Abstract
Since 2002, the world has witnessed major outbreaks of acute respiratory illness by three zoonotic coronaviruses (CoVs), which differ from each other in pathogenicity. Reasons for the lower pathogenicity of SARS-CoV-2 than the other two zoonotic coronaviruses, SARS-CoV and MERS-CoV, are not well understood. We herein compared the codon usage patterns of the three zoonotic CoVs causing severe acute respiratory syndromes and four human-specific CoVs (NL63, 229E, OC43, and HKU1) causing mild diseases. We found that the seven viruses have different codon usages, with SARS-CoV-2 having the lowest effective number of codons (ENC) among the zoonotic CoVs. Human codon adaptation index (CAI) analysis revealed that the CAI value of SARS-CoV-2 is the lowest among the zoonotic CoVs. The ENC and CAI values of SARS-CoV-2 were more similar to those of the less-pathogenic human-specific CoVs. To further investigate adaptive evolution within SARS-CoV-2, we examined codon usage patterns in 3573 genomes of SARS-CoV-2 collected over the initial 4 months of the pandemic. We showed that the ENC values and the CAI values of SARS-CoV-2 were decreasing over the period. The low ENC and CAI values could be responsible for the lower pathogenicity of SARS-CoV-2. While mutational pressure appears to shape codon adaptation in the overall genomes of SARS-CoV-2 and other zoonotic CoVs, the E gene of SARS-CoV-2, which has the highest codon usage bias, appears to be under strong natural selection. Data from the study contribute to our understanding of the pathogenicity and evolution of SARS-CoV-2 in humans.
Keywords: SARS-CoV-2, Pathogenicity, Codon usage bias, Human host adaptation, Adaptive evolution
1. Introduction
The pathogen responsible for Coronavirus Disease 2019 (COVID-19), SARS-CoV-2, was initially discovered at the end of 2019 and has spread across the globe within three months. In the past two decades, there have been two other major outbreaks of lower respiratory diseases caused by zoonotic coronaviruses (CoVs), severe acute respiratory syndrome by SARS-CoV (2002) and Middle East respiratory syndrome by MERS-CoV (2012) (de Wit et al., 2016). At the genome level, SARS-CoV-2 has sequence similarity of 79% with SARS-CoV and 50% with MERS-CoV. In addition, four other human coronaviruses (HCoV), NL63, 229E, OC43 and HKU1, have been circulating worldwide in humans for some time, causing mild, self-limiting respiratory infections in human (Fehr and Perlman, 2015). To enter the host cells SARS-CoV, SARS-CoV-2, and HCoV-NL63 utilize the angiotensin-converting enzyme 2 (ACE-2), HCoV-OC43 and HCoV-HKU1 utilize 9-O-acetylsialic acids, MERS-CoV utilizes dipeptidyl-peptidase 4, and HCoV-229E utilizes aminopeptidase N as the receptor (Fehr and Perlman, 2015; Wan et al., 2020). HCoV-OC43 and HCoV-HKU1 had possibly originated from rodents, while the three zoonotic CoVs, HCoV-NL63, and HCoV-229E are believed to have originated from bats (Cui et al., 2019; Zhou et al., 2020). The zoonotic CoVs potentially used different intermediate hosts in the initial transmission of the pathogen to humans (Cui et al., 2019; Lam et al., 2020; Xiao et al., 2020).
All seven CoVs have similar genomes, with four genes encoding structural proteins, including spike (S), nucleocapsid (N), envelope (E), and membrane (M) proteins (Cui et al., 2019; Forni et al., 2017). Additionally, the CoV genomes encode two open-reading frames (ORFs), ORF1a and ORF1b, which are translated to form large polyproteins 1a and 1b (polyprotein AB in MERS CoV). The polyproteins are processed by proteases to yield several nonstructural proteins, including an RNA-dependent RNA polymerase (RdRp). The RdRp is the essential enzyme in a replicase complex used to transcribe the viral genome (Ulrich et al., 2003).
Although there is high sequence similarity among the zoonotic CoVs, SARS-CoV-2 is spreading more rapidly than SARS-CoV and MERS-CoV. The number of secondary cases resulting from a primary case (reproduction number, R0) before the implementation of control measures is estimated to be 2.0–3.7 for SARS-CoV-2 (Lonergan and Chalmers, 2020). In contrast, the R0 for SARS-CoV and MERS-CoV are 1.7–1.9 and less than 1, respectively (Petrosillo et al., 2020). Therefore, SARS-CoV-2 has higher transmissibility than SARS-CoV and MERS-CoV. In addition, most of the patients infected with SARS-CoV-2 have mild symptoms, with case mortality (around 5%) significantly lower than those infected with SARS-CoV (10%) and MERS-CoV (37%) (Chen, 2020). Reasons for these differences are poorly understood.
Analysis of codon usage patterns can provide insight into the evolution of viruses and their adaption to hosts (Burns et al., 2006; Costafreda et al., 2014). Because the genetic code is redundant in most organisms, during the adaptation to their hosts, viruses can modify codon usage therefore protein synthesis efficiency while maintaining the amino acid sequences, leading to changes in viral replication (Chen et al., 2020). Effective number of codons (ENC) and codon adaptation index (CAI) analyses are frequently used to evaluate the codon usage patterns of viruses (Baha et al., 2019; Belalov and Lukashev, 2013; Khandia et al., 2019; Li et al., 2018b; Stoletzki and Eyre-Walker, 2007; Zhang et al., 2019). Between them, ENC values are measurements of codon preference (Comeron and Aguadé, 1998), while CAI is an indirect measurement of the likely expression efficiency of viral genes in the host (Carbone et al., 2003; Chen et al., 2020).
