Abstract
Objective
Asthma is the most common chronic disease in children. Short-acting bronchodilator medications are the most commonly prescribed asthma treatment worldwide, regardless of disease severity. Puerto Rican children display the highest asthma morbidity and mortality of any US population. Alarmingly, Puerto Rican children with asthma display poor bronchodilator drug response (BDR). Reduced BDR may explain, in part, the increased asthma morbidity and mortality observed in Puerto Rican children with asthma. Gene-environment interactions may explain a portion of the heritability of BDR. We aimed to identify gene-environment interactions associated with BDR in Puerto Rican children with asthma.
Setting
Genetic, environmental, and psycho-social data from the Genes-environments and Admixture in Latino Americans (GALA II) case-control study.
Participants
Our discovery dataset consisted of 658 Puerto Rican children with asthma; our replication dataset consisted of 514 Mexican American children with asthma.
Main Outcome Measures
We assessed the association of pairwise interaction models with BDR using ViSEN (Visualization of Statistical Epistasis Networks).
Results
We identified a non-linear interaction between Native American genetic ancestry and air pollution significantly associated with BDR in Puerto Rican children with asthma. This interaction was robust to adjustment for age and sex but was not significantly associated with BDR in our replication population.
Conclusions
Decreased Native American ancestry coupled with increased air pollution exposure was associated with increased BDR in Puerto Rican children with asthma. Our study acknowledges BDR’s phenotypic complexity, and emphasizes the importance of integrating social, environmental, and biological data to further our understanding of complex disease.
Keywords: Asthma, Minority Health, Precision Medicine, Air Pollution, Health Disparities
Introduction
Asthma is the most common chronic disease in children in the United States.1 Asthma is also the most disparate common disease in the country, with significant disparities in prevalence, morbidity, and mortality between US populations.2 Asthma prevalence is highest in Puerto Ricans (36.5%) compared with European Americans (12.1%), African Americans (13.0%), and Mexican Americans (7.5%).3 Puerto Rican children also have the highest asthma mortality rate in the United States.4 Short-acting bronchodilator medications, such as albuterol, are the first line of treatment for asthma symptoms in the United States regardless of disease severity.5 Bronchodilator drug response (BDR) varies widely across populations. Puerto Rican children with asthma demonstrate the lowest BDR compared with all other groups.6
Variability in BDR may contribute to asthma disparities in morbidity and mortality. BDR is a complex phenotype with an estimated heritability of approximately 28.5%, indicating that variation in BDR is significantly impacted by both genetic and environmental factors.7 Previous research has focused primarily on identifying single biological, environmental, or social factors associated with BDR. While there have been significant discoveries for independent effects associated with BDR, these associations account for only a small portion of the trait’s estimated heritability, leaving the majority of the variation in BDR undefined.8
A growing body of evidence suggests that gene-gene and gene-environment interactions have a significant impact on variation in complex phenotypes such as BDR.9 Comparatively few population-based studies of complex disease have incorporated interaction effects, and the majority of the studies that have attempted to assess interaction effects in pulmonary research have employed regression-based methods aimed at identifying linear, or additive, interactions.10 A growing body of evidence supports the presence and importance of epistatic (non-linear) interactions between genetic and environmental factors in the etiology of complex phenotypes.11,12 However, due to their non-linear nature, epistatic interactions are not reliably captured by the standard regression-based methods characteristic of large genetic/epidemiological studies.
We postulate that a portion of the undefined heritability in BDR could be attributed to the impact of epistatic interactions between biological, environmental, and psychosocial factors. Although Puerto Ricans carry a disproportionate share of the asthma burden in the United States, this population has been historically underrepresented in asthma research, and as a result, has been both understudied and underserved.13 To our knowledge, a comprehensive evaluation of linear and epistatic interactions between genetic, environmental, and social factors on variation in BDR has never been performed in Puerto Rican children with asthma. In this study, we investigate the impact of linear and non-linear interactions between clinical, environmental, and psychosocial factors on BDR in Puerto Rican children with asthma using analytical methods that acknowledge and embrace the complexity of this important asthma-related phenotype.
Methods
Study Participants
The Genes-environment and Admixture in Latino Americans (GALA II) study is an ongoing case-control study that began in 2006 aimed at discovering genetic and environmental factors associated with asthma and asthma-related phenotypes in Latino children. The study design for this cohort is described in detail elsewhere.8 Briefly, self-identified Latino children with and without asthma between the ages of 8-21 years were enrolled in the GALA II study at several clinical sites in the continental United States and Puerto Rico. Biological samples, environmental exposures, and demographic and clinical data were collected at study enrollment. The current study population includes 658 GALA II participants who self-identified as Puerto Rican with complete phenotypic and genetic information. Our replication population includes 514 GALA II participants who self-identified as Mexican American with complete phenotypic and genetic information (Supplemental Table 1, available from corresponding author).
Predictor Variables
Information on socioeconomic status (SES) and experiences of discrimination (EOD) were collected via questionnaire as previously described.14,15 Briefly, a composite score for SES was derived from three socioeconomic factors: mother’s educational attainment, insurance status, and household income weighted by region. Each composite score was then assigned a value of low SES (0), medium SES (1), and high SES (2). For the purposes of this study, EOD data were dichotomized into two categories; no experiences of discrimination (0) and at least one experience of discrimination (1). Body mass index (BMI) was calculated for each participant (BMI = kg/ m2). BMI percentile values were generated using a standard reference population according to the guidelines from the US Centers for Disease Control and Prevention growth charts.16 Obesity is a known asthma risk factor and co-morbidity that has been shown to significantly affect bronchodilator response in pediatric populations.16 BMI percentiles were dichotomized by obesity status (OBESE), where obesity status was defined as BMI ≥ 95th percentile (obese) or BMI <95th percentile (non-obese).
