Abstract
The severity of the new COVID-19 pandemic caused by the SARS-CoV-2 virus is strikingly variable in different global populations. SARS-CoV-2 uses ACE2 as a cell receptor, TMPRSS2 protease, and FURIN peptidase to invade human cells. Here, we investigated 1,378 whole-exome sequences of individuals from the Middle Eastern populations (Kuwait, Qatar, and Iran) to explore natural variations in the ACE2, TMPRSS2, and FURIN genes. We identified two activating variants (K26R and N720D) in the ACE2 gene that are more common in Europeans than in the Middle Eastern, East Asian, and African populations. We postulate that K26R can activate ACE2 and facilitate binding to S-protein RBD while N720D enhances TMPRSS2 cutting and, ultimately, viral entry. We also detected deleterious variants in FURIN that are frequent in the Middle Eastern but not in the European populations. This study highlights specific genetic variations in the ACE2 and FURIN genes that may explain SARS-CoV-2 clinical disparity. We showed structural evidence of the functionality of these activating variants that increase the SARS-CoV-2 aggressiveness. Finally, our data illustrate a significant correlation between ACE2 variants identified in people from Middle Eastern origins that can be further explored to explain the variation in COVID-19 infection and mortality rates globally.
Keywords: COVID-19, SARS-CoV-2, ACE2, TMPRSS2, FURIN, Variants, eQTLs, Middle Eastern populations, Precision medicine, Genomic medicine
COVID-19; SARS-CoV-2; ACE2; TMPRSS2; FURIN; Variants; eQTLs; Middle Eastern populations; Precision medicine; Genomic medicine
1. Introduction
The global pandemic COVID-19 caused by SARS-CoV-2 virus is life-threatening and has become a significant concern to humanity. Notably, the severity of this disease is highly variable in different populations across the world [1, 2]. Since the outbreak, several studies have reported specific factors including age, gender, and pre-existing health conditions that could have contributed to the increased severity of the disease [3, 4, 5, 6]. The genetic susceptibility to COVID-19 has also been explored by scrutinizing Angiotensin converting enzyme 2 (ACE2) genetic variations in different populations. ACE2 is the functional receptor mediating entry of SARS-CoV-2 into the host cells [7], which is facilitated by FURIN cleavage [8, 9, 10]. Transmembrane serine protease 2 (TMPRSS2) is another candidate gene that has been linked to COVID-19 disease [10, 11, 12]. TMPRSS2 expression enhances ACE2-mediated SARS-CoV-2 cell invasion by operating as a co-receptor [12]. The increased cleavage activity of this protease was suggested to diminish viral recognition by neutralizing antibodies and by activating SARS spike (S) protein for virus-cell fusion [13] and facilitates the active binding of SARS-CoV-2 through ACE2 receptor, which is a risk factor for a more serious COVID-19 presentation [10, 11, 12].
The hallmark of the novel SARS-CoV-2, as compared to other SARS viruses, is the presence of a polybasic FURIN cleavage site. FURIN has been reported to facilitate the transport of SARS-CoV-2 into or from the host cell [8, 9, 10, 14]. Notably, a recent study has highlighted the presence of a unique functional polybasic FURIN cleavage consensus site between the two spike subunits S1 and S2 by the insertion of 12 nucleotides encoding PRRA in the S protein of SARS-CoV-2 virus [15]. The FURIN-like cleavage-site is cleaved during virus egress, which primes the S-protein providing a gain-of-function for the efficient spreading of the SARS-CoV-2 among humans [16, 17]. It is, therefore, likely that the presence of a deleterious ACE2, TMPRSS2 and FURIN gene variants may modulate viral infectivity among humans, making some people less vulnerable than others. Recent studies, assessed the genetic variations and eQTL (expression quantitative trait locus) expression profiles in the candidate genes ACE2, TMPRSS2, and FURIN to demonstrate the sex and population-wise differences that may influence the pathogenicity of SARS-CoV-2 [3, 11, 18, 19, 20, 21, 22]. It is to be noted that these studies focused only on the European and East Asian populations. Given the extremely high prevalence of obesity (80%), hypertension (28%) and diabetes (20%) of the population in the Gulf states [23, 24, 25] which are considered as risk factors for mortality from COVID-19 [26, 27], the witnessed low infectivity and mortality rates registered in this area of the world are intriguing. Even though this could be due to various factors that are not well accounted for yet such as testing, hot weather or extreme measures taken early on by some countries, it could also be due to ethnic genetic variations in the ACE2-TMPRSS2-FURIN genes that are key regulators for orchestrating SARS-CoV-2 cellular access.
As a result, it is crucial to study the variation of these candidate genes ACE2, TMPRSS2 and FURIN in Middle Eastern populations to better understand possible natural genetic components that can be responsible for these differences. Here, we present a comprehensive comparative assessment of deleterious or gain of function mutations of ACE2, TMPRSS2, and FURIN that may have influenced disease progression in the Middle Eastern populations compared to European, African and East Asian populations. While these findings are preliminary and based on our genetic data and worldwide public datasets, they present a compelling perception into the role of naturally occurring genetic variants in ACE2, TMPRSS2, and FURIN in different populations. They can shed light on the reported variations in susceptibility or resistance to SARS-CoV-2 infection in different populations and can be available for other scientists to utilize them in precision medicine.
2. Materials and methods
2.1. Genetic data
The whole-exome sequences data of 473 Kuwaitis [28], 800 Iranians [29] and 100 Qataris [30] published previously were used in this analysis. The genetic data of non-Finnish European, East Asian and African American populations were obtained from the gnomAD repository [31], which contain data on a total of 125,748 exomes and 71,702 genomes (https://gnomad.broadinstitute.org/). The minor allele frequencies of the ACE2 variant, rs41303171 (N720D) for different countries were obtained from 1000 Genomes Project phase 3 [32] and ABraOM [33] databases.
2.2. Gene expression data
The expression data for ACE2, TMPRSS2, and FURIN were obtained from the genotype-tissue expression (GTEx) database (https://gtexportal.org/home/). The same database portal was used to extract quantitative trait loci (eQTLs) for the three genes.
2.3. Data on case fatality rate (CFR)
Country level numbers of cases and deaths were obtained from the data available from https://covid.ourworldindata.org/data/owid-covid-data.xlsx (accessed on 7 August 2020) and the CFR was calculated as stated in https://ourworldindata.org/mortality-risk-covid i.e., Case Fatality Rate (CFR in %) = (Number of deaths from disease/Number of diagnosed cases of disease) x 100. The country-wise CFR values are available online at https://www.coronatracker.com/country/kuwait/(accessed on 7 August 2020).
2.4. Statistical analysis
The missense variants were defined as deleterious when predicted to be damaging, probably damaging, disease causing and deleterious by the five algorithms applied, SIFT [34], PolyPhen-2 HumVar, PolyPhen-2 HumDiv [35], MutationTaster [36] and LRT score [37] and/or CADD (Combined Annotation-Dependent Depletion) score of more than 20 [38]. We considered only deleterious variants with minor allele frequency (MAF) less than 1% in the burden analysis. The significance of the differences in MAFs between different populations was calculated using Chi-Square test, using the R software (https://www.r-project.org/). All the p-values presented in the tables are not corrected for multiple testing. P values ≤ 0.05 were considered significant.
2.5. Structural analysis
All the identified ACE2 missense exon variants were mapped, modeled, and analyzed using Pymol modeling software (https://pymol.org/2/). DynaMut web server was used to predict the effect of genetic variants on the stability and flexibility of ACE2 receptor [39, 40].
3. Results
3.1. ACE2 receptor variations in the Middle East
We first examined ACE2 gene variation frequencies in Kuwait, Qatar, East Asia, and Africa where the impact of SARS-CoV-2 has been modest and compared it to Iran, which is moderately affected and then to Europe, the continent with the most deaths per population. Overall, we found human ACE2 gene variations and the probability of loss of function mutations (pLOF = 0.1, C.I. 0.04–0.25) to be low in comparison to ACE (pLOF = 0.87, C.I. 0.71–1.08), which is a gene of similar size, indicating that the ACE2 gene is highly intolerant to loss of function mutations. Additionally, we identified 19 missense variants in the ACE2 gene from Kuwait, Qatar and Iran (Table 1). All were rare variants defined by minor allele frequency (MAF) of less than 1%. The genetic variants included four novel variants from Kuwait, and six from Iran.
Table 1.
