understanding of the specific genes that activate cellular pathways and insights into the modulation of those pathways remains elusive. Recently, in an effort to better understand the pathogenesis of arthrofibrosis at the molecular level, investigators in our laboratory used the RNA-sequencing analysis of posterior capsular tissue to identify deferential gene expression between arthrofibrotic and nonarthrofibrotic knees [65]. Importantly, this study demonstrated that deferentially expressed genes were enriched in ECM organization pathways that participate in cell-to-cell adhesion, wound healing, and post-translational collagenous protein modification [65]. Although this represents significant progress and enables identification of druggable pathways, it requires action after diagnosis. Future endeavors investigating changes at the level of DNA are required to identify patients at risk for arthrofibrosis development to foster the development of preventative strategies.
Taken together, these findings identify specific genes that play a role in fibrotic processes, and identification of variants within these genes holds both therapeutic and diagnostic benefits. For the very first time, cellular direct reprogramming and molecular targets could indeed be a part of novel therapeutic options, also known as theragnostic (a treatment strategy that combines therapeutics with diagnostics). Theragnostics in MSK fibrosis, including arthrofibrosis after TKA, may become a successful component of personalized medicine.
Conclusion
Despite shared genetic variations in both the lung and hand, there remains limited information about genetic variants associated with fibrosis in other MSK regions. This finding establishes the necessity of further studies to elucidate the genetic determinants involved in the knee, shoulder, and other joint fibrotic pathways.
Acknowledgments
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of the National Institutes of Health (NIH) under Award Number AR072597 (Principal Investigator Matthew P. Abdel, MD) and the Anna-Maria and Stephen Kellen Foundation (Principal Investigator Matthew P. Abdel, MD). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Louis Dagneaux, MD, also reports funding from Société Française de Chirurgie Orthopédique et Traumatologique (SOFCOT) and the University of Montpellier (MUSE Explorer#2).
Appendix A
Ovid
Database(s): EBM Reviews—Cochrane Central Register of Controlled Trials August 2019, EBM Reviews—Cochrane Database of Systematic Reviews 2005 to October 3, 2019, Embase 1974 to October 4, 2019, Ovid MEDLINE(R) and Epub Ahead of Print, In-Process & Other Non-Indexed Citations and Daily 1946 to October 4, 2019 Search Strategy:
| # | Searches | Results |
|---|---|---|
| 1 | exp Fibrosis/ge [Genetics] | 1551 |
| 2 | exp Pulmonary Fibrosis/ge [Genetics] | 1419 |
| 3 | exp Endomyocardial Fibrosis/ge [Genetics] | 80 |
| 4 | exp Dupuytren Contracture/ge [Genetics] | 166 |
| 5 | (arthrofibroses or arthrofibrosis or fibroses or fibrosis).ti,ab,hw,kw. | 547,154 |
| 6 | Genetics/ | 704,070 |
| 7 | Genomics/ | 104,087 |
| 8 | exp Genetic Predisposition to Disease/ | 281,969 |
| 9 | (gene or genes or genetic or genetics or genomic or genomics).ti,ab,hw,kw. | 7,915,383 |
| 10 | 6 or 7 or 8 or 9 | 7,915,383 |
| 11 | exp Joint Diseases/ge [Genetics] | 16,780 |
| 12 | exp Joints/ | 668,503 |
| 13 | exp Shoulder Joint/ or exp Shoulder/ | 94,461 |
| 14 | exp Hand/ or exp Hand Joints/ | 175,741 |
| 15 | exp Knee/ or exp Knee Joint/ | 133,234 |
| 16 | exp Heart Diseases/ge [Genetics] | 47,024 |
| 17 | (arthrofibroses or arthrofibrosis or cardiac or dupuytren* or endomyocardial or hand or hands or heart orjoint orjoints or knee or knees or lung or lungs or myocardial or pulmonary or shoulder).ti,ab,hw,kw. | 8,874,025 |
| 18 | 11 and (1 or 5) | 133 |
| 19 | 12 and (1 or (5 and 10)) | 607 |
| 20 | 13 and (1 or (5 and 10)) | 91 |
| 21 | 14 and (1 or (5 and 10)) | 103 |
| 22 | 15 and (1 or (5 and 10)) | 118 |
| 23 | 16 and (1 or 5) | 1933 |