Air pollution exposure was defined as the measure of average lifetime exposure to particulate matter <2.5 μm in size (PM2.5) prior to recruitment gathered from the geographic coordinates of each participant’s residence using the TomTom/Tele ATLAS EZ-locate software (TomTom, Amsterdam, the Netherlands). Coordinates were linked to regional ambient daily air pollution data from the US Environmental Protection Agency Air Quality System. The inverse distance-squared weighted lifetime average of PM2.5 exposure was then calculated from up to four air pollution monitoring stations within 50 km of the participant’s residential geographic coordinates. Median PM2.5 was calculated for each recruitment site. Individual PM2.5 exposure measurements for each study participant were then dichotomized as less than or greater than the site-specific PM2.5 median.
All study participants were previously genotyped using the Axiom® LAT1 array (World Array 4, Affymetrix, Santa Clara, CA) as described elsewhere.17 Global ancestry was determined for each study participant using the ADMIXTURE software package.18 ADMIXTURE was run unsupervised assuming three ancestral populations (African, European, and Native American) to calculate the proportion of global African (AFR), Native American (NAT), and European (EUR) ancestry for each participant. Reference haplotypes were gathered from HapMap phase III CEU (European) and YRI (African). Reference haplotypes for Native American ancestry were derived from 71 Native Americans previously genotyped using the Axiom LAT1 array.19
Outcome Variable: Bronchodilator Drug Response (BDR)
Spirometric testing was performed to collect measurements of forced expiratory volume in one second (FEV1), at the time of study enrollment for all participants, as previously described.20 FEV1 is commonly used to describe baseline lung function in adult and pediatric populations.21 Maximal BDR was calculated according to American Thoracic Society/European Respiratory Society guidelines and was defined as the percent change in FEV1 (∆ FEV1) after treatment with albuterol, calculated as shown in the equation: 20
∆FEV1 = (post bronchodilator-FEV1 - pre bronchodilator-FEV1 / pre bronchodilator- FEV1) x 100%
A clinically meaningful bronchodilator response is defined as ∆FEV1 ≥ 12%.22 BDR was analyzed as a dichotomous variable; individuals were classified as bronchodilator responders (∆FEV1 ≥ 12%) and bronchodilator non-responders (∆FEV1 < 12%).
Outcome Variable: Baseline Lung Function (FEV1 Percent Predicted)
Correct interpretation of FEV1 measurements requires the use of age, sex, and race/ethnicity specific reference values. The Hankinson prediction equation was used to generate predicted FEV1 values for each study participant.23 We then compared actual FEV1 values (measured as described in the preceding subsection) to predicted FEV1 values to determine the percentage of predicted FEV1 actually achieved by each participant. FEV1 percent predicted values were then dichotomized to indicate optimal (actual FEV1 ≥ 80% predicted FEV1) and compromised (measured FEV1 < 80% predicted FEV1) baseline lung function.
Statistics
Visualization of Statistical Epistasis Networks (ViSEN) Interaction Analysis
ViSEN is a non-parametric entropy-based method capable of identifying interactions with and without the presence of single variant (main) effects.24 Previous studies have shown that ViSEN is as powerful as standard regression-based tests in identifying additive effects, and more powerful than standard regression-based methods in identifying non-linear, synergistic, interaction effects.24,25 ViSEN uses a measure from information theory called information gain (IG) to gauge the strength of the association between an interaction model and a specified outcome or phenotype. IG describes the percentage of variation in phenotype class (ie, BDR status) explained by a given interaction. Mutual information (MI) describes the percentage of variation in phenotype class explained by the independent effects of a single variable (main effects). For a given phenotype, ViSEN constructs an interaction network composed of the underlying biological and environmental factors impacting variation in phenotype that is centered on the strongest interaction effects rather than the strongest main effects. ViSEN’s IG calculation explicitly excludes the MI from each of the single variables that contribute to any given interaction model. This ensures that ViSEN identifies only “genuine” synergistic interaction effects, rather than “de facto” interaction effects that are merely artifacts of strong main effects. Permutation of BDR status (n = 1000) was used to create an empirical IG distribution, estimate IG p-values, and correct for multiple testing, as previously described.25 Interactions were deemed statistically significant with the IG P <.05.
Covariate Adjustment for ViSEN Analyses
Age and sex were identified as potential confounders in our study. Therefore, it was necessary to adjust for age and sex in our ViSEN analyses. The ViSEN software, however, is currently unable to directly adjust for covariates. Following a previously published protocol used to incorporate covariates into ViSEN analyses, we employed the local case-control (LCC) subsampling method to adjust for the effects of age and sex.26,27 Local case-control subsampling adjusts for covariates in a dataset by extracting a subset of individuals from the original dataset that consists of only the individuals for which prediction based on the covariates alone does not explain their case-control status well. For this subset of individuals, the impact of the specified covariates on case-control status is effectively removed.
We generated an unadjusted (for covariates) ViSEN interaction network model for BDR in our study population and calculated the IG for all possible pairwise (two variable) interaction models constructed from our six predictor variables; OBESE, SES, EOD, PM2.5, NAT, and AFR. Since ViSEN can accept only discrete variables as predictor and outcome variables, all variables used in ViSEN analyses were recoded as described (Supplementary Table 2, available from corresponding author). We also generated a ViSEN interaction network model for BDR adjusted for age and sex using a subset of our study population (bronchodilator responders = 153 and bronchodilator non-responders = 167) selected as previously described (LCC method) to assess the robustness of significant ViSEN associations identified in the full dataset.
Further Characterization of Significantly Associated Interaction Models Identified by ViSEN
All significant pairwise interaction models associated with BDR identified using ViSEN that remained significant after adjustment for age and sex, underwent additional analyses to further elucidate the nature of the relationship between the specified interaction and BDR. For significantly associated interactions, participants were split into groups defined by the specified interaction, and further described in terms of clinical and demographic factors. If the identified interaction model appeared to be non-linear in nature, descriptive statistics were generated in the R computing environment and used to make pairwise comparisons between groups.