In this study, we performed comparative analyses of codon usage patterns among the three zoonotic CoVs and the four human-specific CoVs to understand the differences in pathogenicity. We also examined the evolution of codon usage in SARS-CoV-2 genomes collected over a 4-month period during the early pandemic.
2. Materials and methods
2.1. Data acquisition
A total of 122 complete CoV genomes (15 from SARS-CoV, 15 from SARS-CoV-2, 25 from MERS-CoV, 15 from HCoV-OC43, 15 from HCoV-HKU1, 15 from HCoV-229E, 15 from HCoV-NL63, 15 from Bat-CoV, and 2 from SARS-CoV-2-related CoV) were downloaded from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/genbank/). After removing low-quality sequences, 86 CoV genomes were used in analyses.
A total of 17,440 SARS-CoV-2 genomes flagged as “collection date from December 1 2019 to April 30 2020” were downloaded from the EpiCoV database of the GISAID Initiative (https://www.epicov.org) as of May 102,020. After filtering to exclude those with lengths less than 29,000 bp, those with Ns or degenerate bases, and those derived from animals, a final dataset of 3573 genomes was used in downstream analyses.
2.2. CoV genome composition analysis
Reference genomes of human and zoonotic CoVs were obtained from GenBank (https://www.ncbi.nlm.nih.gov/genbank/). The open reading frames of the genomes were predicted using Geneious v11.1.5 (https://www.geneious.com/) and annotated using Blastn v2.10.1 (https://blast.ncbi.nlm.nih.gov/Blast.cgi)
2.3. Phylogenetic analysis
The CoV genomes were aligned using MUSCLE v3.8.31 (https://www.ebi.ac.uk/). The substitution model was selected using jModelTest v2.1.10 (Posada, 2008), based on values from the Akaike Information Criterion (AIC), the Bayesian Information Criterion (BIC), and hierarchical likelihood-ratio tests (hLRTs). A maximum likelihood (ML) tree was reconstructed using PhyML (Guindon et al., 2010), with the GTR + GAMMA I + I model and bootstrap value of 1000.
2.4. Nucleotide composition analysis
The frequency of each nucleotide (A, U, G, C), AU, and GC in the genes or genomes was calculated using BioEdit v7.1.3.0 (https://bioedit.software.informer.com/). The nucleotide frequency of the third position of synonymous codons (A3%, U3%, G3%, C3%) was calculated using CodonW (http://codonw.sourceforge.net/). The GC content at the first (GC1S), second (GC2S), and third codon positions (GC3S) was calculated using Emboss explorer (http://codonw.sourceforge.net/), with the GC12S as the mean of GC1S and GC2S. The three termination codons (UAA, UGA, UAG) and the codons AUG and UGG were excluded from these analyses.
2.5. Relative synonymous codon usage (RSCU) analysis
To assess the codon usage patterns without the effect of sequence length, RSCU values for 59 codons (excluding UAA, UGA, UAG, AUG, and UGG) were calculated using DAMBE v7.2.43 (http://dambe.bio.uottawa.ca/). An RSCU value of 1 indicates that the codon is used equally, while codons with RSCU values of >1.6 and < 0.6 are considered as over-represented and under-represented, respectively (Sharp and Li, 1986). The principal component analysis (PCA) implemented in TBtools v0.66831 (http://www.tbtools.com/) was used in the analysis of the RSCU data based on a 59-dimension vector.
2.6. Assessment of effective number of codons (ENC) and selection pressure
The ENC was calculated using the CodonW software v1.4.4 (http://codonw.sourceforge.net/). The ENC values range from 20 (only one synonymous codon is used per amino acid, showing an extreme codon usage bias) to 61 (all synonymous codons are equally used showing no bias) (Wong et al., 2010). In general, ENC values of less than 35 indicate strong codon usage bias (Comeron and Aguadé, 1998). The ENC values generated were plotted against GC3S. If the codon usage bias is merely constrained by mutational pressure, the ENC values would lie on or around the expected standard curve. Values below the expected curve, on the other hand, indicate that natural selection could play a role in shaping the codon usage (Wong et al., 2010). Plots of the expected ENC values against GC3S values were generated as described (Zhang et al., 2019).
2.7. Codon adaptation index (CAI) analysis
CAI analysis was used to further assess the adaptability of the CoV codons to humans. It was calculated using the cai script implemented in Emboss explorer (http://www.bioinformatics.nl/emboss-explorer/). The reference dataset for humans was downloaded from the Codon Usage Database (http://www.kazusa.or.jp/codon/). The CAI value has a range from 0 to 1, with higher CAI values being indicators of better adaptation to humans by the viruses (Sharp and Li, 1987).
