Studies have shown that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can appear in feces within three days of infection, which is much sooner than the time taken for people to develop symptoms and get an official diagnosis (Mallapaty 2020). Therefore, earlier identification in wastewater of the virus’s arrival in a community might limit the health and economic damage caused by the coronavirus disease 2019 (COVID-19).
Despite centuries of technological advances, the COVID-19 pandemic is highlighting the limitations of our ability to control viral spread and infections worldwide. This knowledge gap has induced highly variable and sometimes contradictory government decisions in many countries, such as whether to wear a mask or not, and various strategies for testing, contact tracing and isolation of infected patients. The failure of control strategies has led many countries to lockdowns. In particular, powerful and rapid analytical techniques are actually still missing because virus detection is a key step in controlling viral transmission.
The biomarker approach, which was initially developed in geochemistry to measure the maturity of dead organic matter and to link molecular fossils with biological homologs, has been applied to modern samples such as soils, sediments, wastewaters and plants (Albrecht and Ourisson 1971; Lichtfouse et al. 1994,1997; Bryselbout et al. 1998; Payet et al. 1999; Dsikowitzky and Schwarzbauer 2014; Choi et al. 2020). Here, we discuss the emergence of wastewater epidemiology based on ribonucleic acid (RNA) biomarkers, a discipline linking biomedical and environmental sciences for the detection of pathogens, with focus on analytical challenges.
The rise of viral epidemics and pandemics
In the past decades, human populations have faced many epidemics causing negative impact on public health and economy (Kitajima et al. 2020; Sharma et al. 2020). Virus-related epidemics included the human immunodeficiency virus (HIV), influenza virus (flu), the Ebola virus (EBOV), the Zika virus (ZIKV), the chikungunya virus (Chik), the Middle East respiratory syndrome coronavirus (MERS-CoV) and the severe acute respiratory syndrome coronavirus (SARS-CoV). Among respiratory viruses, coronaviruses (CoV) are a major threat to public health due to easy human-to-human transmissions (Li et al. 2020; Prather et al. 2020). Coronaviruses may also be transmittable via birds (Shi et al. 2020) or other mammalian intermediate hosts. The novel coronavirus SARS-CoV-2 (or COVID-19) pandemic has resulted in more than 61 million infected people with nearly 1.5 million death according to the World Health Organization on November 29, 2020 (World health Organization 2020). COVID-19 remains a challenge to public health, which requires its tracking to implement rational-based virus countermeasures in order to minimize further risk of viral spreading to human population. Knowledge on COVID-19 in environmental samples such as wastewater appears critical to shrink the pandemic, as explained below.
COVID-19 in wastewater
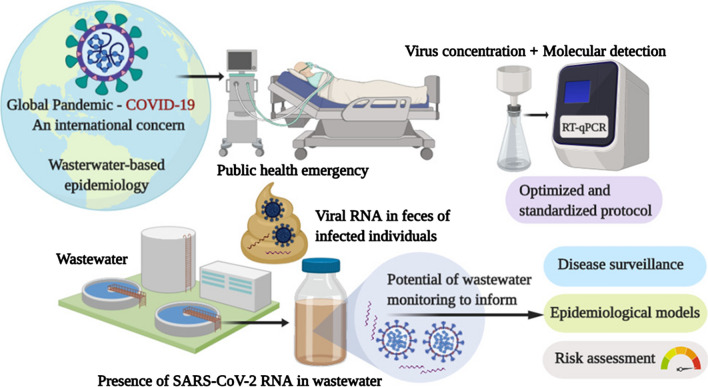
SARS-CoV-2 belongs to the family of Coronaviridae that contains a group of enveloped viruses having a single-stranded RNA genome. The main transmission pathway of SARS-CoV-2 includes person-to-person contact and inhalation of droplets and aerosols (Dinnon et al. 2020; Li et al. 2020). Infected persons shed both viable SARS-CoV-2 and viral RNA through bodily excretion such as sputum, saliva, urine and feces (Arnaud 2020; Daughton 2020; Foladori et al. 2020; Kitajima et al. 2020; Kweinor Tetteh et al. 2020; Peccia et al. 2020; Randazzo et al. 2020). Accordingly, SARS-CoV-2 RNA has been detected in urine, anal and rectal swabs and human feces (Ahmed et al. 2020; Hart and Halden 2020; Jones et al. 2020; Medema et al. 2020; Peccia et al. 2020). The human excreta ultimately enter into the wastewater (Fig. 1, Kitajima et al. 2020). As a consequence, SARS-CoV-2 has been detected in wastewater worldwide (Adelodun et al. 2020; Al Huraimel et al. 2020; Bivins et al. 2020; Daughton 2020; Foladori et al. 2020; Hart and Halden 2020; Kweinor Tetteh et al. 2020; Mallapaty, 2020; Peccia et al. 2020; Randazzo et al. 2020).