Since baseline lung function (FEV1) is one of the strongest clinical predictors of BDR,28 any model with a significant IG from ViSEN analysis using BDR as the outcome was analyzed a second time in ViSEN using FEV1 (dichotomized as described above) as the phenotypic outcome. We performed both unadjusted (full dataset) and adjusted (LCC subset adjusted for age and sex; bronchodilator responders = 86 and bronchodilator non-responders = 94) ViSEN analyses. If the interaction was also found to be significantly associated with FEV1, and remained significant after adjustment for age and sex, mediation analysis was performed to determine if the association with BDR was due to an intermediate association with FEV1. Mediation analysis was performed in the R computing environment using the Robust Mediation (robmed) package.29 Robust mediation analysis used median regression and bootstrapping (n = 5000) to generate direct and indirect effect estimates and 95%CIs. Ps were computed from 95%CIs.30 For mediation analysis, BDR and FEV1 were analyzed as continuous variables. All mediation analyses were performed using data from the full dataset and were adjusted for age and sex.
Results
Study Demographics
Our study included 658 Puerto Rican children and adolescents with physician-diagnosed asthma from the GALA II cohort. Participants were categorized by BDR status (318 bronchodilator responders and 340 bronchodilator non-responders) and demographic variables were compared between groups (Table 1). Significant differences in age (P=.04), baseline lung function (4.08 x 10-15), and exposure to PM2.5 (P=.04) were discovered between bronchodilator responders and non-responders. It should be noted that the average difference in age between bronchodilator responders and non-responders was .3 years (Table 1); while this difference may be statistically significant, it is unlikely to be clinically relevant.
Table 1. Demographics for study population of Puerto Rican children.
| Bronchodilator responders | Bronchodilator non-responders | Pa | ||
| Sample size, N | 318 | 340 | --- | |
| Sex, % female | 40% | 44% | .25c | |
| Age, yrs, mean (SE) | 11.7 (.17) | 12.0 (.15) | .04b | |
| Baseline lung function, median (SE) | 81.28 (.87) | 91.03 (.90) | 4.08 x 10-15 b | |
| Obese | non-obese | 210 | 230 | .72 |
| obese | 108 | 110 | ||
| SES | low | 96 | 106 | .65 |
| medium | 66 | 74 | ||
| high | 156 | 160 | ||
| PM2.5 | <median | 146 | 185 | .04 |
| ≥median | 172 | 155 | ||
| EOD | none = 0 | 277 | 304 | .43 |
| yes ≥ 1 | 41 | 36 | ||
| Global ancestry, % mean (SE) | AFR | 23% (.007) | 22% (.006) | .49b |
| NAT | 10% (.002) | 11% (.003) | .26b | |
| EUR | 67% (.007) | 67% (.006) | .67b | |
SES, socioeconomic status; PM2.5, particulate matter <2.5 microns in diameter; EOD, perceived experience of discrimination; EUR, European ancestry; NAT, Native American ancestry; AFR, African Ancestry; SE, standard error.
a. significant associations. Ps were generated from the Chi-square test unless otherwise indicated.
b. P generated from the Wilcoxon Rank Sum test.
c. P generated from the difference in proportions test.