ACE2 missense variants in the Middle East and gnomAD populations.
| Variant ID | Protein Consequence |
Minor Allele Frequency |
Functional Risk Prediction (Scores) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kuwait | Iran | Qatar | Europe | East Asia | Africa | SIFT | PP2HVAR | PP2HDIV | MUTTASTER | LRT | CADD | ||
| rs372923812 | I741V | 0 | 0 | 0.00655 | 0.00009 | 0 | 0.00005 | T (0.32) | B (0.004) | B (0.001) | N (0.07) | N (0.005) | 0.066 |
| rs4646116 | K26R | 0.00209 | 0 | 0 | 0.00587 | 0.00007 | 0.00095 | T (0.47) | B (0.001) | B (0.0) | N (0.31) | N (0.83) | 8.188 |
| rs200540199 | R716H | 0.00105 | 0 | 0 | 0.00014 | 0 | 0 | D (0.02) | B (0.277) | P (0.771) | N (0.03) | N (0.207) | 11.52 |
| rs769062069 | R708Q | 0 | 0.00188 | 0 | 0.00002 | 0 | 0 | D (0.00) | D (0.935) | D (0.999) | D (0.99) | D (0.00) | 24.9 |
| rs776995986 | R708W | 0.00105 | 0.00584 | 0 | 0.00004 | 0 | 0 | D (0.00) | D (0.997) | D (1.00) | D (1.00) | D (0.00) | 22.9 |
| rs765152220 | D494V | 0 | 0.00063 | 0 | 0.00005 | 0 | 0 | D (0.01) | D (0.926) | D (0.995) | D (1.00) | D (0.00) | 31 |
| rs750145841 | Y199C | 0 | 0.00125 | 0 | 0.00002 | 0 | 0 | T (0.08) | D (0.984) | D (1.0) | D (0.98) | D (0.0004) | 26.2 |
| rs759162332 | Q60R | 0 | 0.00125 | 0 | 0.00003 | 0 | 0 | T (0.21) | B (0.009) | B (0.001) | N (0.20) | N (0.23) | 15.34 |
| rs41303171 | N720D | 0.00314 | 0.00625 | 0.00204 | 0.02521 | 0 | 0.00269 | T (0.07) | B (0.02) | B (0.006) | N (0.20) | N (0.009) | 14.9 |
| X:15580089∗ | I786T | 0 | 0.00188 | 0 | 0 | 0 | 0 | T (0.18) | B (0.088) | B (0.297) | N (0.00) | U (0.001) | 6.519 |
| X:15582247∗ | P737A | 0.00314 | 0 | 0 | 0 | 0 | 0 | D (0.01) | B (0.210) | P (0.489) | N (0.00) | U (0.953) | 2.44 |
| X:15585864∗ | Q661P | 0.00105 | 0 | 0 | 0 | 0 | 0 | D (0.02) | B (0.30) | P (0.812) | N (0.00) | U (0.3) | 22.9 |
| X:15589919∗ | F555L | 0.00105 | 0 | 0 | 0 | 0 | 0 | T (0.09) | B (0.145) | B (0.16) | N (0.00) | U (0.223) | 8.825 |
| X:15591578∗ | V485L | 0 | 0.00188 | 0 | 0 | 0 | 0 | T (0.10) | P (0.56) | B (0.028) | N (0.00) | U (0.63) | 23.7 |
| X:15593877∗ | F452V | 0 | 0.00083 | 0 | 0 | 0 | 0 | D (0.03) | B (0.038) | B (0.189) | N (0.00) | U (0.004) | 26.3 |
| X:15596394∗ | A372G | 0.00209 | 0 | 0 | 0 | 0 | 0 | D (0.05) | B (0.049) | B (0.437) | N (0.00) | U (0.001) | 25.4 |
| X:15599413∗ | T334M | 0 | 0.00188 | 0 | 0 | 0 | 0 | T (0.11) | B (0.026) | B (0.01) | N (0.00) | U (0.084) | 12.87 |
| X:15599422∗ | S331F | 0 | 0.00083 | 0 | 0 | 0 | 0 | D (0.00) | P (0.944) | B (0.006) | N (0.00) | U (0.344) | 24.6 |
| X:15607489∗ | D225G | 0 | 0.00125 | 0 | 0 | 0 | 0 | T (0.37) | B (0.007) | B (0.002) | N (0.00) | U (0.03) | 26.1 |
SIFT (Sorting Intolerant From Tolerant): D = damaging, T = tolerated.
PP2HVAR (PolyPhen-2 Polymorphism Phenotyping v2 HumVar): D = probably damaging, P = possibly damaging, B = benign.
PP2HDIV (PolyPhen-2 Polymorphism Phenotyping v2 HumDiv): D = probably damaging, P = possibly damaging, B = benign.
MUTTASTER (MutationTaster): A = disease causing automatic, D = disease causing, N = polymorphism, P = polymorphism automatic.
LRT (Likelihood Ratio Test): D = deleterious, N = Neutral, U = unknown.
CADD - Combined Annotation Dependent Depletion based scores.
The missense variants were defined as deleterious when predicted to be damaging, probably damaging, disease causing and deleterious by the five algorithms applied (SIFT, PolyPhen-2 HumVar, PolyPhen-2 HumDiv, MutationTaster and LRT score) and/or CADD score of more than 20. We considered only deleterious variants with minor allele frequency less than 1% in the burden analysis.
Denotes Novel variants presented with chromosome: position (based on the human reference genome build GRCh37/hg19).
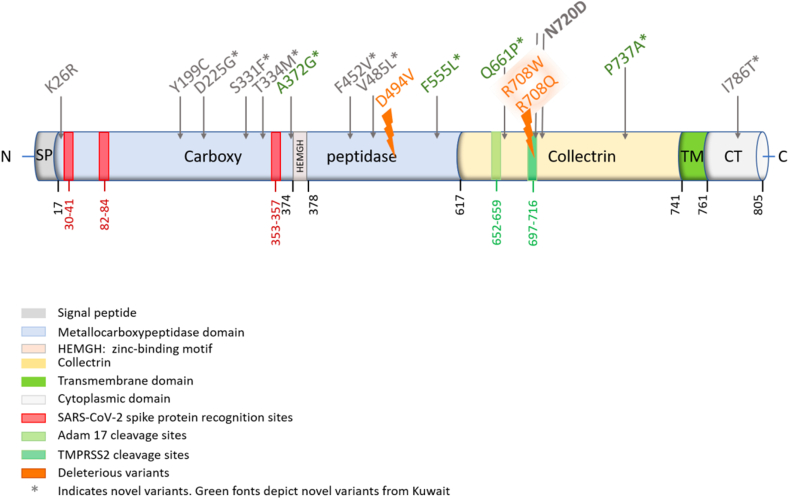
We identified four deleterious variants (rs776995986, rs769062069, rs765152220, and rs750145841), causing R708W, R708Q, D494V, Y199C missense amino acid substitutions in the ACE2 gene using risk prediction tools as described in the methods section (Table 1). All the ACE2 gene deleterious variants were absent from the African, East Asian and Qatari data and were very rare in Europeans but were present at MAF of 0.063–0.5% in the Iranian population (Table 1). This suggests a more protective effect and a significant decrease in the disease burden in Iran compared to Europe (p < 0.05; Table 2). The positions of the ACE2 receptor polymorphisms on the linearized ACE2 protein model are shown in Figures 1 and 3 D-models for the same are shown in Figures S1 and S2. It is noteworthy that none of the ACE2 polymorphisms identified in this study involved the three ACE2 regions known to directly bind the SARS-CoV-2 S-Protein Receptor Binding Domain (RBD), namely amino acids 30–41, 82–84, and 353–357 (Figure 1).
Table 2.
Burden of ACE2 rare variants in the Middle East and gnomAD populations.
| Variant ID | Protein Consequence |
Minor Allele Frequency |
P value |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| KWT | IRN | QAR | EUR | EAS | AFR | KWT vs IRN | KWT vs EUR | IRN vs EUR | ||
| rs776995986 | R708W | 1.05E-03 | 5.64E-03 | 0.00 | 3.79E-05 | 0.00 | 0.00 | 0.1084 | 0.0515 | 0.0005 |
| rs769062069 | R708Q | 0.00 | 1.88E-03 | 0.00 | 1.46E-05 | 0.00 | 0.00 | __ | __ | 0.0005 |
| rs765152220 | D494V | 0.00 | 6.25E-04 | 0.00 | 5.37E-05 | 0.00 | 0.00 | __ | __ | 0.0995 |
| rs750145841 | Y199C | 0.00 | 1.25E-03 | 0.00 | 2.00E-05 | 0.00 | 0.00 | __ | __ | 0.0015 |
The missense variants were defined as deleterious when predicted to be damaging, probably damaging, disease causing and deleterious by the five algorithms applied (SIFT, PolyPhen-2 HumVar, PolyPhen-2 HumDiv, MutationTaster and LRT score) and/or CADD score of more than 20. We considered only deleterious variants with minor allele frequency less than 1% in the burden analysis. P values are calculated using Chi-square test.
KWT-Kuwaitis; IRN-Iranians; QAR-Qataris; EUR-Europeans (non-Finnish); EAS-East Asians; AFR-Africans.