| 24 | (arthrofibroses or arthrofibrosis or fibroses or fibrosis).ti. and 10 and (arthrofibroses or arthrofibrosis or cardiac or dupuytren* or endomyocardial or hand or hands or heart or joint or joints or knee or knees or lung or lungs or myocardial or pulmonary or shoulder).ti. | 8844 |
| 25 | 1 and 17 | 346 |
| 26 | 2 or 3 or 4 or 18 or 19 or 20 or 21 or 22 or 23 or 24 or 25 | 12,326 |
| 27 | (case* adj3 report*).mp,pt. | 4,842,191 |
| 28 | exp Genetic Diseases, Inborn/ | 1,707,136 |
| 29 | (“Alagille Syndrome*” or “alpha 1-Antitrypsin Deficienc*” or “Ataxia Telangiectasia*” or “Autoimmune Lymphoproliferative Syndrome*” or “Brugada Syndrome*” or CADASIL or “Camurati-Engelmann Syndrome*” or “CHARGE Syndrome*” or Cherubism or “Chromosome disorder*” or Ciliopath* or “congenital Adrenal Hyperplasia*” or “congenital Angioedema*” or “congenital Autoinflammatory Disease*” or “congenital Eye Disease*” or “congenital hemolytic Anemia*” or “congenital hypertrophic Cardiomyopath*” or “congenital hypoplastic Anemia*” or “congenital Multiple Lipomatos*” or “Congenital Myasthenic Syndrome*” or “congenital NeoplasticNeoplastic Syndrome*” or “Congenital Pain Insensitivity” or “Costello Syndrome*” or “Cystic Fibros*” or “Donohue Syndrome*” or Dwarfism or “familial Adrenal Hyperplasia*” or “familial Angioedema*” or “familial Autoinflammatory Disease*” or “familial Eye Disease*” or “familial hemolytic Anemia*” or “familial hypertrophic Cardiomyopath*” or “familial hypoplastic Anemia*” or “Familial Multiple Lipomatos*” or “familial Myasthenic Syndrome*” or “familial NeoplasticNeoplastic Syndrome*” or “familial Pain Insensitivity” or “Frasier Syndrome*” or “GATA2 Deficienc*” or “genetic Skin Disease*” or “Graves Ophthalmopath*” or “Hajdu-Cheney Syndrome*” or Hemoglobinopath* or “hereditary Adrenal Hyperplasia*” or “hereditary Angioedema*” or “Hereditary Autoinflammatory Disease*” or “hereditary Eye Disease*” or “hereditary hemolytic Anemia*” or “hereditary hypertrophic Cardiomyopath*” or “hereditary hypoplastic Anemia*” or “hereditary Multiple Lipomatos*” or “hereditary Myasthenic Syndrome*” or “Hereditary NeoplasticNeoplastic Syndrome*” or “hereditary Pain Insensitivity” or “Heredodegenerative disorder*” or “Hyper-IgM Immunodeficiency Syndrome*” or “Inborn Errors of Metabolism” or “inborn Genetic Disease*” or “inherited Blood Coagulation disorder*” or “Kallmann Syndrome*” or “Kartagener Syndrome*” or “Lennox Gastaut Syndrome*” or “Loeys-Dietz Syndrome*” or “Marfan Syndrome*” or “Muscular Dystroph*” or “Nail-Patella Syndrome*” or “Oculocerebrorenal Syndrome*” or “Orofaciodigital Syndrome*” or Osteochondrodysplasia* or “Osteogenesis Imperfecta” or “Pelger-Huet Anomaly” or “Primary hypertrophicOsteoarthropath*” or Pycnodysostos* or “rheumatoid disease*” or scleroderma* or “Werner Syndrome*” or “X-Linked Genetic Disease*” or “Yellow Nail Syndrome*” or “Y-Linked Genetic Disease*”).ti,ab,hw,kw. | 471,956 |
| 30 | exp Atrial Fibrillation/ | 116,660 |
| 31 | “atrial fibrillation*”.ti,ab,hw,kw. | 223,252 |
| 32 | exp Sarcoidosis/ | 58,514 |
| 33 | sarcoidosis.ti,ab,hw,kw. | 70,450 |
| 34 | Myositis/ | 22,687 |
| 35 | myositis.ti,ab,hw,kw. | 37,853 |
| 36 | exp Epigenomics/ | 74,627 |
| 37 | (epigenetic* or epigenomic*).ti,ab,hw,kw. | 208,463 |
| 38 | exp Smoking/ | 520,985 |
| 39 | smoking.ti,ab,hw,kw. | 761,622 |
| 40 | or/27–39 | 7,503,536 |
| 41 | 26 not 40 | 8531 |
| 42 | limit 41 to english language [Limit not valid in CDSR; records were retained] | 8210 |
| 43 | limit 42 to yr=“2000 -Current” | 7798 |
| 44 | (exp animals/ or exp nonhuman/) not exp humans/ | 10,852,970 |
| 45 | ((alpaca or alpacas or amphibian or amphibians or animal or animals or antelope or armadillo or armadillos or avian or baboon or baboons or beagle or beagles or bee or bees or bird or birds or bison or bovine or buffalo or buffaloes or buffalos or “c elegans” or “Caenorhabditis elegans” or camel or camels or canine or canines or carp or cats or cattle or chick or chicken or chickens or chicks or chimp or chimpanze or chimpanzees or chimps or cow or cows or “D melanogaster” or “dairy calf” or “dairy calves” or deer or dog or dogs or donkey or donkeys or drosophila or “Drosophila melanogaster” or duck or duckling or ducklings