Identification of Pairwise Interactions Associated with BDR
ViSEN was used to create an entropy-based interaction network and analyze the relationships between all possible pairwise interactions models (constructed from six predictor variables; PM2.5, EOD, SES, OBESE, AFR, NAT) and BDR (Figure 1). Only one interaction model (PM2.5 x NAT) was significantly associated with BDR after correction for multiple testing (unadjusted ViSEN IG%=.53, P=.03) (Figure 1). The association between PM2.5 x NAT and BDR was robust to adjustment for age and sex (adjusted ViSEN IG%=1.34, P=.02).
Figure 1. Interaction network of variables underlying variation in bronchodilator drug response.
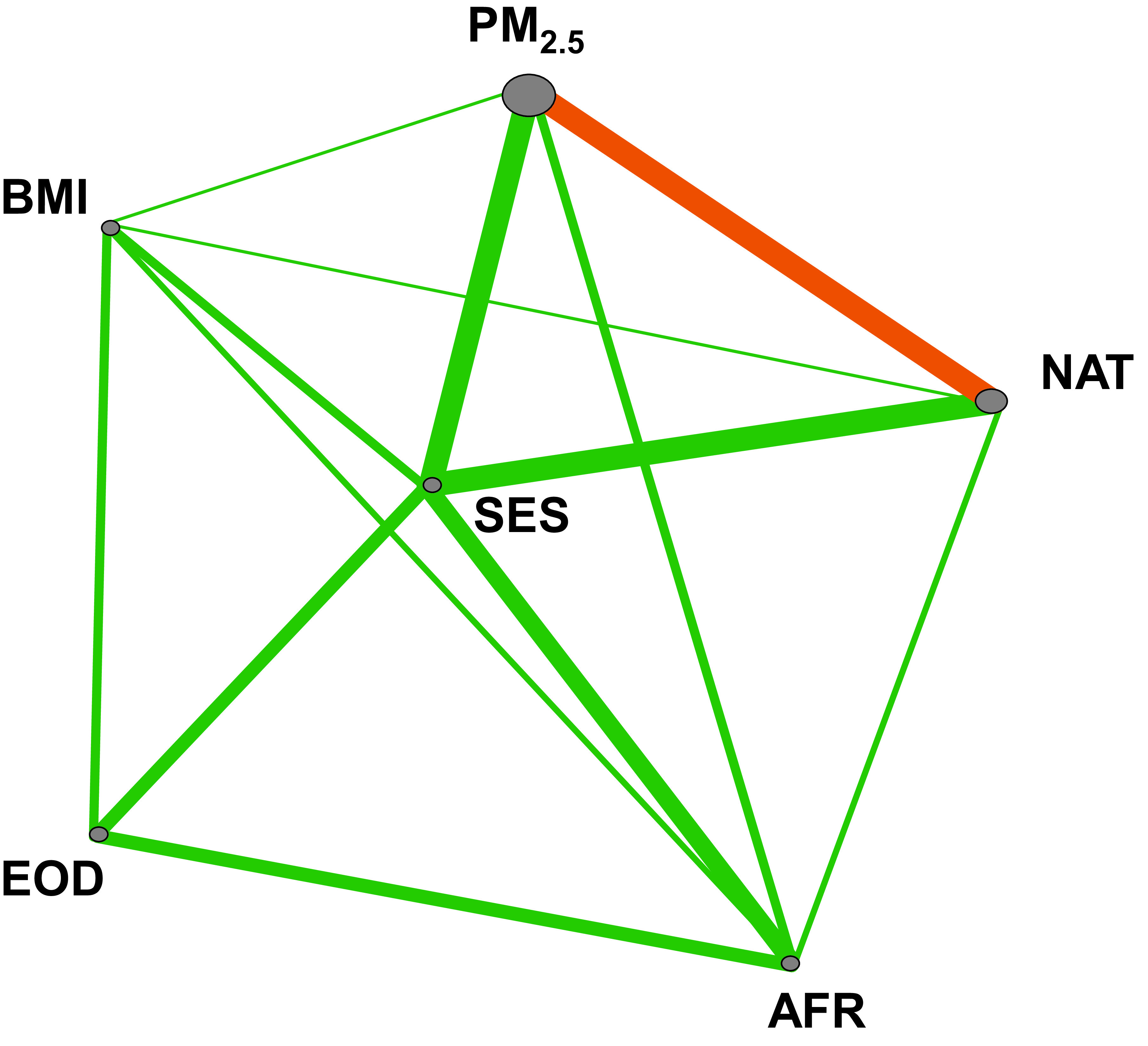
Graphical visualization of an entropy-based interaction network impacting BDR variation constructed by ViSEN. Gray nodes represent individual variables, while the size of a node indicates the strength of the variables independent effect on BDR. Lines represent pairwise interactions between variables; the thickness of the line indicates the strength of the pairwise interaction effect on BDR measured by information gain (IG). Significant pairwise interactions (IG P<.05) are colored in orange. EOD, experience of discrimination; AFR, African ancestry; NAT: Native American ancestry; OBESE: obesity status; SES, socioeconomic status; PM2.5: Particulate matter <2.5 μm in size.
Previous studies have shown that Puerto Ricans have the lowest BDR while the Mexican American population displays the highest BDR among US populations.31 To determine if PM2.5 x NAT interaction effects observed in our Puerto Rican study population were conserved across the BDR phenotypic distribution, we attempted to replicate our ViSEN results in an independent population of 514 Mexican American children. The association between the PM2.5 x NAT interaction and BDR was not significant in Mexican American children (unadjusted ViSEN IG%=.02%, P=.73; age and sex adjusted ViSEN IG%=.23%, P=.43).
Further Characterization of the PM2.5 x NAT Interaction Model
Thorough investigation of the PM2.5 x NAT interaction model revealed that the interaction’s relationship with BDR was non-linear (Figure 2). Only participants in PM2.5 x NAT interaction Group 3 (increased PM2.5 exposure and decreased NAT) demonstrated clinical responsiveness to bronchodilator treatment (Figure 2).
Figure 2. Variation in bronchodilator drug response by PM2.5 x NAT interaction group.
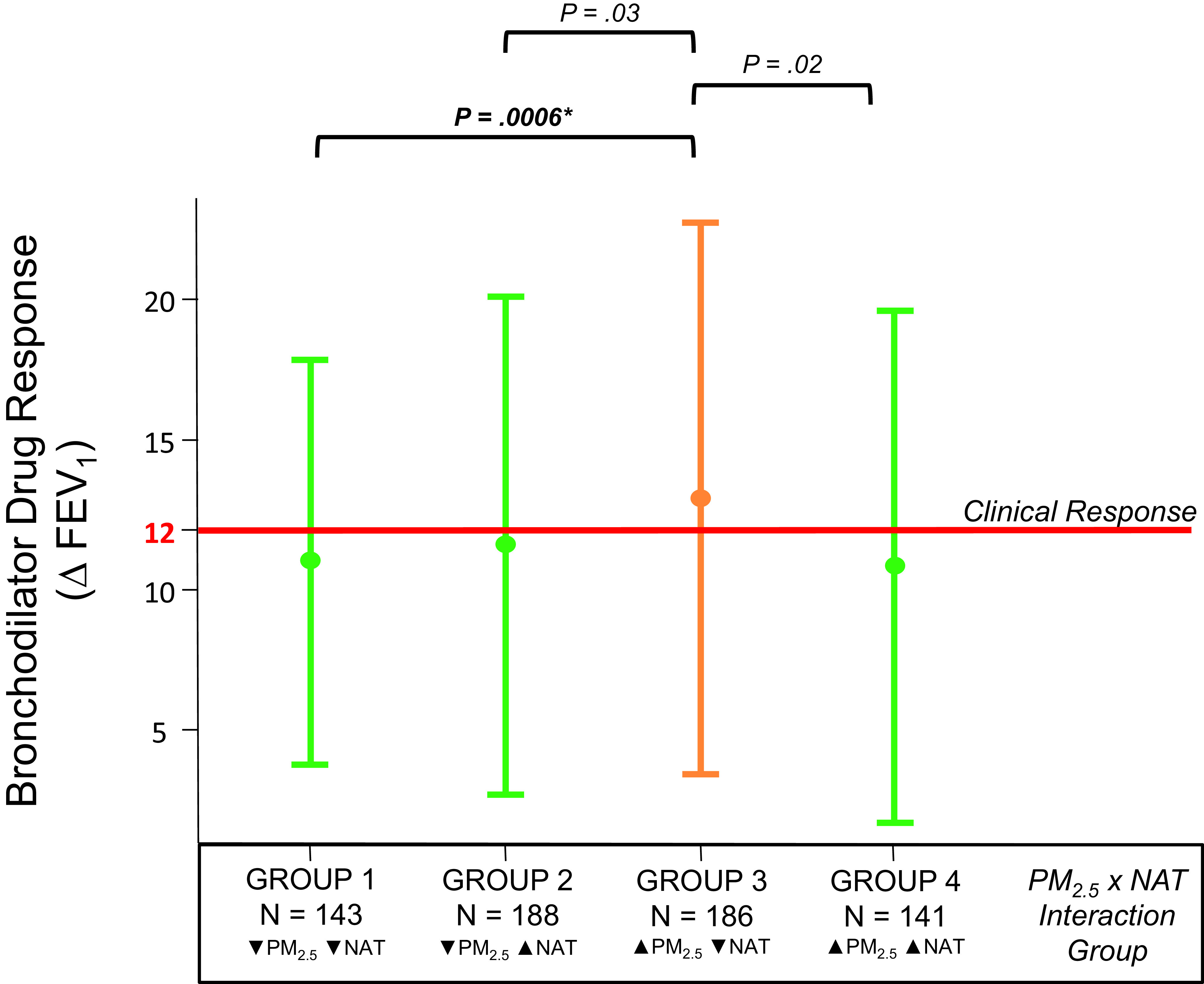
Error plot graph of bronchodilator drug response by PM2.5 x NAT interaction group membership. Vertical lines denote the interquartile range (IQR) of group data; circles indicate the group median. Orange lines indicate groups that achieved clinically significant BDR (∆ FEV1 ≥ 12%). Ps presented here are from the Dunn Test of differences between groups; black brackets indicate the groups being compared. P-values in bold remained significant after correction for Bonferroni correction for multiple testing. ∆ FEV1 is the difference in % of Predicted FEV1 achieved before and after bronchodilator treatment. ▼ means that the group median < study population median; ▲ means that the group median ≥ study population median. PM2.5 denotes exposure to particulate matter, while NAT indicates Native American genetic ancestry. FEV, forced expiratory volume.