Fig. 1.
Release of SARS-CoV-2 into wastewater and wastewater epidemiology.
Adapted from Kitajima et al. (2020). COVID-19 patients shed SARS-CoV-2 viral RNA in feces. The feces in the catchment are converged at the wastewater treatment facility. Wastewater samples are collected. Viral RNA in the wastewater is isolated and analyzed. SARS-CoV-2: severe acute respiratory syndrome coronavirus 2, COVID-19: coronavirus disease 2019, RNA: ribonucleic acid
Wastewater epidemiologic surveillance
Classical clinical diagnostic tests to detect the virus in individuals are useful but are limited by cost, supplies, ratio of tested versus untested people, and by the inability to test a large population urgently, as well as by the complicated process of nasopharyngeal swabbing. Alternatively, wastewater epidemiology, which involves tracking SARS-CoV-2 in wastewater, provides rapid and complementary information to assist public health decisionmakers to minimize risks associated with COVID-19 in the community (Kitajima et al. 2020)(Fig. 1). Indeed, wastewater epidemiology provides a community diagnostic integrating the potential contamination of a large number of individuals. In particular, in regions with limited resources where it is difficult to perform clinical diagnosis, monitoring wastewater may assist in controlling the spread of epidemics (Bivins et al. 2020). Overall, tracing of SARS-CoV-2 through detection at sewer lines and wastewater treatment locations will immediately identify areas of epidemic concerns. Moreover, wastewater epidemiology addresses the social stigma of a community toward the clinical diagnosis of COVID-19 (Murakami et al. 2020).
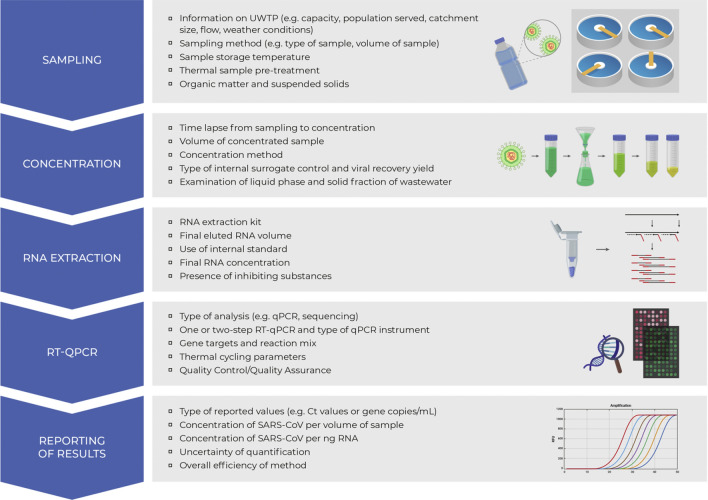
Since research on the identification of SARS-CoV-2 in human excreta and wastewaters has just begun, many unknowns need to be unveiled, such as a solid analytical methodology for isolating, identifying and quantifying the virus. Michael-Kordatou et al. (2020) have recently reviewed factors and conditions to optimize the analytical methodology, with details of protocols including sampling, concentrating, extracting viral RNA, detecting and subsequently quantifying (Fig. 2). In particular, they found that the selection of the sampling method, e.g., grab or composite, the sampling time and the temperature of sample storage is critical. Moreover, organic matter, suspended solids, pretreatment temperature and concentrating the virus from large volumes of wastewater may influence the analysis.
Fig. 2.
Procedure for analysis of viral RNA from SARS-CoV-2 in wastewaters, showing critical steps. RNA: ribonucleic acid, SARS-CoV-2: severe acute respiratory syndrome coronavirus 2, UWTP: urban wastewater treatment plant, RT-qPCR: reverse transcription polymerase chain reaction, q-PCR: quantitative polymerase chain reaction.