or ducks or equid or equids or equine or equines or feline or felines or ferret or ferrets or finch or finches or fish or flatworm or flatworms or fox or foxes or frog or frogs or “fruit flies” or “fruit fly” or “G mellonella” or “Galleria mellonella” or geese or gerbil or gerbils or goat or goats or goose or gorilla or gorillas or hamster or hamsters or hare or hares or heifer or heifers or horse or horses or insect or insects orjellyfish or kangaroo or kangaroos or kitten or kittens or lagomorph or lagomorphs or lamb or lambs or llama or llamas or macaque or macaques or macaw or macaws or marmoset or marmosets or mice or minipig or minipigs or mink or minks or monkey or monkeys or mouse or mule or mules or nematode or nematodes or octopus or octopuses or orangutan or “orang-utan” or orangutans or “orang-utans” or oxen or parrot or parrots or pig or pigeon or pigeons or piglet or piglets or pigs or porcine or primate or primates or quail or rabbit or rabbits or rat or rats or reptile or reptiles or rodent or rodents or ruminant or ruminants or salmon or sheep or shrimp or slug or slugs or swine or tamarin or tamarins or toad or toads or trout or urchin or urchins or vole or voles or waxworm or waxworms or worm or worms or xenopus or “zebra fish” or zebrafish) not (human or humans or patient or patients)).ti,ab,hw,kw. | 9,357,844 |
| 46 | 43 not (44 or 45) | 4075 |
| 47 | limit 46 to (letter or conference abstract or editorial or erratum or note or addresses or autobiography or bibliography or biography or blogs or comment or dictionary or directory or interactive tutorial or interview or lectures or legal cases or legislation or news or newspaper article or overall or patient education handout or periodical index or portraits or published erratum or video-audio media or webcasts) [Limit not valid in CCTR, CDSR,Embase, Ovid MEDLINE(R),Ovid MEDLINE(R) Daily Update, Ovid MEDLINE(R) In-Process,Ovid MEDLINE(R) Publisher; records were retained] | 976 |
| 48 | 46 not 47 | 3099 |
| 49 | remove duplicates from 48 | 2390 |
Scopus
TITLE(arthrofibroses OR arthrofibrosis OR fibroses OR fibrosis)
TITLE-ABS-KEY(gene OR genes OR genetic OR genetics OR genomic OR genomics)
TITLE(arthrofibroses OR arthrofibrosis OR cardiac OR dupuytren* OR endomyocardial OR hand OR hands OR heart OR joint OR joints OR knee OR knees OR lung OR lungs OR myocardial OR pulmonary OR shoulder)
PUBYEAR AFT 1999 AND LANGUAGE(english)
1 and 2 and 3 and 4
TITLE-ABS-KEY((case* W/3 report*) OR “Alagille Syndrome*” OR “alpha 1-Antitrypsin Deficienc*” OR “Ataxia Telangiectasia*” OR “atrial fibrillation*” OR “Autoimmune Lymphoproliferative Syndrome*” OR “Brugada Syndrome*” OR CADASIL OR “Camurati-Engelmann Syndrome*” OR “CHARGE Syndrome*” OR Cherubism OR “Chromosome disorder*” OR Ciliopath* OR “congenital Adrenal Hyperplasia*” OR “congenital Angioedema*” OR “congenital Autoinflammatory Disease*” OR “congenital Eye Disease*” OR “congenital hemolytic Anemia*” OR “congenital hypertrophic Cardiomyopath*” OR “congenital hypoplastic Anemia*” OR “congenital Multiple Lipomatos*” OR “Congenital Myasthenic Syndrome*” OR “congenital NeoplasticNeoplastic Syndrome*” OR “Congenital Pain Insensitivity” OR “Costello Syndrome*” OR “Cystic Fibros*” OR “Donohue Syndrome*” OR Dwarfism OR epigenetic* OR epigenomic* OR “familial Adrenal Hyperplasia*” OR “familial Angioedema*” OR “familial Autoinflammatory Disease*” OR “familial Eye Disease*” OR “familial hemolytic Anemia*” OR “familial hypertrophic Cardiomyopath*” OR “familial hypoplastic Anemia*” OR “Familial Multiple Lipomatos*” OR “familial Myasthenic Syndrome*” OR “familial NeoplasticNeoplastic Syndrome*” OR “familial Pain Insensitivity” OR “Frasier Syndrome*” OR “GATA2 Deficienc*” OR “genetic Skin Disease*” OR “Graves Ophthalmopath*” OR “Hajdu-Cheney Syndrome*” OR Hemoglobinopath* OR “hereditary Adrenal Hyperplasia*” OR “hereditary Angioedema*” OR “Hereditary Autoinflammatory Disease*” OR “hereditary Eye Disease*” OR “hereditary hemolytic Anemia*” OR “hereditary hypertrophic Cardiomyopath*” OR “hereditary hypoplastic Anemia*” OR “hereditary Multiple Lipomatos*” OR “hereditary