We then compared measurements of our five other ViSEN predictor variables (SES, OBESE, EOD, AFR, and baseline lung function) between NAT x PM2.5 interaction groups. Baseline lung function was the only predictor that differed significantly by PM2.5 x NAT interaction status. Pairwise comparisons, using interaction Group 3 as the reference group, revealed that participants in interaction Group 3 had significantly lower baseline lung function (median FEV1 % predicted = 82%) compared to all other groups (Group 1 median FEV1 % predicted = 88%, P=.0006; Group 2 median FEV1 % predicted = 86%, P=.03; Group 4 median FEV1 % predicted = 86%, P=.02) (Figure 2). It should be noted that while we did identify significant differences in baseline lung function between PM2.5 x NAT interaction groups, the interquartile range (IQR) for each interaction group is quite wide. This indicates high variability in FEV1 % predicted between individuals within the same interaction group; this high degree of inter-individual variability is characteristic of FEV1 measurements. Though we have separated individuals into groups defined by PM2.5 and NAT, these groups are by no means homogenous in terms of FEV1 measurements and this should be taken into account when determining the clinical relevance of NAT x PM2.5 group membership.
Baseline lung function (FEV1 % predicted) is one of the strongest clinical predictors of BDR. Specifically, decreased FEV1 % predicted has been associated with increased BDR in children with asthma.28 Given this a priori knowledge, we performed a secondary ViSEN analysis to evaluate the effect of all possible pairwise interactions between our six predictor variables (and variation in FEV1. Once again, the PM2.5 x NAT interaction was the only model significantly associated with our outcome (ViSEN IG%=.53 %, P=.03).
As the PM2.5 x NAT interaction was associated with both BDR and with FEV1, we decided to perform a mediation analysis to explicitly test whether the significant association observed with BDR was merely an artifact of an intermediate association with FEV1. One of the assumptions of mediation analysis is that the effects are additive in nature; to gain more power in our mediation analysis, PM2.5 x NAT groups were recoded prior to mediation analysis to simulate an additive effect as follows: participants with increased PM2.5 and decreased NAT (Group 1) vs all other study participants (Group 2). Mediation analysis uncovered significant direct (P=.03) and indirect (P=.004) effects of the PM2.5 x NAT interaction on BDR (Figure 3). Although the indirect effect of the PM2.5 x NAT interaction on BDR, via the intermediate phenotype of FEV1, was statistically significant, it did not account for the majority of the interactions effect on BDR (33% of the total interaction effect) (Figure 3).
Figure 3. Direct and indirect effects of the PM2.5 x NAT interaction on bronchodilator drug response.