Figure 1.
The positions of the ACE2 receptor polymorphisms on the linearized ACE2 protein. The translated protein contains an N-terminal signal sequence (1–17), single catalytic domain (18–740) with zinc-binding motif (HEMGH 374–378), a transmembrane region (741–761), a small C-terminal cytosolic domain (762–805).
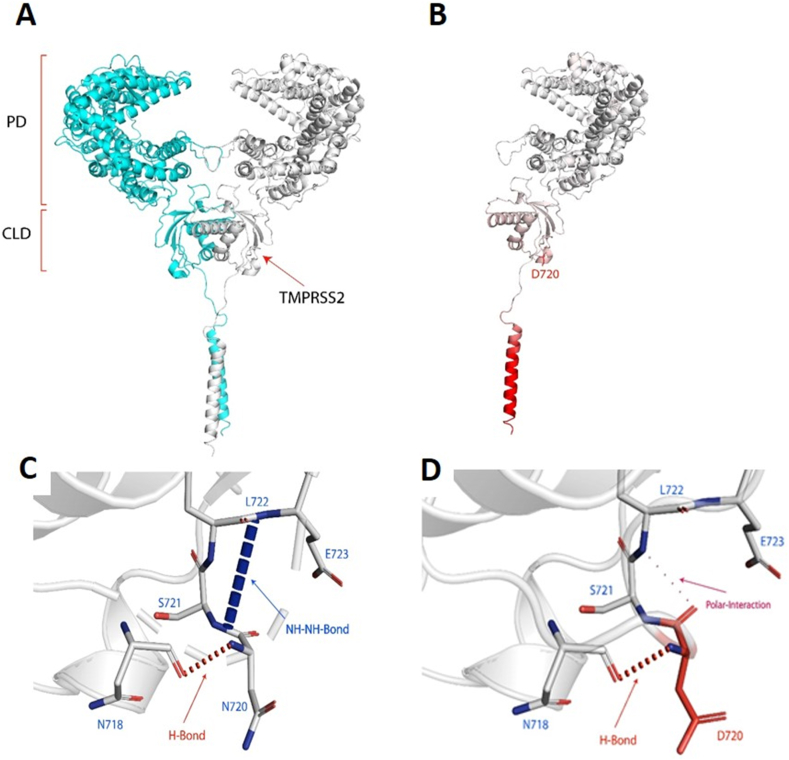
Figure 3.
(A)ACE2 dimer complex (promoter 1 Blue), (promoter 2 white). The peptidase domain (PD) is the site where the SARS-CoV-2 S protein ribosomal domain (RBD) binds. The C-terminal collectrin-like domain (CLD) is the region where the TMPRSS2 cleaves ACE2. (B) Single domain of the ACE2 mutation N720D, the red region of the protein depicted the more flexible region of the protein due to the N720D mutation with a (ΔΔG): -0.470 kcal/mol and ΔΔS: 0.070 kcal mol−1.K−1 (increase of molecule flexibility). (C) Depicts N720 backbone NH forming a H-bond with N718 and E723 forming an NH–NH bond with S721. (D) D720 backbone NH forming a H-bond with N718 and COO- group of D720 forming a weak polar interaction with L722 backbone.
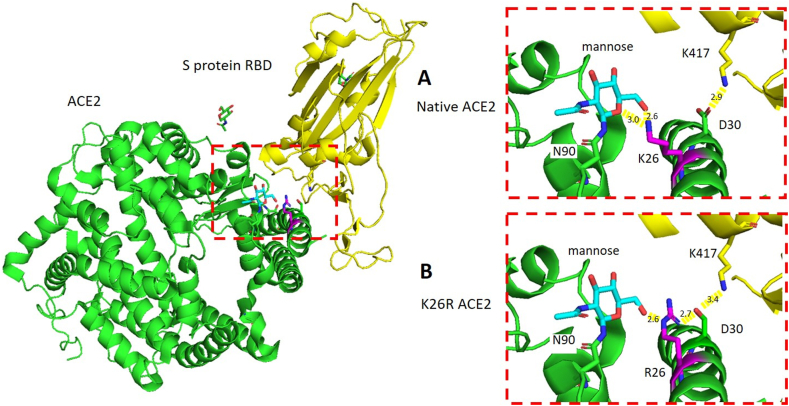
Next, we examined whether natural ACE2 gene variations that increase the affinity of ACE2 to the S-protein or facilitate viral entry/viral load exist more frequently in high-burden compared to low-burden populations. Two such genetic variants existed in our data (Table 1). The first, rs4646116, is a missense variant that changes a lysine amino acid at position 26 to arginine (K26R) (Table 1). The K26, which is just proximal to the first region of the ACE2 receptor involved in S-protein binding, has been shown previously to bind the sterically hindering first mannose in the glycan that is linked to N90 and thus stabilizes the glycan moiety hindering the binding of S-protein RBD to ACE2 [41] (Figure 2A). The missense variant R26 creates a new hydrogen bond with D30, which is then poised to build a salt-bridge with the S-protein RBD K417 that increases the affinity of SARS-CoV-2 to the ACE2 receptor [21] (Figure 2B). Indeed, the ACE2 K26R activating variant was extremely rare in East Asian (MAF = 0.007%), Africans (MAF = 0.095%), but the second most common variant in Europeans with MAF of 0.587% (shown in green fonts in Table 1). The MAF of this variant in the Kuwaiti population was nearly half that of Europeans (MAF = 0.29%), and it was absent from the Qatari and Iranian exome data (Table 1). Our structural modeling supports the notion that K26R is an ACE2 receptor activating variant (Figure 2A, B). Consistent with these findings, using a synthetic human ACE2 mutant library, a recent study reported that the R26 variant increased S-protein binding and susceptibility to the virus significantly [42].
Figure 2.
K26R polymorphism in ACE2 mapped to the structure of ACE2, green; in complex with S protein RBD, yellow (PDB ID: 6VW1). (A) In native ACE2, K26, magenta, forms two H-bonds with the mannose moiety, cyan, of ACE2 N90 which may stabilize the glycan-ACE2 complex. (B) The mutant R26, magenta, forms one H-bond with mannose. A process that could provoke the moiety-ACE2 stability and increases the affinity of the ACE2 a-helix to S protein RBD binding, where, R26 functions as a backbone and interacts with D30 which in turn dignified to build a salt-bridge with the S protein RBD K417, yellow stick.
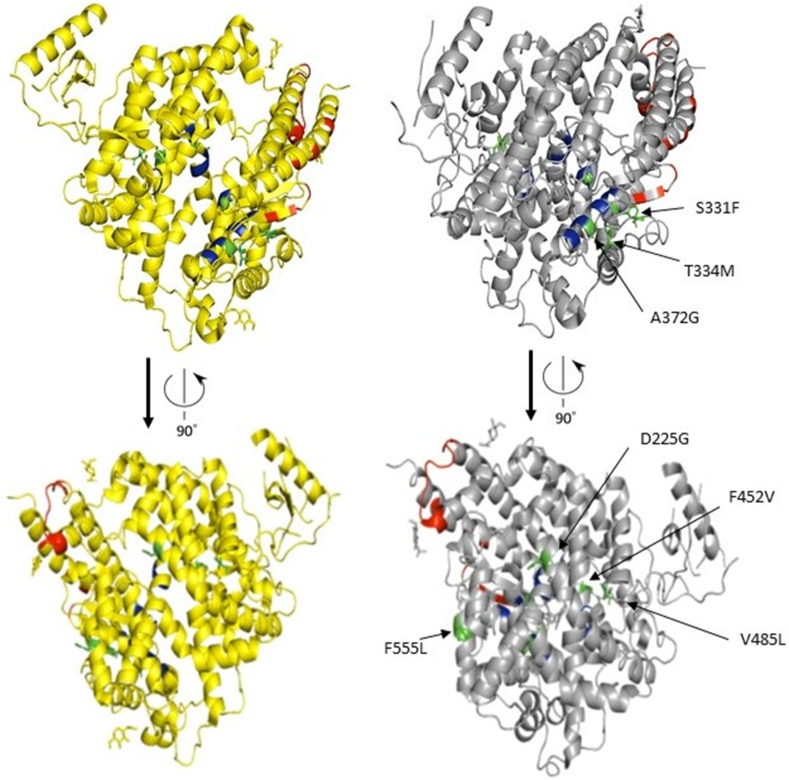
We also subjected novel and known ACE2 variants to structural predictions that may impact the binding of SARS-CoV-2 to the host cells. These changes (A372G, Y199C, D225G, F452V and V485L) were located proximal to the protein residues that mediate its activity. A previous study indicated that the three amino acid (aa) regions 30–41, especially the residue near lysine 31 and tyrosine 41, 82–84 and 353–357 in ACE2 were essential for the binding of S-protein in coronavirus [43]. Our structural estimation based on the 19 ACE2 missense variants identified in Kuwaitis, Qataris and Iranians (Table 1), revealed the location of K26R variant near the aa residues 30–41 and the novel variants, S331F and T334M, adjacent to tyrosine at 353–357. The A372G is adjacent to the zinc-binding domain (HEMGH 374–378) critical for ACE2 enzymatic activity [44]. While Y199C, D225G, F452V, and V485L all fall within the metallocarboxypeptidase domain (aa 18–617) [45] (Figure 1).