3. Results
3.1. Genome composition and phylogenetic relationship of CoVs
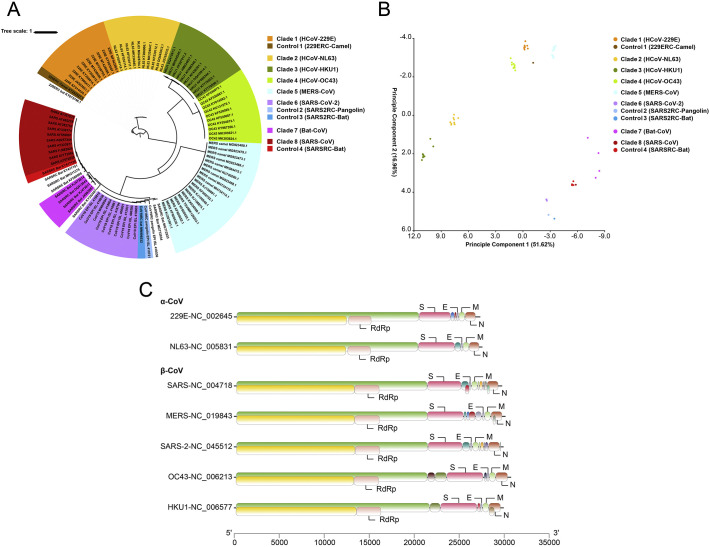
The genomes of CoVs were AT rich (58.3–67.4%). Overall, GC contents of the three zoonotic CoVs were higher than of human-specific CoVs (Table 1 ). Among the eight clades of CoVs analyzed in the study (Fig. 1A), Clade 3 (HCoV-HKU1) had the lowest GC content (32.6%) and the lowest GC content at the third codon position (GC3) (18.9%). In contrast, the highest values of both indices (GC = 41.7% and GC3 = 36.3%) were seen in Clade 7 (Bat-CoV). Specifically among the three zoonotic CoVs, SARS-CoV-2 (Clade 6) had the lowest GC content (39.2%) and the lowest GC3 content (30.1%) (Table 1), while MERS-CoV (Clade 5) had the highest values of both indices (GC = 41.6% and GC3 = 35.6%). Moreover, GC contents of other clades were 35.9%, 37.1%, 39.1%, and 40.9% for Clade 2 (HCoV-NL63), Clade 4 (HCoV-OC43), Clade 1 (HCoV-229E), and Clade 8 (SARS-CoV), respectively. In comparison, the GC3 contents of those clades were 24.7%, 28.3%, 32.4%, and 33.8%, respectively.
Table 1.
The nucleotide compositions and the codon usage indices of the RdRp, S, E, M, and N genes in different CoV groups.
| GC | GC1S | GC2S | GC3S | GC12S | AT | AT3S | A3S | T3S | C3S | G3S | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Clade 1 (HCoV-229E) | 39.1 ± 0 | 45.9 ± 0.1 | 39 ± 0.1 | 32.4 ± 0.1 | 42.4 ± 0 | 60.9 ± 0 | 67.6 ± 0.1 | 20.7 ± 0 | 47 ± 0.1 | 16.4 ± 0.1 | 16 ± 0.1 |
| Control 1 (229ERC-Camel) | 39.1 ± 0 | 46.3 ± 0 | 39.3 ± 0 | 31.9 ± 0 | 42.8 ± 0 | 60.9 ± 0 | 68.1 ± 0 | 21.2 ± 0 | 46.9 ± 0 | 16.4 ± 0 | 15.5 ± 0 |
| Clade 2 (HCoV-NL63) | 35.9 ± 0.1 | 44.7 ± 0.1 | 38.3 ± 0.1 | 24.7 ± 0.2 | 41.5 ± 0.1 | 64.1 ± 0.1 | 75.3 ± 0.2 | 18.6 ± 0.1 | 56.7 ± 0.2 | 11.6 ± 0.1 | 13.1 ± 0.1 |
| Clade 3 (HCoV-HKU1) | 32.6 ± 0.1 | 41.1 ± 0.1 | 37.8 ± 0.1 | 18.9 ± 0.3 | 39.5 ± 0 | 67.4 ± 0.1 | 81.1 ± 0.3 | 20.4 ± 0.2 | 60.7 ± 0.2 | 8.2 ± 0.2 | 10.7 ± 0.1 |
| Clade 4 (HCoV-OC43) | 37.1 ± 0.1 | 44.8 ± 0.1 | 38.4 ± 0.1 | 28.3 ± 0.1 | 41.6 ± 0.1 | 62.9 ± 0.1 | 71.7 ± 0.1 | 22.3 ± 0.1 | 49.4 ± 0.1 | 13 ± 0.1 | 15.3 ± 0.1 |
| Clade 5 (MERS-CoV) | 41.6 ± 0.1 | 48.7 ± 0.1 | 40.5 ± 0.1 | 35.6 ± 0.1 | 44.6 ± 0.1 | 58.4 ± 0.1 | 64.4 ± 0.1 | 19.9 ± 0.1 | 44.5 ± 0.1 | 20.1 ± 0.1 | 15.5 ± 0 |
| Clade 6 (SARS-CoV-2) | 39.2 ± 0 | 47.3 ± 0 | 40 ± 0 | 30.1 ± 0 | 43.7 ± 0 | 60.8 ± 0 | 69.9 ± 0 | 27.1 ± 0 | 42.7 ± 0 | 17.9 ± 0 | 12.2 ± 0 |
| Control 2 (SARS2RC-Pan) | 39 ± 0 | 46.7 ± 0 | 40.1 ± 0 | 30.1 ± 0 | 43.4 ± 0 | 61 ± 0 | 69.9 ± 0 | 27.9 ± 0 | 42 ± 0 | 18.3 ± 0 | 11.8 ± 0 |
| Control 3 (SARS2RC-Bat) | 39.3 ± 0 | 47.5 ± 0 | 40 ± 0 | 30.5 ± 0 | 43.7 ± 0 | 60.7 ± 0 | 69.5 ± 0 | 27.4 ± 0 | 42.1 ± 0 | 18.4 ± 0 | 12.1 ± 0 |
| Clade 7 (Bat-CoV) | 41.7 ± 0.2 | 48.9 ± 0.1 | 40 ± 0.2 | 36.3 ± 0.4 | 44.4 ± 0.1 | 58.3 ± 0.2 | 63.7 ± 0.4 | 25.5 ± 0.5 | 38.2 ± 0.5 | 21.6 ± 0.5 | 14.7 ± 0.5 |
| Clade 8 (SARS-CoV) | 40.9 ± 0 | 48.8 ± 0 | 40.1 ± 0 | 33.8 ± 0.1 | 44.4 ± 0 | 59.1 ± 0 | 66.2 ± 0.1 | 24.7 ± 0 | 41.5 ± 0.1 | 19.9 ± 0 | 13.9 ± 0.1 |
| Control 4 (SARSRC-Bat) | 41.1 ± 0 | 49.1 ± 0 | 40.1 ± 0 | 34 ± 0 | 44.6 ± 0 | 58.9 ± 0 | 66 ± 0 | 24.6 ± 0 | 41.4 ± 0 | 20 ± 0 | 14 ± 0 |
Fig. 1.