Myasthenic Syndrome*” OR “Hereditary NeoplasticNeoplastic Syndrome*” OR “hereditary Pain Insensitivity” OR “Heredodegenerative disorder*” OR “Hyper-IgM Immunodeficiency Syndrome*” OR “Inborn Errors of Metabolism” OR “inborn Genetic Disease*” OR “inherited Blood Coagulation disorder*” OR “Kallmann Syndrome*” OR “Kartagener Syndrome*” OR “Lennox Gastaut Syndrome*” OR “Loeys-Dietz Syndrome*” OR “Marfan Syndrome*” OR “Muscular Dystroph*” OR myositis OR “Nail-Patella Syndrome*” OR “Oculocerebrorenal Syndrome*” OR “Orofaciodigital Syndrome*” OR Osteochondrodysplasia* OR “Osteogenesis Imperfecta” OR “Pelger-Huet Anomaly” OR “Primary hypertrophicOsteoarthropath*” OR Pycnodysostos* OR “rheumatoid disease*” OR sarcoidosis OR scleroderma* OR smoking OR “Werner Syndrome*” OR “X-Linked Genetic Disease*” OR “Yellow Nail Syndrome*” OR “Y-Linked Genetic Disease*”)
5 and not 6
TITLE-ABS-KEY((alpaca OR alpacas OR amphibian OR amphibians OR animal OR animals OR antelope OR armadillo OR armadillos OR avian OR baboon OR baboons OR beagle OR beagles OR bee OR bees OR bird OR birds OR bison OR bovine OR buffalo OR buffaloes OR buffalos OR “c elegans” OR “Caenorhabditis elegans” OR camel OR camels OR canine OR canines OR carp OR cats OR cattle OR chick OR chicken OR chickens OR chicks OR chimp OR chimpanze OR chimpanzees OR chimps OR cow OR cows OR “D melanogaster” OR “dairy calf” OR “dairy calves” OR deer OR dog OR dogs OR donkey OR donkeys OR drosophila OR “Drosophila melanogaster” OR duck OR duckling OR ducklings OR ducks OR equid OR equids OR equine OR equines OR feline OR felines OR ferret OR ferrets OR finch OR finches OR fish OR flatworm OR flatworms OR fox OR foxes OR frog OR frogs OR “fruit flies” OR “fruit fly” OR “G mellonella” OR “Galleria mellonella” OR geese OR gerbil OR gerbils OR goat OR goats OR goose OR gorilla OR gorillas OR hamster OR hamsters OR hare OR hares OR heifer OR heifers OR horse OR horses OR insect OR insects OR jellyfish OR kangaroo OR kangaroos OR kitten OR kittens OR lagomorph OR lagomorphs OR lamb OR lambs OR llama OR llamas OR macaque OR macaques OR macaw OR macaws OR marmoset OR marmosets OR mice OR minipig OR minipigs OR mink OR minks OR monkey OR monkeys OR mouse OR mule OR mules OR nematode OR nematodes OR octopus OR octopuses OR orangutan OR “orang-utan” OR orangutans OR “orang-utans” OR oxen OR parrot OR parrots OR pig OR pigeon OR pigeons OR piglet OR piglets OR pigs OR porcine OR primate OR primates OR quail OR rabbit OR rabbits OR rat OR rats OR reptile OR reptiles OR rodent OR rodents OR ruminant OR ruminants OR salmon OR sheep OR shrimp OR slug OR slugs OR swine OR tamarin OR tamarins OR toad OR toads OR trout OR urchin OR urchins OR vole OR voles OR waxworm OR waxworms OR worm OR worms OR xenopus OR “zebra fish” OR zebrafish) AND NOT (human OR humans or patient or patients))
7 and not 8
DOCTYPE(le) OR DOCTYPE(ab) OR DOCTYPE(ed) OR DOCTYPE(bk) OR DOCTYPE(er) OR DOCTYPE(no) OR DOCTYPE(sh)
9 and not 10
INDEX(embase) OR INDEX(medline) OR PMID(0* OR 1* OR 2* OR 3* OR 4* OR 5* OR 6* OR 7* OR 8* OR 9*)
11 and not 12
Appendix B
Level of Evidence and Quality Assessment According to the Newcastle—Ottawa Quality Assessment Scale.
| Authors, Year | LOE | Selection | Comparability | Exposure |
|---|---|---|---|---|
| Xaubet et al 2003 [20] | III | ** | * | ** |
| Jonsson et al 2013 [21] | III | *** | ** | ** |
| Ng et al 2017 [27] | III | *** | ** | ** |
| Huang et al 2014 [28] | III | *** | ** | ** |
| Shih et al 2010 [29] | III | ** | * | * |
| Fingerlin et al 2016 [30] | III | *** | * | ** |
| Xue etal 2011 [31] | III | **** | * | ** |
| Ojwhang et al 2010 [32] | III | *** | - | ** |
| Cecchini et al 2018 [33] | III | ** | * | ** |
| Anderson et al 2013 [34] | III | *** | ** | ** |
| Bayat et al 2003 [36] | III | *** | * | ** |
| Dolmans etal 2011 [37] | III | *** | * | ** |
| Bayat et al 2003 [38] | III | *** | * | ** |
Newcastle—Ottawa Quality Assessment Scale Case Control Studies
Note: A study can be awarded a maximum of one star for each numbered item within the selection and exposure categories. A maximum of 2 stars can be given for comparability.