3.2. ACE2 N720D receptor variation and structural modeling
The second activating, and by far the most common ACE2 gene variant in Europeans (MAF = 2.52%) and Italians (MAF = 1.6%; detected in 105 of 6984 exomes) [20] was rs41303171, which replaces the amino acid asparagine at position 720 to aspartic acid (N720D) (Green font in Table 1). This ACE2 variant was absent from the East Asian population (13,784 exomes) and was significantly rarer in the Middle East and Africa (Table 1). This particular variant has been reported before, but its clinical relevance was persistently dismissed because the codon 720, being far from ACE2-Spike protein interface, does not appear to be an obvious candidate for ACE2 receptor binding to the S-protein of SARS-CoV-2 [21, 46]. However, we noted that N720D is located 4 amino acids proximal to the TMPRSS2 cleavage site (aa 697–716) as shown in Figure 1. Recent studies demonstrated that TMPRSS2 cleavage of the ACE2 receptor increases SARS-CoV-2 cellular entry [47, 48]. It is not unreasonable to suggest that this activating variant may play a similar role in SARS-CoV-2, rendering people who harbor it more prone to severe infection and higher viral load.
ACE2 cleavage by TMPRSS2 enhances the S-protein viral entry [7]. TMPRSS2 cleaves ACE2 between residues 697 and 716, which is the third and fourth helices in the C-terminal collectrin neck domain of the dimer interface of ACE2 [47]. To further dissect the mechanisms underlying the enhanced ACE2-TMPRSS2 accessibility, we performed structural analysis of the recently published ACE2 domain bound to B0AT1 (PDB ID:6M18) [7] and showed that N720 is located on the same interface of the loop region in close vicinity to the TMPRSS2 cleavage site (Figure 3A). Since loop region of protein are unordered and display conformational dynamics, a mutation close to the cleavage site can affect the binding affinity of TMPRSS2. Therefore, we used DynaMut web server to predict the effect of mutation N720D on the stability and flexibility of ACE2 [39,40]. Whereby, the predicted stability change was (ΔΔG): -0.470 kcal/mol, which indicated the destabilization of ACE2 receptor after the introduction of D720 (Figure 3B). The N720D mutation has resulted in an increase in entropy in the loop region (ΔΔSVib: 0.070 kcal mol−1.K−1) [39] (Figure 3B), depicting a more unstable state, which makes ACE2 more readily cleavable by TMPRSS2.
In addition, we modelled in the various non-covalent interactions for both N720 and mutant D720 and other amino acids in the loop region (Figure 3C, D). In Figure 3C, N720 formed a backbone hydrogen-bond with N718, this conformation also resulted in an NH–NH between E723 and S721. Whereas, with the activating variant D720 (Figure 3D) the back-bone hydrogen bond with N718 is still established, however, the conformational change has resulted in a polar interaction between the backbone D720 COO- group and backbone NH of L722, forming a weak polar interaction (through water-mediated hydrogen bond). As such, the D720 variant altered the conformation of the loop, and the NH–NH bond between E723 and S721 cannot be formed. Whereby, such intermolecular amide interactions are significant for protein stability [49]. Such a change in interatomic interactions between amino acids near the cleavage region can decrease the stability and increase the flexibility of the loop [50], which makes it easier for TMPRSS2 to cleave.
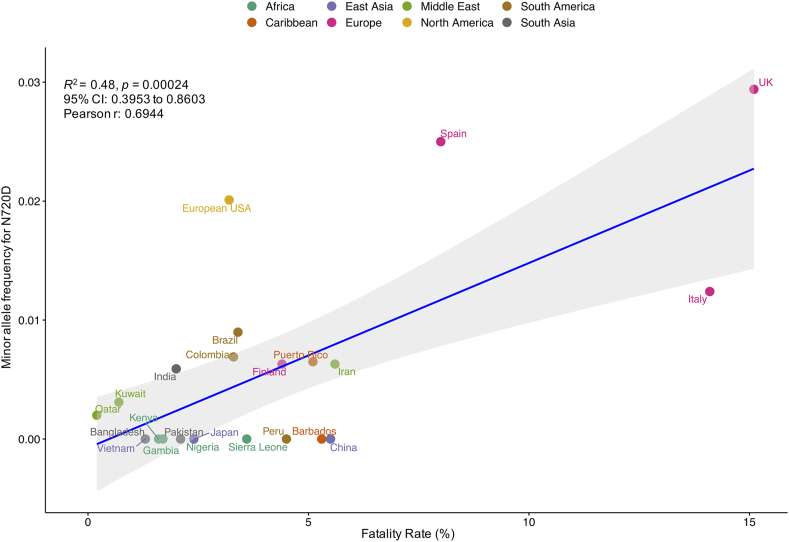
To gain insights that are pathologically and clinically relevant, we then asked whether a correlation exists between the N720D MAF data and mortality rates reported in the corresponding regions. We identified a significant correlation between N720D MAF and case fatality rate reported from countries of different continents (Figure 4) with Pearson's correlation coefficient of 69.44% (p < 0.0003; C.I. 0.3953–0.8603).
Figure 4.
The plot shows a significant correlation between N720D minor allele frequency (MAF) and case fatality rate reported from countries of different continents with Pearson's correlation coefficient of 69.44% (p < 0.0003; C.I. 0.3953–0.8603).
Next, we examined ACE2 eQTL variants in the Middle Eastern populations (Table S1). There were few variants in the Kuwaiti and Iranian populations that influenced ACE2 expression. In the Qatari population the variants modifying ACE2 gene expression were similar in frequencies to Europeans (Table S1) with most upregulating ACE2 expression in the brain and tibial nerve tissues (https://gtexportal.org/home/). Notably, the tibial nerves were affected in diabetic conditions [51], which inflicts a high number of patients in the Middle East. The most downregulating eQTL variant, rs112171234, was present in 20% and 6% of the African and Qatari populations respectively. Overall, the eQTL data pertaining to ACE2 expression were not significantly predictive nor informative.
3.3. TMPRSS2 variation
Five rare and one common deleterious variants were identified in the TMPRSS2 gene in the Middle Eastern population (Figure S3, Tables 2, and S3). Overall, there was no significant conclusion that may be withdrawn from the TMPRSS2 genetic variation data. On the expression level, we discerned four TMPRSS2 eQTL variants, which were detected only in Qataris among the Middle Eastern populations (Table S4). One of these variants, rs6517673 is intronic and downregulates the expression of TMPRSS2 in the prostate. Two of the eQTL variants, rs79391937 and rs79566442, decrease the expression of the TMPRSS2 gene in the thyroid and ovary tissues, respectively. While the variant rs11701542 upregulates the TMPRSS2 gene expression in the testis (https://gtexportal.org/home/). It is worth noting that two-third of the mortality due to COVID-19 disease affects males [18, 52].
3.4. FURIN variation
Genetic variation analysis of the FURIN gene resulted in the identification of 13 known missense variants (Table 3 and Figure S4). Like ACE2, all the identified variants were rare in the Middle Eastern populations. However, unlike the ACE2 gene, no novel variants were observed in FURIN in the Middle Eastern population (Table 3). Among the 13, we detected seven deleterious variants suggesting a possible decrease in FURIN protease function, which can potentially reduce the risk of SARS-CoV-2 in the studied populations. In this context, deleterious FURIN gene variations were observed least in East Asians, Africans, followed by Europeans then Iranians (Table 4). Interestingly, both in Qatar and Kuwait the deleterious FURIN genetic variations were more common, suggesting a possible protective effect against the SARS-CoV-2 (p < 0.05; Table 4). For example, the MAF of R37C, and R81C in Kuwait and Qatar were 0.4%/0.7% and 2.4%/3.4% respectively compared to significantly lower MAF in corresponding variants in Europeans (p < 0.05; Table 4). Together, these data support the premise that FURIN gene variants may play important roles in protection against SARS-CoV-2 in the Middle East. It is worth noting, however, that in Africa FURIN gene variants may not be a contributing factor in viral protection (Table 4). We next sought to determine the extent of FURIN expression in the Middle East. We detected 16 FURIN eQTL variants in the Middle Eastern populations, most of which were reported in the Qatar population (Table 5). We observed a high frequency of the FURIN upregulating variants, rs6226 (93%) and rs8039305 (81%) in the African populations compared to the Middle Eastern, European and East Asian populations (p < 0.05) (Table 5). In the GeneATLAS PheWAS database [53], these frequent variants were significantly related to hypertension (rs6226: P = 1.3569e-09, OR = 1.03; rs8039305: P = 1.7643e-38, OR = 1.05), which was one of the common comorbid conditions associated with the increased risk of SARS-CoV-2 infection in Chinese [54, 55] and American [56] populations.