Genome composition and phylogeny of zoonotic CoVs (SARS-CoV, MERS-CoV, and SARS-CoV-2), human-specific CoVs (HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1) and related viruses. (A) Maximum likelihood tree of CoVs constructed using a GTR + G + I model implemented in PhyML and bootstrapping with 1000 replicates. The observed eight clades and three controls, i.e. Clade 1 (HCoV-229E), Clade 2 (HCoV-NL63), Clade 3 (HCoV-HKU1), Clade 4 (HCoV-OC43), Clade 5 (MERS-CoV), Clade 6 (SARS-CoV-2), Clade 7 (Bat-CoV), Clade 8 (SARS-CoV), Control 1 (229ERC-Camel), Control 2 (SARS2RC-Pangolin), Control 3 (SARS2RC-Bat), and Control 4 (SARSRC-Bat) are represented in orange, yellow, green, light green, sky blue, purple, violet, dark red, brown, light blue, blue, and red, respectively. (B) Outcome of the PCA analysis of RSCU data, with clades and controls colored the same as in A. The PCA was done on RSCU data from CoVs based on a 59-dimension vector. (C) Genome composition of different CoVs. Two-third of the genome from the 5′-terminus encodes a polyprotein, 1ab, which is further cleaved into an RNA-dependent RNA polymerase (RdRp) involved in genome transcription and replication. The other one-third of the genome from the 3′ terminus encodes structural proteins, including spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins.
In the ML analysis, the CoVs under analysis formed eight clades with high bootstrap support: Clade 1 (HCoV-229E), Clade 2 (HCoV-NL63), Clade 3 (HCoV-HKU1), Clade 4 (HCoV-OC43), Clade 5 (MERS-CoV), Clade 6 (SARS-CoV-2), Clade 7 (Bat-CoV), and Clade 8 (SARS-CoV) (Fig. 1A). In addition, the ML tree showed that the genome KT253327 was genetically related to HCoV-229E, the genome KY417150 was genetically related to SARS-CoV, and the pangolin CoV EPIISL410721 and the bat CoV MN996532 were genetically related to SARS-CoV-2. Therefore, KT253327 was used as the control for HCoV-229E, KY417150 as the control for SARS-CoV, and EPIISL410721 and KY417150 as controls for SARS-CoV-2 in other analyses in the study. In PCA analysis of the RSCU data (Table S1), all clades formed separate clusters together with their controls (Fig. 1B).
Comparison of the predicted coding regions of the three zoonotic CoVs and four human-specific CoVs showed that they possessed a similar genomic organization (Fig. 1C). At least six common coding regions were predicted, including 1ab, 1a, S, E, M, and N (Fig. 1C). The lengths and the order of these genes were similar among the seven CoVs.
3.2. Codon usage bias in CoVs
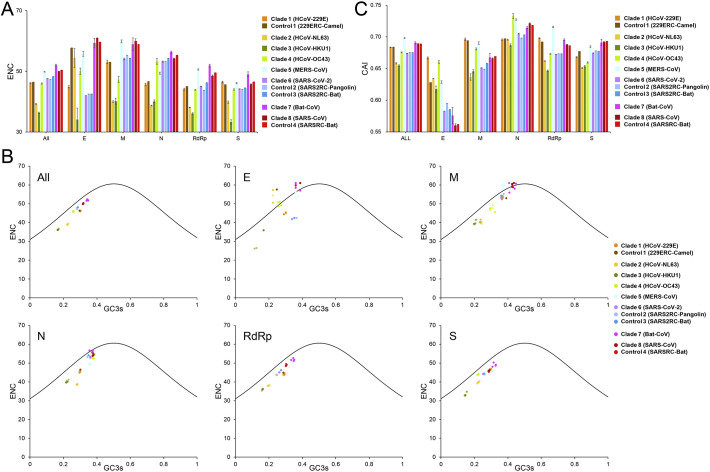
In the ENC analysis of the CoV genomes, the values generated were all higher than 35, indicating low codon usage bias by CoVs. Among the eight major phylogenetic clades, the highest ENC value was 52.02 ± 0.30 from the Clade 7 (Bat-CoVs), indicating that these CoVs had the lowest codon usage bias among the lineages. In contrast, the lowest ENC value of human-specific CoVs was 36.27 ± 0.22 from Clade3 (HCoV-HKU1). Among the three zoonotic CoVs, the lowest ENC value was 47.60 ± 0.02 from Clade 6 (SARS-CoV-2), suggesting that SARS-CoV-2 uses a narrower set of synonymous codons (Fig. 2A). This was also the case in ENC analysis of individual genes, especially those from the E gene. In the latter, the ENC value was 42.00 ± 0.00 in Clade 6 (SARS-CoV-2), compared with 61.00 ± 0.00 and 55.71 ± 0.81 in Clades 8 (SARS-CoV) and 5 (MERS-CoV), respectively (Fig. 2A).
Fig. 2.