Selection
-
1
Is the case definition adequate?
- yes, with independent validation Ø
- yes, eg, record linkage or based on self-reports
- no description.
Appendix C
Genes with Unknown or Nonspecific Fibrotic Function From the Literature Review.
| Lung (Idiopathic Pulmonary Fibrosis) | Hand (Dupuytren’s Disease) |
|---|---|
|
AHNAK2, ATP8A1, C1orf162, CTD-2245O6.1, F11, FMO5, LNX2, LRP2, MOP-1, NECAB1, OBFC1, PARN, PDL1M3, PHACTR1, PRUNE2, PTAFR, RP13–870H17.3, RSAD2, SDR16C5, hTERT, TERC, TERT, TMEM38A, TXLNGY, WARS |
ALDH2, ATL1, BOP1, C8orf34, CFDP1, CHST6, CYCSP14, DHDH, DUXA, GNB1, HSD17B7, L1NC01230, LOC100129495, LOC100131502, LOC100132514, LOC100132626, LOC100132627, LOC100505718, LOC727945, M1R8079, M1RLET7BHG, OR10D3P, OR8F1P, RAB31, SM1M2, SULF1, SUMO4 |
-
2Representativeness of the cases
- consecutive or obviously representative series of cases Ø
- potential for selection biases or not stated.
-
3Selection of controls
- community controls Ø
- hospital controls
- no description.
-
4Definition of controls
- no history of disease (endpoint) Ø
- no description of source.
Comparability
- Comparability of cases and controls on the basis of the design or analysis
- study controls for _______________ (Select the most important factor.) Ø
- study controls for any additional factor Ø (This criteria could be modified to indicate specific control for a second important factor.)
Exposure
- Ascertainment of exposure
- secure record (eg, surgical records) Ø
- structured interview where blind to case/control status Ø
- interview not blinded to case/control status
- written self-report or medical record only
- no description.
- Same method of ascertainment for cases and controls
- yes Ø
- no.
- Nonresponse rate
- same rate for both groups Ø
- nonrespondents described
- rate different and no designation.
Footnotes
Investigation was performed at the Mayo Clinic, Rochester, MN.
One or more of the authors of this paper have disclosed potential or pertinent conflicts of interest, which may include receipt of payment, either direct or indirect, institutional support, or association with an entity in the biomedical field which may be perceived to have potential conflict of interest with this work. For full disclosure statements refer to https://doi.org/10.1016/j.arth.2020.05.070.
References
- [1].Gulati A, Jabbour A, Ismail TF, Guha K, Khwaja J, Raza S, et al. Association of fibrosis with mortality and sudden cardiac death in patients with nonischemic dilated cardiomyopathy. JAMA 2013;309:896–908. 10.1001/jama.2013.1363. [DOI] [PubMed] [Google Scholar]
- [2].Dulai PS, Singh S, Patel J, Soni M, Prokop LJ, Younossi Z, et al. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: systematic review and meta-analysis. Hepatology 2017;65:1557–65. 10.1002/hep.29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sgalla G, Biffi A, Richeldi L. Idiopathic pulmonary fibrosis: diagnosis, epidemiology and natural history. Respirology 2016;21:427–37. 10.1111/resp.12683. [DOI] [PubMed] [Google Scholar]
- [4].Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med 2018;378: 1811–23. 10.1056/NEJMra1705751. [DOI] [PubMed] [Google Scholar]
- [5].Dibenedetti DB, Nguyen D, Zografos L, Ziemiecki R, Zhou X. Prevalence, incidence, and treatments of Dupuytren’s disease in the United States: results from a population-based study. Hand 2011;6:149–58. 10.1007/s11552-010-9306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol 2008;214: 199–210. 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rockey DC, Bell PD, Hill JA. Fibrosis-a common pathway to organ injury and failure. N Engl J Med 2015;372:1138–49. 10.1056/NEJMra1300575. [DOI] [PubMed] [Google Scholar]
- [8].Friedman SL. Clarity and challenges in tissue fibrosis In: Nakao K, Minato N, Uemoto S, editors. Innov. Med Tokyo: Springer Japan; 2015. p. 187–94. [PubMed] [Google Scholar]
- [9].Friedman SL, Sheppard D, Duffield JS, Violette S. Therapy for fibrotic diseases: nearing the starting line. Sci Transl Med 2013;5 10.1126/scitranslmed.3004700 167sr1–167sr1 [DOI] [PubMed] [Google Scholar]
- [10].Schwartz DA. Idiopathic pulmonary fibrosis is a genetic disease involving mucus and the peripheral airways. Ann Am Thorac Soc 2018;15:S192–7. 10.1513/AnnalsATS.201802-144AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tibbo ME, Limberg AK, Salib CG, Ahmed AT, van Wijnen AJ, Berry DJ, et al. Acquired idiopathic stiffness after total knee arthroplasty: a systematic review and meta-analysis. J Bone Joint Surg Am 2019;101:1320–30. 10.2106/JBJS.18.01217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cheuy VA, Foran JRH, Paxton RJ, Bade MJ, Zeni JA, Stevens-Lapsley JE. Arthrofibrosis associated with total knee arthroplasty. J Arthroplasty 2017;32: 2604–11. 10.1016/j.arth.2017.02.005. [DOI] [PubMed] [Google Scholar]