Table 3.
FURIN known missense variants in the Middle East and gnomAD populations.
| Variant ID | Protein Consequence | Minor Allele Frequency |
Functional Risk Prediction (Scores) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kuwait | Iran | Qatar | Europe | East Asia | Africa | SIFT | PP2HVAR | PP2HDIV | MUTTASTER | LRT | CADD | |||
| rs201172453 | R37C | 0.00418 | 0.00375 | 0.00700 | 0.00007 | 0 | 0.00016 | D (0.01) | P (0.586) | D (0.995) | N (0.000) | N (0.052) | 22.7 | |
| rs749761700 | R37H | 0.00105 | 0 | 0 | 0.00001 | 0 | 0 | T (1.00) | B (0.001) | B (0.001) | N (0.000) | N (0.052) | 0.496 | |
| rs16944971 | A43V | 0.00628 | 0.00188 | 0.00500 | 0.00255 | 0 | 0.07835 | T (0.25) | B (0.007) | B (0.010) | P (0.411) | N (0.007) | 11.82 | |
| rs148110342 | R81C | 0.02406 | 0.00750 | 0.03400 | 0.00159 | 0 | 0.00016 | T (0.17) | P (0.586) | D (0.989) | D (0.995) | N (0.051) | 23 | |
| rs751909359 | R86Q | 0 | 0.00070 | 0 | 0.00003 | 0 | 0 | T (0.31) | B (0.028) | B (0.244) | D (0.999) | D (0.000) | 22.3 | |
| rs116359616 | V109M | 0.00105 | 0 | 0 | 0.00002 | 0 | 0.02318 | D (0.03) | B (0.010) | B (0.083) | D (0.635) | N (0.168) | 4.625 | |
| rs143276283 | Q399R | 0.00209 | 0.00063 | 0 | 0.00006 | 0 | 0 | T (1.00) | B (0.002) | B (0.000) | D (0.967) | N (0.002) | 18.69 | |
| rs750446344 | V580I | 0.00314 | 0 | 0 | 0.00004 | 0 | 0 | T (0.23) | B (0.007) | B (0.010) | N (0.000) | N (0.835) | 0.211 | |
| rs752639409 | R637Q | 0 | 0.00070 | 0 | 0 | 0 | 0.00006 | T (0.58) | B (0.051) | B (0.200) | N (0.003) | N (0.031) | 21.3 | |
| rs761541008 | R677W | 0 | 0.00070 | 0 | 0.00005 | 0 | 0 | T (0.05) | B (0.332) | P (0.947) | N (0.046) | D (0.000) | 27.4 | |
| rs193268286 | S685P | 0 | 0 | 0 | 0.00004 | 0 | 0 | T (0.09) | D (0.994) | D (0.999) | D (0.999) | D (0.000) | 26.6 | |
| rs760368022 | H712Y | 0.00105 | 0 | 0 | 0.00001 | 0 | 0 | D (0.01) | B (0.089) | B (0.367) | N (0.016) | N (0.032) | 19.66 | |
| rs35641241 | R745Q | 0 | 0 | 0 | 0.00118 | 0.00102 | 0.00008 | T (0.13) | B (0.375) | D (0.989) | N (0.149) | N (0.021) | 23.4 | |
SIFT (Sorting Intolerant From Tolerant): D = damaging, T = tolerated.
PP2HVAR (PolyPhen-2 Polymorphism Phenotyping v2 HumVar): D = probably damaging, P = possibly damaging, B = benign.
PP2HDIV (PolyPhen-2 Polymorphism Phenotyping v2 HumDiv): D = probably damaging, P = possibly damaging, B = benign.
MUTTASTER (MutationTaster): A = disease causing automatic, D = disease causing, N = polymorphism, P = polymorphism automatic.
LRT (Likelihood Ratio Test): D = deleterious, N = Neutral, U = unknown.
CADD - Combined Annotation Dependent Depletion based scores.
The missense variants were defined as deleterious when predicted to be damaging, probably damaging, disease causing and deleterious by the five algorithms applied (SIFT, PolyPhen-2 HumVar, PolyPhen-2 HumDiv, MutationTaster and LRT score) and/or CADD score of more than 20. We considered only deleterious variants with minor allele frequency less than 1% in the burden analysis.
Table 4.
Burden of FURIN rare variants in the Middle East and gnomAD populations.
| Variant ID | Protein Consequence |
Minor Allele Frequency |
P value |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KWT | IRN | QAR | EUR | EAS | AFR | KWT vs IRN | KWT vs QAR | KWT vs EUR | KWT vs AFR | IRN vs QAR | IRN vs EUR | IRN vs AFR | QAR vs EUR | QAR vs AFR | EUR vs EAS | EUR vs AFR | EAS vs AFR | |||
| rs201172453 | R37C | 0.004 | 0.004 | 0.007 | 7.0E-05 | 0 | 1.6E-04 | 1 | 1 | 0.0005 | 0.0005 | 1 | 0.0005 | 0.0005 | 0.0120 | 0.0610 | __ | 0.2724 | __ | |
| rs148110342 | R81C | 0.024 | 0.008 | 0.034 | 1.6E-03 | 0 | 1.6E-04 | 0.0015 | 0.4468 | 0.0005 | 0.0005 | 0.0055 | 0.0010 | 0.0005 | 0.0005 | 0.0005 | __ | 0.0005 | __ | |
| rs751909359 | R86Q | 0 | 0.001 | 0 | 3.0E-05 | 0 | 0 | __ | __ | __ | __ | __ | 0.0645 | __ | __ | __ | __ | __ | __ | |
| rs752639409 | R637Q | 0 | 0.001 | 0 | 0 | 0 | 6.0E-05 | __ | __ | __ | __ | __ | __ | 0.1709 | __ | __ | __ | __ | __ | |
| rs761541008 | R677W | 0 | 0.001 | 0 | 5.0E-05 | 0 | 0 | __ | __ | __ | __ | __ | 0.1000 | __ | __ | __ | __ | __ | __ | |
| rs35641241 | R745Q | 0 | 0 | 0 | 1.2E-03 | 1.0E-03 | 8.0E-05 | __ | __ | __ | __ | __ | __ | __ | __ | __ | 0.5857 | 0.0005 | 0.0005 | |
| rs193268286 | S685P | 0 | 0 | 0 | 4.0E-05 | 0 | 0 | __ | __ | __ | __ | __ | __ | __ | __ | __ | __ | __ | __ | |
The missense variants were defined as deleterious when predicted to be damaging, probably damaging, disease causing and deleterious by the five algorithms applied (SIFT, PolyPhen-2 HumVar, PolyPhen-2 HumDiv, MutationTaster and LRT score) and/or CADD score of more than 20. We considered only deleterious variants with minor allele frequency less than 1% in the burden analysis. P values are calculated using Chi-square test.
KWT-Kuwaitis; IRN-Iranians; QAR-Qataris; EUR-Europeans (non-Finnish); EAS-East Asians; AFR-Africans.
Table 5.
FURIN eQTL variants in the Middle East and gnomAD populations.