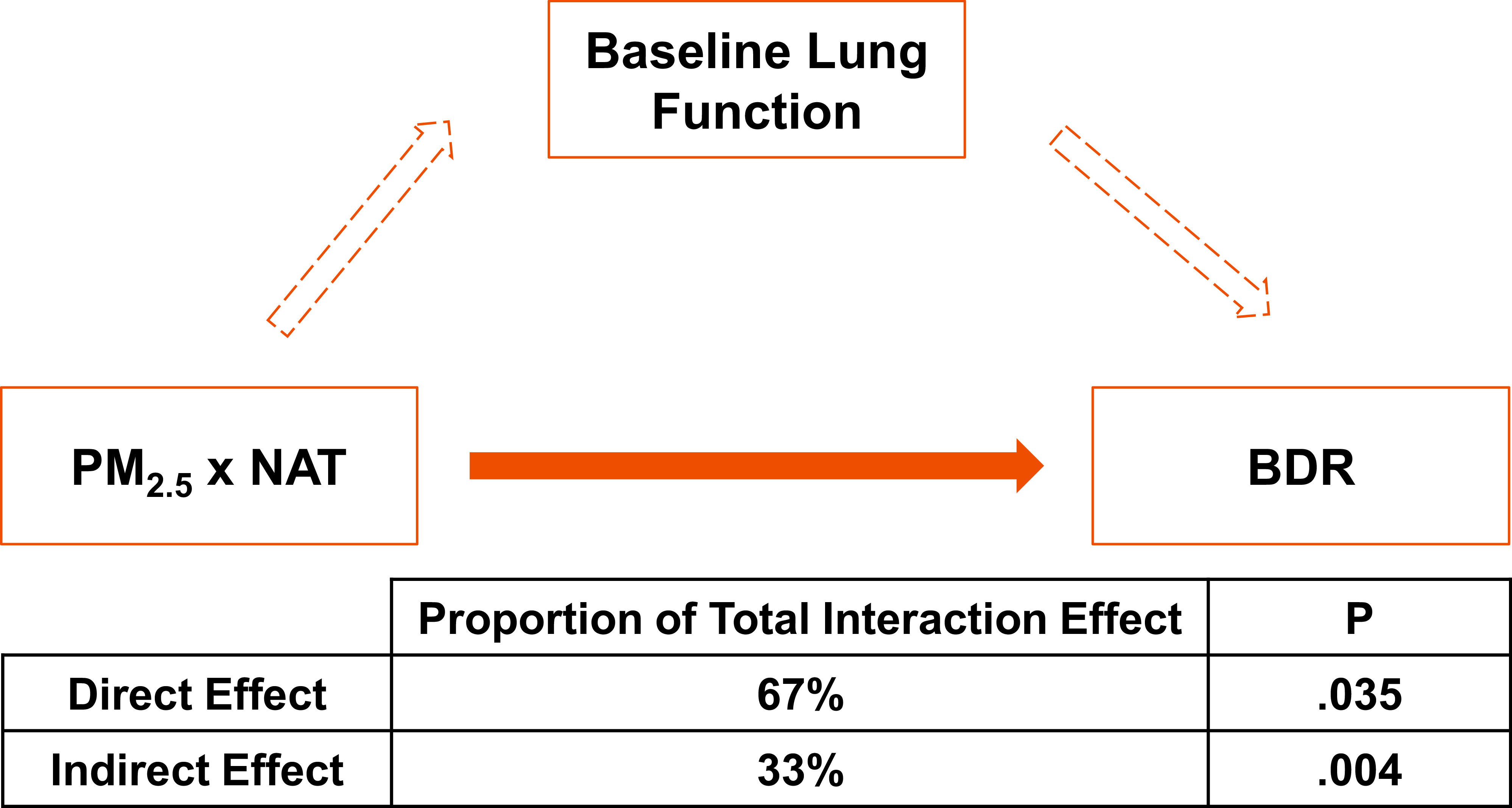
Graphical representation of the effect of PM2.5 x NAT on BDR. The solid orange arrow indicates a direct effect of PM2.5 x NAT on BDR; dotted orange arrows indicate indirect effect of PM2.5 x NAT on BDR. The proportion of the total effect of PM2.5 x NAT on BDR due to direct and indirect effects, and the corresponding P, were calculated via robust mediation analysis. Effect proportions and corresponding P presented above are based on bootstrap effect estimates (n=5000). NAT, Native American ancestry; PM2.5, particulate matter <2.5 μm in size; BDR, bronchodilator drug response.
Discussion
Increased asthma prevalence, morbidity, and mortality, coupled with a documented decreased responsiveness to albuterol, the most commonly prescribed bronchodilator medication in the United States, underscore the disproportionately heavy disease burden carried by the Puerto Rican population. Investigating the complex network of linear and non-linear interactions between biological, demographic, and environmental factors impacting variation in BDR, and focusing our efforts on patient populations with the highest risk of adverse outcomes, is an essential step toward addressing asthma health disparities.
Acknowledging recent evidence that the uni-disciplinary strategies historically favored in biomedical research may not suffice when disease etiology is complex, we performed a comprehensive trans-disciplinary study of BDR in Puerto Rican children with asthma that brought together clinical, biological, environmental, and psycho-social data.32 Our study identified a non-linear interaction between air pollution exposure (PM2.5) and Native American ancestry (NAT) that was significantly associated with BDR in a population of Puerto Rican children with asthma. We found that Puerto Rican children with asthma who had increased air pollution exposure (PM2.5) and decreased Native American ancestry displayed increased BDR due, in part, to the decreased baseline lung function characteristic of this group. Of the four groups defined by the PM2.5 x NAT interaction model, only one (Group 3; N = 186) had a median BDR that crossed the threshold of clinically relevant response to bronchodilator treatment, meaning that the majority of our study population (N = 472) may be non-responsive to bronchodilator treatment. These metrics are potentially valuable to clinicians as a means of identifying patients less likely to be responsive to bronchodilator treatment.
Recent studies highlight the existence of quantile-specific effects for gene-environment interactions.33 Owing to this observation, we chose our replication population from the opposite end of the BDR phenotypic distribution (Mexican Americans) as our study populations (Puerto Ricans). Our failure to replicate the significant interaction effect between PM2.5 x NAT and BDR discovered in Puerto Ricans in a population of Mexican Americans indicates that our interaction may be BDR quantile-specific and/or population-specific. Further studies incorporating diverse populations spanning the BDR phenotypic spectrum are needed to further characterize and validate the impact of the PM2.5 x NAT interaction on variation in BDR.
Limitations
It is worth emphasizing that we discovered a significant association between the PM2.5 x NAT interaction and BDR using ViSEN, an entropy-based method that employs information gain (IG), a metric from information theory typically used in machine learning. If we had employed the standard regression-based interaction analysis methods characteristic of genetic and epidemiological studies, then the non-linear association between the PM2.5 x NAT interaction and BDR may have remained hidden. While our study yielded positive results, it should be noted that there are three potential limitations inherent with the use of ViSEN in identifying interaction effects that should be carefully considered by researchers interested in using this method in studies of complex traits.
ViSEN is currently only able to directly analyze dichotomous outcomes; therefore, in order to analyze continuous outcomes in ViSEN, they must first be collapsed into a ranked form (ie, dichotomized) manually prior to input into the program. Dichotomization of continuous outcomes may result in some loss of information and could lead to over-simplification of the resulting interaction models. In the current study, we were able to counter this limitation by using a biologically relevant threshold (clinical response to BDR) to define BDR responders and non-responders thereby increasing the interpretability of the resulting interaction models.