- [13].Stang A Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- [14].Skutek M, Elsner H-A, Slateva K, Mayr H-O, Weig T-G, van Griensven M, et al. Screening for arthrofibrosis after anterior cruciate ligament reconstruction: analysis of association with human leukocyte antigen. Arthroscopy 2004;20: 469–73. [DOI] [PubMed] [Google Scholar]
- [15].Alhamad EH, Cal JG, Shakoor Z, Almogren A, AlBoukai AA. Cytokine gene polymorphisms and serum cytokine levels in patients with idiopathic pulmonary fibrosis. BMC Med Genet 2013;14:66 10.1186/1471-2350-14-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bournazos S, Grinfeld J, Alexander KM, Murchison JT, Wallace WA, McFarlane P, et al. Association of FcgammaRIIa R131H polymorphism with idiopathic pulmonary fibrosis severity and progression. BMC Pulm Med 2010;10:51 10.1186/1471-2466-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jiang H, Hu Y, Shang L, Li Y, Yang L, Chen Y. Association between MUC5B polymorphism and susceptibility and severity of idiopathic pulmonary fibrosis. Int J Clin Exp Pathol 2015;8:14953–8. [PMC free article] [PubMed] [Google Scholar]
- [18].Korthagen NM, van Moorsel CHM, Barlo NP, Kazemier KM, Ruven HJT, Grutters JC. Association between variations in cell cycle genes and idiopathic pulmonary fibrosis. PLoS One 2012;7:e30442 10.1371/journal.pone.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Noth I, Zhang Y, Ma S-F, Flores C, Barber M, Huang Y, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med 2013;1:309–17. 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Xaubet A, Marin-Arguedas A, Lario S, Ancochea J, Morell F, Ruiz-Manzano J, et al. Transforming growth factor-β1 gene polymorphisms are associated with disease progression in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2003;168:431–5. 10.1164/rccm.200210-1165OC. [DOI] [PubMed] [Google Scholar]
- [21].Jonsson T, Gudmundsson KG, Bjarnadottir K, Hjalmarsdotti IB, Gudmundsson S, Arngrimsson R. Association of HLA-DRB1*01 with Dupuytren’s disease. Scand J Rheumatol 2013;42:45–7. 10.3109/03009742.2012.713982. [DOI] [PubMed] [Google Scholar]
- [22].Kishore A, Zizkova V, Kocourkova L, Petrkova J, Bouros E, Nunes H, et al. Association study for 26 candidate loci in idiopathic pulmonary fibrosis patients from four European populations. Front Immunol 2016;7:274 10.3389/fimmu.2016.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lorenzo-Salazar JM, Ma S-F, Jou J, Hou P-C, Guillen-Guio B, Allen RJ, et al. Novel idiopathic pulmonary fibrosis susceptibility variants revealed by deep sequencing. ERJ Open Res 2019;5 10.1183/23120541.00071-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Peljto AL, Zhang Y, Fingerlin TE, Ma S-F, Garcia JGN, Richards TJ, et al. Association between the MUC5B promoter polymorphism and survival in patients with idiopathic pulmonary fibrosis. JAMA 2013;309:2232–9. 10.1001/jama.2013.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med 2011;364:1503–12. 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet 2013;45:613–20. 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ng M, Thakkar D, Southam L, Werker P, Ophoff R, Becker K, et al. A genome-wide association study of Dupuytren disease reveals 17 additional variants implicated in fibrosis. Am J Hum Genet 2017;101:417–27. 10.1016/j.ajhg.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Huang SK, Scruggs AM, McEachin RC, White ES, Peters-Golden M. Lung fibroblasts from patients with idiopathic pulmonary fibrosis exhibit genome-wide differences in DNA methylation compared to fibroblasts from non-fibrotic lung. PLoS One 2014;9:e107055 10.1371/journal.pone.0107055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shih BB, Tassabehji M, Watson JS, McGrouther AD, Bayat A. Genome-wide high-resolution screening in Dupuytren’s disease reveals common regions of DNA copy number alterations. J Hand Surg 2010;35:1172–83. 10.1016/j.jhsa.2010.03.006. [DOI] [PubMed] [Google Scholar]