| Variant ID | Gene | Tissue | P GTEx |
NES GTEx |
Minor Allele Frequency |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| KWT | IRN | QAR | EUR | EAS | AFR | |||||
| rs117976310 | S2VB, LOC101926928 | Adrenal Gland | 3.0E-06 | -1.1 | NA | NA | 0.01 | 0.02 | 0.00 | 0.002 |
| rs12904679 | BLM, FURIN | Esophagus - Mucosa | 3.1E-06 | -0.2 | NA | NA | 0.12 | 0.12 | 0.003 | 0.02 |
| rs4932373 | FES | Esophagus - Mucosa | 6.6E-19 | 0.28 | NA | NA | 0.30 | 0.33 | 0.09 | 0.13 |
| rs2238332 | BLM | Pituitary | 2.7E-06 | -0.17 | NA | NA | 0.36 | 0.36 | 0.33 | 0.70 |
| rs8039305∗ | FURIN | Esophagus - Mucosa | 1.7E-17 | 0.23 | NA | 0.41 | 0.49 | 0.48 | 0.17 | 0.81 |
| rs6226∗ | FURIN | Artery - Tibial | 1.6E-09 | 0.12 | 0.72 | 0.65 | 0.69 | 0.68 | 0.53 | 0.93 |
| rs12904055 | CRTC3 | Brain - Putamen (basal ganglia) | 1.1E-05 | 0.2 | NA | NA | 0.24 | 0.22 | 0.17 | 0.31 |
| rs80160059 | SV2B | Colon - Sigmoid | 9.0E-06 | 0.54 | NA | NA | 0.06 | 0.001 | 0.00 | 0.11 |
| rs2071410 | FURIN | Esophagus - Mucosa | 1.7E-20 | 0.28 | NA | 0.26 | 0.27 | 0.33 | 0.06 | 0.18 |
| rs8026133 | LOC101926928, SLCO3A1 | Minor Salivary Gland | 1.4E-06 | 0.83 | NA | NA | 0.12 | 0.01 | 0.09 | 0.47 |
| rs2227935 | BLM | Esophagus - Mucosa | 2.9E-10 | 0.32 | 0.05 | 0.03 | 0.03 | 0.07 | 0.0004 | 0.07 |
| rs16944923 | BLM, FURIN | Skin - Sun Exposed (Lower leg) | 9.0E-06 | 0.12 | NA | NA | 0.13 | 0.14 | 0.20 | 0.05 |
| rs28385078 | BLM | Esophagus - Mucosa | 5.7E-10 | 0.32 | NA | NA | 0.02 | 0.07 | 0.00 | 0.03 |
| rs116449376 | BLM, FURIN | Esophagus - Mucosa | 1.5E-09 | 0.34 | NA | NA | 0.02 | 0.06 | 0.00 | 0.04 |
| rs7495370 | BLM, FURIN | Whole Blood | 5.6E-07 | 0.072 | NA | NA | 0.64 | 0.57 | 0.78 | 0.47 |
| rs7165790 | BLM | Heart - Atrial Appendage | 4.4E-07 | -0.12 | NA | NA | 0.34 | 0.00 | 0.001 | 0.00 |
P GTEx- P values reported in GTEx database; NES-normalized effect size in GTEx database; ∗P values calculated using Chi-square test.
KWT-Kuwaitis; IRN-Iranians; QAR-Qataris; EUR-Europeans (non-Finnish); EAS-East Asians; AFR-Africans. NA denotes Not Available in exome data.
4. Discussion
At the time of revising this article during December 2020, SARS-CoV-2 has infected 77,394,940 and killed about 1,703,164 people worldwide [1]. The case fatality rates (CFR) varied significantly around the world, with the highest reported in France, United Kingdom, Italy, Belgium, and Spain reaching 15.3%, 15%, 14.1%, 13.6% and 7.9%, respectively (https://ourworldindata.org/mortality-risk-covid; accessed on 7 August 2020). Whereas, the CFRs were distinctly lower in Qatar, Kuwait, South Africa and Japan at 0.2%, 0.7%, 1.8% and 2.4%, respectively and moderately higher in Iran (5.6%) and China (5.5%) that reported the highest CFR in Asia (https://ourworldindata.org/mortality-risk-covid; accessed on 7 August 2020). The lower infection and CFRs in developing countries obviously cannot be attributed to increased number of tests or better medical services than what is already provided in Europe. Moreover, prior knowledge of the SARS-CoV-2 RNA genome revealed by whole genome sequencing of the virus, performed by us and others, has shown remarkable genetic similarities among the virus [57] circulating between Qatar, Kuwait and Europe, which was attributed to traveling and repatriation between these countries [Manuscript in preparation; GISAID [57]]. For these reasons, it is not unrealistic to propose that the differences observed in CFR among countries may be attributed to host/ethnic germline variations in genes involved in SARS-CoV-2 processing and cellular entry or exit. Therefore, we screened the genetic variations and eQTL expression of the SARS-CoV-2 candidate genes, ACE2, TMPRSS2 and FURIN in three Middle Eastern populations: Kuwaiti, Iranian, and Qatari and compared them to available MAF of the continental populations including Europeans, East Asians and Africans from the gnomAD database [31].
Through comprehensive survey, we identified novel and common missense variants at differential frequencies in the study populations. Among the detected nine known ACE2 missense variants, we observed that N720D (rs41303171) and K26R (rs4646116) were the most frequent in the European datasets [21]. Moreover, recent structural predictions by Stawiski and colleagues revealed that the K26R missense variant enhanced the affinity of ACE2 for SARS-CoV-2 whereas N720D had little involvement in the SARS-CoV-2 S-protein interaction [21]. Our structural investigation suggests that the ACE2 receptor N720D variant may enhance TMPRSS2 activation and subsequent viral entry. Interestingly, the prevalence of the most ACE2 activating variant N720D (rs41303171) was highest among Europeans (2.5%), Iranians (0.6%) when compared to Kuwaitis (0.3%), Qataris (0.2%) and other global populations (0.4%) and MAF of this variant significantly correlated with the CFRs (p < 0.0003) in the corresponding countries (Figure 4). However, further functional assessments are required to confirm our predictions. Notably, a recent Iranian population based study missed this activating variant N720D despite its prevalence in the Iranome database and claimed no association between the ACE2 variants and COVID-19 disease severity or mortality among Iranians [58]. Albeit, the study was limited to 45 patients, which is insufficient to establish significant conclusions. While another recent study highlighted the impact of the activating variant N720D (rs41303171) on epidemiological disparities, especially in Europeans when compared to other global populations [59] and stands in support of our observations.
In the context of the occurrence of rare deleterious ACE2 variants, while the MAF of R708W in Kuwait is 0.105%, which may indicate again a protective role in the Kuwaiti population, the absence of this and the other four deleterious variants from Africa, East-Asia [31], Qatar [30] does not explain the lower disease burden in these countries. Similarly, we did not observe a significant difference in the burden of novel deleterious variants comparing the Kuwaiti population with Europeans (Table 2), although they may play a protective role against COVID-19 locally. The functional roles of the two Kuwait-specific and the two Iran-specific ACE2 deleterious novel variants Q661P (MAF = 0.1%), A372G (MAF = 0.2%) and V485L (MAF = 0.18%), F452V (MAF = 0.08%) respectively are yet to be experimentally determined.
Further, our structural estimations of the ACE2 novel variants predicted the following, (i) Q661P is close to the aa region 652–659, which is important for cleavage by the metalloprotease ADAM17; (ii) R716H and the two deleterious variants R708W and R708Q are located within residues 697–716 essential for cleavage by TMPRSS11D and TMPRSS2. In fact, the mutation of arginine -such as R708- and lysine residues within amino acid residues 697-716 markedly reduced ACE2 cleavage by TMPRSS2 [60]. However, it should be mentioned that sequentially distant aa residues in the ACE2 receptor can be seen brought structurally proximal to each other to create active sites for catalysis [45]. We illustrated this in Figure 5 to show that the novel aa changes (colored green) are proximal to the protein residues that mediate its activity (colored blue and red). Further studies are needed to directly assess the functional aspects of the reported missense variants. We urge the international community to assess ACE2 variation differences among people with mild/asymptomatic disease versus patients presenting with severe respiratory distress syndrome.
Figure 5.
ACE2 protein structure (open form, yellow; and closed form with substrate, gray). The active site residues are coded in red color. The zinc binding residues are coded in blue color. The identified novel amino acid changes in the Middle Eastern populations are coded in green color. The novel changes are proximal to the protein residues that mediate its activity.
Interestingly, there was not a single individual in the Middle East with ACE2 receptor variations in amino acids known to be crucial for SARS-CoV-2 S-protein binding (K31, E35, D38, M82, K353) [61]. This may indicate that natural immunity conferred by the ACE2 receptor variations is lacking or extremely rare. The rareness is evident from a recent comprehensive study that scrutinized ACE2 gene variations in more than 290,000 individuals from 400 different worldwide population groups and in which only eight rare ACE2 gene missense variants (K31R, E35K, E37K, D38V, N33I, H34R, Y83H, and Q388L) with reduced binding to the S-protein of SARS-CoV-2 were reported [21]. Notably, this study has disproved the claim of Cao and colleagues on the absence of such protective ACE2 variants in human populations [18].
It is to be pointed out that there is no explicit published experimental evidence for our proposition that Lys26Arg increases virus entry and N720D increases cleavage of ACE2 which also has a positive effect on virus entry. However, there are sufficient suggestive evidences, as indicated below. A recent study [62] mined the whole-exome sequencing data of 6,930 Italian control individuals (available in the Network of Italian Genomes) from five different centers and identified the ACE2 variants, K26R and N720D along with G211R as common missense changes with a potential to interfere with protein structure and stabilization. Another recent work [18] also listed these two variants as hotspot variants that could make some people differentially susceptible to SARS-CoV-2 infection. We demonstrated through structural modelling that the K26R change creates a new hydrogen bond with D30, which is then poised to build a salt-bridge with the S-protein RBD K417 that increases the affinity of SARS-CoV-2 to the ACE2 receptor [21] (Figure 2B). The work by Stawiski and colleagues (2020) has also revealed that the K26R variant enhanced the affinity of ACE2 for SARS-CoV-2. As mentioned earlier, a recent study using a synthetic human ACE2 mutant library has reported that the R26 variant increased S-protein binding and susceptibility to the virus significantly [42]. As regards N720D change, our data presents a novel prediction that the ACE2 receptor N720D variant may enhance TMPRSS2 activation and subsequent viral entry. The amino acid substitution can make a change in interatomic interactions between amino acids near the cleavage region and subsequently can decrease the stability and increase the flexibility of the loop, which makes it easier for TMPRSS2 to cleave. Benetti and colleagues (2020) also concluded that this N720D residue is located close to the cleavage sequence of TMPRSS2 and is likely to affect the cleavage-dependent virion intake. Though there exists no direct experimental evidence, the reported novel proposition will lead to future studies dealing with TMPRSS2 enzyme activity which is important for coronavirus spread and pathogenesis in the infected host.