Adapted from Michael-Kordatou et al. (2020) with permission from Elsevier
Extracting the viral RNA without damage to its molecular structure is particularly difficult because, contrary to ‘ideal’ biological samples, wastewater is a very complex organo-mineral mixture that contains compounds which could adsorb, react with and degrade RNA (Limsawat and Ohgaki 1997; Lu et al. 2020). The key challenge is in selecting a suitable procedure that breaks virus particles without altering nucleic acids to achieve a maximum recovery or yield. Quantitative reverse transcription polymerase chain reaction (RT-qPCR) is usually applied to detect and quantify SARS-CoV-2. This technique relies on the presence of SARS-CoV-2 specific RNA in the sample detected by the TaqMan assay, i.e., the 5′ nuclease assay, which has been established for samples of clinical origins. The use of this assay for environmental sample requires additional considerations such as proper controls of similar matrices. In the technique, consistency in the combinations of assay sets can be applied in the study.
In conclusion, systematic and continuous monitoring using wastewater epidemiology can be applied to give early warning to the existence of infections in the community to guide public health authorities and drive the public health counter measures. Interestingly, wastewater, which was previously considered as unwanted junk matter, appears now as a gold mine of information, chemicals and energy in the context of the future circular economy.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Virender K. Sharma, Email: vsharma@tamu.edu
Chetan Jinadatha, Email: Chetan.Jinadatha@va.gov.
Eric Lichtfouse, Email: eric.lichtfouse@gmail.com.
Etienne Decroly, Email: etienne.decroly@gmail.com.
Jacques van Helden, Email: Jacques.van-Helden@univ-amu.fr.
References
- Adelodun B, Ajibade FO, Ibrahim RG, Bakare HO, Choi K. Snowballing transmission of COVID-19 (SARS-CoV-2) through wastewater: Any sustainable preventive measures to curtail the scourge in low-income countries? Sci Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.140680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O'Brien JW, Choi PM, Kitajima M, Simpson SL, Li J, Tscharke B, Verhagen R, Smith WJM, Zaugg J, Dierens L, Hugenholtz P, Thomas K, V., Mueller JF, First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Huraimel K, Alhosani M, Kunhabdulla S, Stietiya MH. SARS-CoV-2 in the environment: Modes of transmission, early detection and potential role of pollutions. Sci Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.140946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht P, Ourisson G. Biogenic Substances in Sediments and Fossils. Angew Chem. 1971;10:209–225. doi: 10.1002/anie.197102091. [DOI] [PubMed] [Google Scholar]
- Arnaud CH. Following SARS-CoV-2 down the drain. Chem Eng News. 2020;98(45):16–20. doi: 10.47287/cen-09845-feature1. [DOI] [Google Scholar]
- Bivins A, North D, Ahmad A, Ahmed W, Alm E, Been F, Bhattacharya P, Bijlsma L, Boehm AB, Brown J, Buttiglieri G, Calabro V, Carducci A, Castiglioni S, Gurol ZC, Chakraborty S, Costa F, Curcio S, de los Reyes, Francis L, Vela JD, Farkas K, Fernandez-Casi X, Gerba C, Gerrity D, Girones R, Gonzalez R, Haramoto E, Harris A, Holden PA, Islam MT, Jones DL, Kasprzyk-Hordern B, Kitajima M, Kotlarz N, Kumar M, Kuroda K, La Rosa G, Malpei F, Mautus M, McLellan SL, Medema G, Meschke JS, Mueller J, Newton RJ, Nilsson D, Noble RT, van Nuijs A, Peccia J, Perkins TA, Pickering AJ, Rose J, Sanchez G, Smith A, Stadler L, Stauber C, Thomas K, van der Voorn T, Wigginton K, Zhu K, Bibby K. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ Sci Technol. 2020;54(13):7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Bryselbout C, Henner P, Lichtfouse E. Fossil fuel biomarkers in plant waxes as pollution parameters. Sci Total Environ. 1998;222:201–204. doi: 10.1016/S0048-9697(98)00297-6. [DOI] [Google Scholar]
- Choi PM, O’Brien JW, Tscharke BJ, Mueller JF, Thomas KV. Samanipour S (2020) Population socioeconomics predicted using wastewater. Environ Sci Technol Lett. 2020;7(8):567–572. doi: 10.1021/acs.estlett.0c00392. [DOI] [Google Scholar]