Codon usage bias in CoVs and related viruses. (A) Mean ENC values of genomes and major genes of CoVs. The error bars are the standard deviation of the means. Clade 1 (HCoV-229E), Clade 2 (HCoV-NL63), Clade 3 (HCoV-HKU1), Clade 4 (HCoV-OC43), Clade 5 (MERS-CoV), Clade 6 (SARS-CoV-2), Clade 7 (Bat-CoV), Clade 8 (SARS-CoV), Control 1 (229ERC-Camel), Control 2 (SARS2RC-Pangolin), Control 3 (SARS2RC-Bat), and Control 4 (SARSRC-Bat) are represented in orange, yellow, green, light green, sky blue, purple, violet, dark red, brown, light blue, blue, and red, respectively. (B) Results of the ENC-GC3S plot analysis of genomes and five major genes (E, M, N, RdRp, and S genes) of CoVs. The color dots represent the observed ENC-GC3S values of the individual groups (colored the same as in A). (C) Human codon adaptation indices (CAI) of the genomes and the five major genes of CoVs. The error bars are the standard deviation of the means (colored the same as in A).
3.3. Selection pressure in SARS-CoV-2
To explore whether mutational pressure, natural selection, or they both shaped the codon usage in CoVs, the ENC values were plotted against the GC3S. The plots generated with data from full genomes and individual genes showed that the points clustered mostly on or near the expected curves, indicating that mutational pressure was largely responsible for the codon usage bias in CoVs (Fig. 2B). For the E gene, data from Clade 6 (SARS-CoV-2) and its relative controls (Controls 2 & 3), however, clustered under the expected curve (Fig. 2B), indicating that natural selection shaped codon usage of this gene in SARS-CoV-2.
3.4. Adaptation to human codon usage by CoVs
We further compared the extent of adaptation to human codon usage by CoVs using CAI analysis. The results obtained suggested that among the eight phylogenetic groups, Clade 5 (MERS-CoV) had the highest CAI value (0.698), followed by Clade 7 (bat-CoVs; 0.690), Clade 8 (SARS-CoV; 0.689), Clade 1 (HCoV-229E; 0.683), Clade 4 (HCoV-OC43; 0.676), Clade 6 (SARS-CoV-2; 0.674), Clade 2 (HCoV-NL63; 0.658), and Clade 3 (HCoV-HKU1; 0.655) (Fig. 2C). The difference among clades was significant (p < 0.01). Among the three zoonotic CoVs, the lower CAI value in SARS-CoV-2 supports the suggestion that the gene expression of SARS-CoV-2 could be less efficient than that of SARS-CoV and MERS-CoV.
3.5. Codon usage of SARS-CoV-2 over time
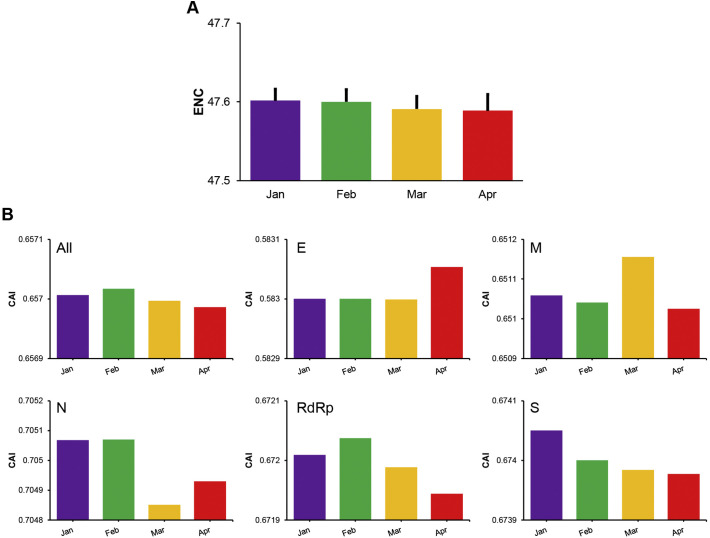
We calculated the ENC values of SARS-CoV-2 isolates collected over a four-month period to evaluate if codon usage changed in SARS-CoV-2. The ENC values decreased gradually during the period, being 47.601 ± 0.014, 47.600 ± 0.015, 47.591 ± 0.016, and 47.588 ± 0.020 for January, February, March, and April 2020, respectively, indicating the codon usage bias of SARS-CoV-2 was increasing slowly over time (Fig. 3A).
Fig. 3.
Codon usage of SARS-CoV-2 over a 4-month period in 2020. (A) Mean ENC values of the genomes of SARS-CoV-2 collected in each month. The error bars are the standard deviation of the means. (B) Human codon adaptation indices (CAI) of the genomes and five genes (E, M, N, RdRp, and S genes) in SARS-CoV-2.
In the longitudinal evaluations of changes in adaptation to human codon usage by SARS-CoV-2, the CAI values of the viral genomes decreased gradually over the four months (p > 0.05), indicating that the likely efficiency of gene expression of SARS-CoV-2 in the human host could be decreasing. This was also the case for most individual viral genes. For example, the CAI values decreased from 0.67200 to 0.67194, 0.67405 to 0.67398, and 0.70507 to 0.70493 for the RdRp, S, and N, genes over the 4-month period, respectively (p < 0.01) (Fig. 3B). For the M gene, the CAI values also decreased from 0.65106 to 0.65102 over the 4 months, although there was a spike in March when the CAI value reached 0.65115 (p < 0.01). In contrast, CAI values of the E gene remained stable during the first three months and increased from 0.58300 to 0.58305 in April (p = 0.1).
We further evaluated the role of mutational pressure in the adaptive evolution of SARS-CoV-2. The ENC-GC3S plots generated with data from the entire genome and individual genes showed that the points mostly clustered on or near the expected curves (Fig. S1). For the E gene, however, the points were dispersed under the expected curve (Fig. S1), indicating that unlike other genes, the E gene was further enduring natural selection during the 4-month period.