Recently [63], examined natural amino acid replacements at ACE2 regions binding to SARS-COV-2 spike protein across 22 mammals. Though pigs and dogs, which are not effectively infected by SARS-COV-2, have only a few changes in the binding site, they exhibit relatively low levels of ACE2 in the respiratory tract. They further showed that bats and pangolins, which are now thought as confirmed carriers of viruses closely related to SARS-CoV-2, exhibit up to seven amino acid replacements in ACE2 residues binding to SARS-CoV-2 spike protein. Further examining the intermediate hosts, they reported that none of the ACE2 proteins of any of the discussed intermediate hosts seems to be especially equipped to attach to the spike protein of SARS-CoV-2. Of the two ACE2 amino acid changes that we report, the N720D was not examined in this article; though the article examines changes at the other amino acid position namely 26, the change that we report Lys26Arg has not been seen in any of the mammals – it is a more or less invariable position showing only K26N and K26Q changes in two mammals.
The hypothesis, that we proposed initially, is that ethnic genetic variations in the ACE2-TMPRSS2-FURIN genes may act as key regulators for orchestrating SARS-CoV-2 cellular access. It has been reported in literature that systematic identification of the genetic determinants of COVID-19 pathway (such as ACE2 and TMPRSS2 polymorphisms) can guide personalized treatments in this pandemic and may even explain the related epidemiologic observations on susceptibility, severity, and clinical outcome, including both virus and host factors. The reported ACE2 deleterious variations are rare or less frequent and hence might not solely attribute to the overall infection rate. Genetic variation is one of the many risk factors that contribute to the overall clinical outcome; there are other contributors, such as age, gender, and pre-existing health conditions [27, 54, 55], that would act in conjunction with the genetic risk and bring about the inter-individual variability in outcome. Notable pre-existing health conditions that lead to severe outcomes in COVID-19 patients are diabetes and hypertension. Public Health England recently reported that hospital admission rates of ethnic minorities (including Indian, Black, and Arab populations) are three times higher than the majority white British population in the United Kingdom. The economists Platt and Warwick discussed the likely reasons for the ethnic inequalities in terms of disparities in age, gender and geographic profiles of ethnic groups Occupational exposure and economic vulnerability [64]. The impact of COVID-19 on a population also depends on the extent and timeliness of prevention efforts which certainly differ from country to country. There is also resistance from societies to adopt social distancing and isolation especially from countries with high socioeconomical status. However, contributions from genetic factors cannot be ruled out. Some of the eQTL variants for the considered genes in this study and missense variants are at higher frequency in African populations than at European population – for example, the FURIN eQTL variant rs8026133 (Table 5) is 47% in Africa, 12% in Qatar and is only 1% in European population. Thus, a large plethora of genetic variants from the different molecular components of COVID-19 pathway need to be considered. It is also possible that other epigenetic factors can also play a role including the socioeconomic status of those individuals amongst many others.
The deleterious variations that we report in ACE2 are rare or less frequent and hence might not attribute to the overall infection or mortality rate. Genetic variation is one of the many risk factors that contribute to the overall clinical outcome; there are other contributors, such as comorbid health conditions, that would act in conjunction with the genetic risk and bring about the inter-individual variability in outcome. The other factors contributing to the clinical outcome include age, gender and pre-existing health conditions [27, 54, 55]. Notable pre-existing health conditions that lead to severe outcomes in COVID-19 patients are diabetes and hypertension. The patients with type 2 diabetes show high prevalence, severity of disease and mortality during SARS-CoV-2 infection (see the commentary by [65]. COVID-19 patients with worse glycemic control evidenced worse prognosis. Notably, as demonstrated by Marfella and colleagues (2020), COVID-19 patients with hyperglycemia could experience a reduced effect of tolicizumab therapy, that is anti-IL6 drug administered for the ICU admitted patients. Thus, hyperglycemia could result in an unfavorable effect on hospital admission, clinical outcomes and drug therapy [66, 67]. Another most common comorbidity and cause of death for COVID-19 patients is hypertension. The severe acute respiratory syndrome is caused by the binding of the virus to the target epithelial lung cells through angiotensin converting enzyme 2 (ACE2). Thus, hypertensive patients with COVID-19 could have worse prognosis. The ACE2 pathway and anti-hypertensive therapies play a central role in COVID-19 [68, 69]. Indeed, the common finding coming from these observations is that the endothelial dysfunction, that is mainly seen in COVID-19 patients with comorbidities, causes severe inflammation and endothelial damage leading to intravascular coagulopathy (DIC) and higher rate of death [70].
In case of TMPRSS2 gene, a deleterious variant rs12329760 (V197M) was frequent in Middle East and global populations from gnomAD (Table S2) and 1000 Genomes Project Phase 3 [71] datasets. Two different in silico investigations predicted the impact of this variant in viral entry into the host cell. One suggested that this variant produces a largest de novo pocket protein, which might affect the TMPRSS2 structure [72] and the other reported decreased stability of the protein in the presence of this variant [73]. However, Vargas-Alarcón and colleagues (2020) have illustrated no possible effect of this variant on the TMPRSS2 post-translational modifications.
Additionally, we showed high prevalence of frequent deleterious FURIN variants in the Middle Eastern populations, especially Kuwait and Qatar, which indicated decreased FURIN protease activity that further reduces the risk of viral infection. As such, our results suggest a possible protective role of FURIN gene variants against the SARS-CoV-2 infection in the studied Middle Eastern populations. However, the data presented here add another variable that needs to be urgently addressed. It is not unreasonable to suggest that European human genetic variation presents optimal access for SARS-CoV-2 infection, whereas Iranian genetic background presents sub-optimal opportunities for the infection, and finally, Kuwaitis and Qataris have the least natural inductive genetic background in the ACE2 and FURIN proteins combined for viral entry.
5. Conclusions
In summary, we report the differential occurrence of gene variants in the Middle Eastern populations that enrich the knowledge on the genetic susceptibility of different global populations to the novel SARS-CoV-2 virus. Especially that in the context of a pandemic, rare variants that increase or decrease disease susceptibility become quantitatively important since millions of people may be infected. The decreased burden of rare deleterious variants and the increased MAF in variants that activate the ACE2 receptor in the European population suggest an inherent susceptibility to SARS-CoV-2 infection and vice versa in the Middle Eastern population. We understand that other confounding factors like SARS-CoV-2 testing, socioeconomical status, the availability of proper medical services and the higher burden of other diseases may be important contributors to the disparities seen in mortality rates around the world. It is imperative that more comprehensive studies should be conducted as more patient data emerges from different parts of the world. Our data are preliminary and highlight the urgent need to correlate patients' medical/infection history to their genetic variants in order to predict in a more accurate and personalized way how human genetic variations may influence this pandemic.
We note that our findings are preliminary based on the computational (in silico) analyses, therefore, further experimental validations are required to understand the ramification of the reported gene variants on the entry of SARS-CoV-2 virus into the host cells.
Declarations
Author contribution statement
Hamad Ali, Mohamed Abu-Farha, Rasheed Ahmad, Jehad Abubaker: Analyzed and interpreted the data.
Fahd Al-Mulla: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Anwar Mohammad, Ashraf Al Madhoun, Dania Haddad: Performed the experiments.
Muthukrishnan Eaaswarkhanth: Analyzed and interpreted the data; Wrote the paper.
Sumi Elsa John, Rasheeba Nizam, Arshad Channanath, Thangavel Alphonse Thanaraj: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by a Coronavirus emergency resilience grant from Kuwait Foundation for the Advancement of Sciences (#PN20-93MM-02).