- Daughton CG. Wastewater surveillance for population -wide Covid-19: The present and future. Sci Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon KH, Leist SR, Schafer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Jr, Hou YJ, Adams LE, Gully KL, Brown AJ, Huang E, Bryant MD, Choong IC, Glenn JS, Gralinski LE, Sheahan TP, Baric RS. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020 doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dsikowitzky L, Schwarzbauer J. Industrial organic contaminants: identification, toxicity and fate in the environment. Environ Chem Lett. 2014;12:371–386. doi: 10.1007/s10311-014-0467-1. [DOI] [Google Scholar]
- Foladori P, Cutrupi F, Segata N, Manara S, Pinto F, Malpei F, Bruni L, La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: What do we know. A review: Sci Total Environ; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart OE, Halden RU. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities and challenges. Sci Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Baluja MQ, Graham DW, Corbishley A, McDonald JE, Malham SK, Hillary LS, Connor TR, Gaze WH, Moura IB, Wilcox MH, Farakas K. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci Total Environ. 2020;749:141364. doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M, Ahmed W, Bibby K, Carducci A, Gerba CP, Hamilton KA, Haramoto E, Rose JB. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweinor Tetteh E, Opoku Amankwa M, Armah EK, Rathilal S. Fate of COVID-19 Occurrences in Wastewater Systems: Emerging Detection and Treatment Technologies-A Review. Water. 2020 doi: 10.3390/w12102680. [DOI] [Google Scholar]
- Li Y, Zhang R, Zhaoa J, Molina MJ. Understanding transmission and intervention for the COVID-19 pandemic in the United States. Sci Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.141560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtfouse E, Albrecht P, Behar F, Hayes JM. A molecular and isotopic study of the organic matter from the Paris basin, France. Geochim Cosmochim Acta. 1994;58:209–221. doi: 10.1016/0016-7037(94)90458-8. [DOI] [PubMed] [Google Scholar]
- Lichtfouse E, Bardoux G, Mariotti A, Balesdent J, Ballentine DC, Macko SA. Molecular, 13C, and 14C evidence for the allochthonous and ancient origin of C16–C18 n-alkanes in modern soils. Geochim Cosmochim Acta. 1997;61:1891–1898. doi: 10.1016/S0016-7037(97)00021-5. [DOI] [Google Scholar]
- Limsawat S, Ohgaki S (1997) Fate of liberated viral RNA in wastewater determined by PCR. Applied Environ. Microbiol. 63, 2932–2933. https://aem.asm.org/content/63/7/2932 [DOI] [PMC free article] [PubMed]
- Lu D, Huang Z, Luo J, Zhang X, Sha S. Primary concentration - The critical step in implementing the wastewater based epidemiology for the COVID-19 pandemic: A mini-review. Sci Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.141245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580:176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- Medema G, Heijnen L, Elsinga C, Italiaander R, Brouwer A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Enviorn Sci Technol Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Michael-Kordatou I, Karaolia P, Fatta-Kassinos D. Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification. Journal of Environmental Chemical Engineering. 2020 doi: 10.1016/j.jece.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Hata A, Honda R, Watanabe T. Letter to the Editor: Wastewater-Based Epidemiology Can Overcome Representativeness and Stigma Issues Related to COVID-19. Environ Sci Technol. 2020 doi: 10.1021/acs.est.0c02172. [DOI] [PubMed] [Google Scholar]
- Payet C, Bryselbout C, Morel JL, Lichtfouse E. Fossil fuel biomarkers in sewage sludges: environmental significance. Naturwissenschaften. 1999;86:484–488. doi: 10.1007/s001140050659. [DOI] [PubMed] [Google Scholar]
- Peccia J, Zulli A, Brackney DE, Grubaugh ND, Kaplan EH, Casanovas-Massana A, Ko AI, Malik AA, Wang D, Wang M, Warren JL, Weinberger DM, Arnold W, Omer SB. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather KA, Marr LC, Schooley RT, McDiarmid MA, Wilson ME, Milton DK. Airborne transmission of SARS-CoV-2. Science. 2020 doi: 10.1126/science.abf0521. [DOI] [PubMed] [Google Scholar]
- Randazzo W, Truchado P, Cuevas-Ferrando E, Simon P, Allende A, Sanchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VK, Jinadatha C, Lichtfouse E. Environmental chemistry is most relevant to study coronavirus pandemics. Environ Chem Lett. 2020 doi: 10.1007/s10311-020-01017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, Zhao Y, Liu P, Liang L, Cui P, Wang J, Zhang X, Guan Y, Tan W, Wu G, Chen H, Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020 doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (Access November 29, 2020) Coronavirus Disease (COVID-19) Dashboard.