4. Discussion
SARS-CoV-2 is the third zoonotic CoV responsible for a major epidemic after SARS-CoV and MERS-CoV but has spread at unprecedented speed across the globe. There are four other more common coronaviruses causing mild, self-limiting respiratory symptoms in humans, including HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1. While these CoVs differ from each other in pathogenicity and transmissibility, the reasons for the differences are poorly understood (Chen, 2020). In this study, we have performed comparative analyses of codon usage among them. We found that the seven viruses had different GC and GC3S contents indicating different codon usages. Among the zoonotic CoVs, SARS-CoV-2 had the lowest ENC and CAI values, suggesting that SARS-CoV-2 is poorly adapted to human codon usage. However, these values are similar to those of the less-pathogenic human-specific CoVs. In addition, we showed that the ENC values and the CAI values of SARS-CoV-2 were decreasing over first four months of the COVID-19 pandemic, which indicates that the codon usage bias of SARS-CoV-2 is increasing during the period, leading to further reduced pathogenicity.
The three zoonotic CoVs and the four human-specific CoVs appear to differ in codon usage. In this study, we compared the nucleotide compositions and the RSCU patterns among the eight groups of phylogenetically related CoVs. They all have AT-rich genomes, in agreement with the previous studies of CoVs (Gu et al., 2004; Jenkins and Holmes, 2003; Kandeel et al., 2020). While, the genomes of human-specific CoVs have higher AT content than zoonotic CoVs, SARS-CoV-2 had the highest AT content among zoonotic CoVs. In correlation with the previous knowledge of CoV genome composition, CoVs frequently use A- and U-ended codons (Chen et al., 2017; Dilucca et al., 2020). Human-specific CoVs have higher A/U frequency at the third codon position than zoonotic CoVs, indicating that the codon usage of zoonotic CoVs is different from the human-specific CoVs. In addition, SARS-CoV-2 has the highest A/U frequency at the third codon position among the three zoonotic CoVs. This indicates that the codon usage of SARS-CoV-2 is different from the other zoonotic CoVs. These were supported by the result of the PCA analysis of RSCU data, which placed these CoVs in different clusters in agreement with their phylogenetic relationships.
Among the three zoonotic CoVs, SARS-CoV-2 has the most codon preference. Although the ENC values of the zoonotic CoV and human-specific CoV genomes were all higher than 35, human-specific CoVs displayed higher codon usage biases than the zoonotic CoVs, and SARS-CoV-2 displayed a higher codon usage bias than the other zoonotic CoVs. In human-pathogenic viruses, high codon preference has generally been associated with reduced host range (Kumar et al., 2018). Thus far, the four human-specific CoVs are known to infect only humans (Fehr and Perlman, 2015). On the contrary, other than humans, SARS-CoV-2 can infect cats and ferrets (Shi et al., 2020). While the sequence characteristics of the receptor in the host is crucial in determining the susceptibility of animals to CoVs, the codon preference by SARS-CoV-2 could potentially make some animals inefficient hosts. Whether SARS-CoV-2 might have a narrower host range than the other two zoonotic CoVs remains to be determined.
SARS-CoV-2 appears to be poorly adapted to human codon usage compared with other zoonotic CoVs. Among the three zoonotic CoVs, SARS-CoV-2 had the lowest human codon CAI value, indicating that it has the lowest adaptation to the human host. Thus, it probably has the lowest likely efficiency of gene expression among the three zoonotic CoVs, as seen in comparative analyses of other viruses (Kumar et al., 2018; Zhang et al., 2019). This appears to contradict the finding in one study (Dilucca et al., 2020), but is in agreement with the finding in another (Kandeel et al., 2020). On the other hand, the four human-specific CoVs have lower human codon CAI values than the zoonotic CoVs, and among the human-specific CoVs, the CAI values of HCoV-229E and HCoV-OC43 higher than that of HCoV-NL63 and HCoV-HKU1. These findings indicate that HCoV-229E and HCoV-OC43 have higher adaptation to the human host than the other two. Over the years, the prevalence of human-specific CoVs in adults and infants is in the following order: HCoV-OC43, HCoV-229E, HCoV-NL63, and HCoV-HKU1 (Bouvier et al., 2018; Zeng et al., 2018). While these four CoVs cause only mild, self-limiting respiratory infections in humans, HCoV-OC43 infected patients have a significantly higher rate of acute respiratory tract infections than HCoV-HKU1 infected patients (Gerna et al., 2007). Therefore, the outcome of the CAI analysis suggests that SARS-CoV-2 might have the lowest translation efficiency of viral proteins in humans among zoonotic CoVs, contributing to its lower pathogenicity than SARS-CoV and MERS-CoV (Chen, 2020; Wang et al., 2020). In other RNA viruses, codon bias has been associated with reduced virulence (Diaz-San Segundo et al., 2016; Goñi et al., 2012; Li et al., 2018a; van Weringh et al., 2011).
Mutational pressure probably plays a major role in shaping the codon usage in SARS-CoV-2. In ENC-GC3S plots, most values clustered on or around the expected curves for all zoonotic and human-specific CoVs, which is an indication of the dominant role of mutational pressure in shaping codon usage of viruses (He et al., 2016). The E gene of SARS-CoV-2, however, appears to be evolved differently from others. Compared with other zoonotic CoVs, the E gene of SARS-CoV-2 has the lowest ENC value. In all zoonotic CoVs, the E gene also has CAI values much lower CAI values to human codons than other genes. This suggests that the E gene of SARS-CoV-2 has a higher codon preference than other genes. Although mutational pressure plays a dominant role in shaping codon usage of all zoonotic CoVs, the E gene in SARS-CoV-2 appears to be under natural selection, as its ENC-GC3S values were dispersed under the expected curve. The function of the E protein is not clear. It has been suggested to play an important role in viral assembly, budding, envelope formation, and pathogenesis (Schoeman and Fielding, 2019).