Data availability statement
Data included in article/supplementarymaterial/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali H. Outcomes of COVID-19: disparities by ethnicity. Infect. Genet. Evol. 2020:104639. doi: 10.1016/j.meegid.2020.104639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alshukry A. Clinical characteristics of coronavirus disease 2019 (COVID-19) patients in Kuwait. PloS One. 2020;15 doi: 10.1371/journal.pone.0242768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alahmad B., Al-Shammari A.A., Bennakhi A., Al-Mulla F., Ali H. Fasting blood glucose and COVID-19 severity: nonlinearity matters. Diabetes Care. 2020;43:3113–3116. doi: 10.2337/dc20-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abu-Farha M. Impact of diabetes in patients diagnosed with COVID-19. Front. Immunol. 2020;11:576818. doi: 10.3389/fimmu.2020.576818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ali H. Clinical characteristics and outcomes of diabetic cOVID-19 patients in Kuwait. medRxiv. 2020 2020.08.20.20178525. [Google Scholar]
- 7.Yan R. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xi J. Virus strain of a mild COVID-19 patient in Hangzhou representing a new trend in SARS-CoV-2 evolution related to furin cleavage site. medRxiv. 2020 doi: 10.1080/22221751.2020.1781551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coutard B. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukassen S. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020 doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asselta R., Paraboschi E.M., Mantovani A., Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. medRxiv. 2020 doi: 10.18632/aging.103415. 2020.3.30.20047878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann M. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glowacka I. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haddad D. SARS-CoV-2: proof of recombination between strains and emergence of possibly more virulent ones. medRxiv. 2020 2020.11.11.20229765. [Google Scholar]
- 15.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Coupanec A. Cleavage of a neuroinvasive human respiratory virus spike glycoprotein by proprotein convertases modulates neurovirulence and virus spread within the central nervous system. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao Y. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y., Shan K., Qian W. 2020. Asians and Other Races Express Similar Levels of and Share the Same Genetic Polymorphisms of the SARS-CoV-2 Cell-Entry Receptor. [Google Scholar]
- 20.Renieri A. ACE2 variants underlie interindividual variability and susceptibility to COVID-19 in Italian population. medRxiv. 2020 doi: 10.1038/s41431-020-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stawiski E.W. Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. bioRxiv. 2020 [Google Scholar]
- 22.Chen J. 2020. Individual Variation of the SARS-CoV2 Receptor ACE2 Gene Expression and Regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alnohair S. Obesity in gulf countries. Int. J. Health Sci. 2014;8:79–83. doi: 10.12816/0006074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khalil A.B. Diabesity in the Arabian gulf: challenges and opportunities. Oman Med. J. 2018;33:273–282. doi: 10.5001/omj.2018.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alkandari A. The prevalence of pre-diabetes and diabetes in the Kuwaiti adult population in 2014. Diabetes Res. Clin. Pract. 2018;144:213–223. doi: 10.1016/j.diabres.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 26.Yang X. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou F. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.John S.E. Assessment of coding region variants in Kuwaiti population: implications for medical genetics and population genomics. Sci. Rep. 2018;8:16583. doi: 10.1038/s41598-018-34815-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fattahi Z. A catalog of genomic variations in the Iranian population. Hum. Mutat. 2019;40:1968–1984. doi: 10.1002/humu.23880. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez-Flores J.L. Exome sequencing identifies potential risk variants for mendelian disorders at high prevalence in Qatar. Hum. Mutat. 2014;35:105–116. doi: 10.1002/humu.22460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karczewski K.J. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auton A. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naslavsky M.S. Exomic variants of an elderly cohort of Brazilians in the ABraOM database. Hum. Mutat. 2017;38:751–763. doi: 10.1002/humu.23220. [DOI] [PubMed] [Google Scholar]
- 34.Ng P.C., Henikoff S.S.I.F.T. Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adzhubei I.A. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46:W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pires D.E.V., Ascher D.B., Blundell T.L. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014;42:W314–W319. doi: 10.1093/nar/gku411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demogines A., Farzan M., Sawyer S.L. Evidence for ACE2-utilizing coronaviruses (CoVs) related to severe acute respiratory syndrome CoV in bats. J. Virol. 2012;86:6350–6353. doi: 10.1128/JVI.00311-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Procko E. The sequence of human ACE2 is suboptimal for binding the S spike protein of SARS coronavirus 2. bioRxiv. 2020 [Google Scholar]
- 43.Li W. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarke N.E., Turner A.J. Angiotensin-converting enzyme 2: the first decade. Int. J. Hypertens. 2012;2012:307315. doi: 10.1155/2012/307315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Towler P. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson W.T., Evans D.M., An J., Jones S.J.M. ACE 2 coding variants: a potential X-linked risk factor for COVID-19 disease. bioRxiv. 2020 [Google Scholar]
- 47.Heurich A. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shulla A. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eberhardt E.S., Raines R.T. Amide-amide and amide-water hydrogen bonds: implications for protein folding and stability. J. Am. Chem. Soc. 1994;116:2149–2150. doi: 10.1021/ja00084a067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahmad S., Kumar V., Ramanand K.B., Rao N.M. Probing protein stability and proteolytic resistance by loop scanning: a comprehensive mutational analysis. Protein Sci. 2012;21:433–446. doi: 10.1002/pro.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishibashi F. Elasticity of the tibial nerve assessed by sonoelastography was reduced before the development of neuropathy and further deterioration associated with the severity of neuropathy in patients with type 2 diabetes. J. Diabetes Investig. 2016;7:404–412. doi: 10.1111/jdi.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan C., Li K., Ding Y., Lu W.L., Wang J. ACE2 expression in kidney and testis may cause kidney and testis damage after 2019-nCoV infection. medRxiv. 2020 doi: 10.3389/fmed.2020.563893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Canela-Xandri O., Rawlik K., Tenesa A. An atlas of genetic associations in UK Biobank. Nat. Genet. 2018;50:1593–1599. doi: 10.1038/s41588-018-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang C. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu C. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Richardson S. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York city area. JAMA. 2020 doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ardeshirdavani A. Clinical population genetic analysis of variants in the SARS-CoV-2 receptor ACE2. medRxiv. 2020 [Google Scholar]
- 59.Khayat A.S. ACE2 polymorphisms as potential players in COVID-19 outcome. medRxiv. 2020 doi: 10.1371/journal.pone.0243887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ko C.-J. Androgen-induced TMPRSS2 activates matriptase and promotes extracellular matrix degradation, prostate cancer cell invasion, tumor growth, and metastasis. Cancer Res. 2015;75:2949–2960. doi: 10.1158/0008-5472.CAN-14-3297. [DOI] [PubMed] [Google Scholar]
- 61.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benetti E. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur. J. Hum. Genet. 2020 doi: 10.1038/s41431-020-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhai X. Comparison of severe acute respiratory syndrome coronavirus 2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J. Virol. 2020;94 doi: 10.1128/JVI.00831-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Platt L., Warwick R. The Institute for Fiscal Studies; 2020. Are Some Ethinic Groups More Vulnerable to COVID-19 than Others? [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sardu C., Gargiulo G., Esposito G., Paolisso G., Marfella R. Impact of diabetes mellitus on clinical outcomes in patients affected by Covid-19. Cardiovasc. Diabetol. 2020;19:76. doi: 10.1186/s12933-020-01047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sardu C. Outcomes in patients with hyperglycemia affected by COVID-19: can we do more on glycemic control? Diabetes Care. 2020;43:1408–1415. doi: 10.2337/dc20-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marfella R. Negative impact of hyperglycaemia on tocilizumab therapy in Covid-19 patients. Diabetes Metab. 2020 doi: 10.1016/j.diabet.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sardu C. Could anti-hypertensive drug therapy affect the clinical prognosis of hypertensive patients with COVID-19 infection? Data from centers of southern Italy. J. Am. Heart Assoc. 2020 doi: 10.1161/JAHA.120.016948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bosso M. The two faces of ACE2: the role of ACE2 receptor and its polymorphisms in hypertension and COVID-19. Mol Ther Methods Clin Dev. 2020;18:321–327. doi: 10.1016/j.omtm.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sardu C. Hypertension, thrombosis, kidney failure, and diabetes: is COVID-19 an endothelial disease? A comprehensive evaluation of clinical and basic evidence. J. Clin. Med. Res. 2020;9 doi: 10.3390/jcm9051417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vargas-Alarcón G., Posadas-Sánchez R., Ramírez-Bello J. Variability in genes related to SARS-CoV-2 entry into host cells (ACE2, TMPRSS2, TMPRSS11A, ELANE, and CTSL) and its potential use in association studies. Life Sci. 2020;260:118313. doi: 10.1016/j.lfs.2020.118313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paniri A., Hosseini M.M., Akhavan-Niaki H. First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS-CoV-2 among different populations. J. Biomol. Struct. Dyn. 2020;1–18 doi: 10.1080/07391102.2020.1767690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vishnubhotla R. Genetic variants in TMPRSS2 and Structure of SARS-CoV-2 spike glycoprotein and TMPRSS2 complex. Cold Spring Harbor Lab. 2020 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementarymaterial/referenced in article.