Another potential limitation of ViSEN is that the program requires complete phenotypic data for each participant. Reductions in sample size due to missingness in the dataset, coupled with potential decreases in sample size following the LCC subsampling method to correct for confounders (if necessary), may result in a substantial loss of statistical power. This may mean that some true interaction effects may not pass correction for multiple testing in studies with modest sample sizes.
The third limitation of the ViSEN program is its inability to provide directionality to the interaction effects that it identifies (ie, whether membership in a particular interaction group correlates positively or negatively with a given outcome). To compensate for this limitation in the current study, we have supplemented ViSEN analysis with additional data visualization and analysis (pairwise tests, mediation analysis) to provide more information about the potential impact of our identified interaction on BDR response (Figure 2, Figure 3).
Conclusions
Our study is a representation of the novel discoveries that can result from transdisciplinary team-based research that embraces the complexity of human disease. It is our hope that future studies will embrace diversity of study populations, types of data, and scientific expertise for the improvement of patient care and health outcomes for all patients, particularly in those areas, such as asthma and asthma-related phenotypes like BDR, that display significant disparities between populations.
Acknowledgments
Research Involving Human Participants
A total of 658 participants from the GALA II study were used to generate the results described in this article. The GALA II study was approved by the institutional review board of the University of California San Francisco (Parnassus Panel) (IRB# 11-05424; Reference # 273089) in accordance with the ethical standards of the IRB and the Helsinki Declaration of 1975, as revised in 2000.
All participants aged 18 years at the time of study enrollment provided their written consent to participate in this study. Parents of participants aged <18 years provided their written assent for the participation of their children in this study.
The authors acknowledge the families and patients for their participation and thank the numerous health care providers and community clinics for their support and participation in the GALA II studies. In particular, the authors thank GALAII study coordinator Sandra Salazar. The authors would also like to acknowledge the GALAII co-investigators, collaborators, advisors, and recruiters involved with subject recruitment, sample processing and quality control: Duanny Alva, MD, Gaby Ayala-Rodriguez, Lisa Caine, Elizabeth Castellanos, Jaime Colon, Denise DeJesus, Blanca Lopez, Brenda Lopez, MD, Louis Martos, Vivian Medina, Juana Olivo, Mario Peralta, Esther Pomares, MD, Jihan Quraishi, Johanna Rodriguez, Shahdad Saeedi, Dean Soto, Ana Taveras, Luisa N. Borrell, DDS, PhD, Emerita Brigino-Buenaventura, MD, Adam Davis, MA, MPH, Michael A. LeNoir, MD, Kelley Meade, MD, Fred Lurmann, MS and Harold J. Farber, MD, MSPH.
This work was supported in part by the Sandler Family Foundation, the American Asthma Foundation, the RWJF Amos Medical Faculty Development Program, Harry Wm. and Diana V. Hind Distinguished Professor in Pharmaceutical Sciences II, National Institutes of Health R01HL117004, R01HL128439, R01HL135156, 1X01HL134589, R01HL141992, R01HL141845, National Institute of Health and Environmental Health Sciences R01ES015794, R21ES24844, the National Institute on Minority Health and Health Disparities P60MD006902, RL5GM118984, R01MD010443, and R56MD013312, the Tobacco-Related Disease Research Program under Award Number 24RT-0025, 27IR-0030 and the National Human Genome Research Institute U01HG009080. The research effort of J.M. was additionally supported by a diversity supplement of NIMHD R01MD010443. The research effort of K.L.K was additionally supported by NHLBI R01HL135156-S1, the Gordon and Betty Moore Foundation grant GBMF3834, and the Alfred P. Sloan Foundation grant 2013-10-27 to UC Berkeley through the Moore-Sloan Data Sciences Environment initiative at the Berkeley Institute for Data Science (BIDS). The research effort of M.J.W. was additionally supported by an NHLBI Research Career Development (K) Award K01HL140218. The research effort of M.G.C was additionally supported by NIH MARC U-STAR grant T34GM008574 at San Francisco State University.
References
- 1.Adams PF, Hendershot GE, Marano MA; Centers for Disease Control and Prevention/National Center for Health Statistics . Current estimates from the National Health Interview Survey, 1996. Vital Health Stat 10. 1999;(200):1-203. [PubMed] [Google Scholar]
- 2.Oh SS, White MJ, Gignoux CR, Burchard EG. Making Precision Medicine Socially Precise. Take a Deep Breath. Am J Respir Crit Care Med. 2016;193(4):348-350. 10.1164/rccm.201510-2045ED [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barr RG, Avilés-Santa L, Davis SM, et al. Pulmonary Disease and Age at Immigration among Hispanics. Results from the Hispanic Community Health Study/Study of Latinos. Am J Respir Crit Care Med. 2016;193(4):386-395. 10.1164/rccm.201506-1211OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akinbami LJ, Moorman JE, Bailey C, et al. Trends in asthma prevalence, health care use, and mortality in the United States, 2001-2010. NCHS Data Brief. 2012;(94):1-8. [PubMed] [Google Scholar]
- 5.Palmer LJ, Silverman ES, Weiss ST, Drazen JM. Pharmacogenetics of asthma. Am J Respir Crit Care Med. 2002;165(7):861-866. 10.1164/ajrccm.165.7.2109096 [DOI] [PubMed] [Google Scholar]
- 6.Naqvi M, Thyne S, Choudhry S, et al. Ethnic-specific differences in bronchodilator responsiveness among African Americans, Puerto Ricans, and Mexicans with asthma. J Asthma. 2007;44(8):639-648. 10.1080/02770900701554441 [DOI] [PubMed] [Google Scholar]