Over the initial four months of the COVID-19 pandemic, SARS-CoV-2 appears to have increased codon preference. The ENC values of SARS-CoV-2 genomes collected over the 4-month period gradually decreased, indicating the codon usage preference has been increasing in SARS-CoV-2. This contradicts somewhat with the recent report indicating that nearly 80% of the recurrent mutations in SARS-CoV-2 produced non-synonymous changes at the protein level, possibly suggesting that adaptation of the virus to humans is occurring (van Dorp et al., 2020). However, the reduced adaptation of the virus to human codons is collaborated in the present study by the decreasing CAI values over the 4-month period. The reduced ENC and CAI values are both suggesting that pathogenicity of SARS-CoV-2 in humans could be decreasing. In contrast, the E gene of the virus, which is the only gene under natural selection (indicated by ENC-GC3S plot), seems to be adapting to human codons. Further studies of the E gene are needed to elucidate its role in the adaptative evolution of SARS-CoV-2.
In conclusion, results of the study suggest that the three zoonotic CoVs have different extent of adaptation to the human host. Among them, SARS-CoV-2 has the lowest ENC and CAI values, which are more similar to those from the human-specific CoVs. This indicates that it has the most codon preference and likely the lowest efficiency of gene expression in human among the three zoonotic CoVs. These data help to explain the lower pathogenicity of SARS-CoV-2 than SARS-CoV and MERS-CoV. In addition, the decreasing ENC and the CAI values of SARS-CoV-2 during the first four months of the COVID-19 epidemic suggest the codon usage bias in SARS-CoV-2 has been increasing, which could contribute to the reduced pathogenicity of the virus over time. The E gene of the virus appears to be evolved differently from other genes in the genome. It has the highest codon usage bias and is under natural selection.
Author contributions
YF and LX conceived and designed the experiments. WH analyzed the data. WH, YF, and LX wrote the manuscript. All authors approved the final manuscript.
Funding
This work was supported in part by the Emergency Projects of COVID-19 of the Department of Agriculture and Rural Affairs of Guangdong Province, Projects of COVID-19 Control and Prevention of the Department of Education of Guangdong Province (2020KZDZX1033), and Innovation Team Project of Guangdong University (2019KCXTD001).
Declaration of Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2021.104736.
Appendix A. Supplementary data
Supplementary material
References
- Baha S., Behloul N., Liu Z., Wei W., Shi R., Meng J. Comprehensive analysis of genetic and evolutionary features of the hepatitis E virus. BMC Genomics. 2019;20:790. doi: 10.1186/s12864-019-6100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belalov I.S., Lukashev A.N. Causes and implications of codon usage bias in RNA viruses. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier M., Chen W.J., Arnold J.C., Fairchok M.P., Danaher P.J., Lalani T., Malone L., Mor D., Ridoré M., Burgess T.H., Millar E.V. Species-specific clinical characteristics of human coronavirus infection among otherwise healthy adolescents and adults. Influenza Other Respir. Viruses. 2018;12:299–303. doi: 10.1111/irv.12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C.C., Shaw J., Campagnoli R., Jorba J., Vincent A., Quay J., Kew O. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J. Virol. 2006;80:3259–3272. doi: 10.1128/jvi.80.7.3259-3272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone A., Zinovyev A., Képès F. Codon adaptation index as a measure of dominating codon bias. Bioinformatics. 2003;19:2005–2015. doi: 10.1093/bioinformatics/btg272. [DOI] [PubMed] [Google Scholar]
- Chen J. Pathogenicity and transmissibility of 2019-nCoV-A quick overview and comparison with other emerging viruses. Microbes Infect. 2020;22:69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Xu Q., Yuan X., Li X., Zhu T., Ma Y., Chen J.L. Analysis of the codon usage pattern in Middle East respiratory syndrome coronavirus. Oncotarget. 2017;8:110337–110349. doi: 10.18632/oncotarget.22738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Wu P., Deng S., Zhang H., Hou Y., Hu Z., Zhang J., Chen X., Yang J.R. Dissimilation of synonymous codon usage bias in virus-host coevolution due to translational selection. Nat. Ecol. Evol. 2020;4:589–600. doi: 10.1038/s41559-020-1124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J.M., Aguadé M. An evaluation of measures of synonymous codon usage bias. J. Mol. Evol. 1998;47:268–274. doi: 10.1007/pl00006384. [DOI] [PubMed] [Google Scholar]
- Costafreda M.I., Pérez-Rodriguez F.J., D’Andrea L., Guix S., Ribes E., Bosch A., Pintó R.M. Hepatitis A virus adaptation to cellular shutoff is driven by dynamic adjustments of codon usage and results in the selection of populations with altered capsids. J. Virol. 2014;88:5029–5041. doi: 10.1128/jvi.00087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-San Segundo F., Medina G.N., Ramirez-Medina E., Velazquez-Salinas L., Koster M., Grubman M.J., de los Santos T. Synonymous deoptimization of foot-and-mouth disease virus causes attenuation in vivo while inducing a strong neutralizing antibody response. J. Virol. 2016;90:1298–1310. doi: 10.1128/jvi.02167-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilucca M., Forcelloni S., Georgakilas A.G., Giansanti A., Pavlopoulou A. Codon usage and phenotypic divergences of SARS-CoV-2 genes. Viruses. 