- [30].Fingerlin TE, Zhang W, Yang IV, Ainsworth HC, Russell PH, Blumhagen RZ, et al. Genome-wide imputation study identifies novel HLA locus for pulmonary fibrosis and potential role for auto-immunity in fibrotic idiopathic interstitial pneumonia. BMC Genet 2016;17:74 10.1186/s12863-016-0377-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Xue J, Gochuico BR, Alawad AS, Feghali-Bostwick CA, Noth I, Nathan SD, et al. The HLA class II allele DRB1*1501 is over-represented in patients with idiopathic pulmonary fibrosis. PLoS One 2011;6:e14715 10.1371/journal.pone.0014715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ojwang JO, Adrianto I, Gray-McGuire C, Nath SK, Sun C, Kaufman KM, et al. Genome-wide association scan of Dupuytren’s disease. J Hand Surg Am 2010;35:2039–45. 10.1016/j.jhsa.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cecchini MJ, Hosein K, Howlett CJ, Joseph M, Mura M. Comprehensive gene expression profiling identifies distinct and overlapping transcriptional profiles in non-specific interstitial pneumonia and idiopathic pulmonary fibrosis. Respir Res 2018;19:153 10.1186/s12931-018-0857-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Anderson ER, Ye Z, Caldwell MD, Burmester JK. SNPs previously associated with Dupuytren’s disease replicated in a North American cohort. Clin Med Res 2014;12:133–7. 10.3121/cmr.2013.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dolmans GH, de Bock GH, Werker PM. Dupuytren diathesis and genetic risk. J Hand Surg Am 2012;37:2106–11. 10.1016/j.jhsa.2012.07.017. [DOI] [PubMed] [Google Scholar]
- [36].Bayat A, Stanley JK, Watson JS, Ferguson MWJ, Ollier WER. Genetic susceptibility to Dupuytren’s disease: transforming growth factor beta receptor (TGFbR) gene polymorphisms and Dupuytren’s diseaseq n.d.:6. Br J Plast Surg 2003;56:328–33. [DOI] [PubMed] [Google Scholar]
- [37].Dolmans GH, Werker PM, Hennies HC, Furniss D, Festen EA, Franke L, et al. Wnt signaling and Dupuytren’s disease. N Engl J Med 2011;365:307–17. 10.1056/NEJMoa1101029. [DOI] [PubMed] [Google Scholar]
- [38].Bayat A, Watson JS, Stanley JK, Ferguson MWJ, Ollier WER. Genetic susceptibility to Dupuytren disease: association of Zf9 transcription factor gene. Plast Reconstr Surg 2003;111:2133–9. 10.1097/01.PRS.0000060531.98180.32. [DOI] [PubMed] [Google Scholar]
- [39].Wijnen AJ, Westendorf JJ. Epigenetics as a new frontier in orthopedic regenerative medicine and oncology. J Orthop Res 2019;37:1465–74. 10.1002/jor.24305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Boucher RC. Idiopathic pulmonary fibrosis — a sticky business. N Engl J Med 2011;364:1560–1. 10.1056/NEJMe1014191. [DOI] [PubMed] [Google Scholar]
- [41].Hobbs BD, Putman RK, Araki T, Nishino M, Gudmundsson G, Gudnason V, et al. Overlap of genetic risk between interstitial lung abnormalities and idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2019;24:24 10.1164/rccm.201903-0511OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Moore C, Blumhagen RZ, Yang IV, Walts A, Powers J, Walker T, et al. Resequencing study confirms that host defense and cell senescence gene variants contribute to the risk of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2019;200:199–208. 10.1164/rccm.201810-1891OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Staats KA, Wu T, Gan BS, O’Gorman DB, Ophoff RA. Dupuytren’s disease susceptibility gene, EPDR1, is involved in myofibroblast contractility. J Dermatol Sci 2016;83:131–7. 10.1016/j.jdermsci.2016.04.015. [DOI] [PubMed] [Google Scholar]
- [44].Bulgen DY, Hazleman BL, Voak D. HLA-B27 and frozen shoulder. Lancet 1976;1:1042–4. 10.1016/s0140-6736(76)92219-4. [DOI] [PubMed] [Google Scholar]
- [45].Noy S, Dekel S, Orgad S, Efter T, Mizrachi Y, Gazit E. HLA-B27 and frozen shoulder. Tissue Antigens 1981;17:251 10.1111/j.1399-0039.1981.tb00695.x. [DOI] [PubMed] [Google Scholar]
- [46].Rizk TE, Pinals RS. Histocompatibility type and racial incidence in frozen shoulder. Arch Phys Med Rehabil 1984;65:33–4. [PubMed] [Google Scholar]
- [47].Seignalet J, Sany J, Caillens JP, Lapinski H. Lack of association between HLA-B27 and frozen shoulder. Tissue Antigens 1981;18:364 10.1111/j.1399-0039.1981.tb01405.x. [DOI] [PubMed] [Google Scholar]
- [48].Prodromidis AD, Charalambous CP. Is there a genetic predisposition to frozen shoulder?: A systematic review and meta-analysis. JBJS Rev 2016;4:1 10.2106/JBJS.RVW.O.00007. [DOI] [PubMed] [Google Scholar]
- [49].Vasakova M, Striz I, Slavcev A, Jandova S, Dutka J, Terl M, et al. Correlation of IL-1alpha and IL-4 gene polymorphisms and clinical parameters in idiopathic pulmonary fibrosis. Scand J Immunol 2007;65:265–70. [DOI] [PubMed] [Google Scholar]