- 7.McGeachie MJ, Stahl EA, Himes BE, et al. Polygenic heritability estimates in pharmacogenetics: focus on asthma and related phenotypes. Pharmacogenet Genomics. 2013;23(6):324-328. 10.1097/FPC.0b013e3283607acf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishimura KK, Galanter JM, Roth LA, et al. Early-life air pollution and asthma risk in minority children. The GALA II and SAGE II studies. Am J Respir Crit Care Med. 2013;188(3):309-318. 10.1164/rccm.201302-0264OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10(6):392-404. 10.1038/nrg2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burchard EG, Oh SS, Foreman MG, Celedón JC. Moving toward true inclusion of racial/ethnic minorities in federally funded studies. A key step for achieving respiratory health equality in the United States. Am J Respir Crit Care Med. 2015;191(5):514-521. 10.1164/rccm.201410-1944PP 10.1164/rccm.201410-1944PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore JH. A global view of epistasis. Nat Genet. 2005;37(1):13-14. 10.1038/ng0105-13 [DOI] [PubMed] [Google Scholar]
- 12.Moore JH, Williams SM. Epistasis and its implications for personal genetics. Am J Hum Genet. 2009;85(3):309-320. 10.1016/j.ajhg.2009.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538(7624):161-164. 10.1038/538161a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thakur N, Oh SS, Nguyen EA, et al. Socioeconomic status and childhood asthma in urban minority youths. The GALA II and SAGE II studies. Am J Respir Crit Care Med. 2013;188(10):1202-1209. 10.1164/rccm.201306-1016OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thakur N, Barcelo NE, Borrell LN, et al. Perceived Discrimination Associated With Asthma and Related Outcomes in Minority Youth: the GALA II and SAGE II Studies. Chest. 2017;151(4):804-812. 10.1016/j.chest.2016.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGarry ME, Castellanos E, Thakur N, et al. Obesity and bronchodilator response in black and Hispanic children and adolescents with asthma. Chest. 2015;147(6):1591-1598. 10.1378/chest.14-2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galanter JM, Gignoux CR, Torgerson DG, et al. Genome-wide association study and admixture mapping identify different asthma-associated loci in Latinos: the Genes-environments & Admixture in Latino Americans study. J Allergy Clin Immunol. 2014;134(2):295-305. 10.1016/j.jaci.2013.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655-1664. 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pino-Yanes M, Thakur N, Gignoux CR, et al. Genetic ancestry influences asthma susceptibility and lung function among Latinos. J Allergy Clin Immunol. 2015;135(1):228- 235. https://doi.org/ 10.1016/j. jaci.2014.07.053 PMID:25301036 [DOI] [PMC free article] [PubMed]
- 20.Drake KA, Torgerson DG, Gignoux CR, et al. A genome-wide association study of bronchodilator response in Latinos implicates rare variants. J Allergy Clin Immunol. 2014;133(2):370-378. 10.1016/j.jaci.2013.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bateman ED, Hurd SS, Barnes PJ, et al. Global strategy for asthma management and prevention: GINA executive summary. Eur Respir J. 2008;31(1):143-178. 10.1183/09031936.00138707 [DOI] [PubMed] [Google Scholar]
- 22.Pellegrino R, Viegi G, Brusasco V, et al. Interpretative strategies for lung function tests. Eur Respir J. 2005;26(5):948-968. 10.1183/09031936.05.00035205 [DOI] [PubMed] [Google Scholar]
- 23.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159(1):179-187. 10.1164/ajrccm.159.1.9712108 [DOI] [PubMed] [Google Scholar]
- 24.Hu T, Chen Y, Kiralis JW, Moore JH. ViSEN: methodology and software for visualization of statistical epistasis networks. Genet Epidemiol. 2013;37(3):283-285. 10.1002/gepi.21718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magaña J, Contreras MG, Keys KL, et al. Pairwise and higher-order epistatic interactions have a significant impact on bronchodilator drug response in African American Youth with Asthma. bioRxiv. 2020:2020.03.04.977066. https://doi. org/ 10.1101/2020.03.04.977066 [DOI] [PMC free article] [PubMed]
- 26.Fithian W, Hastie T. Local case-control sampling: efficient subsampling in imbalanced data sets. Ann Stat. 2014;42(5):1693-1724. 10.1214/14-AOS1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore JH, Olson RS, Schmitt P, Chen Y, Manduchi E. How computational experiments can improve our understanding of the genetic architecture of common human diseases. Artif Life. 2020;26(1):23-37. 10.1162/artl_a_00308 [DOI] [PubMed] [Google Scholar]
- 28.Coverstone AM, Bacharier LB, Wilson BS, et al. Clinical significance of the bronchodilator response in children with severe asthma. Pediatr Pulmonol. 2019;54(11):1694-1703. 10.1002/ppul.24473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alfons A. Robmed: (Robust) Mediation Analysis. 2020. Last accessed April 7, 2020 from https://CRAN.R-project.org/ package=robmed.
- 30.Altman DG, Bland JM. How to obtain the P value from a confidence interval. BMJ. 2011;343(aug08 1):d2304. https://doi. org/ 10.1136/bmj.d2304 PMID:22803193 [DOI] [PubMed]
- 31.Burchard EG, Avila PC, Nazario S, et al. ; Genetics of Asthma in Latino Americans (GALA) Study . Lower bronchodilator responsiveness in Puerto Rican than in Mexican subjects with asthma. Am J Respir Crit Care Med. 2004;169(3):386-392. 10.1164/rccm.200309-1293OC [DOI] [PubMed] [Google Scholar]
- 32.Ciesielski TH, Aldrich MC, Marsit CJ, Hiatt RA, Williams SM Transdisciplinary approaches enhance the production of translational knowledge. Transl Res. 2017;182:123- 134. https://doi.org/ 10.1016/j. trsl.2016.11.002 PMID:27893987 [DOI] [PMC free article] [PubMed]
- 33.Williams PT. Gene-environment interactions due to quantile-specific heritability of triglyceride and VLDL concentrations. Sci Rep. 2020;10(1):4486. 10.1038/s41598-020-60965-9 10.1038/s41598-020-60965-9 [DOI] [PMC free article] [PubMed] [Google Scholar]