2020;12 doi: 10.3390/v12050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerna G., Percivalle E., Sarasini A., Campanini G., Piralla A., Rovida F., Genini E., Marchi A., Baldanti F. Human respiratory coronavirus HKU1 versus other coronavirus infections in Italian hospitalised patients. J. Clin. Virol. 2007;38:244–250. doi: 10.1016/j.jcv.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goñi N., Iriarte A., Comas V., Soñora M., Moreno P., Moratorio G., Musto H., Cristina J. Pandemic influenza A virus codon usage revisited: biases, adaptation and implications for vaccine strain development. Virol. J. 2012;9:263. doi: 10.1186/1743-422x-9-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W., Zhou T., Ma J., Sun X., Lu Z. Analysis of synonymous codon usage in SARS coronavirus and other viruses in the Nidovirales. Virus Res. 2004;101:155–161. doi: 10.1016/j.virusres.2004.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- He B., Dong H., Jiang C., Cao F., Tao S., Xu L.A. Analysis of codon usage patterns in Ginkgo biloba reveals codon usage tendency from A/U-ending to G/C-ending. Sci. Rep. 2016;6:35927. doi: 10.1038/srep35927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G.M., Holmes E.C. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res. 2003;92:1–7. doi: 10.1016/s0168-1702(02)00309-x. [DOI] [PubMed] [Google Scholar]
- Kandeel M., Ibrahim A., Fayez M., Al-Nazawi M. From SARS and MERS CoVs to SARS-CoV-2: moving toward more biased codon usage in viral structural and nonstructural genes. J. Med. Virol. 2020;92:660–666. doi: 10.1002/jmv.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandia R., Singhal S., Kumar U., Ansari A., Tiwari R., Dhama K., Das J., Munjal A., Singh R.K. Analysis of Nipah virus codon usage and adaptation to hosts. Front. Microbiol. 2019;10:886. doi: 10.3389/fmicb.2019.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Kulkarni D.D., Lee B., Kaushik R., Bhatia S., Sood R., Pateriya A.K., Bhat S., Singh V.P. Evolution of codon usage bias in henipaviruses is governed by natural selection and is host-specific. Viruses. 2018;10:604. doi: 10.3390/v10110604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., Li W.J., Jiang B.G., Wei W., Yuan T.T., Zheng K., Cui X.M., Li J., Pei G.Q., Qiang X., Cheung W.Y., Li L.F., Sun F.F., Qin S., Huang J.C., Leung G.M., Holmes E.C., Hu Y.L., Guan Y., Cao W.C. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Li P., Ke X., Wang T., Tan Z., Luo D., Miao Y., Sun J., Zhang Y., Liu Y., Hu Q., Xu F., Wang H., Zheng Z. Zika virus attenuation by codon pair deoptimization induces sterilizing immunity in mouse models. J. Virol. 2018;92:e00701–e00718. doi: 10.1128/jvi.00701-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Wang H., Wang S., Xing G., Zhang C., Zhang W., Liu J., Zhang J., Su S., Zhou J. Insights into the genetic and host adaptability of emerging porcine circovirus 3. Virulence. 2018;9:1301–1313. doi: 10.1080/21505594.2018.1492863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan M., Chalmers J.D. Estimates of the ongoing need for social distancing and control measures post-“lockdown” from trajectories of COVID-19 cases and mortality. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.01483-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020;26:729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P.M., Li W.H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986;24:28–38. doi: 10.1007/bf02099948. [DOI] [PubMed] [Google Scholar]
- Sharp P.M., Li W.H. The codon adaptation index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletzki N., Eyre-Walker A. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol. Biol. Evol. 2007;24:374–381. doi: 10.1093/molbev/msl166. [DOI] [PubMed] [Google Scholar]
- Ulrich K., Wehner S., Bekaert M., Di Paola N., Dilcher M., Muir K.F., Taggart J.B., Matejusova I., Weidmann M. Molecular epidemiological study on infectious pancreatic necrosis virus isolates from aquafarms in Scotland over three decades. J. Gen. Virol. 2003;99:1567–1581. doi: 10.1099/jgv.0.001155. [DOI] [PubMed] [Google Scholar]
- van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C.C.S., Boshier F.A.T., Ortiz A.T., Balloux F. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weringh A., Ragonnet-Cronin M., Pranckeviciene E., Pavon-Eternod M., Kleiman L., Xia X. HIV-1 modulates the tRNA pool to improve translation efficiency. Mol. Biol. Evol. 2011;28:1827–1834. doi: 10.1093/molbev/msr005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. (e00127–20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E.H., Smith D.K., Rabadan R., Peiris M., Poon L.L. Codon usage bias and the evolution of influenza A viruses. Codon usage biases of influenza virus. BMC Evol. Biol. 2010;10:253. doi: 10.1186/1471-2148-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.J., Li N., Guo Y., Li X., Shen X., Zhang Z., Shu F., Huang W., Li Y., Zhang Z., Chen R.A., Wu Y.J., Peng S.M., Huang M., Xie W.J., Cai Q.H., Hou F.H., Chen W., Xiao L., Shen Y. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Zeng Z.Q., Chen D.H., Tan W.P., Qiu S.Y., Xu D., Liang H.X., Chen M.X., Li X., Lin Z.S., Liu W.K., Zhou R. Epidemiology and clinical characteristics of human coronaviruses OC43, 229E, NL63, and HKU1: a study of hospitalized children with acute respiratory tract infection in Guangzhou, China. Eur. J. Clin. Microbiol. Infect. Dis. 2018;37:363–369. doi: 10.1007/s10096-017-3144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhang L., He W., Zhang X., Wen B., Wang C., Xu Q., Li G., Zhou J., Veit M., Su S. Genetic evolution and molecular selection of the HE gene of influenza C virus. Viruses. 2019;11 doi: 10.3390/v11020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material