- [50].Ahn M-H, Park B-L, Lee S-H, Park S-W, Park J-S, Kim D-J, et al. A promoter SNP rs4073T>A in the common allele of the interleukin 8 gene is associated with the development of idiopathic pulmonary fibrosis via the IL-8 protein enhancing mode. Respir Res 2011;12:73 10.1186/1465-9921-12-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Barlo NP, van Moorsel CHM, Korthagen NM, Heron M, Rijkers GT, Ruven HJT, et al. Genetic variability in the IL1RN gene and the balance between interleukin (IL)-1 receptor agonist and IL-1beta in idiopathic pulmonary fibrosis. Clin Exp Immunol 2011;166:346–51. 10.1111/j.1365-2249.2011.04468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Pantelidis P, Fanning GC, Wells AU, Welsh KI, Du Bois RM. Analysis of tumor necrosis factor-alpha, lymphotoxin-alpha, tumor necrosis factor receptor II, and interleukin-6 polymorphisms in patients with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 2001;163:1432–6. [DOI] [PubMed] [Google Scholar]
- [53].Vasakova M, Striz I, Slavcev A, Jandova S, Kolesar L, Sulc J. Th1/Th2 cytokine gene polymorphisms in patients with idiopathic pulmonary fibrosis. Tissue Antigens 2006;67:229–32. [DOI] [PubMed] [Google Scholar]
- [54].Whittington HA, Freeburn RW, Godinho SIH, Egan J, Haider Y, Millar AB. Analysis of an IL-10 polymorphism in idiopathic pulmonary fibrosis. Genes Immun 2003;4:258–64. [DOI] [PubMed] [Google Scholar]
- [55].Chen AF, Lee YS, Seidl AJ, Abboud JA. Arthrofibrosis and large joint scarring. Connect Tissue Res 2019;60:21–8. 10.1080/03008207.2018.1517759. [DOI] [PubMed] [Google Scholar]
- [56].Abdel MP, Morrey ME, Barlow JD, Grill DE, Kolbert CP, An KN, et al. Intraarticular decorin influences the fibrosis genetic expression profile in a rabbit model of joint contracture. Bone Joint Res 2014;3:82–8. 10.1302/2046-3758.33.2000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Morrey ME, Abdel MP, Riester SM, Dudakovic A, van Wijnen AJ, Morrey BF, et al. Molecular landscape of arthrofibrosis: microarray and bioinformatic analysis of the temporal expression of 380 genes during contracture genesis. Gene 2017;610:15–23. 10.1016/j.gene.2017.01.025. [DOI] [PubMed] [Google Scholar]
- [58].Abdel MP, Morrey ME, Barlow JD, Kreofsky CR, An K-N, Steinmann SP, et al. Myofibroblast cells are preferentially expressed early in a rabbit model of joint contracture. J Orthop Res 2012;30:713–9. 10.1002/jor.21588. [DOI] [PubMed] [Google Scholar]
- [59].Salib CG, Reina N, Trousdale WH, Limberg AK, Tibbo ME, Jay AG, et al. Inhibition of COX-2 pathway as a potential prophylaxis against arthrofibrogenesis in a rabbit model of joint contracture. J Orthop Res 2019;37:2609–20. 10.1002/jor.24441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kim Y-S, Kim J-M, Lee Y-G, Hong O-K, Kwon H-S, Ji J-H. Intercellular adhesion molecule-1 (ICAM-1, CD54) is increased in adhesive capsulitis. J Bone Joint Surg Am 2013;95 10.2106/JBJS.K.00525. e18–1–8. [DOI] [PubMed] [Google Scholar]
- [61].Issa K, Rifai A, Boylan MR, Pourtaheri S, McInerney VK, Mont MA. Do various factors affect the frequency of manipulation under anesthesia after primary total knee arthroplasty? Clin Orthop Relat Res 2015;473:143–7. 10.1007/s11999-014-3772-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zmistowski B, Restrepo C, Kahl LK, Parvizi J, Sharkey PF. Incidence and reasons for nonrevision reoperation after total knee arthroplasty. Clin Orthop Relat Res 2011;469:138–45. 10.1007/s11999-010-1558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Clement ND, Merrie KL, Weir DJ, Holland JP, Deehan DJ. Asynchronous bilateral total knee arthroplasty: predictors of the functional outcome and patient satisfaction for the second knee replacement. J Arthroplasty 2019;34:2950–6. 10.1016/j.arth.2019.06.056. [DOI] [PubMed] [Google Scholar]
- [64].Meehan JP, Monazzam S, Miles T, Danielsen B, White RH. Postoperative stiffness requiring manipulation under anesthesia is significantly reduced after simultaneous versus staged bilateral total knee arthroplasty. J Bone Joint Surg Am 2017;99:2085–93. 10.2106/JBJS.17.00130. [DOI] [PubMed] [Google Scholar]
- [65].Bayram B, Limberg AK, Salib CG, Bettencourt JW, Trousdale WH, Lewallen EA, et al. Molecular pathology of human knee arthrofibrosis defined by RNA sequencing. Genomics 2020. 10.1016/j.ygeno.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


